Abstract
Get3, Get4, and Get5 in Saccharomyces cerevisiae participate in the insertion of tail-anchored proteins into the endoplasmic reticulum membrane. We elucidated the interaction between Get4 and Get5 and investigated their interaction with Get3 and a tetratricopeptide repeat-containing protein, Sgt2. Based on co-immunoprecipitation and crystallographic studies, Get4 and Get5 formed a tight complex, suggesting that they constitute subunits of a larger complex. In contrast, although Get3 interacted physically with the Get4-Get5 complex, low amounts of Get3 co-precipitated with Get5, implying a transient interaction between Get3 and Get4-Get5. Sgt2 also interacted with Get5, although the amount of Sgt2 that co-precipitated with Get5 varied. Moreover, GET3, GET4, and GET5 interacted genetically with molecular chaperone YDJ1, suggesting that chaperones might also be involved in the insertion of tail-anchored proteins.
Keywords: Protein Structure, Protein Targeting, Protein-Protein Interactions, X-ray Crystallography, Yeast, GET, Guided Entry of Tail-anchored Proteins, Post-translational Protein Targeting
Introduction
Protein transport in cells is an elaborate process that requires extensive cooperation among various biochemical pathways. In Saccharomyces cerevisiae, the Hsp70 homologs Ssa1/Ssa2 and Hsp40/DnaJ-related protein Ydj1 participate in protein transport to the endoplasmic reticulum (ER)4 and mitochondria (1). Moreover, several tetratricopeptide repeat (TPR)-containing proteins have been shown to interact with Ssa1. Each TPR motif composed of 34 amino acids forms an antiparallel pair of α-helices with equal length (2). Some Ssa1-interacting TPR-containing proteins are also involved in the targeting of intracellular proteins in S. cerevisiae. For example, Sti1 (3) and Cns1 (4) interact with Ssa1 to stimulate its ATP hydrolytic activity, and Tom70 (5) interacts with Ssa1 and Hsc82 (Hsp90 in yeast) to facilitate protein translocation into mitochondria. Another TPR-containing protein, Sgt2, co-immunoprecipitates with Ssa1/Ssa2, and yeast SGT2 interacts genetically with YDJ1, which encodes a molecular chaperone, when yeast are under stress (6). Sgt2 is thought to be a homolog of the vertebrate small glutamine-rich TPR-containing protein, SGT (7). Recently, SGT was implicated in several processes related to cell growth, including androgen receptor signaling (8) and mitotic cell division (9); however, it is unclear whether Sgt2 has similar functions in yeast.
Recently, Hillenmeyer et al. (10) used small molecules to probe the growth fitness of yeast strains in deletion collections. In this type of assay, genes that show a similar fitness profile for various chemicals may be scored and clustered together. MDY2 and YOR164C are the two genes that have the highest score for homozygous co-sensitivity with SGT2. The N-terminal domain of Sgt2 interacts with Mdy2 (6), but details of the interaction between Sgt2 and Yor164c have not been elucidated. Interestingly, the genes showing co-sensitivity with MDY2 and YOR164C are quite similar and include GET3. In S. cerevisiae, GET complexes, including Get1-Get2 and Get3, take part in the insertion of a subset of tail-anchored (TA) proteins into the ER membrane (11, 12). These TA proteins insert into the membrane using a single transmembrane segment in the C-terminal region, leaving the N-terminal functional domain in the cytosol. Jonikas et al. (13) reported that Yor164c and Mdy2 also participate in the insertion of TA proteins. They appear to play a role upstream of Get3. Accordingly, YOR164C and MDY2 were renamed as GET4 and GET5, respectively (13). In addition, FLAG-tagged Get3 has the capacity to precipitate Get4 (Yor164c) and Get5 (Mdy2) (13), but it is unclear whether Get3 interacts physically with Get4 and Get5 or whether this association is mediated by another protein(s).
Data obtained from large scale yeast two-hybrid assays (14) and mass spectrometry analyses of protein complexes (15, 16) have revealed that the four proteins, Sgt2, Get5, Get4, and Get3, interact with one another, although the details of the interactions have not been elucidated. We therefore investigated the interactions among these proteins and determined the crystal structure of full-length Get4 complexed with the N-terminal domain of Get5. In addition, the molecular chaperon YDJ1 was also shown here to genetically interact with several GET complexes members. The results of our functional and structural studies led to a deeper understanding of the machinery for TA protein targeting into ER membrane.
EXPERIMENTAL PROCEDURES
Yeast Strains
S. cerevisiae strains in the S288C genetic background (BY4741, BY4742, and single-deletion mutants) used in this study were purchased from Research Genetics/Open Biosystems, and the double mutant, get4Δydj1Δ, was generated by first crossing the appropriate haploid strain followed by sporulation and tetrad dissection. The wild-type yeast strain in the W303 background (MATa ura3-1 ade2-1 his3-11 leu2-3 trp1-1 can1-100) was provided by Dr. J.-Y. Leu (Institute of Molecular Biology, Academia Sinica), and the mutants were generated by replacing the genes with the knaMX4 cassette (17). Growth and manipulation of yeast were carried out using the procedures described by Amberg et al. (18) unless otherwise specified.
Construction of Plasmids for Protein Expression and Yeast Two-hybrid Assays
Plasmids were constructed by standard molecular cloning and recombinant DNA technologies. Briefly, to construct plasmids of GET4, its coding sequence with 288 nucleotides upstream and 338 nucleotides downstream was obtained from the genome of S. cerevisiae using PCR. After introducing an NdeI site at the N terminus of the coding sequence, the DNA was cloned into the pET-15b (Novagen). The plasmid obtained, Get4/15b, was used to express recombinant Get4 for the production of antibodies (6). To express Get4 and Get5 simultaneously, the coding sequences of GET4 and GET5 were inserted into MCS2 and MCS1 of the vector pRSFDuet-1 (Novagen), respectively. The plasmid (Get4-Get5)/pRSFDuet-1 can be used to simultaneously express the His6-tagged Get4 and untagged Get5. The expression plasmids containing GET3 were constructed by amplifying its coding sequence using PCR from the genomic DNA of S. cerevisiae. During the amplification, a NdeI site was introduced at the 5′-end of the coding sequence, and a XhoI site was introduced after the stop codon. The NdeI-XhoI fragment obtained was cloned into the pET-15b vector using the same two restriction sites. The resulting plasmid, Get3/15b, was used to express the Get3 recombinant protein in E. coli.
For the yeast two-hybrid assays, a fragment containing the coding sequence of GET4 was isolated and was cloned into pACT2 vector (Clontech), and the fragment containing the coding sequence of GET5 was cloned into pAS2-1 (Clontech), respectively. The SGT2-contaning plasmids with pAS2-1 as backbone were obtained previously (6). To generate the plasmid containing the coding sequence for the first 110 amino acids of GET5, we digested Mdy2/15b (6) with StuI and EcoRV. The digested plasmid was allowed to self-ligate, and the NdeI-XhoI fragment containing the Get5-(1–110) was excised and ligated with pAS2-1 using NdeI and SalI sites. The NcoI-XhoI fragment was also obtained and was inserted into the pACT2 vector using the same two sites. The plasmids containing fragments of GET5 corresponding to the coding sequences for amino acids 69–153 and the one with amino acids 69–212 were engineering by amplifying these fragments using PCR with Mdy2/15b as template. During the process, appropriate restriction sites were introduced at the 5′-end and the 3′-end. Subsequently, they were cloned into both pACT2 and pAS2-1 vectors. To engineer the plasmids containing the coding sequence of Get4-(1–148) and Get4-(149–312), the corresponding DNA fragments were amplified by PCR and ligated with pAS2-1 using NdeI and XhoI sites.
Yeast Mating Assay
All mutants used were the a mating type. They were mated with tester strain 4275 (MATα ade8) in the SK1 genetic background (provided by Dr. T.-F. Wang, Academia Sinica). BY4742 (MATα) was also used in this assay as a negative control. Briefly, overnight cultures were diluted 30-fold in YPD medium and cultured at 30 °C for 3 h. Then, 5 × 106 cells from each sample and 107 cells of the 4275 tester strain were mixed, washed once with water, and diluted with 1 ml of water. The cells were allowed to settle onto a 0.22-μm GSWP nitrocellulose membrane (25 mm in diameter) using a glass microanalysis filter holder (Millipore). The filters were placed on YPAD plates (1% yeast extract, 2% peptone, 2% glucose, 0.002% adenine, 2% agar) and incubated for 3 h at 30 °C. The cells were subsequently recovered by washing with 2 ml of water. They were serially diluted to 105-fold and spread either on synthetic complete plates lacking adenine to quantify the input cell number or on synthetic dextrose plates (with adenine) for the estimation of successful mating events. The plates were incubated at 30 °C for 2–3 days. Mating efficiency was calculated as the ratio of successful mating events to the input cell number.
Purification of Recombinant Proteins
Expression of recombinant Get4-Get5 complex was induced by adding isopropyl-β-d-thiogalactopyranoside (final concentration 1 mm) to bacterial cultures harboring the plasmid (Get4-Get5)/pRSFDuet-1. The cells were collected by centrifugation and suspended in 0.1 culture volume of homogenization buffer (20 mm Tris-HCl, pH 7.9, 500 mm NaCl, 5 mm imidazole) with protease inhibitors (EDTA-free, Roche Applied Science). After the addition of glass beads (150–212 μm; Sigma) to 1 g/ml suspension, the mixtures were homogenized by vortexing (maximum speed, five times, 30 s each). Then, 5 volumes of buffer with protease inhibitors were added to each homogenate. The mixtures were resuspended and centrifuged. The Get4-Get5 complex in the supernatants was purified using Ni2+-NTA resin (Qiagen). Recombinant Get3 was purified using an identical procedure. Alternatively, the cells were homogenized using a Microfluidizer (Microfluidics) under high pressure, and the lysate was centrifuged at 40,000 × g for 1 h at 4 °C. The supernatant was then loaded onto a HisTrapTM HP column (GE Healthcare) and eluted with an imidazole gradient (5–300 mm in buffer containing 20 mm Tris-HCl, pH 7.9, and 500 mm NaCl). Fractions containing the Get4-Get5 complex were pooled and further purified by size exclusion chromatography on a 16/60 Superdex 200 column (GE Healthcare) equilibrated with a buffer containing 20 mm Tris-HCl, pH 7.9, and 300 mm NaCl. The purified protein complex was concentrated to 10 mg/ml and stored at 4 °C. After 90 days in storage at 4 °C, the remaining stable complex containing full-length Get4 and Get5-(2–59) was purified with a 16/60 Superdex 200 size exclusion column (GE Healthcare) with the same buffer.
Preparation of Yeast Cytosol for Immunoprecipitation
Overnight cultures (A600 nm = 5–8) were harvested after the addition of NaN3 to 0.1%. The pellets were washed once with ice-cold water and resuspended in phosphate-buffered saline (2× volume over packed cells) containing protease inhibitors. The mixtures were homogenized with glass beads (425–600 μm, Sigma). After homogenization, 20 volumes of phosphate-buffered saline with protease inhibitors were added, and the diluted homogenates were mixed briefly by vortexing. The suspensions were then centrifuged at 20,000 × g for 15 min, and each supernatant was collected and subjected to high speed centrifugation (100,000 × g, 30 min). Each clear supernatant (cytosol) was then incubated at 25 °C for 50 min with anti-Get5 antibodies that had been coupled to protein A-Sepharose (6). The resin was then washed with phosphate-buffered saline, and the protein bound to the resin was eluted with 20 mm HCl in water. Each eluate was lyophilized, and samples of the dry residues were solubilized with SDS sample buffer for gel electrophoresis.
Crystallization of Get4-Get5N Complex and Structure Determination
For crystallization trials, the Get4-Get5N complex was first concentrated to 6 mg/ml in buffer containing 20 mm Tris-HCl, pH 7.9, and 300 mm NaCl with a Centricon concentrator (Millipore). The hanging drop vapor diffusion method was then carried out at 298 K by mixing 1 μl of Get4-Get5N with an equal volume of crystal screening solutions. Rod-shaped crystals appeared after 1 day under the condition containing 200 mm diammonium hydrogen citrate and 20% (v/v) polyethylene glycol 3350. Because no structure similar to Get4 and Get5 could be used for molecular replacement phasing, heavy atom derivatives were tried for solving the phase problem using the multiwavelength anomalous dispersion method. After an extensive search, one useful mercury derivative was obtained by the co-crystallization method with 0.1 mm tetrakis(acetoxymercury)methane using a modified reservoir condition containing 300 mm diammonium hydrogen citrate and 18% (v/v) polyethylene glycol 3350. The crystals belong to space group P21 and contain four Get4-Get5N molecules per asymmetric unit. The cryogenic multiwavelength anomalous dispersion data collection was conducted on an Area Detector Systems Corp. Quantum-315 charge-coupled device detector using a synchrotron radiation x-ray source at beamline BL13B1 of the National Synchrotron Radiation Research Center (NSRRC) in Taiwan. Two energies were chosen near the absorption peak and the edge of mercury atoms in Get4-Get5N, 1.0050 and 1.0085 Å, which corresponded to the maximum f″ and the minimum f′, respectively. A remote energy was selected as reference wavelength at 0.8550 Å. X-ray diffraction data integration and scaling were performed using the HKL2000 package (19). The redundancy-independent merging R-factor (Rr.i.m.) and the precision-indicating merging R-factor (Rp.i.m.) were calculated using the program RMERGE (20, 21). SOLVE (22) was used to locate the only mercury site and generate the initial multiwavelength anomalous dispersion phases at 2.50 Å resolution. The extension of initial phases to 1.99 Å and the preliminary auto-model building were carried out by RESOLVE (23). XtalView (24) was used for examining electron density maps and manually model building. Further refinement was performed by using CNS (25) and Refmac5 (26). After completion, the R-factor of final model for all reflections above 2 σ between 25.59 and 1.99 Å resolution was refined to 17.5%, and an Rfree value of 20.6% was obtained using 4.8% randomly distributed reflections. The Ramachandran plot showed that 93.8% of residues lie within the most favored regions, 6.2% lie in the additional allowed regions, and no residues were found in generously allowed and disallowed regions.
Mass Spectrometry Analysis
Protein bands of interest were excised from Coomassie Brilliant Blue-stained gels, digested with trypsin (20 μg/ml), and then spotted onto Bruker's prespotted α-cyano-4-hydroxycinnamic acid target. The masses of the fragments were acquired using an Autoflex III matrix-assisted laser desorption/ionization-time of flight/time of flight mass spectrometer (Bruker). The spectra were first subjected to fingerprinting search with MASCOT (version 2.2.1) using the National Center for Biotechnology Information (NCBI) data base. The identity of the proteins was confirmed with peptide sequencing using the same instrument.
Other Methods
The yeast two-hybrid assays were carried out as described (27) except that in some cases we used yeast strain Y190 with GET4 or GET5 deleted using the kanMX4 cassette (17). To generate the double mutant, get3Δydj1:: natkM, we first crossed strain get3Δ in BY4741 with strain BY4742. The diploids heterozygous for YDJ1 were obtained by replacing one copy of the gene with the natMX cassette. To engineer the cassette, we first isolated the HindIII/EcoRV fragment from the pAG25 plasmid (17). The fragment was inserted into YDJ1 between the same two restriction sites in plasmid. Subsequently, the diploids heterozygous for both GET3 and YDJ1 were sporulated for tetrad dissection.
RESULTS
Get5 Mediates the Interaction of Sgt2 with Get4
Tandem affinity purification-tagged Sgt2 has the capacity to pull down Get4, Ssa1/Ssa2, and Sse1/Sse2 (15). Moreover, based on homozygous co-sensitivity for yeast deletion strains (10), Get4 (Yor164c) has the highest correlation score as an interactor of Sgt2. Therefore, we first considered whether Sgt2 might physically interact with Get4; indeed, yeast two-hybrid assays demonstrated this interaction (Fig. 1A). Further investigation demonstrated that the N-terminal region of Sgt2 was necessary and sufficient for interaction with Get4 (Fig. 1A). Interestingly, the same N-terminal region of Sgt2 has been demonstrated to interact with Get5 (Mdy2) (6). To confirm the physical interaction between Get4 and Sgt2, we purified recombinant proteins, including Get4 fused with glutathione S-transferase (GST). Surprisingly, GST·Get4 failed to bind Sgt2 in a pulldown assay. On the other hand, the physical interaction between GST·Get4 and Get5 was evident (supplemental Fig. S1), suggesting that the observed yeast two-hybrid interaction between Sgt2 and Get4 could have been mediated by Get5. Based on this hypothesis, Sgt2 and Get4 should not interact in yeast strains lacking GET5. Indeed, Sgt2 and Get4 did not interact in the GET5 deletion strain (Fig. 1B). Thus, it is likely that the apparent two-hybrid interaction between Sgt2 and Get4 is mediated by Get5.
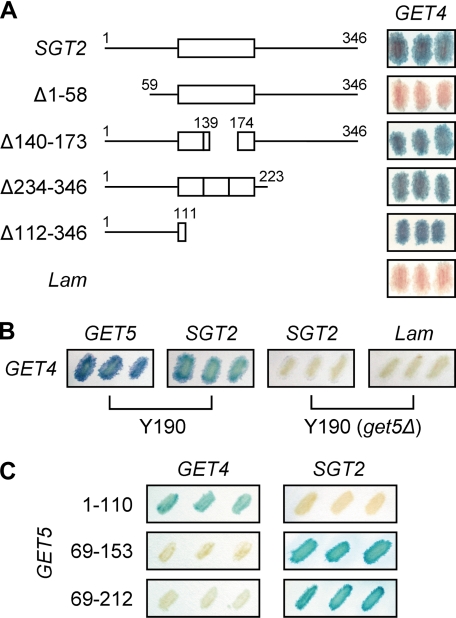
FIGURE 1.
Yeast two-hybrid assays to assess the interaction between different pairs of proteins among Sgt2, Get4, and Get5. A, interaction of SGT2 and its fragments (in pAS2-1 vector) with GET4 (in pACT2 vector) was determined, and the blue filters are shown. Human lamin C-(66–230) in pAS2-1 (Lam) was used as a control. B, interaction of GET4 with SGT2 was determined using yeast strain Y190 and Y190 with GET5 deleted; the blue filters are shown. C, interaction of GET4 and SGT2 with fragments of GET5 (in either pAS2-1 or pACT2) was determined, and the blue filters are shown. The ubiquitin-like domain of Get5 ranges from residues 74–173.
Based on the assumption that Get5 mediates the interaction between Sgt2 and Get4, we constructed GET5 truncation mutants for use in yeast two-hybrid assays. The results showed that the N-terminal region of Get5 interacted with Get4 (Fig. 1C, also see Fig. 5A below), whereas fragments containing the Get5 ubiquitin-like domain were sufficient for interaction with Sgt2 (Fig. 1C).
FIGURE 5.
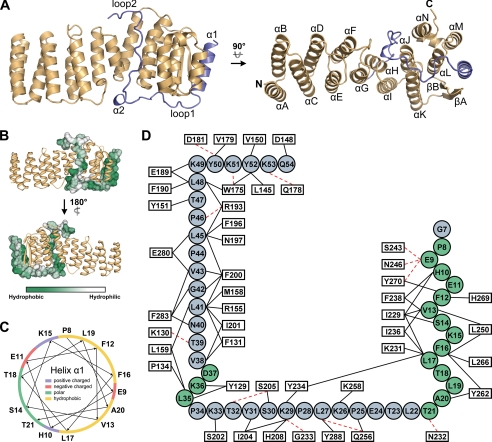
Crystal structure of Get4-Get5N. A, ribbon diagrams for the Get4-Get5N complex structure. The structure of Get4 in the complex (gold) and Get5N (blue) is shown. The left and right panels are the front orthogonal view and top orthogonal view, respectively. B, hydrophobicity of the Get5N surface. The surface hydrophobicity of Get5N is shown by a color gradient from green (hydrophobic) to white (hydrophilic). Get4 is shown as the gold ribbons. C, helical plot of the α1 in Get5N. The basic, acidic, polar, and hydrophobic residues are indicated in blue, red, green, and yellow, respectively. D, interactions between Get5N and Get4. Residues of Get5N and Get4 are displayed by circles and rectangles, respectively. The α-helices and loops of Get5N are colored in green and blue, respectively. Residues participating in intermolecular interactions are represented by black solid lines (hydrophobic interactions) or red dotted lines (hydrogen bonds).
Cells Lacking GET4 or GET5 Show Identical Phenotypes
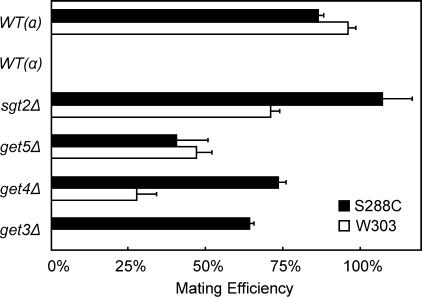
Because Sgt2, Get4, and Get5 show co-fitness in chemical genomic assays (10), they might perform a similar, if not identical, function in yeast. If so, one would expect that deletion of these genes would result in similar phenotypes. Deletion of GET5 results in a decrease in mating efficiency (28); therefore, we examined whether deletion of SGT2 or GET4 might also have such an effect. The mutants were generated from yeast cells with two distinct genetic backgrounds, and they were mated with the tester strains at a ratio of 1:2. The mating efficiency was then quantified. Similar to get5Δ cells, the get4Δ strain showed a reduction in mating efficiency with yeast cells having the W303 or S288C genetic background (Fig. 2). For sgt2Δ strains, however, a decrease in mating efficiency was only observed with W303 cells, suggesting that the sgt2Δ strains behaved differently from the get5Δ and get4Δ strains.
FIGURE 2.
Effect of deletions of GET or SGT2 genes on mating efficiency. The mating efficiency of the mutants having the W303 or S288C genetic background was quantified. Data represent the means ± S.E. of three independent determinations. WT, wild type.
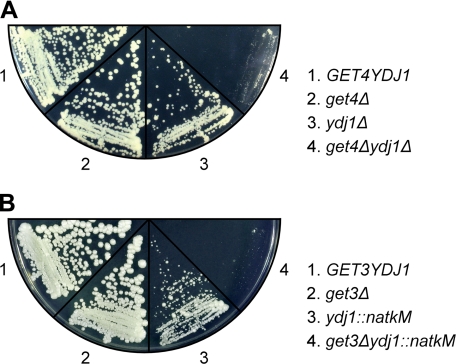
The genetic interaction between SGT2 and YDJ1 can be observed in yeast cells under stress; under normal growth conditions, however, GET5 interacts genetically with YDJ1 (6). We therefore tested whether GET4 interacts genetically with YDJ1. Mutants, including get4Δydj1Δ, were first generated through sporulation and were verified by immunoblotting. These mutants were then subjected to growth fitness assays. As shown in Fig. 3A, get4Δ had no growth defect, and ydj1Δ had a slow growth phenotype. Notably, however, get4Δydj1Δ showed a much more severe growth defect than ydj1Δ, clearly indicating that GET4 interacts genetically with YDJ1. Taken together, these analyses show that Get4 and Get5 are closely related in a functional sense, whereas the function of Sgt2 is more distantly related to these proteins.
FIGURE 3.
Genetic interaction of YDJ1 with GET4 and GET5. Diploid yeast strains heterozygous for YDJ1 and GET4 (A) or for YDJ1 and GET3 (B) were sporulated, and tetrads were dissected. Growth fitness assays were performed on tetrads with all four possible combinations. Here, an equal number of cells was streaked onto YPD plates and allowed to grow at 30 °C for 3–4 days before being photographed.
Get4 and Get5 Form a Tight Complex in the Cytosol
Although we demonstrated that Get5 has the capacity to interact with both Sgt2 and Get4, there are conflicting reports about whether these proteins are located in the same cellular compartment in yeast. Fleischer et al. (29) reported that Get5 is associated with ribosomes, indicating that Get5 should be located in the cytosol. On the other hand, by examining the cellular location of Get5 fused with green fluorescent protein (GFP), Hu et al. (28) concluded that Get5 was predominantly found in the nucleus. However, using an approach identical to that adopted by Hu et al. (28), Jonikas et al. (13) recently demonstrated that Get5·GFP was indeed cytosolic. To clarify these discrepancies, we examined the subcellular localization of endogenous Get5 by immunofluorescence microscopy. Our result (supplemental Fig. S2) suggested that Get5 is predominantly located in the cytosol. Because Sgt2 and Get4 are also cytosolic, these three proteins would indeed have the capacity to interact with each other in vivo.
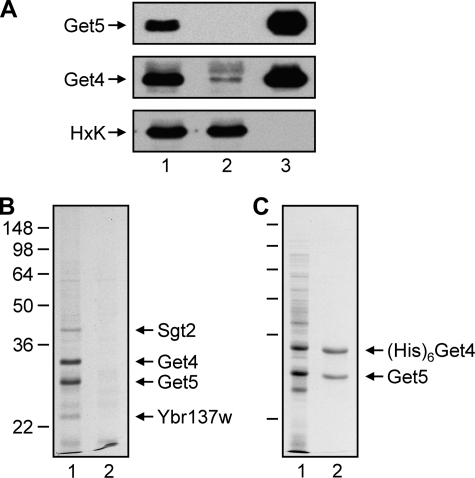
The next question was whether Sgt2, Get4, and Get5 form a complex in yeast. We prepared a cytosolic fraction from yeast cultured in rich medium, and this fraction was incubated with anti-Get5 antibodies coupled to protein A-Sepharose. The protein bound to the antibodies was then eluted with acid for analysis. Under our experimental conditions, most (if not all) of the Get5 in the cytosolic fraction was immunoprecipitated by anti-Get5 (Fig. 4A). The immunoprecipitate was subjected to SDS gel electrophoresis (Fig. 4B), which revealed two major proteins and a few minor ones that were subsequently identified by mass spectrometry. The two major proteins were Get5 and Get4, which were always present in similar, if not identical, amounts. Based on the Saccharomyces Genome Database (SGD), the number of copies per cell for Get4 (5.4 × 103) and Get5 (6.5 × 103) is similar. Indeed, most of Get4 also precipitated with Get5 (Fig. 4A). The minor bands found in the anti-Get5 immunoprecipitate were identified as Sgt2 and Ybr137c. They were always present at much lower levels than Get5 and Get4, and the amount of these minor proteins varied from experiment to experiment (data not shown). These results clearly suggest that Get4 and Get5 form a tight complex in yeast cells, and Sgt2 associates transiently with this complex.
FIGURE 4.
Get4 and Get5 form a complex. A, yeast lysates were prepared and utilized for immunoprecipitation with anti-Get5. The immunoprecipitates were subjected to gel electrophoresis and then blotted with anti-Get5, anti-Get4, and anti-hexokinase (HxK). Lanes 1, 2, and 3 represent the lysate diluted 1:2000, flow-through diluted 1:2000, and protein bound to antibodies diluted 1:200, respectively. B, the protein immunoprecipitated with anti-Get5 was subjected to gel electrophoresis, and the gel was stained with Coomassie Brilliant Blue. Lanes 1 and 2 represent immunoprecipitates from wild-type and get5Δ lysates, respectively. C, Get5 co-purifies with His-tagged Get4 using Ni2+-NTA resin. Lanes 1 and 2 represent the soluble fraction of the bacterial lysates and the purified protein, respectively.
Crystal Structure of Get4-Get5N Complex
To obtain a crystal structure of the Get4-Get5 complex, we co-expressed His-tagged Get4 and untagged Get5 in Escherichia coli and purified the protein. Get5 co-purified with Get4 on Ni2+-NTA resin, and the amount of the two proteins was similar (Fig. 4C). Our attempts to use the purified protein complex for crystallization met with limited success because of apparent protein degradation upon storage at 4 °C. After 3 months of storage at 4 °C, two major stable fragments remained. Mass spectrometry and N-terminal sequencing revealed that they were the full-length Get4 and the N-terminal fragment of Get5 (Get5N, residues 2–59). These two polypeptides remained associated in a complex in solution as determined by size exclusion chromatography (data not shown). The Get4-Get5N complex was further purified to homogeneity for crystallization trials. The single crystals we obtained were used for structural determination. The statistics of data collection and structure refinement are shown in Table 1.
TABLE 1.
Data collection and refinement statistics
| Data collection | |||
| Space group | P21 | ||
| Cell dimensions | |||
| a (Å) | 48.28 | ||
| b (Å) | 118.77 | ||
| c (Å) | 168.38 | ||
| β (°) | 95.17 |
| Peak | Inflection | Remote | |
|---|---|---|---|
| Wavelength | 1.0050 | 1.0085 | 0.8550 |
| Resolution (Å) | 2.15 | 1.99 | 1.99 |
| Redundancy | 6.3 (6.0)a | 3.1 (3.0) | 3.1 (3.0) |
| I/σ(I) | 19.5 (3.9) | 15.3 (3.7) | 12.4 (2.4) |
| Completeness (%) | 99.7 (99.9) | 99.3 (99.1) | 98.9 (96.9) |
| Rmerge (%)b | 9.0 (47.2) | 7.3 (33.0) | 9.6 (49.2) |
| Rr.i.m. (%)c | 9.5 (51.3) | 8.7 (40.1) | 11.4 (59.1) |
| Rp.i.m. (%)d | 3.8 (21.0) | 4.9 (22.2) | 6.4 (32.8) |
| Refinement | |||
| Resolution (Å) | 25.59-1.99 | ||
| No. of reflections | 203797 | ||
| Rwork/Rfree (%) | 17.5/20.6 | ||
| No. of atoms | |||
| Protein | |||
| Get4 | 9288 | ||
| Get5N | 1515 | ||
| Mercury ion | 4 | ||
| Water | 1202 | ||
| B-factors | |||
| Protein | |||
| Get4 | 21.1 | ||
| Get5N | 24.4 | ||
| Mercury ion | 49.2 | ||
| Water | 35.1 | ||
| r.m.s.e deviations | |||
| Bond lengths (Å) | 0.005 | ||
| Bond angles (°) | 1.0 | ||
a Numbers in parentheses refer to the highest resolution shell.
b Rmerge = ΣhΣi|Ihi − 〈Ih〉|/ΣhΣiIhi.
c Rr.i.m. = Σh[N/(N − 1)]½Σi|Ihi − 〈Ih〉|/ΣhΣiIhi.
d Rp.i.m. = Σh[1/(N − 1)]½Σi|Ihi − 〈Ih〉|/ΣhΣiIhi.
e r.m.s., root mean square.
A ribbon diagram of the structure at 1.99 Å resolution is shown in Fig. 5A. The complex had an overall oblong shape: about 80 Å in length, 30 Å in width, and 40 Å in pitch. The organization of the secondary structures of Get4 and Get5N are given in supplemental Fig. S3. Get4 contains 14 α-helices (helix A to helix N) and is composed of several helix-turn-helix motifs; an antiparallel β-sheet (βA and βB) lies between helices K and L. Get5N in the complex has an N-terminal α-helix (α1) followed by two long loops (loop 1 and loop 2) with a short α-helix (α2) in between, and it straddles the C-terminal bisection of the elongated Get4 structure. Get5N α1 is located in the cleft formed by βA, βB, and helices L and M of Get4. Loop 1 lies across the bottom of helices J, K, and L of Get4, and helix α2 makes a turn in Get5N that allows loop 2 to fit into the concavity on the Get4 surface comprised of helices H, I, J, and N.
The structure also showed that Get4 and Get5N associate mainly through hydrophobic interactions (Fig. 5B). The nonpolar surface of the amphipathic helix α1 (Fig. 5C) of Get5N is lodged in the hydrophobic cleft on Get4 formed by βA, βB, helix L, and helix M through extensive hydrophobic interactions and a few hydrogen bonds. Loops 1 and 2 of Get5N also interact with Get4 mainly through hydrophobic interactions. These binding behaviors imply that Get4 and Get5 are not likely to dissociate in the cytosol, which is hydrophilic in nature. Moreover, there is an abnormally high density of prolyl residues (6 of 48) in Get5N, five of which are in the loop regions. Because of the inherent inflexibility of prolyl residues, the long loops of Get5N may be able to adopt more rigid conformations to fit into the complementary positions on the Get4 surface. A schematic representation summarizing the interaction of Get5N with Get4 is given in Fig. 5D.
The continuous antiparallel α-helices in Get4 structure suggest an architectural similarity to the TPR proteins. However, the structural comparison between Get4 and the TPR domain of human SGT (30) reveals that Get4 does not behave in a right-handed helical conformation, as do regular TPR proteins (supplemental Fig. S4). The varied numbers of amino acid residues in each antiparallel α-helix pair of Get4 also overrule the relatedness to the TPR family (supplemental Fig. S3). Nevertheless, the structural feature of Get4 may still suggest a possible evolutionary relationship to proteins containing helix-turn-helix repeats and provide hints for a similar interaction manner with other proteins.
Interaction of the Get4-Get5 Complex with Get3
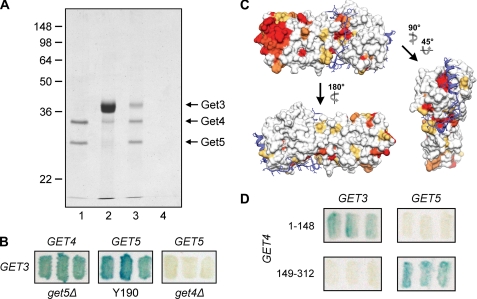
Homozygous co-fitness data (10) show that GET4 and GET5 have a high correlation with GET3, and deletion of GET3 reduced mating efficiency (Fig. 2). Moreover, Get4 and Get5 have been shown to take part in the insertion of TA proteins into the ER membrane, and they function upstream of Get3 (13). Because GET3 was found to interact with GET4 in a large scale two-hybrid analysis (14) and because FLAG-tagged Get3 has the capacity to pull down Get4 and Get5 (13), we next tested whether Get3 and Get4 interact physically. We first purified the Get4-Get5 complex and mixed it with purified Get3. The mixture was then immunoprecipitated with anti-Get5. The result clearly demonstrated that Get3 was pulled down by the Get4-Get5 complex (Fig. 6A). We next determined whether Get3 interacts specifically with Get4 or Get5 or with both proteins. Yeast two-hybrid assays (Fig. 6B) demonstrated that both Get4 and Get5 appeared to interact with Get3. Because deletion of GET4 abolished the interaction of GET5 with GET3, however, the apparent interaction between GET5 and GET3 (31) might be mediated by GET4. These results suggest that in the Get4-Get5 complex, either only Get4 interacts with Get3 or the interaction of Get5 with Get3 is dependent on structural aspects of Get5 that are only manifested when it is bound to Get4.
FIGURE 6.
Interaction of the Get4-Get5 complex with Get3. A, recombinant Get4-Get5 complex (lane 1) was incubated with purified Get3 (lane 2), and the mixtures were immunoprecipitated with anti-Get5. Get3 co-precipitated in the presence of the Get4-Get5 complex (lane 3) but not in its absence (lane 4). B, yeast two-hybrid interactions of GET3 with GET4 in Y190(get5Δ) and of GET3 with GET5 in Y190 and Y190(get4Δ) are shown. C, conserved residues on the Get4 surface. Residue conservation in Get4 determined by sequence alignment (supplemental Fig. S5) is shown by a surface diagram with a color gradient from yellow (70% conserved) to red (100% identical). Get5N (blue) is shown as ribbons with stick side chains. D, the yeast two-hybrid assays were used to dissect the interaction of Get4 with Get3 and Get5. The N-terminal region (residues 1–148) and C-terminal region (residues 149–312) of Get4 are responsible for interacting with Get3 and Get5, respectively.
Get4 is expressed ubiquitously across many species, and thus we chose some of the most conserved Get4 homologs from different species for sequence alignment (supplemental Fig. S5), and a color gradient from yellow (70% similarity) to red (100% identity) was assigned to the residues on the surface of Get4 (Fig. 6C). Besides the relatively conserved Get5N-binding troughs, Get4 contains a highly conserved region located distal to the Get5N-interacting surface. This conservation of surface charge suggested that the N-terminal region of Get4 is responsible for interacting with the conserved Get3. To test this possibility, we generated deletion plasmids of GET4 for the yeast two-hybrid assay. As expected, the C-terminal domain of Get4 interacted with Get5, but the results also clearly showed that the N-terminal domain of Get4 interacted with Get3 (Fig. 6D).
GET3 Interacts Genetically with the Molecular Chaperone YDJ1
Using Hsp70 inhibitors, Rabu et al. (32) have demonstrated in vitro that in vertebrates, membrane insertion of a subset of TA proteins depends on the cytosolic Hsc70/Hsp40 chaperone system. Thus, we employed a genetic approach in yeast to investigate the possible involvement of chaperones in this process. Because we previously demonstrated a synthetic interaction of GET4 and GET5 with YDJ1, we wanted to determine whether GET3 also interacted genetically with YDJ1. Diploids with heterozygous mutations in both GET3 and YDJ1 were generated, and the tetrads were dissected. After verifying the mutations by immunoblotting, we examined the growth phenotypes of the mutants. As shown in Fig. 3B, get3Δ cells had no growth defect in rich medium, and ydj1::natkM cells displayed a slow growth phenotype. Moreover, the growth of get3Δydj1::natkM was severely retarded, demonstrating that GET3 interacts genetically with YDJ1. This result is consistent with the hypothesis that in yeast, there might be another chaperone-dependent pathway for the insertion of TA proteins into membranes.
DISCUSSION
Our results clearly show that Get4 and Get5 in S. cerevisiae form a tight complex to carry out their function(s). This conclusion is supported by the observation that both Get4 and Get5 are completely immunoprecipitated from yeast cytosol by anti-Get5 and that Get4 and Get5 also co-purify from extracts prepared from bacteria co-expressing the two proteins. In both cases, the stoichiometry of the recovered proteins was essentially 1:1, in agreement with the data deposited in SGD, indicating that the number of copies of these two proteins in yeast is virtually identical. The crystal structure of the complex formed by Get4 and Get5N clearly indicates that these proteins associate primarily via hydrophobic interactions. Although two other proteins, Sgt2 and Ybr137w, also have the capacity to associate with Get4 or Get5, the amount of these proteins associated with this complex varied and was not stoichiometric. Therefore, their association with the Get4-Get5 complex is probably dynamic, and Get4 and Get5 are likely the only two subunits of the complex. The quaternary structure of this molecule, however, remains to be determined.
The structure of Get4-Get5N also suggests that Get4 may play an important role in the stabilization of Get5 in yeast cells. Based on the structure, the N-terminal domain of Get5 would be highly exposed in the absence of Get4. Because of the flexible long loops and the amphipathic helix α1, Get5 would conceivably be easily cleaved by proteases or abnormally aggregated unless it was associated with Get4. If so, then deletion of GET4 should significantly accelerate the turnover of Get5. Indeed, deletion of GET4 brought about a pronounced reduction in the level of Get5 (supplemental Fig. S6) without affecting the level of Get3 and Sgt2 (data not shown). In vertebrates, however, the situation may be different. Homologs of Get3 and Get4 have been identified based on amino acid sequence data. The Get3 homolog, Ansa-1, participates in the insertion of TA proteins into membranes (32). Vertebrate homologs of Get4 have also been identified (33), but their function(s) has not been elucidated. The vertebrate homolog of Get5 remains elusive, and no protein has been found to have amino acid sequence similarity with the N-terminal region of Get5. There are at least two simple interpretations for this observation. First, it may be that only the three-dimensional structure, and not the sequence, of the putative vertebrate equivalent of Get5 needs to be conserved; i.e. the sequences of the yeast and putative vertebrate proteins may have diverged to the extent that the search algorithm may not recognize any existing similarity between them. Second, the vertebrate equivalent of Get5 may not associate with Get4, eliminating the need for sequence conservation (or even maintenance) of the N-terminal region. It is also possible that the putative vertebrate Get5 may be associated with some as yet unidentified component or it may act alone. Further work is needed to clarify these hypotheses.
Get1, Get2, and Get3 are involved in the insertion of TA proteins into the ER membrane (12). More recently, Jonikas et al. (13) demonstrated that Get4 and Get5 also participate in the membrane insertion of TA proteins and that Get4 and Get5 function upstream of Get3. Our present data demonstrate that the Get4-Get5 complex interacts physically with Get3. Although the experiments were carried out using recombinant proteins in the absence of TA proteins, the results nevertheless imply that Get4-Get5 may have the capacity to transfer TA proteins to Get3 without any additional GET protein. In intact cells, however, the association of the Get4-Get5 complex with Get3 may be transient because Get3 does not co-precipitate in any significant amount with the Get4-Get5 complex in yeast cytosol. Moreover, Get5 is associated with ribosomes (29). It is possible that nascent TA proteins emerging from ribosomes may be quickly transferred to the Get4-Get5 complex(es). Therefore, the number of essential components upstream of Get4-Get5 for this process may be quite limited. These upstream components, if any, might be important for specificity because certain TA proteins are inserted into the mitochondrial outer membrane instead of the ER (12).
Sgt2 is one of the major proteins associated with the Get4-Get5 complex. However, the role of Sgt2 in the insertion of TA proteins is unclear. On one hand, the phenotypes of sgt2Δ cells differ from those of get3Δ, get4Δ, and get5Δ cells. For example, for yeast strains having the S288C genetic background, deletion of GET3, GET4, or GET5 reduces mating efficiency, but deletion of SGT2 does not affect the mating process. It is possible that reduction in mating efficiency for those strains may originate from a defect in TA protein insertion because some TA proteins (e.g. Prm3) are important for the signaling involved in the mating process (34). If so, the lack of an effect of SGT2 deletion on mating efficiency may imply that the product of this gene is not an essential component for the insertion of TA proteins into the ER membrane. On the other hand, Sgt2 may associate with the Get4-Get5 complex in vivo, suggesting that these three proteins may be somewhat interdependent with respect to their various functions. Similarly, chemical genomics analysis results (10) also imply that SGT2 may be functionally linked to GET4 and GET5. Moreover, Sgt2 interacts physically with molecular chaperone Ssa1 (6). Because chaperones may play a role in the insertion of TA proteins into the ER membrane, Sgt2 might also be involved in, but may not be an essential component of, that process.
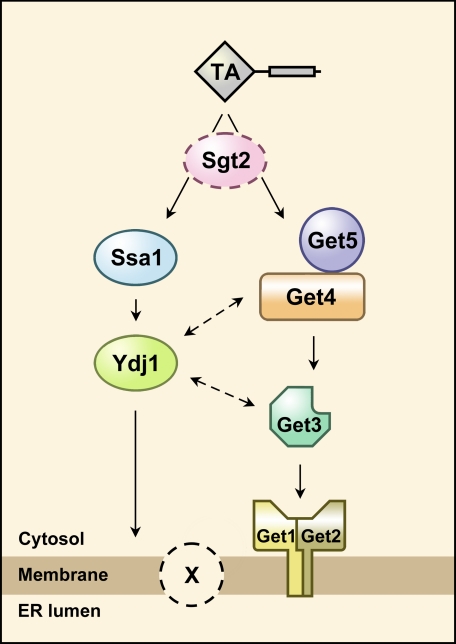
Yeast strains with GET3, GET4, or GET5 deleted do not show any defect in growth fitness in rich medium. This observation suggests that there might be another pathway for the insertion of TA proteins into the ER membrane. However, this putative pathway may have lower target-membrane specificity because certain TA proteins are transported to an incorrect location in the GET3 deletion strain (12). Similar to vertebrates (32), this additional pathway may be chaperone-dependent. We therefore examined the synthetic interaction between YDJ1 and GET3 as well as GET4. Indeed, both GET3 and GET4 show synthetic interaction with YDJ1. It has also been shown that YDJ1 and GET5 interact genetically (6). It cannot be ruled out that the GET protein complexes might physically interact with chaperones and that the chaperones improve the efficiency of TA protein transfer mediated by the GET complexes. Nevertheless, the simplest interpretation of this result is that besides the pathway involving GET protein complexes, there is a parallel chaperone-dependent pathway for the insertion of TA proteins. A simple working model is given in Fig. 7, in which we hypothesize that a membrane component (X) participates in this process because it is unclear whether the Get1-Get2 complex is sufficient for proper insertion of TA proteins into membranes.
FIGURE 7.
Model for the insertion of tail-anchored proteins. Besides the known pathway involving GET proteins, there is a chaperone-dependent pathway for the insertion of tail-anchored proteins. However, the role of Sgt2, if any, in this process needs to be elucidated. It also remains to be determined whether there are additional membrane proteins (indicated by X) participating in the insertion of tail-anchored proteins.
Acknowledgments
We thank Miranda Loney for comments on the manuscript, and we thank Hui-Chen Yan for the plasmid (Get4-Get5)/pRSFDuet-1 and help with the yeast two-hybrid assays. We are grateful for access to synchrotron radiation beamlines BL13B1 and BL13C1 at the National Synchrotron Radiation Research Center in Taiwan and BL12B2 at SPring-8 in Japan.
This work was supported by National Science Council Grants NSC96-2311-B-001-035-MY3 (to C. W.) and NSC-95-2311-B-001-064 (to C.-D. H.) and grants from the Academia Sinica Taiwan, ROC.
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 2WPV) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- ER
- endoplasmic reticulum
- TPR
- tetratricopeptide repeat
- TA
- tail-anchored
- Ni2+-NTA
- Ni2+-nitrilotriacetic acid
- GET
- Golgi ER trafficking
- SGT
- small glutamine-rich TPR-containing protein.
REFERENCES
- 1.Becker J., Walter W., Yan W., Craig E. A. (1996) Mol. Cell. Biol. 16, 4378–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Andrea L. D., Regan L. (2003) Trends Biochem. Sci. 28, 655–662 [DOI] [PubMed] [Google Scholar]
- 3.Wegele H., Haslbeck M., Reinstein J., Buchner J. (2003) J. Biol. Chem. 278, 25970–25976 [DOI] [PubMed] [Google Scholar]
- 4.Hainzl O., Wegele H., Richter K., Buchner J. (2004) J. Biol. Chem. 279, 23267–23273 [DOI] [PubMed] [Google Scholar]
- 5.Young J. C., Hoogenraad N. J., Hartl F. U. (2003) Cell 112, 41–50 [DOI] [PubMed] [Google Scholar]
- 6.Liou S. T., Cheng M. Y., Wang C. (2007) Cell Stress Chaperones 12, 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kordes E., Savelyeva L., Schwab M., Rommelaere J., Jauniaux J. C., Cziepluch C. (1998) Genomics 52, 90–94 [DOI] [PubMed] [Google Scholar]
- 8.Buchanan G., Ricciardelli C., Harris J. M., Prescott J., Yu Z. C., Jia L., Butler L. M., Marshall V. R., Scher H. I., Gerald W. L., Coetzee G. A., Tilley W. D. (2007) Cancer Res. 67, 10087–10096 [DOI] [PubMed] [Google Scholar]
- 9.Winnefeld M., Grewenig A., Schnölzer M., Spring H., Knoch T. A., Gan E. C., Rommelaere J., Cziepluch C. (2006) Exp. Cell Res. 312, 2500–2514 [DOI] [PubMed] [Google Scholar]
- 10.Hillenmeyer M. E., Fung E., Wildenhain J., Pierce S. E., Hoon S., Lee W., Proctor M., St Onge R. P., Tyers M., Koller D., Altman R. B., Davis R. W., Nislow C., Giaever G. (2008) Science 320, 362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabu C., Schmid V., Schwappach B., High S. (2009) J. Cell Sci. 122, 3605–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuldiner M., Metz J., Schmid V., Denic V., Rakwalska M., Schmitt H. D., Schwappach B., Weissman J. S. (2008) Cell 134, 634–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonikas M. C., Collins S. R., Denic V., Oh E., Quan E. M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J. S., Schuldiner M. (2009) Science 323, 1693–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 16.Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. (2006) Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 17.Goldstein A. L., McCusker J. H. (1999) Yeast 15, 1541–1553 [DOI] [PubMed] [Google Scholar]
- 18.Amberg D. C., Bruke D. J., Strathern J. N. (2005) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 19.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 20.Weiss M. S. (2001) J. Appl. Crystallogr. 34, 130–135 [Google Scholar]
- 21.Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 22.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terwilliger T. C. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McRee D. E. (1999) J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 25.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 26.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 27.Liu F. H., Wu S. J., Hu S. M., Hsiao C. D., Wang C. (1999) J. Biol. Chem. 274, 34425–34432 [DOI] [PubMed] [Google Scholar]
- 28.Hu Z., Potthoff B., Hollenberg C. P., Ramezani-Rad M. (2006) J. Cell Sci. 119, 326–338 [DOI] [PubMed] [Google Scholar]
- 29.Fleischer T. C., Weaver C. M., McAfee K. J., Jennings J. L., Link A. J. (2006) Genes Dev. 20, 1294–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutta S., Tan Y. J. (2008) Biochemistry 47, 10123–10131 [DOI] [PubMed] [Google Scholar]
- 31.Yu H., Braun P., Yildirim M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., Hao T., Rual J. F., Dricot A., Vazquez A., Murray R. R., Simon C., Tardivo L., Tam S., Svrzikapa N., Fan C., de Smet A. S., Motyl A., Hudson M. E., Park J., Xin X., Cusick M. E., Moore T., Boone C., Snyder M., Roth F. P., Barabási A. L., Tavernier J., Hill D. E., Vidal M. (2008) Science 322, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabu C., Wipf P., Brodsky J. L., High S. (2008) J. Biol. Chem. 283, 27504–27513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandes J. M., Macqueen D. J., Lee H. T., Johnston I. A. (2008) Genomics 91, 315–325 [DOI] [PubMed] [Google Scholar]
- 34.Shen S., Tobery C. E., Rose M. D. (2009) Mol. Biol. Cell 20, 2438–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]