Abstract
Lipoate-protein ligase A (LplA) catalyzes the attachment of lipoic acid to lipoate-dependent enzymes by a two-step reaction: first the lipoate adenylation reaction and, second, the lipoate transfer reaction. We previously determined the crystal structure of Escherichia coli LplA in its unliganded form and a binary complex with lipoic acid (Fujiwara, K., Toma, S., Okamura-Ikeda, K., Motokawa, Y., Nakagawa, A., and Taniguchi, H. (2005) J Biol. Chem. 280, 33645–33651). Here, we report two new LplA structures, LplA·lipoyl-5′-AMP and LplA·octyl-5′-AMP·apoH-protein complexes, which represent the post-lipoate adenylation intermediate state and the pre-lipoate transfer intermediate state, respectively. These structures demonstrate three large scale conformational changes upon completion of the lipoate adenylation reaction: movements of the adenylate-binding and lipoate-binding loops to maintain the lipoyl-5′-AMP reaction intermediate and rotation of the C-terminal domain by about 180°. These changes are prerequisites for LplA to accommodate apoprotein for the second reaction. The Lys133 residue plays essential roles in both lipoate adenylation and lipoate transfer reactions. Based on structural and kinetic data, we propose a reaction mechanism driven by conformational changes.
Keywords: Enzymes/Catalysis, Enzymes/Structure, Methods/X-ray Crystallography, Organisms/Bacteria, Protein/Protein-Protein Interactions, Protein/Structure, Vitamins and Cofactors, Enzyme Catalysis, Adenylate-forming Enzyme, Lipoic Acid
Introduction
Lipoic acid is a disulfide-containing cofactor widely distributed among living organisms. It acts as a prosthetic group after covalently attaching to the acyltransferase (E2) subunit of the α-ketoacid dehydrogenase multienzyme complexes and to H-protein of the glycine cleavage system via amide linkage between the ϵ-amino group of a specific lysine residue of these proteins and the carboxyl group of lipoic acid (1–4). The lipoyllysine arm plays a central role in the reactions of complexes, shuttling the reaction intermediate and the reducing equivalents between active sites of the complexes.
The lipoate-protein ligase A (LplA)4 catalyzes the lipoate attachment reaction (5) by a two-step reaction as follows.
Lipoic acid is activated to a high energy lipoyl-5′-AMP (lipoyl-AMP) intermediate at the expense of ATP by Reaction 1 (lipoate adenylation reaction), and then the lipoyl moiety of the intermediate is transferred to the lysine residue of the apoprotein by Reaction 2 (lipoate transfer reaction).
The structure of Escherichia coli LplA in its unliganded form is composed of a large N-terminal domain (residues 1–244) and a small C-terminal domain (residues 253–337) with a lipoic acid binding site in the N-terminal domain (6). Analysis using the DALI program (7) showed that the LplA had the closest structural similarity to the central domain of the E. coli biotin-protein ligase (BirA) (8) except for LplA homologues, with a root mean square (r.m.s.) deviation of 3.5 Å for the alignment of 139 of 292 Cα atoms. BirA consists of three domains, and the central domain catalyzes the attachment of biotin to biotin-dependent enzymes by a two-step reaction similar to that of LplA. An evolutionary relationship between LplA, BirA, and class II aminoacyl-tRNA synthetase has been suggested by Wood et al. (9) on the basis of structural analysis. On the other hand, LplA from Thermoplasma acidophilum is composed of only an N-terminal domain (10, 11), and a second protein, LplB, is required for lipoyl-adenylate formation (12). Three structures of T. acidophilum LplA (the LplA in its unliganded form, the LplA·ATP complex, and the LplA·lipoyl-AMP complex) have been determined. Protein folding of the three structures and the geometry of the substrate binding site are essentially the same and are similar to that of the N-terminal domain of E. coli LplA, except for the presence of a long loop (residues 124–138) in the T. acidophilum enzyme (10).
We have also determined the structure of bovine lipoyltransferase in complex with lipoyl-AMP, which is a mammalian homologue of LplA (13). The lipoyltransferase has no ability to catalyze the lipoate adenylation reaction despite high amino acid sequence identity of more than 30% with E. coli LplA. Therefore, it uses lipoyl-AMP as a substrate and catalyzes the transfer of lipoyl moiety from the substrate to lipoate-dependent apoproteins. The structure of the lipoyltransferase consists of a large N-terminal domain and a small C-terminal domain, having endogenous lipoyl-AMP at the active site in the N-terminal domain (13). The fold of the two domains is similar to that of the respective domain of E. coli LplA in the unliganded form; however, the overall structure of the lipoyltransferase adopts a more stretched conformation than that of LplA because of rotation of the C-terminal domain by about 180° relative to the N-terminal domain. Therefore, we speculated a conformational change of E. coli LplA upon binding of the lipoyl-AMP intermediate.
The two-step reaction mechanism of LplA resembles the feature of the adenylate-forming enzyme superfamily, which is characterized by the two-step reaction mechanism, implying the formation of an acyl- or aryl-adenylate intermediate from ATP and a carboxylate substrate. The family comprises three subfamilies: firefly luciferases (14, 15), acyl- and aryl-CoA synthetases (16–19), and the adenylation domain of non-ribosomal peptide synthetases (20–22). Despite the low sequence identity between subfamilies, they show structural similarity, consisting of a large N-terminal domain and a small C-terminal domain and having an active center at the interface between the two domains. LplA, biotin-protein ligase, and aminoacyl-tRNA synthetase may be added to the adenylate-forming enzyme superfamily in the context of their two-step reaction mechanism, comprising other subfamilies. Recently, the conformational change of some superfamily enzymes along with the progress of the reaction has been shown by comparing the structure in a reaction stage with that in other reaction stages of the enzyme, including different enzymes (16–19, 23). For example, it was demonstrated that the C-terminal domain of acyl- or aryl-CoA synthetase rotates to the adenylate-forming conformation, triggered by binding of the first substrate, ATP or carboxylate, and returns to the thioester-forming conformation by binding of the third substrate, CoA (17, 19). The significance of domain rotation is the reconfiguration of a single active site for multiple catalytic steps.
To our knowledge, E. coli LplA is the only LplA whose two consecutive overall activities have been demonstrated without an aid of other proteins. However, little is known about the reaction mechanism catalyzed by the enzyme. To understand the reaction mechanism and to examine whether the reaction stage-specific domain rearrangement occurs, we determined two LplA structures: LplA·lipoyl-AMP binary complex and LplA·octyl-5′-AMP·apoH-protein ternary complex. These structures represent a post-lipoate adenylation stage and a pre-lipoate transfer stage, respectively. We found three large scale conformational changes in the E. coli LplA structure upon completion of the lipoate adenylation reaction, which enabled LplA to accommodate apoprotein for the second reaction. Lys133 directly plays key roles in both lipoate adenylation and lipoate transfer reactions. On the basis of the structural and kinetic data, we propose a reaction mechanism catalyzed by E. coli LplA coordinated with conformational changes. We also determined the structure of bovine lipoyltransferase in its unliganded form and found that rotation of the C-terminal domain did not occur upon binding lipoyl-AMP in bovine lipoyltransferase, which was different from E. coli LplA.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
We prepared E. coli wild-type and mutant LplAs (6) and E. coli wild-type and mutant apoH-proteins (24), as described previously. Mutations LplA-K133A, -N121A, and -D122A and apoH-protein-K64A (apoH-K64A) were introduced using the QuikChange kit (Stratagene) according to the manufacturer's instructions, employing a sense and antisense primer set (supplemental Table S1). Mutation LplA-H149A was introduced using the In-Fusion PCR cloning kit (Clontech) with a set of primers. Mutations were verified by DNA sequencing.
Assay of Activities
The overall reaction activity of wild-type and mutant LplAs was determined as described previously at constant concentrations of R-(+)-lipoic acid (30 μm), ATP-Mg (2 mm), and apoH-protein (4 μm) (6). Lipoate transfer activity was determined as for overall reaction activity but replacing lipoic acid and ATP-Mg with R-(+)-lipoyl-AMP (16 μm).
Lipoate adenylation activity was determined in a reaction mixture of 0.25 ml containing 4.21 nmol (0.16 mg) of LplA, 20 mm potassium phosphate, pH 7.0, 2 mm ATP, 2 mm MgCl2, and 0.03 mm R-(+)-lipoic acid. The reaction was initiated by adding lipoic acid, carried out at 0 °C for 15 s, and terminated by adding 0.04 ml of 0.5% trifluoroacetic acid. The produced lipoyl-AMP was immediately separated by high performance liquid chromatography (HPLC) on a C18 column (2.1 × 250 mm; VYDAC 218TP52) with an acetonitrile gradient of 0–80% using 0.1% trifluoroacetic acid (medium A) and 100% acetonitrile (medium B). The amount of lipoyl-AMP was calculated using chemically synthesized standard lipoyl-AMP (25).
Synthesis of Octanol-5′-AMP (Octyl-AMP)
We synthesized octyl-AMP from AMP and octanol as described previously (26). Octyl-AMP was purified by chromatography on the HiPrep DEAE FF column (GE Healthcare) as described (27) and desalted by HPLC on a C18-120T column (4.6 × 250 mm; Tosoh, Japan) with an acetonitrile gradient of 10–70% using medium A and B as described for lipoyl-AMP analysis. A mass of octyl-AMP obtained in the positive ion mode (QToF-2, Waters/Micromass) was consistent with the expected mass of 460.20 (MH+).
Crystallization and Data Collection
We prepared LplA·lipoyl-AMP binary complex by incubating LplA in a mixture of 1 mm ATP, 1 mm MgCl2, 0.15 mm R-(+)-lipoate, and 40 mm potassium phosphate buffer, pH 7.0, at 25 °C for 10 min and then purified the complex with the HiTrap desalting column (GE Healthcare) developed with 20 mm Tris-Cl, pH 7.5, 0.03 m NaCl, 1 mm dithiothreitol, and 0.01 mm p-amidinophenylmethanesulfonyl fluoride hydrochloride.
We also prepared the LplA·lipoyl-AMP·apoH-K64A ternary complex by adding a 1.3-fold molar excess of apoH-K64A to the binary complex mixture and further incubating the mixture for 20 min. The ternary complex was purified by chromatography on the Superdex 200 pg column (GE Healthcare), developing with the same medium used for the desalting column. The ternary complex was eluted at the position ahead of the LplA monomer on the column and isolated.
All crystals were generated by the hanging drop vapor diffusion method at 20 °C. Crystals of the LplA·lipoyl-AMP binary complex were obtained by mixing the binary complex solution (6 mg/ml) with an equal volume of reservoir solution of 1.6 m Li2SO4, 0.01 m MgSO4, and 0.05 m sodium cacodylate, pH 6.2. On the following day, microseeding was carried out, employing binary complex crystals those were accidentally generated from LplA·lipoyl-AMP·apoH-K64A ternary complex solution with the same reservoir solution. Crystals were flash-frozen in liquid nitrogen.
Next, we attempted to isolate the LplA·octyl-AMP·apoH-protein ternary complex by gel filtration; however, this was unsuccessful, presumably because of the lower affinity of octyl-AMP for LplA than that of lipoyl-AMP. Crystals of the ternary complex were obtained by mixing the protein solution of LplA and apoH-protein (1:1 molar ratio, 16 mg/ml total) containing 1 mm octyl-AMP directly with the reservoir solution of 16.2% polyethylene glycol 3350, 0.045 m MgSO4, 0.045 m NaCl, 1 mm NiCl2, 0.01 m Tris-Cl, pH 8.5, and 2% polyethylene glycol monomethyl ether 2000. The crystals were frozen after soaking in the same reservoir solution containing 14% (v/v) ethylene glycol.
Crystals of apoH-protein were generated by mixing the protein solution (11.8 mg/ml) with an equal volume of reservoir solution containing 2.3 m ammonium sulfate, 0.1 m sodium citrate, pH 4.9, and 3% (v/v) 2-methyl-2,4-pentanediol. The crystals were frozen after soaking in the same reservoir solution containing 22% (v/v) glycerol.
Because our previously determined structure of recombinant bovine lipoyltransferase expressed in E. coli contained endogenous lipoyl-AMP at the active site, we removed lipoyl-AMP to prepare unliganded lipoyltransferase. For this purpose, purified recombinant lipoyltransferase was incubated with a 3-fold molar excess of bovine apoH-protein in a reaction mixture containing 40 mm potassium phosphate, pH 7.8, 0.2 mm dithiothreitol, and 0.01 mm MnCl2 at 25 °C for 20 min to expend lipoyl-AMP for the lipoylation of apoH-protein, and then unliganded lipoyltransferase was isolated by chromatography on the HiPrep DEAE FF column with a linear gradient of 0–0.5 m NaCl in 20 mm potassium phosphate, pH 7.4, 0.5 mm dithiothreitol, and 0.01 mm p-amidinophenylmethanesulfonyl fluoride hydrochloride. Crystals of unliganded lipoyltransferase were generated as described for the complex with lipoyl-AMP (13), except that the protein solution was mixed with 0.7 volume of the reservoir solution.
Diffraction data sets were collected at 100 K using synchrotron radiation on beamline BL-17A at the Photon Factory (Ibaraki, Japan) for LplA·lipoyl-AMP crystals and on beamline BL44XU at SPring-8 (Hyogo, Japan) for the other crystals. Diffraction data were processed and scaled with the HKL-2000 program suite (28). Data collection statistics are summarized in Table 1.
TABLE 1.
Data collection and structural refinement statistics
| Parameters | Values |
|||
|---|---|---|---|---|
| LplA·lipoyl-AMP | LplA·octyl-AMP·apoH-protein | E. coli apoH-protein | Unliganded bovine lipoyltransferase | |
| Data collection | ||||
| Space group | P6122 | P212121 | C2 | P6122 |
| Cell dimensions | ||||
| a, b, c (Å) | 102.3, 102.3, 221.3 | 70.0, 102.0, 159.9 | 46.5, 52.0, 46.8 | 91.2, 91.2, 232.2 |
| α, β, γ (degrees) | 90, 90, 120 | 90, 90, 90 | 90, 93.1, 90 | 90, 90, 120 |
| Wavelength (Å) | 1.0000 | 1.0000 | 0.9000 | 0.9000 |
| Resolution range (Å) | 50.00–2.05 (2.12–2.05)a | 50.00–3.00 (3.11–3.00) | 30.00–1.30 (1.35–1.30) | 50.00–3.45 (3.51–3.45) |
| Rmerge (%)b | 12.2 (47.2) | 7.0 (48.4) | 5.3 (47.3) | 9.6 (45.1) |
| I/σ (I) | 38.1 (8.2) | 19.7 (2.0) | 32.5 (3.0) | 44.0 (6.5) |
| Completeness (%) | 100 (100) | 98.1 (97.0) | 97.9 (96.9) | 100 (100) |
| No. of observed reflections | 941,504 | 97,400 | 92,325 | 110,246 |
| No. of unique reflections | 44,005 (4,299) | 23,803 (2,322) | 26,800 (2,627) | 8,169 (402) |
| Redundancy | 21.4 (20.3) | 4.1 (3.8) | 3.4 (3.1) | 13.5 (14.0) |
| Wilson B value (Å2) | 27.5 | 73.1 | 18.7 | 74.3 |
| Structural refinement | ||||
| Resolution (Å) | 20.00-2.05 | 20.00-3.1 | 15.00-1.30 | 20.00-3.44 |
| No. of reflections | 41,478 | 19,375 | 25,438 | 7,452 |
| Rc/Rfreed (%) | 17.5/20.4 | 22.8/27.8 | 17.6/20.5 | 24.2/31.8 |
| No. of atoms | ||||
| Protein | 2,663 | 7,238 | 963 | 2,600 |
| Ligande | 34 | 62 | ||
| Water | 506 | 76 | ||
| Ion | 21 | |||
| B-factors (Å2) | ||||
| Protein | 16.8 | 48.5 | 17.9 | 73.0 |
| Ligand | 12.3 | 41.0 | ||
| Water | 31.1 | 29.9 | ||
| Ion | 58.4 | |||
| r.m.s. deviation | ||||
| Bond length (Å) | 0.010 | 0.003 | 0.011 | 0.015 |
| Bond angle (degrees) | 1.119 | 0.636 | 1.234 | 1.741 |
| Ramachandran plot | ||||
| Most favored (%) | 92.7 | 84.1 | 88.5 | 73.2 |
| Additional allowed (%) | 7.0 | 15.2 | 10.6 | 25.8 |
| Generously allowed (%) | 0.3 | 0.6 | 0.9 | 1.0 |
| Disallowed (%) | 0.0 | 0.1 | 0.0 | 0.0 |
a Numbers in parentheses represent statistics in the highest resolution shell.
b Rmerge = ΣΣj|〈I(h)〉 − I(h)j|/ΣΣj|〈I(h)〉, where 〈I(h)〉 is the mean intensity of symmetry-equivalent reflections.
c R = Σ‖Fo| − |Fc‖/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors for data used for refinement, respectively.
d Rfree = Σ‖Fo| − |Fc‖/Σ|Fo| for 7.5% of the data (bovine lipoyltransferase) and 5% of the data (the others) not used at any stage of structural refinement.
e Ligand, lipoyl-AMP or octyl-AMP.
Structure Solution and Refinement
The structures of all proteins were determined by the molecular replacement method with MOLREP (29) in the CCP4 suite (30) using the structures of bovine lipoyltransferase (13) (Protein Data Bank code 2E5A) for E. coli LplA and the unliganded bovine lipoyltransferase and pea H-protein (31) (Protein Data Bank code 1HPC) for E. coli apoH-protein. After initial model building using COOT (32), more than 10 rounds of manual model revision and structural refinement were performed with REFMAC5 (33) for all structures using weighted individual atomic B-factors. TLS refinement was carried out for low resolution crystals; however, the Rfree value for bovine lipoyltransferase was not reduced enough despite cumulative refinement because of the low resolution. Refinement statistics are summarized in Table 1. Clear electron densities for lipoyl-AMP and octyl-AMP were observed and could be modeled into densities. The final models were evaluated by the Ramachandran plot defined by PROCHECK (34). The preparation of figures and superposition of structures were carried out using PyMOL (DeLano Scientific LLC). The electrostatic potential on the molecular surface was calculated using APBS (35). The interface area between proteins was calculated with PISA (36). Interactions between LplA and a ligand or apoH-protein were analyzed by LIGPLOT (37).
RESULTS
Structure of LplA·Lipoyl-AMP Complex
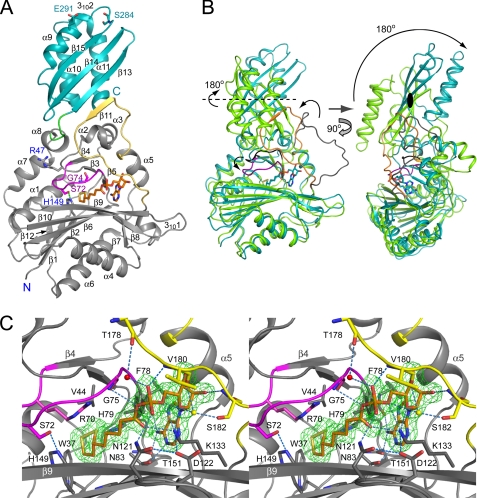
The structure of the LplA·lipoyl-AMP binary complex was solved by the molecular replacement method using the structure of bovine lipoyltransferase in complex with lipoyl-AMP as a search model because molecular replacement using the structure of E. coli unliganded LplA (Protein Data Bank code 1X2G) was unsuccessful. The reason for this became apparent when the binary structure was solved as described below. Data collection and refinement statistics are given in Table 1. Similar to the unliganded LplA, LplA in complex with lipoyl-AMP consists of a large N-terminal domain (residues 1–246) and a small C-terminal domain (residues 251–337) connected by a four-residue linker (247–250) (Fig. 1A). The respective domain has the same fold as that of the unliganded form; however, the overall structure of the complex is largely different from that of the unliganded form, adopting a rather stretched conformation (Fig. 1B).
FIGURE 1.
Structure of the LplA·lipoyl-AMP complex. A, overall structure of the LplA·lipoyl-AMP complex. LplA is shown in ribbon form. N- and C-terminal domains and the connecting loop of LplA are in gray, cyan, and green, respectively. The lipoate-binding loop (residues 69–76) and the adenylate-binding loop (residues 165–184) are highlighted in magenta and yellow, respectively. The hydrogen bond between Ser72 and His149 is shown by a dashed line in green. The secondary structural elements, N and C termini, and some residues shown in stick form are labeled. Lipoyl-AMP is shown in stick form in orange (bond) and atom colors. B, superposition of structures of the LplA·lipoyl-AMP complex (in cyan) and unliganded LplA (in green; Protein Data Bank code 1X2G, molC). Lipoate-binding and adenylate-binding loops in the complex are highlighted in magenta and orange, respectively, and those in unliganded LplA are in black and gray, respectively. A black ellipse and dashed line represent the 2-fold rotational symmetry axis of the C-terminal domain. C, stereo view of the LplA active site. The Fo − Fc omit electron density map for lipoyl-AMP contoured at 3σ is shown in green. Lipoyl-AMP and some residues of LplA are shown in stick form. The colors are the same as in A. An ordered water molecule is in red. Mg2+ introduced from solvent is in yellow. Hydrogen bonds are represented by dashed lines in blue.
In this structure, we observed strong electron densities for the lipoyl-AMP intermediate, making it possible to assign R-(+)-lipoyl-AMP in the densities (Fig. 1C). Lipoyl-AMP adopts a U-shaped conformation at the bifurcated active site sandwiched between the residue 69–76 loop (hereafter referred to as the “lipoate-binding loop”) and the large β-sheet in the N-terminal domain. The characteristic sequence motif RRXXGGG in LplAs comprises the lipoate-binding loop (Fig. 1C and supplemental Fig. S1). The AMP moiety interacts with LplA through numerous hydrogen bonds and hydrophobic interactions (Fig. 1C and supplemental Fig. S2). N1 and N6 of the adenine ring form hydrogen bonds with the side chain of Thr151 and with the side chain of Asn83 and the main chain oxygen atom of Phe78, respectively. O2* forms a hydrogen bond with the main chain nitrogen atom of Ser182. O4* and O5* form hydrogen bonds with the ϵ-amino group of Lys133. The phosphate group interacts with the side chain of Asn121, the main chain nitrogen and oxygen atoms of Val180, and the nitrogen atom of Gly75. The carbonyl oxygen atom of the lipoyl moiety forms a hydrogen bond with the ϵ-amino group of Lys133. The aliphatic part and dithiolane ring of the lipoyl moiety is varied in the hydrophobic pocket, composed of hydrophobic and aromatic residues, such as Trp37, Val44, Arg70, Val77, His79, Gly136, Ser137, His149, and Gly150, interacting through van der Waals interactions. The dithiolane ring stacks on the face of the imidazole ring of His149. The residues interacting with lipoyl-AMP are well conserved among LplAs (supplemental Fig. S1).
The structure reveals a series of three large scale conformational changes upon binding lipoyl-AMP, which is formed from lipoic acid and ATP. First, the loop from residue 165 to 184 (hereafter referred to as the “adenylate-binding loop”) stays away from the active site (in one of three molecules in the asymmetric unit) or is partially disordered (residues 176–182 in two of three molecules in the asymmetric unit) in the unliganded LplA structure (6) (Fig. 1B, left). However, in the lipoyl-AMP bound form, the loop covers the adenylate moiety and interacts with it through many hydrogen bonds and hydrophobic interactions. The loop is further stabilized by the formation of a β-sheet between β11 and β13 (Fig. 1A). Second, the lipoate-binding loop is pulled down to the lipoyl-AMP side by van der Waals interactions as mentioned above, resulting in cleavage of the hydrogen bond between the main chain nitrogen atom of Gly74 and the main chain oxygen atom of Ser284 and in the formation of a hydrogen bond between Ser72 and His149 (Fig. 1, A and C). Thus, the bound lipoyl-AMP is shielded from the solvent by the two loops, exposing only carbonyl and phosphate groups on the molecular surface (similar to the structure shown in Fig. 3A). This structure can account for the high affinity of LplA for lipoyl-AMP. The LplA·lipoyl-AMP complex can be isolated by column chromatography, as described under “Experimental Procedures.”
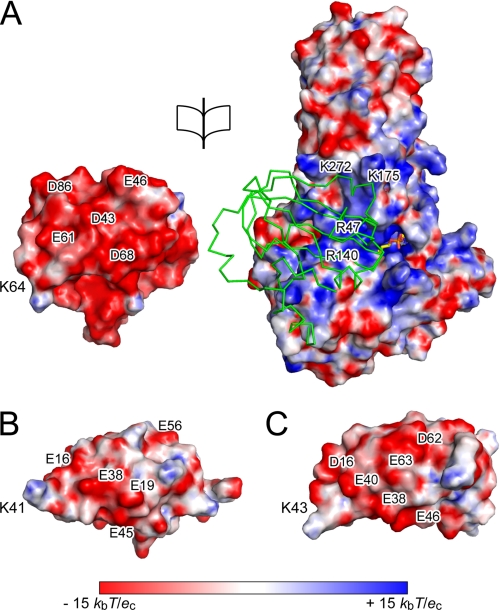
FIGURE 3.
Electrostatic potential on the molecular surface. A, open view of the electrostatic potential on the interface between LplA (right) and apoH-protein (left). The Cα trace of apoH-protein in complex with LplA is shown in green (right). The visible part of octyl-AMP is shown in stick form in yellow (bond) and atom colors. B and C, electrostatic potential on the surface of the lipoyl domain of the E. coli pyruvate dehydrogenase complex (Protein Data Bank code 1QJO) and the α-ketoglutarate dehydrogenase complex (Protein Data Bank code 1PMR) are shown, respectively. Lys41 (K41) and Lys43 (K43) are lysine residues to be lipoylated. Negatively and positively charged surfaces are shown in red and blue, respectively. Some charged residues are labeled.
The third conformational change is a rotation of the C-terminal domain by about 180° relative to the N-terminal domain as compared with that of unliganded LplA (Fig. 1B, right). This rotation is likely to be triggered by the second conformational change, which also induces rearrangement of the region from the β3 strand to α2 helix, including cleavage of the charge-charge interaction between the N-terminal domain (residue Arg47) and the C-terminal domain (residue Glu291) (Fig. 1A); as a consequence, the free C-terminal domain rotates. The conformation is further stabilized by new hydrogen bonds between Ala335 and Lys170 and between Arg337 and Thr57 and charge-charge interactions between the C-terminal group of Arg337 and Lys170 and between Arg337 and Glu61. This is the first observation of the large scale conformational changes of LplA.
Structure of E. coli ApoH-Protein
Prior to determination of the structure of the LplA·octyl-AMP·apoH-protein ternary complex, we determined the structure of E. coli apoH-protein in its free form to compare the structure with that in complex with LplA·octyl-AMP, because the structure of E. coli H-protein has not been solved. The structure was determined by the molecular replacement method using the coordinates of pea H-protein (31) (Table 1). The two H-proteins share amino acid sequence identity of 46.5% (supplemental Fig. S3A). The E. coli apoH-protein exhibits a somewhat flattened α/β-structure sandwiched with two β-sheets, a twisted six-stranded β-sheet (β1, β2, β3, β9, β8, and β5), and a small three-stranded β-sheet (β4, β6, and β7), with five α-helices surrounding the β-sheets (supplemental Fig. S3B). Lys64, which accepts lipoic acid, is located in the β6-β7 hairpin loop.
Structure of LplA·Octyl-AMP·ApoH-Protein Ternary Complex
The structure of the LplA·lipoyl-AMP binary complex represents a snapshot of the post-lipoate adenylation intermediate state. Next, we attempted to obtain a snapshot of a complex that represents the pre-lipoate transfer intermediate state, the LplA·lipoyl-AMP·apoH-protein ternary complex, using apoH-protein as an acceptor of lipoic acid. When LplA·lipoyl-AMP complex was incubated with apoH-protein, the lipoyl moiety of lipoyl-AMP was transferred to apoH-protein, forming lipoylated H-protein and unliganded LplA (data not shown), and the ternary complex was not formed because of the conformation of the unliganded LplA as described below. We therefore used octyl-AMP instead of lipoyl-AMP. Because octyl-AMP lacks the carbonyl group, which is involved in amide linkage formation with the lysine residue of apoprotein, the transfer reaction does not occur. We could generate crystals of the LplA·octyl-AMP·apoH-protein ternary complex and report here the first structure of LplA in complex with a lipoate acceptor protein.
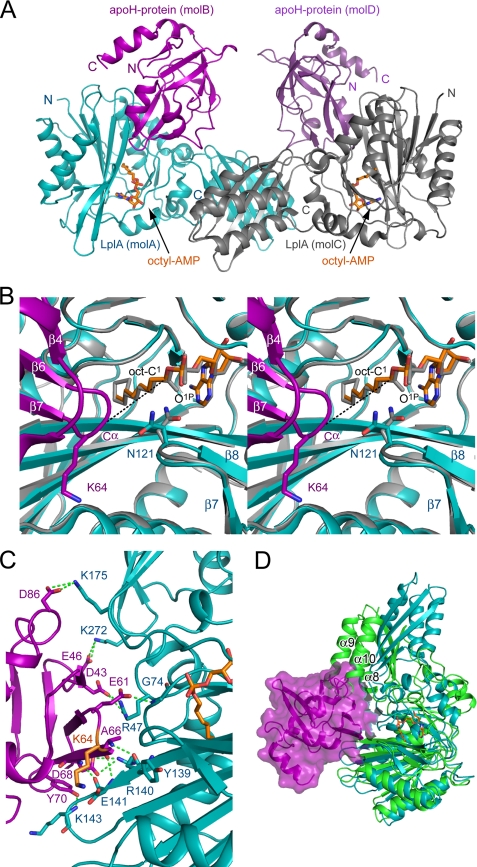
The structure of the LplA·octyl-AMP·apoH-protein ternary complex was determined by the molecular replacement method using coordinates of the LplA·lipoyl-AMP complex and apoH-protein determined above (Table 1). The structure is composed of 2:2 heterotetramer proteins in the asymmetric unit (Fig. 2A). Each pair of the heterodimer is associated via a non-crystallographic 2-fold symmetric interface between β-sheets in the C-terminal domain.
FIGURE 2.
Structure of the LplA·octyl-AMP·apoH-protein ternary complex. A, ribbon form of the 2:2 heterotetramer protein structure in the asymmetric unit. LplAs (molA and molC) are shown in cyan and gray, respectively. ApoH-proteins (molB and molD) are shown in purple and violet-purple, respectively. Octyl-AMP is shown in stick form in orange (bond) and atom colors. B, the active site of the ternary complex (molA·molB). The ternary complex (colored as in A) is superimposed onto the LplA·lipoyl-AMP binary complex (in gray). Octyl-AMP (bond, orange), lipoyl-AMP (gray), Asn121 of LplA in the ternary complex (bond, cyan) and the binary complex (bond, gray), and Lys64 (bond, purple) of apoH-protein are shown in stick form. The distance between Cα of Lys64 of apoH-protein and the C1 atom of the octyl moiety of octyl-AMP is shown by a dashed line in black (12.7 Å). C, interaction between LplA (cyan) and apoH-protein (purple). Lys64 of apoH-protein (bond, orange) and hydrogen-bonding residues are shown in stick forms. Hydrogen bonds are represented by dashed lines in green. D, superimposition of the ternary complex (molA·molB, colored as in A) onto the unliganded LplA (green). The surface structure of the apoH-protein is shown in 40% transparence. Some helices of unliganded LplA are labeled.
In individual components of the complex, structures of LplA·octyl-AMP in the ternary complex (molA and molC) have not changed at this stage by binding to apoH-protein (Fig. 2B and supplemental Fig. S4A) and are well superimposed onto that of the LplA·lipoyl-AMP binary complex determined above with r.m.s. deviations of 0.51 Å (for 301 Cα atoms of molA) and 0.47 Å (for 315 Cα atoms of molC). However, the pattern of hydrogen bond formation between LplA and octyl-AMP or LplA and apoH-protein in the ternary complex displays heterogeneity between molA·molB and molC·molD (supplemental Table S2 and Fig. S4B). As shown in Fig. 2B, the geometry of the active site of the ternary complex is quite similar to that of the binary complex except for two points: (i) the hydrogen bond between the carbonyl oxygen and Lys133 of LplA disappears in the ternary complex because the octyl-AMP compound lacks the carbonyl group, and (ii) the O1P atom becomes free because of the conformational change of the β7-β8 loop, including Asn121.
The N-terminal serine residue of apoH-protein (molB and molD) in the ternary complex has not been assigned because of the missing electron density for the residue. The Cα traces of molB and molD are well superimposed onto that of the free form with an r.m.s. deviation of 0.72 Å (for 121 Cα atoms of molB and molD); however, β6 and β7 strands in molB and molD are shifted to the contact surface with LplA (supplemental Fig. S3B, right side), showing a maximum shift by 2.7 Å at the Cα atom of Ala66 in molD from that in the free form.
In terms of the interaction between LplA and apoH-protein, they interact with contact surface areas of 748.4 Å2 (between molA and molB) and 714.6 Å2 (between molC and molD). ApoH-protein interacts with LplA, forming extensive hydrogen bonds and hydrophobic interactions. Most of the hydrogen-bonding residues in apoH-protein are acidic residues, such as Asp43, Glu46, Glu61, Asp68, and Asp86, whereas many basic amino acid residues of LplA, Arg47, Arg140, Lys143, Lys175, and Lys272, contribute to the hydrogen-bonding interactions (Fig. 2C and supplemental Table S2). The result suggests that the substrate specificity of LplA for the lipoate acceptor protein is determined by charge complementarity between LplA and the apoprotein. In E. coli, the E2 subunit of pyruvate dehydrogenase and α-ketoglutarate dehydrogenase complexes also accepts lipoic acid as a prosthetic group on the lipoyl domain. The molecular surface of apoH-protein and lipoyl domains is predominantly covered with negative charges (Fig. 3). Negatively charged patches are well complemented with the positively charged patch on the LplA surface. The glutamate residue of apoH-protein, Glu61, which is located on the N-terminal side, three residues from the lysine residue to be lipoylated and conserved among various lipoyl domains, has been demonstrated to be essential for the lipoate attachment reaction using bovine lipoyltransferase (25). The glutamate residues of the lipoyl domains (Glu38 (E38) in Fig. 3B and Glu40 (E40) in Fig. 3C) are located in a similar position relative to the lysine residue (Lys41 (K41) and Lys43 (K43), respectively) on the surface of the lipoyl domains, showing the possibility for the glutamate residue of these domains also to form a hydrogen bond with Gly74 of LplA (Fig. 2C and supplemental Table S2).
Superposition of the LplA·octyl-AMP·apoH-protein ternary complex onto the unliganded LplA shows that the N-terminal region of the α8 helix and the region from the C-terminal half of the α9 helix to the N terminus of the α10 helix of unliganded LplA cause steric clashes with apoH-protein (Fig. 2D). This striking finding indicates that rotation of the C-terminal domain is essential for apoH-protein to approach the LplA for the lipoate transfer reaction.
Alternatively, we introduced a mutation in apoH-protein at Lys64 that accepts lipoate (apoH-K64A) and prepared the LplA·lipoyl-AMP·apoH-K64A ternary complex. Because the lipoyl transfer reaction cannot happen with mutant apoH-K64A, the ternary complex is trapped in the pre-lipoyl transfer intermediate state. The crystal structure of the LplA·lipoyl-AMP·apoH-K64A ternary complex is almost the same as that of the LplA·octyl-AMP·apoH-protein complex, including LplA, lipoyl-AMP, and the contact regions of apoH-protein with LplA; however, the structure could not be determined completely because of insufficient electron density of the distal regions of apoH-protein in the complex.
Structure of Bovine Lipoyltransferase in Its Unliganded Form
The rotation of the C-terminal domain of E. coli LplA prompted us to examine whether bovine lipoyltransferase also undergoes conformational change upon binding of lipoyl-AMP. Because previously determined bovine lipoyltransferase structure contained endogenous lipoyl-AMP (13), in this study, we determined the structure of lipoyltransferase in its unliganded form after removing endogenous lipoyl-AMP. The overall structure of the unliganded lipoyltransferase is similar to that in complex with lipoyl-AMP, except for the disordering of the adenylate-binding loop (residues 169–186), the ordering of α10 helix (residues 318–329), and the shift of α11 helix (supplemental Fig. S5). This result indicates that rotation of the C-terminal domain does not occur upon binding lipoyl-AMP in bovine lipoyltransferase.
Kinetic Analyses
Next, we explored the residues essential for the lipoate transfer reaction. Some residues present around Lys64 of apoH-protein and the C1 atom of the octyl moiety of octyl-AMP were substituted with alanine residue, and activities of the overall lipoate attachment reaction, lipoate adenylation, and lipoate transfer reactions were determined. The lipoate adenylation reaction by wild-type LplA was very fast, but comparison with those of mutants was possible by incubating the reaction mixture at 0 °C for a very short time (supplemental Fig. S6 and Table 2).
TABLE 2.
Specific activities of wild-type and mutant LplAs
| LplA | Overall reaction activitya (percentage)c | Lipoate adenylation reaction activityb (percentage)c | Lipoate transfer reaction activitya (percentage)c |
|---|---|---|---|
| nmol/min/mg protein | nmol | nmol/min/mg protein | |
| Wild type | 236.2 ± 20.5 (100%) | 4.21 (100%) | 238.8 ± 5.8 (100%) |
| K133A | <0.03 (0.01%) | 0.01 (0.2%) | 6.06 ± 1.05 (2.5%) |
| N121A | 0.45 ± 0.05 (0.19%) | 0.06 (1.4%) | 57.9 ± 1.31 (24.2%) |
| D122A | 0.32 ± 0.03 (0.14%) | 0.17 (4.0%) | 9.48 ± 1.05 (4.0%) |
| S72A | 169.1 ± 3.9 (71.6%) | 2.84 (67.9%) | 156.2 ± 12.1 (65.4%) |
| H149A | 56.4 ± 2.6 (23.9%) | 4.09 (97.0%) | 119.6 ± 10.1 (50.1%) |
a Overall and lipoate transfer activities are presented as the mean of three separate experiments with S.D.
b The lipoate adenylation activity is presented as nmol of lipoyl-AMP synthesized by the incubation at 0 °C for 15 min using 4.21 nmol (0.16 mg) of LplA (supplemental Fig. S6).
c Numbers in parentheses represent percentage activities of that of wild-type LplA.
Lys133 is strictly conserved among LplAs (supplemental Fig. S1) and forms hydrogen bonds with the carbonyl group and the ribose moiety of lipoyl-AMP. The K133A mutation almost completely abolished the overall reaction activity (0.01% of that of wild type) and caused marked reduction in lipoate adenylation and lipoate transfer activities (0.2 and 2.5% of that of wild type, respectively) (Table 2). This result indicates that Lys133 plays essential roles in both lipoate adenylation and lipoate transfer reactions.
Asp122 is also conserved among LplAs but does not interact with lipoyl-AMP. Since Asp122 interacts with Asn121 in the binary complex (Fig. 1C), we examined the possibility that Asp122 is involved in abstraction of a proton from the nucleophilic ϵ-amino group of the lysine residue of apoprotein via Asn121. Abstraction of the proton is important in the lipoate transfer reaction to facilitate nucleophilic attack on the carbonyl carbon atom by the ϵ-amino group. The D122A mutation resulted in a marked reduction in the overall, lipoate adenylation, and lipoate transfer reaction activities (0.14, 4, and 4% of those of wild type, respectively); however, the mutation of N121A affected only the lipoate adenylation activity and consequently the overall reaction activity (1.4 and 0.19% of those of wild-type, respectively) but retained a significant lipoate transfer activity (24.2%). These results indicate that Asn121 is not involved in the lipoate transfer reaction; therefore, the proton transfer to Asp122 through Asn121 is impossible. Because Asp122 forms hydrogen bonds with the side chain of Lys133, it might contribute indirectly to the reactions, supporting conformations of Lys133 required for both lipoate adenylation and lipoate transfer reactions. On the other hand, Asn121 probably contributes to the formation of lipoyl-AMP by interacting with the phosphate group of ATP.
Ser72 and His149 form a hydrogen bond in lipoyl-AMP-bound LplA (Fig. 1C). Although their mutations did not cause significant reduction in three reaction activities, their Km values for ATP and lipoic acid increased to 28- and 2.3-fold, respectively, in S72A and 15- and 5.8-fold, respectively, in H149A relative to those of wild-type (data not shown); therefore, Ser72 and His149 seem to stabilize the fixation of ATP and lipoic acid at the active site. In conclusion, only Lys133, among the residues examined, is directly involved in the lipoate transfer reaction.
DISCUSSION
We have presented two structures of E. coli LplA in this study: the LplA·lipoyl-AMP binary complex, which represents the post-lipoate adenylation state, and the LplA·octyl-AMP·apoH-protein ternary complex, which mimics the pre-lipoate transfer state. LplA undergoes large scale conformational changes upon completion of the lipoate adenylation reaction, which are essential to bind apoprotein for the second reaction. To speculate upon the sequence of conformational changes, the following observations are worth considering. The structure of the LplA·lipoate binary complex determined previously was the same as that of unliganded LplA, and bound lipoate was shifted from the correct position at the active site (6). Incorrect localization seems to be a result of the open conformation of the lipoate-binding loop. The binding of ATP to the “adenylate-forming enzymes” often induces the disorder-to-order change of the adenylate-binding loop (9, 14); therefore, ATP seems to be bound first at the active site, or both substrates are bound almost simultaneously. Conformational changes of the adenylate-binding loop and the lipoate-binding loop are induced by interactions with the substrates. The formation of lipoyl-AMP strengthens the interactions between lipoyl-AMP and the two loops, resulting in cleavage of the bond between Gly74 and Ser284 and in additional rearrangements in the region from the β3 strand to α2 helix, which induce cleavage of the charge-charge interaction between Arg47 and Glu291; consequently, the free C-terminal domain rotates.
In contrast to these conformational changes of E. coli LplA, T. acidophilum LplA does not undergo any large scale conformational change upon formation of lipoyl-AMP. Its lipoate-binding loop adopts a closed conformation in the unliganded form, and the adenylate-binding loop is still disordered upon formation of lipoyl-AMP (10). Because T. acidophilun LplA consists of only the N-terminal domain, rotation of the C-terminal domain cannot occur. E. coli LplA shows functional and structural similarities with the biotin-protein ligases as mentioned above. In the ligase from E. coli (BirA), ordering of the biotin-binding loop and the adenylate-binding loop upon the binding of biotinol-5′-AMP has been reported, accompanying the slight rotation of the C-terminal domain by 3° (9). On the other hand, in the biotin-protein ligase from Pyrococcus horikoshii, ordering of the active site loop occurs upon binding of biotinyl-5′-AMP, and a small local conformational change in the C-terminal domain (residues 225–C terminus 235) is caused in the next stage by binding of the apo-biotinyl domain to make a space for the second reaction (38, 39). However, LplA in complex with lipoyl-AMP does not undergo additional large scale conformational changes upon binding of apoH-protein.
The lipoate transfer reaction is initiated by nucleophilic attack of the ϵ-amino group of Lys64 in apoH-protein on the carbonyl carbon atom (C10) of lipoyl-AMP. Although Lys64 is depicted in Fig. 2B, its side chain is probably moving freely in the complex because of a lack of electron density for the side chain. Whereas the average length of the side chain of Lys64 (Cα atom to Nζ atom) is estimated to be about 6 Å, the distance between the Cα atom of Lys64 and the C1 atom of the octyl moiety of octyl-AMP is determined to be 12.7 Å (Fig. 2B, dashed line) (molA·molB) or 11.7 Å (molC·molD). Therefore, the distance is too far for the ϵ-amino group to attack the C1 atom even if the side chain of Lys64 points toward the C1 atom, which corresponds to the C10 atom of lipoyl-AMP. Although the reason for this distance gap is not yet understood, similar observations have been reported in the structure of biotin-protein ligase (39) and acetyl-CoA synthetase (16). When true substrates (ATP, lipoic acid, and apoH-protein) are employed, some deformation should occur in the β6-β7 loop, including Lys64, to make the interatomic distance between the Nζ atom of Lys64 and the C10 atom of lipoyl-AMP within 3 Å.
Lys133 of LplA contributes to both lipoate adenylation and lipoate transfer reactions. The contribution of Lys133 to the second reaction is different from that of the essential lysine residue in firefly luciferase (40) and propionyl-CoA synthetase (27), where the lysine residues are located in the C-terminal domain and contribute only to the first adenylation reaction, bringing two substrates into a vicinal position; however, the lysine residues are about 20 Å away from the active site in the second reaction by rotation of the C-terminal domain (16, 17). Because Lys133 of E. coli LplA is involved also in the second reaction, the significance of the rotation of the C-terminal domain is quite different from that of acyl-CoA synthetase subfamily enzymes.
After completion of the lipoate transfer reaction, LplA returns to the unliganded form. Conformational changes during the returning phase are probably not required for the release of products because the structure of bovine lipoyltransferase in its unliganded form, which represents the post- product release state, has not changed from that of the lipoyl-AMP-bound form. This observation means that the products can be released without conformational change of the lipoate binding loop and rotation of the C-terminal domain and may suggest that the stretched conformation of unliganded lipoyltransferase relates to the inability of this enzyme to catalyze the lipoate adenylation reaction. The largely open active site found in E. coli unliganded LplA is necessary for the binding of ATP and lipoate.
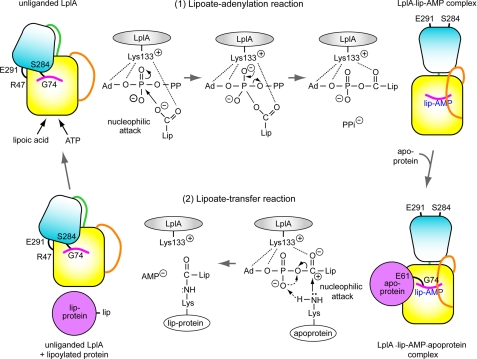
Based on the structures and kinetic data, we propose a reaction mechanism of the lipoate attachment catalyzed by E. coli LplA (Fig. 4). Unliganded LplA has the C-terminal domain adopting a bending conformation. In the first lipoate adenylation reaction, Lys133 assembles ATP and lipoic acid at the active site, forming hydrogen bonds with the carboxyl group of lipoic acid and oxygen atoms of the ribose moiety of ATP. Numerous residues surrounding the active site, shown in Fig. 1C, support this assembling. These interactions enable a nucleophilic attack by the carboxyl group of lipoic acid on the α-phosphorus atom of ATP, which leads to the pentacoordinated transition state. The P–O bond is then cleaved, producing lipoyl-AMP intermediate bound to LplA and pyrophosphate. Upon completion of the first reaction, the C-terminal domain rotates by about 180°, adopting a stretched conformation.
FIGURE 4.
Structure-based proposal for a reaction mechanism catalyzed by LplA. The mechanism of the two successive reactions is presented with schematic illustrations of the conformational change of E. coli LplA. N- and C-terminal domains and the connecting loop of LplA are shown in yellow and gradient cyan and in green, respectively. Apoprotein is shown in magenta. Adenylate and lipoate binding loops are in orange and magenta, respectively. Ad, adenosine moiety of ATP; Lip, aliphatic tail and a dithiolane ring of lipoic acid. The dotted line, arrow, and arrow with dashed line represent the hydrogen bond, electron transfer, and proton transfer, respectively. Details of the mechanism are described under “Discussion.”
In the second lipoate transfer reaction, the interaction between Lys133 and the carbonyl oxygen atom of the lipoyl moiety stimulates the polarization of the carbonyl group, which accelerates nucleophilic attack by the ϵ-amino group of the lysine residue of the apoprotein. This nucleophilic attack is further facilitated by a speculative substrate-assisted proton relay mechanism, in which a proton is accepted from the ϵ-amino group of apoprotein and donated to the departing phosphate oxygen (O3P) through the non-bridging phosphate oxygen (O1P). The C–O bond is then cleaved, forming lipoylated protein and AMP. This proton relay mechanism is similar to the substrate-assisted proton shuttling mechanism in the peptidyl transfer reaction by the ribosome, where the 2′-OH group of the A76 ribose moiety of the P-site peptidyl-tRNA picks up a proton from the attacking α-amino group of the A-site aminoacyl-tRNA, and donates a proton to the departing 3′-oxygen. The O–C bond is then cleaved, accompanying peptidyl transfer (41). The structures and kinetic data of the mutant LplAs support this reaction mechanism: (i) kinetic analyses show that Lys133 plays key roles in both lipoate adenylation and lipoate transfer reactions; (ii) in the structure of the pre-lipoate transfer state, the O1P atom becomes free (Fig. 2B); (iii) an amino acid residue that can abstract the proton from the ϵ-amino group of the lysine residue is not found at the active site.
After the release of products, lipoate-binding and adenylate-binding loops might leave the active site. Disruption of the β-sheet between the β11 and β13 strands will trigger the return to the unliganded LplA structure.
E. coli LplA is an intriguing enzyme that undergoes conformational changes coordinating with the progress of the reaction. Although various reaction stage-specific conformational changes of adenylate-forming enzymes have been reported, the rotation of the C-terminal domain of E. coli LplA is unique with respect to the rotation degree, the stage at which rotation occurs, and the significance of the rotation. The movements of lipoate- and adenylate-binding loops, but not the disorder-to-order change, are also characteristic features of the conformational changes. It is likely that the adenylate-forming enzyme undergoes the subfamily-specific domain rearrangements with the progress of the reaction.
Supplementary Material
Acknowledgments
We thank Dr. Lixy Yamada for help with the mass spectrometric analysis of octyl-AMP and ASTA Medica (Germany) for the generous gift of R-(+)-lipoate.
This work was supported by Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research 18570108 (C) and 19370041 (B) and by the National Projects on Protein Structural and Functional Analyses of the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
We dedicate this article to Dr. Yutaro Motokawa, who contributed to our research and died during preparation of this paper.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S6.
The atomic coordinates and structure factors (codes 3A7R, 3A7A, 3A7L, and 3A7U) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- LplA
- lipoate-protein ligase A
- lipoyl-AMP
- lipoyl-5′-adenylate
- octyl-AMP
- octanol-5′-adenylate
- HPLC
- high performance liquid chromatography
- r.m.s.
- root mean square.
REFERENCES
- 1.Reed L. J., Hackert M. L. (1990) J. Biol. Chem. 265, 8971–8974 [PubMed] [Google Scholar]
- 2.Perham R. N. (1991) Biochemistry 30, 8501–8512 [DOI] [PubMed] [Google Scholar]
- 3.Fujiwara K., Okamura-Ikeda K., Motokawa Y. (1986) J. Biol. Chem. 261, 8836–8841 [PubMed] [Google Scholar]
- 4.Fujiwara K., Okamura-Ikeda K., Motokawa Y. (1992) J. Biol. Chem. 267, 20011–20016 [PubMed] [Google Scholar]
- 5.Morris T. W., Reed K. E., Cronan J. E., Jr. (1994) J. Biol. Chem. 269, 16091–16100 [PubMed] [Google Scholar]
- 6.Fujiwara K., Toma S., Okamura-Ikeda K., Motokawa Y., Nakagawa A., Taniguchi H. (2005) J. Biol. Chem. 280, 33645–33651 [DOI] [PubMed] [Google Scholar]
- 7.Holm L., Sander C. (1993) J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 8.Wilson K. P., Shewchuk L. M., Brennan R. G., Otsuka A. J., Matthews B. W. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 9257–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood Z. A., Weaver L. H., Brown P. H., Beckett D., Matthews B. W. (2006) J. Mol. Biol. 357, 509–523 [DOI] [PubMed] [Google Scholar]
- 10.Kim D. J., Kim K. H., Lee H. H., Lee S. J., Ha J. Y., Yoon H. J., Suh S. W. (2005) J. Biol. Chem. 280, 38081–38089 [DOI] [PubMed] [Google Scholar]
- 11.McManus E., Luisi B. F., Perham R. N. (2006) J. Mol. Biol. 356, 625–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen Q. H., Cronan J. E. (2009) J. Biol. Chem. 284, 21317–21326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujiwara K., Hosaka H., Matsuda M., Okamura-Ikeda K., Motokawa Y., Suzuki M., Nakagawa A., Taniguchi H. (2007) J. Mol. Biol. 371, 222–234 [DOI] [PubMed] [Google Scholar]
- 14.Conti E., Franks N. P., Brick P. (1996) Structure 4, 287–298 [DOI] [PubMed] [Google Scholar]
- 15.Nakatsu T., Ichiyama S., Hiratake J., Saldanha A., Kobashi N., Sakata K., Kato H. (2006) Nature 440, 372–376 [DOI] [PubMed] [Google Scholar]
- 16.Gulick A. M., Starai V. J., Horswill A. R., Homick K. M., Escalante-Semerena J. C. (2003) Biochemistry 42, 2866–2873 [DOI] [PubMed] [Google Scholar]
- 17.Reger A. S., Wu R., Dunaway-Mariano D., Gulick A. M. (2008) Biochemistry 47, 8016–8025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hisanaga Y., Ago H., Nakagawa N., Hamada K., Ida K., Yamamoto M., Hori T., Arii Y., Sugahara M., Kuramitsu S., Yokoyama S., Miyano M. (2004) J. Biol. Chem. 279, 31717–31726 [DOI] [PubMed] [Google Scholar]
- 19.Kochan G., Pilka E. S., von Delft F., Oppermann U., Yue W. W. (2009) J. Mol. Biol. 388, 997–1008 [DOI] [PubMed] [Google Scholar]
- 20.Conti E., Stachelhaus T., Marahiel M. A., Brick P. (1997) EMBO J. 16, 4174–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yonus H., Neumann P., Zimmermann S., May J. J., Marahiel M. A., Stubbs M. T. (2008) J. Biol. Chem. 283, 32484–32491 [DOI] [PubMed] [Google Scholar]
- 22.May J. J., Kessler N., Marahiel M. A., Stubbs M. T. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12120–12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jogl G., Tong L. (2004) Biochemistry 43, 1425–1431 [DOI] [PubMed] [Google Scholar]
- 24.Okamura-Ikeda K., Ohmura Y., Fujiwara K., Motokawa Y. (1993) Eur. J. Biochem. 216, 539–548 [DOI] [PubMed] [Google Scholar]
- 25.Fujiwara K., Okamura-Ikeda K., Motokawa Y. (1996) J. Biol. Chem. 271, 12932–12936 [DOI] [PubMed] [Google Scholar]
- 26.Khorana H. G. (1959) J. Am. Chem. Soc. 81, 4657–4660 [Google Scholar]
- 27.Horswill A. R., Escalante-Semerena J. C. (2002) Biochemistry 41, 2379–2387 [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 29.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 30.Collaborative Computational Project 4 (1994) Acta Crystallogr. D 50, 760–76315299374 [Google Scholar]
- 31.Pares S., Cohen-Addad C., Sieker L. C., Neuburger M., Douce R. (1995) Acta Crystallogr. D 51, 1041–1051 [DOI] [PubMed] [Google Scholar]
- 32.Emsley P., Cowtan K. (2004) Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 34.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 35.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 37.Wallace A. C., Laskowski R. A., Thornton J. M. (1995) Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 38.Bagautdinov B., Kuroishi C., Sugahara M., Kunishima N. (2005) J. Mol. Biol. 353, 322–333 [DOI] [PubMed] [Google Scholar]
- 39.Bagautdinov B., Matsuura Y., Bagautdinova S., Kunishima N. (2008) J. Biol. Chem. 283, 14739–14750 [DOI] [PubMed] [Google Scholar]
- 40.Branchini B. R., Murtiashaw M. H., Magyar R. A., Anderson S. M. (2000) Biochemistry 39, 5433–5440 [DOI] [PubMed] [Google Scholar]
- 41.Beringer M., Rodnina M. V. (2007) Mol. Cell 26, 311–321 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.