Abstract
Podocyte structural and transcriptional phenotype plasticity characterizes glomerular injury. Transcriptional activity of WT1 (Wilm's tumor 1) is required for normal podocyte structure and is repressed by the podocyte adherens junction protein, WTIP (WT1 interacting protein). Here we show that WTIP translocated into podocyte nuclei in lipopolysaccharide (LPS)-treated mice, a model of transient nephrotic syndrome. Cultured podocytes, which stably expressed an epitope-tagged WTIP, were treated with LPS. Imaging and cellular fractionation studies demonstrated that WTIP translocated from podocyte cell contacts into nuclei within 6 h and relocalized to cell contacts within 24 h after LPS treatment. LPS-stimulated WTIP nuclear translocation required JNK activity, which assembled a multiprotein complex of the scaffolding protein JNK-interacting protein 3 and the molecular motor dynein. Intact microtubule networks and dynein activity were necessary for LPS-stimulated WTIP translocation. Podocytes expressing sh-Wtip change morphology and demonstrate altered actin assembly in cell spreading assays. Stress signaling pathways initiate WTIP nuclear translocation, and the concomitant loss of WTIP from cell contacts changes podocyte morphology and dynamic actin assembly, suggesting a mechanism that transmits changes in podocyte morphology to the nucleus.
Keywords: Cell/Differentiation, Cytoskeleton/Tubulin, Nucleus/Nuclear Transport, Signal Transduction/Protein Kinases/MAP, Tissue/Organ Systems/Kidney, LIM Domain Protein, WTIP, Chronic Kidney Disease
Introduction
Normal glomerular filtration barrier function requires wild type differentiation of the highly specialized glomerular epithelial cell, the podocyte. Podocytes express three distinct domains: cell body, primary processes, and secondary foot processes (FPs)2 that express slit diaphragms (SDs), which are highly specialized cell-cell contacts (1). FPs are highly dynamic structures that reorganize within minutes through actin cytoskeletal rearrangement (2). Glomerular diseases are characterized by a persistent simplification in podocyte domain structure with a loss of FPs, a phenotype described as FP effacement (3, 4). Mutations affecting specific podocyte SD proteins such as CD2AP, nephrin (5), podocin (6), and α-actinin-4 (7) alter actin dynamics and result in FP effacement, emphasizing the central role for the actin cytoskeleton in regulating podocyte cytoarchitecture. Alteration in podocyte morphology (8, 9) is contemporaneous with the onset of heavy proteinuria and associated with genome-wide changes in transcript abundance (10). The molecular mechanisms that regulate changes in morphology through transcription are incompletely understood. Given its unique microenvironment with exposure to hemodynamic forces and high flow of ultrafiltrate, we reasoned that podocytes express intracellular molecules that transmit morphological changes into the nucleus. Supporting this concept, nuclear relocation of the nephrin and a CD2AP-binding protein, dendrin, translates changes in SD integrity into podocyte apoptosis (11).
WTIP is a LIM domain-containing protein with a nuclear export signal that we previously characterized as a scaffold molecule that linked the SD protein complex to the actin cytoskeleton and, when its nuclear export is blocked, repressed WT1 transcriptional activity (12). Based on data in this and other studies (12–14), we hypothesized that WTIP functions as molecular messenger that choreographs changes in podocyte FP morphology and transcriptional activity and proposed the following model of WTIP function. Normally, the WTIP nuclear export signal excludes it from the nucleus, and an-as-yet-to-be-defined sequence or structural motif(s) targets WTIP to podocyte cell contacts. After podocyte injury, WTIP shuttles into the nucleus where it is retained and alters gene expression. Nuclear retention of WTIP results in its loss from podocyte cell contacts, changing actin dynamics and contributing to foot process effacement.
The experiments in this paper provide support for two aspects of this model. First, we phenotyped WTIP expression patterns in LPS-treated mice and defined the mechanisms of WTIP translocation in LPS-stimulated podocytes. LPS induces transient proteinuria in mice by destabilization of the actin cytoskeleton (15), and LPS treatment of cultured podocytes recapitulates FP actin reorganization and cell contact disassembly. We show that LPS activated JNK to assemble a dynein-based motor that ferried WTIP to the nucleus on MTs. Second, we show that knockdown of WTIP changed podocyte morphology and actin cytoskeletal dynamics.
EXPERIMENTAL PROCEDURES
Materials
Human Podocyte Lines
A conditionally immortalized human podocyte cell line has been generated by Drs. Michael Ross and Leslie Bruggeman. The cells were maintained in RPMI 1640 medium (Cambrex, Walkersville, MD) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 mg/ml streptomycin. As previously described, the podocytes were maintained in 5% CO2 at 33 °C (permissive conditions). To induce differentiation, the podocytes were plated on type I collagen-coated coverslips or tissue culture plastic and maintained at 37 °C (nonpermissive conditions) for 10 days. Phenotypic characterization is described in the supplemental “Results,” and supplemental Fig. S1A demonstrates positive immunostaining for synaptopodin and WT1 (supplemental Fig. S1A).
Podocytes Expressing TCN-inducible WTIP
Human podocytes stably expressing TCN-inducible WTIP epitope-tagged with V5 (GEC-WTIP-V5) were generated using a ViraPower T-Rex lentiviral expression system and Gateway Technology vectors, according to the manufacturer's protocol (Invitrogen). Briefly, 3 μg of each lentiviral vector, pLenti4/TO/V5-DEST carrying the WTIP gene with a C-terminal V5 epitope tag (WTIP-V5) and the pLenti6/TR containing the TetR gene, together with 9 μg of the ViraPower packaging mix, were transfected into 293FT cells using Lipofectamine 2000 reagent (Invitrogen). The DNA-Lipofectamine 2000 complexes diluted in Opti-MEM I medium (Invitrogen) were allowed to form for 20 min at room temperature before addition to 293FT cells. The cells were maintained for 24 h at 37 °C and 5% CO2 before removing the medium containing the DNA-Lipofectamine 2000 complexes and replacing with Dulbecco's modified Eagle's medium (10% fetal bovine serum, 2 mm l-glutamine, 0.1 mm nonessential amino acids, 1% penicillin/streptomycin, and 1 mm sodium pyruvate). The resulting retroviral particles were harvested by removing medium 72 h after transfection and used to generate a podocyte cell line stably expressing both transgenes. To induce WTIP-V5 expression, 1 μg/ml of TCN (Calbiochem, La Jolla, CA) was added to the cell culture medium for 24 h. Preliminary time course experiments established that WTIP-V5 expression was steady state by 24 h (supplemental Fig. S1).
LPS Treatment of Cultured Podocytes
GEC-WTIP-V5 were preincubated with 1 μg/ml TCN (24 h) and were exposed to LPS (1 μg/ml) for varying time points (30 min to 72 h). Each set of experiments was repeated in triplicate.
LPS-induced FP Effacement and Albuminuria in Mice
Immunostaining
Cultured podocytes on sterile glass coverslips coated with type I collagen were washed in Dulbecco's phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde (10 min, room temperature), washed three times for 10 min with PBS, and permeabilized with 0.2% Triton-X-100 in PBS for 5 min on ice. After blocking with 5% normal goat serum, 2% bovine serum albumin, and 0.2% fish gelatin for 1 h at 37 °C, the cells were incubated with primary antibodies: FITC-V5 1:500 (1 h, room temperature), ZO-1 1:50 (1 h, room temperature), β-catenin 1:50 (1 h, room temperature), and FITC-tubulin 1:100 (1 h, room temperature). In addition, incubation with rhodamine-phalloidin 1:300 (1 h, room temperature) was performed. Subsequently, the coverslips were washed and incubated with Texas Red donkey anti-mouse secondary antibody (1:300; 20 min, room temperature). The coverslips were washed and mounted in anti-fade, aqueous medium containing 4′,6 diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA) on standard glass slides. Confocal images were obtained with the Leica TCS SP2 confocal system (Leica Microsystems, Wetzlar, Germany), using a 63× water immersion lens. WTIP distribution patterns were scored as cytoplasmic/membrane if the intensity was greater in the cytoplasm/membrane than in the nucleus, as nuclear if the intensity was greater in the nucleus than in the cytoplasm, and as equal if no difference was observed between nucleus and cytoplasm, using a published technique of quantification (16, 17). The digital images were processed and grouped using Adobe Photoshop version 7.0.1 (Adobe Systems, San Jose, CA) and Auto-Quant software (Media Cybernetics, Bethesda, MD).
Murine kidneys were harvested and snap-frozen according to standard protocols, and 6-μm-thick sections were fixed in 4% paraformaldehyde for 5 min. For immunofluorescent labeling, sections were washed once with PBS and incubated in blocking solution (5% normal goat serum, 30 min, room temperature) followed by 0.5% SDS for 2 min. Afterward, the sections were incubated with anti-WTIP for 1 h at room temperature followed by anti-WT1 or anti-synaptopodin for 1 h at room temperature. The slides were then washed three times with PBS for 10 min followed by incubation with appropriate secondary antibodies conjugated with fluorochrome Texas Red or FITC for 1 h at room temperature to visualize the antigen-antibody complexes.
Co-localization Analysis
Confocal microscopic images were analyzed for co-localization of WTIP and nuclear marker TOPRO-3 by use of ImageJ (W. S. Rasband, National Institutes of Health, Bethesda, MD, 1997–2004). For details of co-localization analysis, see the supplemental “Materials and Methods.”
Cellular Fractionation of Cytosolic and Nuclear Compartments
The podocytes were incubated for 6 or 24 h in the presence or absence of LPS (1 μg/ml) and TCN (1 μg/ml) and collected by scraping in PBS plus protease inhibitor mixture tablet (Roche Applied Science). The cytoplasmic and nuclear fractions were prepared using NE-PER nuclear and cytoplasmic extraction reagents (Pierce), following the manufacturer's instructions. The extraction buffers follow a classical method of low to high salt for nuclear and cytoplasmic separation. To the cytoplasmic extraction the following inhibitors were added: 0.75 mm phenylmethylsulfonyl fluoride, protease inhibitor mixture tablet (Roche Applied Science), 10 mm β-glycerophosphate, and 0.2 mm sodium orthovanadate. The nuclear extract solution was supplemented with 2 mm phenylmethylsulfonyl fluoride and the above inhibitors listed for the cytoplasmic extracts. The protein concentrations were determined using the Bio-Rad DC protein assay kit. The purity of the cell fractions was determined by Western blotting with RhoGDI, a marker for cytoplasmic fraction and histone H3 and/or lamin B2 for nuclear compartments.
Immunoprecipitation and Immunoblotting
RNA Isolation and Reverse Transcription-PCR
Cell Spreading Assay
Mouse podocytes stably expressing sh-EMP or sh-Wtip, as described in the supplemental “Methods and Methods,” were trypsinized, washed, and pelleted. 1 × 105 cells in complete medium were reseeded onto six-well plates containing collagen type I-coated coverslips (5 μg/ml). The plates were incubated at 37 °C for the time periods specified in the results. The cells were fixed using 4% paraformaldehyde and stained with rhoadmine-phalloidin and nuclear dye TOPRO-3. The images were captured using a Leica SP2 confocal microscope at 40× magnification. Quantification of cell spreading by calculation of average cell diameter was assessed using Leica Quantify software.
Statistical Analysis
The data from all of the experimental groups were expressed as the means ± S.E. An unpaired Student's t test was used to compare differences between control and experimental groups. Statistical significance was defined as p < 0.05.
RESULTS
Podocyte Injury with LPS Promotes Translocation of WTIP-V5 from Cell Junctions to the Nucleus
We developed a human podocyte cell line that expressed WTIP with a V5 C-terminal epitope tag (GEC-WTIP-V5) in response to TCN treatment and focused our efforts on defining the mechanism by which LPS induced the translocation of WTIP-V5 into podocyte nuclei. An inducible expression system was developed to mitigate any effect of WTIP overexpression during the 10–14 days required for podocyte differentiation. GEC-WTIP-V5 were induced to differentiate using standard procedures and then stimulated for 24 h with TCN to induce WTIP-V5 (supplemental Fig. S1, A and B) prior to LPS treatment. A 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay was used to determine whether LPS treatment caused GEC-WTIP-V5 cytotoxicity. No difference was observed in cell viability between LPS-treated and untreated control podocytes (supplemental Fig. S1C). In untreated cells, WTIP-V5 co-localized with the adherens junction markers β-catenin and ZO-1 (supplemental Fig. S1D).
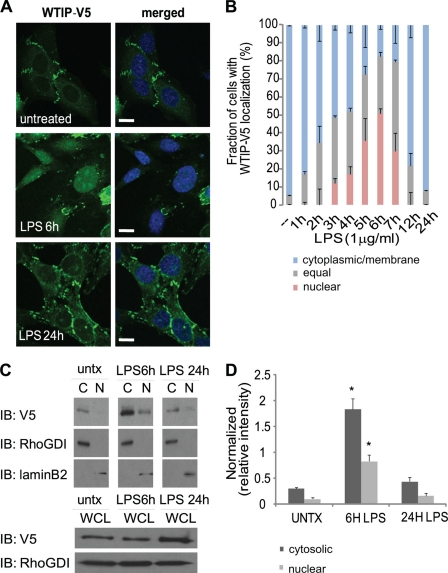
After LPS treatment, WTIP-V5 shifted from sites of cell-cell contacts (Fig. 1A, top panels) into the cytosol and nucleus, as shown by co-localization with the nuclear dye TOTO-3 (Fig. 1A, middle panels). WTIP-V5 had returned to cell-cell contacts 24 h after LPS stimulation (Fig. 1A, bottom panels). A more detailed time course (supplemental Fig. S2) demonstrated that WTIP-V5 began shifting from podocyte cell contacts as early as 1 h after LPS stimulation and localized in intracellular plaques. Intranuclear localization of WTIP-V5 was observed as early as 3 h after LPS treatment. Maximal nuclear translocation of WTIP-V5 occurred 6 h after LPS treatment (supplemental Fig. S2). Following LPS treatment, WTIP-V5 intracellular distribution patterns were quantified by scoring podocytes at each time point using published criteria (as described under “Experimental Procedures”). The percentage of cells with nuclear and/or equal (nuclear + cytoplasmic) WTIP-V5 staining is as follows: 5 ± 0.4% (base line), 48 ± 2.9% (3 h LPS), 52 ± 7.9% (4 h LPS), 73 ± 12.2% (5 h LPS), 83 ± 7.7% (6 h LPS), 82 ± 10.1% (7 h LPS), 21 ± 7.2% (12 h LPS), and 8 ± 0.3% (24 h LPS) (Fig. 1B). Therefore LPS podocyte injury induces WTIP-V5 nuclear translocation in a reversible manner that paralleled the time course of WTIP translocation to podocyte nuclei observed in vivo in LPS-treated mice (see Fig. 6C).
FIGURE 1.
Translocation of WTIP-V5 to the nucleus following LPS treatment. A, GEC-WTIP-V5 cells incubated with or without LPS (1 μg/ml, 6 and 24 h) at 37 °C were fixed and immunostained for FITC-V5 to visualize WTIP localization. The nuclei were visualized by nuclear dye TOTO-3. Scale bars, 10 μm. B, quantification of WTIP-V5 accumulation over time. At least 150 cells from three separate experiments were imaged randomly and scored for localization of WTIP-V5. The bars represent the means ± S.E. (n = 3). C, in the upper three panels, GEC-WTIP-V5 cells were fractionated into cytosolic and nuclear compartments (untreated, LPS 6 and 24 h) and analyzed by immunoblot (IB). Specificity of the fractionation was assessed by blotting for RhoGDI (cytosolic) and lamin B2 (nuclear). The lower two panels show that WTIP-V5 abundance in whole cell lysates (WCL) does not change after LPS stimulation. The RhoGDI and laminB2 immunoblots document equivalent protein loading. D, densitometric analysis of WTIP-V5 expression in cytosolic and nuclear fractions was done by normalizing WTIP-V5 to WTIP-V5 in whole cell lysates. The bars represent the means ± S.E. (n = 3). *, p < 0.05, Student's t test.
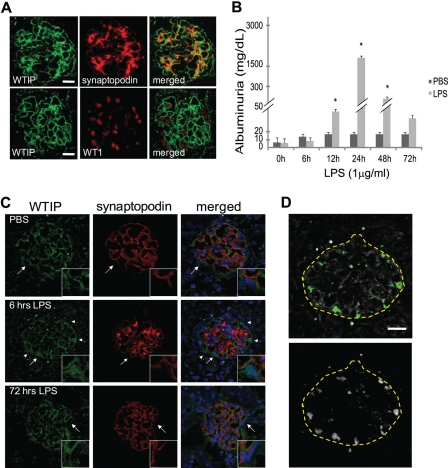
FIGURE 6.
Endogenous Wtip co-localizes with the podocyte actin binding protein synaptopodin and shifts to podocyte nuclei in LPS-injected mice. A, glomerular co-staining of kidney cross sections of Wtip and the podocyte foot process actin binding protein, synaptopodin, or podocyte nuclear marker WT1. Scale bars, 10 μm. B, albuminuria was assessed by dipstick as described under “Experimental Procedures.” The bars represent the means ± S.E. (n = 25 mice in total; n = 11, PBS-injected; n = 14, LPS-injected). *, p < 0.05, Student's t test. C, immunofluorescence staining for Wtip (green, left column), synaptopodin (red, middle column), and merge with nuclear dye TOPRO-3 (blue, right column) in sections of kidneys from mice injected with LPS or PBS. The insets represent zoomed images of glomerular areas, indicated by arrows. The arrowheads highlight regions of nuclear translocation of Wtip. D, using ImageJ (see “Experimental Procedures”), two unbiased co-localization analyses of immunofluorescence images (shown in C) demonstrate that Wtip localizes within nuclei at 6 h after LPS, whose anatomic location is consistent with WTIP-V5 translocation into podocyte nuclei. Yellow dashed lines represent outlines of the glomerulus. Co-localization analysis using the co-localization threshold plug-in displays co-localized pixels pseudocolored green (top panel). The Intensity correlation analysis was used to confirm co-localization threshold analysis, and co-localized pixels were pseudocolored white (bottom panel). Scale bar, 10 μm.
Aliquots of LPS-treated podocytes (shown in Fig. 1A) were fractionated into nuclear and cytoplasmic compartments, equalized for total protein, and probed with anti-V5 antibody. WTIP-V5 accumulated in the nuclear fraction 6 h after LPS treatment and returned to the cytosolic fraction 24 h after LPS treatment (Fig. 1C, top three panels). Immunoblotting for cytosolic protein RhoGDI and nuclear protein lamin B2 demonstrated stringent subcellular fractionation (Fig. 1C). WTIP-V5 abundance in cytosolic extracts increased 6 h after LPS treatment. This increase reflects a shift of WTIP-V5 from membrane/cytoskeletal fractions into cytosol rather than de novo synthesis, because immunoblots of the podocyte lysates used to generate the cytosolic and nuclear fractions demonstrate that LPS did not increase WTIP abundance (Fig. 1C, 6 h, lower panels). Fig. 1D quantifies WTIP translocation between podocyte compartments in three separate experiments. Taken together, both cellular fractionation and confocal images demonstrate that LPS treatment of podocytes stimulates a reversible translocation of WTIP-V5 into nuclei.
To determine whether the translocation of WTIP was a specific effect in response to LPS, we evaluated the effects of various stimuli of proteinuric glomerular disease in vivo on WTIP localization in cultured podocytes. After treatment with LPS (1 μg/ml, 6 h), puromycin aminonucleoside (100 μg/ml, 24 h), ultraviolet C (50 mJ/m2), or H2O2 (50 μm, 6 h), green fluorescent protein-WTIP translocated into nuclei (supplemental Fig. S3). These data suggest WTIP transit into podocyte nuclei is a universal response to injury.
WTIP-V5 Translocation to the Nucleus Requires JNK Activation
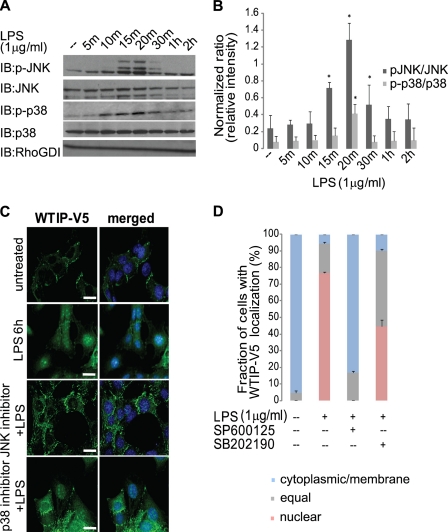
Previous reports have demonstrated that LPS stimulation activates MAPKs, in particular JNK and p38 (18). LPS rapidly activated both JNK and p38 following in cultured podocytes as assayed by immunoblotting with phosphospecific antibodies (Fig. 2A) and quantitated by densitometry (Fig. 2B). In addition, JNK activation was demonstrated in the various podocyte toxins that cause WTIP nuclear translocation by immunoblot analysis (supplemental Fig. S3B). To evaluate whether JNK and/or p38 activation is necessary for the translocation of WTIP-V5 to the nucleus, we incubated GEC-WTIP-V5 with either the JNK inhibitor SP600125 or the p38 inhibitor SB202190 for 1 h prior to LPS treatment for 6 h. WTIP-V5 localized in a plasma membrane pattern in untreated cells and shifted into podocyte nuclei after LPS treatment (Fig. 2C). WTIP-V5 remained localized predominately at the cell contacts in LPS-treated cells preincubated with the JNK inhibitor SP600125 (Fig. 2C). In contrast, WTIP-V5 translocated to the nucleus and/or cytosol after treatment with the p38 inhibitor SB202190 and LPS (Fig. 2C, bottom panels). The pattern in these cells was indistinguishable from podocytes treated with LPS alone. Quantification (Fig. 2D), as described above, demonstrated the percentage of cells with nuclear and/or equal WTIP-V5 staining: 5 ± 0.3% (base line), 94 ± 1.7% (LPS), 91 ± 9.5% (SB202190 + LPS), and 17 ± 2.4% (SP600125 + LPS). The specificity of inhibitors SP600125 and SB202190 was demonstrated by immunoblot (supplemental Fig. S4A) and densitometric analysis (supplemental Fig. S4B). Therefore, JNK, not p38, activation is necessary for the nuclear translocation of WTIP-V5 following LPS treatment.
FIGURE 2.
WTIP-V5 translocation to the nucleus requires JNK activation. A, activation of phosphorylated JNK and p38 by LPS in GEC-WTIP-V5. GEC-WTIP-V5 cells were treated with LPS (1 μg/ml), and whole cell lysates were harvested at the indicated time points (0–2 h) post-treatment. Protein was resolved by SDS-PAGE and immunoblot (IB) analysis was performed. The blots were probed with phosphospecific antibodies for p38 and JNK and then stripped and reprobed with antibodies for total JNK, total p38, or RhoGDI. B, densitometric analysis revealed that the levels of the phosphorylated p38 (p-p38) were low in unstimulated podocytes but increased significantly (p < 0.05) 20 min after LPS treatment. JNK was phosphorylated (pJNK) within 15–30 min following LPS treatment. Total JNK and p38 expression did not change in response to LPS, demonstrating that the change in JNK and p38 phosphorylation was not a result of changes in kinase abundance. RhoGDI expression demonstrates equivalent protein loading between lanes. LPS treatment in GEC-WTIP-V5 resulted in a statistically significant increase in both JNK and p38 phosphorylation (p < 0.05). Densitometric analysis was performed by normalizing phosphorylated p38 or JNK to total levels of p38 or JNK, respectively. The bars represent the means ± S.E. (n = 3). *, p < 0.05, Student's t test. C, GEC-WTIP-V5 cells of untreated control, LPS (1 μg/ml, 6 h), JNK inhibitor + LPS (SP600125, 10 μm, 1 h), or p38 inhibitor + LPS (SB202190, 10 μm, 1 h). The cells were fixed and immunostained with FITC-V5 to visualize WTIP localization. Scale bars, 10 μm. D, quantification of WTIP-V5 accumulation. At least 150 cells from three separate experiments were imaged randomly and scored for localization of WTIP-V5. The bars represent the means ± S.E. (n = 3).
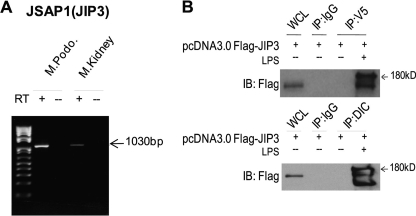
The JNK Scaffolding Protein JIP3 Linked the Dynein Motor Subunit, Dynein Intermediate Chain (DIC), and WTIP-V5 in a Multiprotein Complex
Neuronal injury induces transport of signaling molecules from cytosol and/or plasma membrane into nucleus (19, 20), and JNK scaffolding proteins, JIPs are required for optimal JNK activity. After sciatic nerve injury in the mouse, JIP3 (JSAP1) assembles into a multiprotein complex containing the motor dynein for retrograde transport from axon to cell body (21). We hypothesized that a similar mechanism for damage signaling mediated LPS-stimulated translocation of WTIP into podocyte nuclei. The JIP3 message was identified by reverse transcription-PCR in both mouse kidney and cultured podocytes (Fig. 3A). Co-precipitation experiments using GEC-WTIP-V5 transiently transfected with FLAG-tagged JIP3 showed that WTIP-V5 forms a protein complex with JIP3 following LPS injury but not in vehicle-treated controls (Fig. 3B, top panel). The observed doublet is consistent with published data demonstrating that JIP3 is phosphorylated when JNK signaling complexes are assembled and active (see “Discussion”). JIP3 and DIC co-localize in transfected CV-1 cells, suggesting JIP3 interaction with a dynein/dynactin retrograde motor system (22). In LPS-treated GEC-WTIP-V5 transiently transfected with the FLAG-JIP3 construct, WTIP-V5 co-precipitated with endogenous DIC (Fig. 3B, bottom panel). These results suggest that FLAG-JIP3/DIC/WTIP-V5 form a multiprotein complex following LPS injury in the podocyte.
FIGURE 3.
LPS injury promotes the multiprotein complex formation of JIP3/DIC/WTIP-V5. A, JIP3 transcript expression in both podocyte (M.Podo.) and kidney (M.Kidney) was demonstrated by reverse transcription (RT)-PCR analysis. B, expression of FLAG-JIP3 was confirmed by whole cell lysate of transiently transfected GEC-WTIP-V5 cells and immunoblot (IB) analysis with FLAG M2 antibody 1:7500. Immunoprecipitation experiments were performed with either V5 or DIC monoclonal antibodies (mouse IgG control) in untreated or LPS-treated (1 μg/ml) GEC-WTIP-V5 cells. WCL, whole cell lysates.
Dynein Motor Activity Is Required for WTIP-V5 Transport to the Nucleus
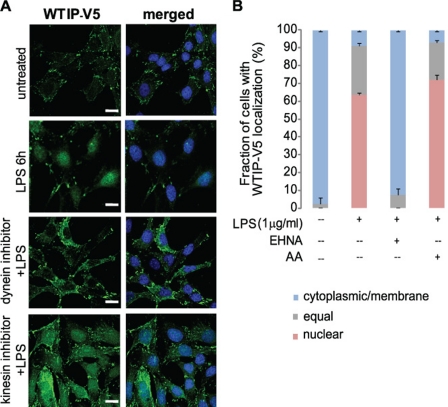
Demonstration of the FLAG-JIP3/DIC/WTIP-V5 complex suggests that the MT motor protein, dynein, transports WTIP to the nucleus. Consistent with our model, dynein motors mediate retrograde transport from the cell periphery to the minus end of the MT at the microtubule organizing center. In contrast to dyneins, kinesins mediate antegrade transport. Both dynein and kinesin MT motor protein superfamilies use ATP-derived energy to directionally transport intracellular cargoes between the cell periphery and cell body (23–25). To document that retrograde translocation of WTIP-V5 toward the nucleus required dynein, not kinesin, activity, WTIP-V5 distribution was assessed in GEC-WTIP-V5 treated with LPS and either the dynein ATPase inhibitor erythro-9-(2-hyroxy-3-nonyl) adenine (EHNA) or the kinesin ATPase inhibitor aurintricarboxylic acid (AA) (26). EHNA (1 mm, 30 min) blocked LPS-induced nuclear accumulation of WTIP-V5, whereas AA (10 μm, 30 min) had no inhibitory effect on WTIP-V5 accumulation in podocyte nucleus after LPS stimulation (Fig. 4A). WTIP-V5 localization was quantified following LPS, EHNA and LPS, or AA and LPS treatment and compared with untreated controls (Fig. 4B). The percentage of cells with nuclear and/or equal WTIP-V5 staining: 5 ± 1.2% (base line), 91 ± 2.5% (LPS), 5 ± 1.4% (EHNA + LPS), and 92 ± 4.0% (AA + LPS). Thus, dynein activity is necessary for nuclear translocation of WTIP-V5 after LPS-mediated injury.
FIGURE 4.
WTIP-V5 translocation to the nucleus requires the dynein motor complex. A, GEC-WTIP-V5 cells treated with LPS (1 μg/ml, 6 h), kinesin ATPase inhibitor + LPS (AA, 10 μm, 30 min), dynein ATPase inhibitor + LPS (EHNA, 1 mm, 30 min), or untreated controls and fixed and immunostained for FITC-V5 for WTIP-V5 localization. Scale bars, 15 μm. B, quantification of WTIP-V5 accumulation. At least 150 cells from three separate experiments were imaged randomly and scored for localization of WTIP-V5. The bars represent the means ± S.E. (n = 3).
LPS-induced Nuclear Translocation of WTIP-V5 Requires an Intact MT Network
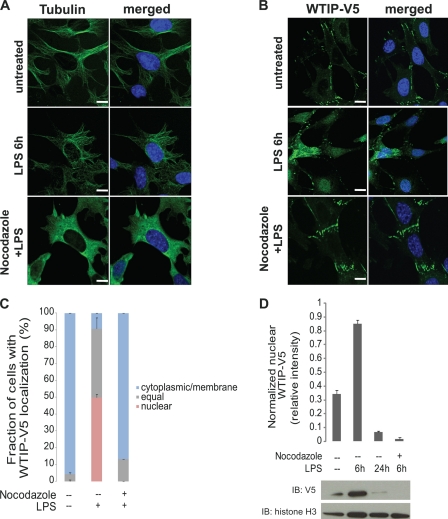
Because dynein motors walk along MTs and WTIP-V5 assembles with JIP3 and the dynein motor subunit DIC, we next examined the role of the podocyte MT network in the nuclear translocation of WTIP-V5. Comparison of MT networks in untreated and LPS-treated GEC-WTIP-V5 by immunostaining with FITC-anti-tubulin antibody showed no difference in MT networks (Fig. 5A). However, preincubation with the MT disruption agent nocodazole (10 μm, 1 h) completely disrupted the MT network (Fig. 5A). In GEC-WTIP-V5 preincubated with nocodazole followed by LPS treatment, WTIP-V5 remained localized at cell contacts, similar to untreated controls (Fig. 5B, top and bottom panels). In contrast, WTIP-V5 shifted from the sites of podocyte cell-cell contacts to the cytoplasm and nucleus after 6 h of LPS treatment when MTs remained intact (Fig. 5B, middle panels). Quantification of WTIP-V5 localization was determined in podocytes treated with LPS alone or with nocodazole and LPS and compared with vehicle-treated controls (Fig. 5C). The percentages of cells with nuclear and/or equal WTIP-V5 staining were 4 ± 1.0% (base line), 91 ± 10.5% (LPS), and 13 ± 0.1% (nocodazole + LPS). WTIP-V5 nuclear and cytoplasmic localization was slightly greater in nocodazole-pretreated, LPS-stimulated cells compared with control podocytes, suggesting that WTIP-V5 can transit to nuclei through MT-independent pathways. Furthermore, nuclear extracts following LPS treatment (6 and 24 h) with or without nocodazole were prepared and analyzed by immunoblot for WTIP-V5. After 6 h of LPS treatment, WTIP-V5 accumulated in the nucleus (Fig. 5D). In contrast, depolymerization of the MT network by nocodazole treatment prior to LPS treatment reduced WTIP-V5 accumulation in the nucleus (Fig. 5D). Histone H3, a resident nuclear protein, demonstrated loading of equivalent amounts of nuclear protein. Densitometric quantification is shown in Fig. 5D.
FIGURE 5.
MT depolymerization impairs LPS-induced nuclear translocation of WTIP-V5. A, MT networks in GEC-WTIP-V5 cells. MT networks were visualized in GEC-WTIP-V5 cells in untreated control, LPS (1 μg/ml, 6 h), or nocodazole (10 μm, 20 min) followed by LPS treatment by immunostaining for FITC-tubulin. Scale bars, 8 μm. B, GEC-WTIP-V5 cells treated with LPS (1 μg/ml, 6 h), nocodazole + LPS (10 μm, 20 min), or untreated controls and fixed and immunostained for FITC-V5. Scale bars, 10 μm. C, quantification of WTIP-V5 accumulation. At least 150 cells from three separate experiments were imaged randomly and scored for localization of WTIP-V5. The bars represent the means ± S.E. (n = 3). D, nuclear extracts of GEC-WTIP-V5 cells were prepared from untreated control, LPS (1 μg/ml, 6 h or 24 h), or nocodazole (10 μm, 20 min) followed by LPS (1 μg/ml, 6 h). Detection of histone H3, a resident nuclear protein, was used as a nuclear loading control. Densitometric analysis was performed normalizing WTIP-V5 to histone H3 expression. Nuclear extracts of GEC-WTIP-V5 cells were prepared from untreated control, LPS (1 μg/ml, 6 or 24 h), or nocodazole (10 μm, 20 min) followed by LPS (1 μg/ml, 6 h). Detection of histone H3, a resident nuclear protein, was used as a nuclear loading control. IB, immunoblot.
Endogenous Wtip Transits to Podocyte Nuclei in Vivo in a Model of LPS-induced Nephrotic Syndrome
We previously demonstrated that Wtip transcripts are expressed by podocyte precursors in developing mouse kidney (12). To determine the renal expression pattern of Wtip in post-natal kidney, we generated an affinity-purified polyclonal rabbit anti-Wtip antibody (see supplemental text and supplemental Fig. S5 for antibody characterization) and assessed Wtip subcellular localization in vivo using podocyte domain marker proteins (Fig. 6A). Wtip expression in mouse kidney localized to the podocyte FP domains, as demonstrated by its co-localization with the actin-associated protein synaptopodin (Fig. 6A, top panel). In contrast, Wtip did not co-localize with WT1, a marker of podocyte nuclei (Fig. 6B, bottom panel). The predominance of Wtip immunostaining was identified in the glomerulus, not in the tubulointerstitium, consistent with our hypothesis that Wtip expression in kidney is restricted to podocytes. To determine whether Wtip translocates to podocyte nuclei in vivo after glomerular injury, we injected LPS (1 μg/ml) intraperitoneally into 3-week-old wild type C57BL/6 mice and analyzed albuminuria and podocyte Wtip localization with the glomerulus. Control animals received PBS. LPS injection caused albuminuria within 24 h (p < 0.05), whereas albumin excretion did not change significantly from base line in mice injected with PBS (p > 0.05; Fig. 6B). As previously reported (15), LPS-induced albuminuria was transient and returned to base-line levels within 72 h after LPS injection (Fig. 6B). Immunofluorescent staining of kidney sections demonstrated that Wtip immunostaining pattern dramatically changed following LPS injection (Fig. 6C, first column) when compared with untreated and PBS-injected controls (Fig. 6, A and C). The linear staining pattern of Wtip observed in the PBS-injected control transitioned to a more diffuse cytosolic and nuclear pattern after 6 h of LPS injection (Fig. 6C, top and middle row), approximately the time we observed an increase in urinary albumin excretion in vivo (Fig. 6B). The zoomed image insets of glomeruli (Fig. 6C) stained with anti-Wtip (green channel), anti-synaptopodin (red channel), and nuclear dye TOPRO-3 (blue channel) demonstrate localization of Wtip within the nucleus 6 h after LPS injection, compared with PBS-injected controls. WTIP immunofluorescence has clearly shifted from its base-line localization with synaptopodin, into the nucleus at that time point. Given the proximity of the nucleus to synaptopodin, the nucleus is considered to be within the podocyte. WTIP has reassociated with synaptopodin by 72 h. Fig. 6D demonstrates co-localization of Fig. 6C at 6 h, using two different unbiased techniques of analysis available within the ImageJ program, intensity correlation analysis (top panel) and co-localization threshold (bottom panel). Note that both analytic methods identified identical regions of co-localized pixels whose anatomic location is consistent with podocyte nuclei. Like albuminuria, the in vivo distribution pattern of Wtip following LPS-induced injury is reversible, and the base-line staining pattern, similar to that of synaptopodin, is re-established 72 h after LPS injection (Fig. 6C, 72 h).
Wtip Affects Podocyte Spreading
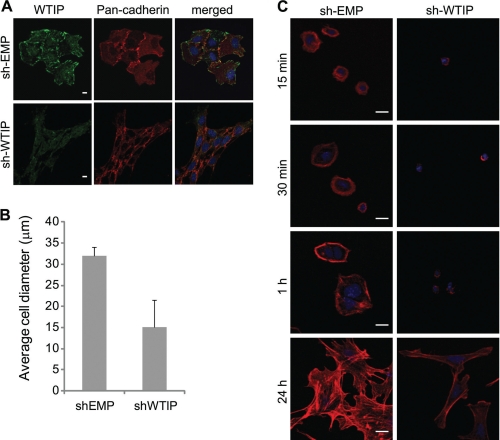
Our model proposes that WTIP retention in podocyte nuclei would deplete WTIP from podocyte cell contacts and promote foot process effacement and actin cytoskeletal rearrangement. To test this aspect of the hypothesis, mouse podocytes with stable knockdown of Wtip (sh-Wtip) and negative control (sh-EMP) were generated to identify the cellular function of Wtip. We initially observed podocyte morphology with pan-cadherin immunostaining. The sh-Wtip cells had altered morphology and junction formation compared with sh-EMP control cells (Fig. 7A). These observations led us to consider the effects of Wtip on cell adhesion and F-actin assembly as measured by cell spreading on collagen type I. The proportion of enlarged sh-EMP cells was significantly increased compared with sh-Wtip cells, as assessed by average cell diameter 24 h after reseeding (Fig. 7B). Fig. 7C demonstrates by visualization of rhodamine-phalloidin that in the early stages of spreading on collagen, sh-Wtip cells have a distinct altered morphology, as compared with the sh-EMP negative control cells. In particular, the sh-EMP cells with endogenous expression of Wtip, spread with an enhanced rate and appear larger and contain increased numbers and lengths of cell protrusions, an effect that persists for several hours after reseeding (Fig. 7C, 24 h). Wtip knockdown appears to have an effect on the degree of cell spreading over time. Therefore, Wtip appears to be necessary for proper cell spreading and actin assembly on collagen. These data, together with the localization of Wtip, would suggest a role for Wtip in the regulation of actin dynamics and/or foot process cytoarchitecture in vivo.
FIGURE 7.
Endogenous Wtip affects the rate of mouse podocyte spreading. Mouse podocytes stably expressing sh-Wtip or negative control sh-EMP vectors were plated onto collagen-coated coverslips and allowed to adhere and spread for the times indicated. A, sh-Wtip or sh-EMP cells were fixed and stained for Wtip (green), pan-cadherin (red), and nuclear dye TOPRO-3 (blue). Scale bars, 5 μm. B, histogram comparing the average cell diameter of sh-EMP and sh-Wtip cells after 24 h of reseeding onto collagen-coated coverslips. C, cells were fixed and stained with rhodamine-phalloidin and nuclear dye TOPRO-3 to visualize endogenous actin stress fibers and nuclei. Scale bars, 10 μm.
DISCUSSION
Podocyte phenotype plasticity is well documented. Electron microscopic analyses of podocyte ultrastructure before and after the resolution of nephrotic syndrome demonstrate that podocytes with FP effacement and simplified morphology re-elaborate the complex cytoarchitecture characteristic of normal podocyte differentiation state. We now define a molecular pathway for WTIP translocation to the nucleus following an environmental signal for a podocyte phenotype switch. Taken in aggregate, our current and previously published data (12, 27) support a model in which WTIP dissociates from cultured podocyte adherens junctions and translocates into the nucleus to regulate nuclear functions such as transcriptional activity. We have now demonstrated, in this model of transient nephrotic syndrome, that WTIP reversibly transits to podocyte nuclei in vivo, providing a mechanism for regulating nuclear functions in response to injury.
The podocyte differentiation state is critical for normal glomerular filtration barrier assembly and function. Glomerular disease is characterized by disassembly of the SD proteins (5, 28, 29), resulting in the reorganization of the podocyte cytoskeleton and changes in gene expression (30, 31). Therefore, scaffolding proteins like WTIP, which interact directly or indirectly with the actin cytoskeleton, become available for transport by motor proteins to regulate nuclear functions. In this way, cytoskeleton-driven changes in podocyte morphology can “signal” necessary alterations in gene expression. For WTIP, we have defined the regulatory mechanisms by which WTIP transits to and accumulates in the podocyte nucleus. However the changes after injury in the macromolecular complexes that allow the release of WTIP from podocyte cell-cell junctions and the specific nuclear functions of WTIP in vivo need to be experimentally defined. Nuclear translocation of other podocyte proteins has been shown to regulate podocyte phenotype. The CD2AP- and nephrin-binding protein dendrin senses changes in SD integrity, shifts into the nucleus, and modulates podocyte survival under pathological conditions (11). The ZHX (zinc fingers and homeoboxes) family proteins are sequestered in podocyte cytosol but migrate into the nucleus and change gene expression with the development of proteinuria (32). WT1, a target of WTIP in the nucleus (12), regulates podocyte phenotype. Interestingly another WT1 binding partner, WTX, a Wilm's tumor suppressor gene, shuttles between cytoplasm and distinct nuclear substructures implicated in transcription and modulates WT1 activity (33). Similar to WTIP, WTX is expressed in the condensing metanephric mesenchyme and in early podocyte precursors (12, 34), raising the hypothesis that WTX and WTIP may coordinately regulate WT1 activity.
In this study, we used LPS as a probe to initiate podocyte injury. LPS causes transient nephrotic syndrome when injected in sublethal doses in mice and, in vitro, changes podocyte actin cytoskeletal assembly. In podocytes, LPS activates the Toll-like receptor TLR-4, which triggers the reorganization of the kidney filtration apparatus, leading to podocyte FP effacement and proteinuria (15, 35). LPS treatment of podocytes stimulates the MAPKs JNK and p38, but WTIP transit to the nucleus only required JNK activity. MAPK activation is enhanced by scaffold proteins that efficiently assemble the necessary upstream kinases (36–38). In macrophages, JIP3 constitutively associates through its N-terminal domain with TLR-4 and enhances LPS-stimulated JNK activation as a scaffold for activating kinases (39). JIPs also link cargo molecules to motor proteins. JIP3 is expressed in cultured podocytes and mouse kidney and co-precipitated with WTIP in LPS-stimulated podocytes. JIP3 is phosphorylated in vivo (40, 41), and stimulation-dependent phosphorylation of JIP3 facilitates formation of a functional JNK signaling module. In our studies, FLAG-JIP3 co-precipitated with WTIP from LPS-stimulated cells FLAG-JIP3 as a doublet, consistent with stimulation-dependent phosphorylation of JIP3. JIP3 phosphorylation may facilitate the formation of a WTIP-based, injury surveillance complex.
Both podocytes and neurons have highly compartmentalized cytoskeletons with MT-based thick processes with branching morphology and thin actin-based projections (i.e. podocyte FPs and dendritic spines) (1, 42). Similar to neurons, information transfer from podocyte cell-cell contacts to podocyte nuclei requires long range transmission of biochemical signals. If WTIP hitchhikes on JIP3, its movement from the glomerular filtration barrier into the nucleus requires a cytoskeleton pathway and a motor protein to provide the engine. The Drosophila JIP3 orthologue, Sunday Driver, undergoes retrograde axonal transport following nerve injury (21). In injured mammalian neurons, retrograde transport of activated JNK and JIP3 to the cell body is driven by a dynactin-dynein complex (21). After LPS-induced podocyte injury, WTIP and JIP3 form a multi-protein complex with a subunit of the dynein motor protein, DIC. Dynein is composed of two identical heavy chains and several associated chains. Dynein processive motion is a property of the heavy chains. DIC regulates attachment of the motor to the appropriate cargo, making DIC a logical partner for WTIP. Using inhibitors of motor protein ATPase activity (24), dynein, but not kinesin motor activity, and intact microtubules were required for WTIP transit to the nucleus. JIP3 also has been co-localized with the dynein heavy chain (21) and with DIC in transfected CV-1 cells (22), suggesting that JIP3 links WTIP to dynein for translocation from podocyte adherens junctions to the nucleus.
In eukaryotic cells, signaling pathways in subcellular compartments must be integrated dynamically for a cell to respond appropriately to information flow from its microenvironment. Molecules that choreograph cytoskeleton-based changes in cell morphology and nuclear function have been described in other systems. WTIP is a member of the Ajuba LIM protein family (Ajuba, WTIP, LIMD1) (13, 14). Similar to WTIP, Ajuba family LIM proteins contain 3′ C-terminal LIM protein interaction domains and unique pre-LIM regions, and link cell adhesive complexes to cytoskeleton (43), and shuttle into nucleus to regulate the activity of specific transcription factors. In mammalian cells, Ajuba functions as a scaffold for assembly of macromolecular transcriptional repressor complexes at target promoters (44–46). In vivo, Xenopus Ajuba LIM proteins, including Xenopus WTIP, serve as Snail/Slug co-repressors and regulate gene expression necessary for Xenopus neural crest development (14). In a similar manner, we believe that WTIP regulates podocyte phenotype changes in response to the microenvironment of the glomerular filtration barrier. We believe that changes in actin dynamics initiate this process, a hypothesis that will be the focus of further experimentation.
Supplementary Material
Acknowledgments
We thank Dr. Roger Davis (University of Massachusetts Medical School, Worcester, MA) for the generous gift of pcDNA3.0-FLAG-JIP3 construct. We thank Dr. Greg Longmore (Washington University School of Medicine, St. Louis, MO) for the generous gift of the anti-LIMD1 antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants DK-07470, P50 DK-054178, DK-064719, and F30 DK083897.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. S1–S5.
- FP
- foot process
- LPS
- lipopolysaccharide
- MT
- microtubule
- TCN
- tetracycline
- SD
- slit diaphragm
- PBS
- phosphate-buffered saline
- FITC
- fluorescein isothiocyanate
- MAPK
- mitogen-activated protein kinase
- JNK
- c-Jun N-terminal kinase
- DIC
- dynein intermediate chain
- JIP
- JNK-interacting protein
- EHNA
- erythro-9-(2-hyroxy-3-nonyl) adenine
- AA
- aurintricarboxylic acid.
REFERENCES
- 1.Pavenstädt H., Kriz W., Kretzler M. (2003) Physiol. Rev. 83, 253–307 [DOI] [PubMed] [Google Scholar]
- 2.Cortes P., Méndez M., Riser B. L., Guérin C. J., Rodríguez-Barbero A., Hassett C., Yee J. (2000) Kidney Int. 58, 2452–2461 [DOI] [PubMed] [Google Scholar]
- 3.Smoyer W. E., Mundel P. (1998) J. Mol. Med. 76, 172–183 [DOI] [PubMed] [Google Scholar]
- 4.Somlo S., Mundel P. (2000) Nat. Genet. 24, 333–335 [DOI] [PubMed] [Google Scholar]
- 5.Patrakka J., Tryggvason K. (2007) Trends Mol. Med. 13, 396–403 [DOI] [PubMed] [Google Scholar]
- 6.Saleem M. A., Ni L., Witherden I., Tryggvason K., Ruotsalainen V., Mundel P., Mathieson P. W. (2002) Am. J. Pathol. 161, 1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michaud J. L., Lemieux L. I., Dubé M., Vanderhyden B. C., Robertson S. J., Kennedy C. R. (2003) J. Am. Soc. Nephrol. 14, 1200–1211 [DOI] [PubMed] [Google Scholar]
- 8.Kerjaschki D. (2001) J. Clin. Invest. 108, 1583–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh J., Reiser J., Mundel P. (2004) Pediatr. Nephrol. 19, 130–137 [DOI] [PubMed] [Google Scholar]
- 10.Hauser P. V., Perco P., Mühlberger I., Pippin J., Blonski M., Mayer B., Alpers C. E., Oberbauer R., Shankland S. J. (2009) Nephron Exp. Nephrol. 112, e43–e58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asanuma K., Campbell K. N., Kim K., Faul C., Mundel P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10134–10139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srichai M. B., Konieczkowski M., Padiyar A., Konieczkowski D. J., Mukherjee A., Hayden P. S., Kamat S., El-Meanawy M. A., Khan S., Mundel P., Lee S. B., Bruggeman L. A., Schelling J. R., Sedor J. R. (2004) J. Biol. Chem. 279, 14398–14408 [DOI] [PubMed] [Google Scholar]
- 13.Rico M., Mukherjee A., Konieczkowski M., Bruggeman L. A., Miller R. T., Khan S., Schelling J. R., Sedor J. R. (2005) Am. J. Physiol. Renal Physiol 289, F431–F441 [DOI] [PubMed] [Google Scholar]
- 14.Langer E. M., Feng Y., Zhaoyuan H., Rauscher F. J., 3rd, Kroll K. L., Longmore G. D. (2008) Dev. Cell 14, 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reiser J., von Gersdorff G., Loos M., Oh J., Asanuma K., Giardino L., Rastaldi M. P., Calvaresi N., Watanabe H., Schwarz K., Faul C., Kretzler M., Davidson A., Sugimoto H., Kalluri R., Sharpe A. H., Kreidberg J. A., Mundel P. (2004) J. Clin. Invest. 113, 1390–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vartiainen M. K., Guettler S., Larijani B., Treisman R. (2007) Science 316, 1749–1752 [DOI] [PubMed] [Google Scholar]
- 17.Kremer B. E., Adang L. A., Macara I. G. (2007) Cell 130, 837–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arndt P. G., Suzuki N., Avdi N. J., Malcolm K. C., Worthen G. S. (2004) J. Biol. Chem. 279, 10883–10891 [DOI] [PubMed] [Google Scholar]
- 19.Gunstream J. D., Castro G. A., Walters E. T. (1995) J. Neurosci. 15, 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindwall C., Kanje M. (2005) Mol. Cell. Neurosci. 29, 269–282 [DOI] [PubMed] [Google Scholar]
- 21.Cavalli V., Kujala P., Klumperman J., Goldstein L. S. (2005) J. Cell Biol. 168, 775–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowman A. B., Kamal A., Ritchings B. W., Philp A. V., McGrail M., Gindhart J. G., Goldstein L. S. (2000) Cell 103, 583–594 [DOI] [PubMed] [Google Scholar]
- 23.Verhey K. J. (2007) Curr. Biol. 17, R804–R806 [DOI] [PubMed] [Google Scholar]
- 24.Batut J., Howell M., Hill C. S. (2007) Dev. Cell 12, 261–274 [DOI] [PubMed] [Google Scholar]
- 25.Goldstein L. S., Yang Z. (2000) Annu. Rev. Neurosci. 23, 39–71 [DOI] [PubMed] [Google Scholar]
- 26.Lalli G., Gschmeissner S., Schiavo G. (2003) J. Cell Sci. 116, 4639–4650 [DOI] [PubMed] [Google Scholar]
- 27.Rico M., Mukherjee A., Konieczkowski M., Bruggeman L. A., Miller R. T., Khan S., Schelling J. R., Sedor J. R. (2005) Am. J. Physiol. Renal Physiol. 289, F431–F441 [DOI] [PubMed] [Google Scholar]
- 28.Benzing T. (2004) J. Am Soc. Nephrol. 15, 1382–1391 [DOI] [PubMed] [Google Scholar]
- 29.Ronco P. (2007) J. Clin. Invest. 117, 2079–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hauser P. V., Perco P., Mühlberger I., Pippin J., Blonski M., Mayer B., Alpers C. E., Oberbauer R., Shankland S. J. (2009) Nephron Exp. Nephrol. 112, e43–e58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mundel P., Shankland S. J. (2002) J. Am Soc. Nephrol. 13, 3005–3015 [DOI] [PubMed] [Google Scholar]
- 32.Liu G., Clement L. C., Kanwar Y. S., Avila-Casado C., Chugh S. S. (2006) J. Biol. Chem. 281, 39681–39692 [DOI] [PubMed] [Google Scholar]
- 33.Rivera M. N., Kim W. J., Wells J., Stone A., Burger A., Coffman E. J., Zhang J., Haber D. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8338–8343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rivera M. N., Kim W. J., Wells J., Driscoll D. R., Brannigan B. W., Han M., Kim J. C., Feinberg A. P., Gerald W. L., Vargas S. O., Chin L., Iafrate A. J., Bell D. W., Haber D. A. (2007) Science 315, 642–645 [DOI] [PubMed] [Google Scholar]
- 35.Sever S., Altintas M. M., Nankoe S. R., Möller C. C., Ko D., Wei C., Henderson J., del Re E. C., Hsing L., Erickson A., Cohen C. D., Kretzler M., Kerjaschki D., Rudensky A., Nikolic B., Reiser J. (2007) J. Clin. Invest. 117, 2095–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cowan K. J., Storey K. B. (2003) J. Exp. Biol. 206, 1107–1115 [DOI] [PubMed] [Google Scholar]
- 37.Jeon Y. J., Choi J. S., Lee J. Y., Yu K. R., Ka S. H., Cho Y., Choi E. J., Baek S. H., Seol J. H., Park D., Bang O. S., Chung C. H. (2008) Mol. Biol. Cell 19, 5116–5130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrison D. K., Davis R. J. (2003) Annu. Rev. Cell Dev. Biol. 19, 91–118 [DOI] [PubMed] [Google Scholar]
- 39.Matsuguchi T., Masuda A., Sugimoto K., Nagai Y., Yoshikai Y. (2003) EMBO J. 22, 4455–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelkar N., Gupta S., Dickens M., Davis R. J. (2000) Mol. Cell. Biol. 20, 1030–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takino T., Nakada M., Miyamori H., Watanabe Y., Sato T., Gantulga D., Yoshioka K., Yamada K. M., Sato H. (2005) J. Biol. Chem. 280, 37772–37781 [DOI] [PubMed] [Google Scholar]
- 42.Rastaldi M. P., Armelloni S., Berra S., Calvaresi N., Corbelli A., Giardino L. A., Li M., Wang G. Q., Fornasieri A., Villa A., Heikkila E., Soliymani R., Boucherot A., Cohen C. D., Kretzler M., Nitsche A., Ripamonti M., Malgaroli A., Pesaresi M., Forloni G. L., Schlöndorff D., Holthofer H., D'Amico G. (2006) FASEB J. 20, 976–978 [DOI] [PubMed] [Google Scholar]
- 43.Kadrmas J. L., Beckerle M. C. (2004) Nat. Rev. Mol. Cell Biol. 5, 920–931 [DOI] [PubMed] [Google Scholar]
- 44.Ayyanathan K., Peng H., Hou Z., Fredericks W. J., Goyal R. K., Langer E. M., Longmore G. D., Rauscher F. J., 3rd (2007) Cancer Res. 67, 9097–9106 [DOI] [PubMed] [Google Scholar]
- 45.Montoya-Durango D. E., Velu C. S., Kazanjian A., Rojas M. E., Jay C. M., Longmore G. D., Grimes H. L. (2008) J. Biol. Chem. 283, 32056–32065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanungo J., Pratt S. J., Marie H., Longmore G. D. (2000) Mol. Biol. Cell 11, 3299–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.