Abstract
To investigate the role of microRNAs in regulating oligodendrocyte (OL) differentiation and myelination, we utilized transgenic mice in which microRNA processing was disrupted in OL precursor cells (OPCs) and OLs by targeted deletion of Dicer1. We found that inhibition of OPC-OL miRNA processing disrupts normal CNS myelination, and that OPCs lacking mature miRNAs fail to differentiate normally in vitro. We identified three miRNAs, miR-219, miR-138, and miR-338, that are induced 10–100x during OL differentiation; the most strongly induced of these, miR-219, is necessary and sufficient to promote OL differentiation, and partially rescues OL differentiation defects caused by total miRNA loss. miR-219 directly represses the expression of PDGFRα, Sox6, FoxJ3, and ZFP238 proteins, all of which normally help to promote OPC proliferation. Together, these findings show that miR-219 plays a critical role in coupling differentiation to proliferation arrest in the OL lineage, enabling the rapid transition from proliferating OPCs to myelinating OLs.
INTRODUCTION
Myelination has evolved in vertebrates to electrically insulate axons to promote rapid, energy efficient action potential propagation. In the central nervous system (CNS), the production of multilamellar myelin sheaths is performed by oligodendrocytes (OLs). The location and timing of CNS myelination is regulated in large part by controlling the progression of OL differentiation, as the expression of markers of OL maturity is rapidly followed by the initiation of myelination in proximal axon tracts (Baumann and Pham-Dinh, 2001). Differentiation from a proliferating, morphologically simple OL precursor cell (OPC) into a postmitotic OL capable of myelinating axons is a complex process requiring the coordinated regulation of many genes. Remarkably, the initiation of OL differentiation is very tightly linked to the cessation of OPC proliferation (Barres and Raff, 1994; Raff et al., 1998; Temple and Raff, 1985). In fact, tight association between cessation of proliferation and initiation of differentiation is observed during the development of many cell types (Buttitta and Edgar, 2007; Kitzmann and Fernandez, 2001; Politis et al., 2008; Truong and Khavari, 2007). Simply arresting proliferation by targeted disruption of cell cycle components, however, is often insufficient to promote the program of OL differentiation (Casaccia-Bonnefil and Liu, 2003). How is it, then, that the repression of proliferative genetic programs and the induction of terminal differentiation programs are so tightly coordinated?
MicroRNAs (miRNAs) are small, non-coding RNA molecules expressed by many eukaryotic organisms. These small miRNAs are generated from longer hairpin-loop RNA sequences, which are trimmed to functional 19–21 mers by successive cleavages by the obligate miRNA processing enzymes Drosha and Dicer1, then incorporated into an RNA-induced silencing complex (RISC) (Bartel, 2004). When a RISC-loaded miRNA recognizes a complementary sequence in the 3′ untranslated region (UTR) of a protein-coding messenger RNA (mRNA), it generally either represses RNA translation, or directly promotes the degradation of the associated mRNA (Wu and Belasco, 2008). Because complementarity to the core (a.k.a. “seed”) 7–8 base pair miRNA recognition sequence within a mRNA transcript is sufficient for miRNA-induced silencing, and because miRNAs can silence mRNAs to which they are only partially complementary, single miRNA molecules are predicted to be capable of influencing the expression of hundreds of different targeted genes (Bartel, 2009).
Several labs have demonstrated the importance of miRNAs in regulating vertebrate development. Transgenic mice that do not produce mature miRNAs due to the loss of functional Dicer1 die embryonically (Bernstein et al., 2003). Using Cre-mediated recombination, lineage-specific disruptions of Dicer1 have also been performed. These have demonstrated that mature miRNAs are required for the normal differentiation and function of several different cell types, including neurons (Cuellar et al., 2008; Davis et al., 2008; Harfe et al., 2005; Lynn et al., 2007). We were therefore interested in determining whether miRNAs play a similarly important role in regulating OL differentiation.
By utilizing mice in which Cre recombinase was expressed from either the OL lineage transcription factor 2 (Olig2) or 2′,3′-cyclic nucleotide 3′ phosphodiesterase (CNP) gene locus, we were able to disrupt a floxed Dicer1 allele within cells of the OPC/OL lineage. Here we show that mice lacking Dicer1 function in their OPCs and OLs display a shivering phenotype, characteristic of classic CNS dysmyelination defects (Baumann and Pham-Dinh, 2001). Normal development of CNS myelination is disrupted in these animals at both the histological and ultrastructural levels. Furthermore, OPCs isolated from these animals are unable to differentiate normally in vitro. To determine which miRNAs may be critical to OL differentiation, we investigated the comparative expression of miRNAs in OPCs and OLs. We found that one of the most highly OL-specific miRNAs, miR-219, was specifically induced by mitogen withdrawal, was both necessary and sufficient to promote the program of OL differentiation, and was also able to partially rescue the differentiation defect caused by the loss of miRNA production in OPCs. To investigate the mechanisms by which miR-219 promotes OL differentiation, we confirmed that miR-219 directly represses the expression of four of its predicted targets, platelet-derived growth factor receptor alpha (PDGFRα), SRY-box containing gene 6 (Sox6), forkhead box J3 (FoxJ3), and zinc finger protein 238 (ZFP238), all of which are involved in promoting OPC proliferation and inhibiting OL differentiation (Barres et al., 1994a; Besnard et al., 1987; Stolt et al., 2006). These data provide evidence that the induction of miRNA expression, and miR-219 in particular, upon the withdrawal of mitogens serves to link the initiation of OL gene expression with the rapid inhibition of OPC proliferation, thereby promoting rapid and coordinated OL differentiation and subsequent myelin formation.
RESULTS
Loss of Dicer1 function in OPCs and OLs leads to a delay in CNS myelination
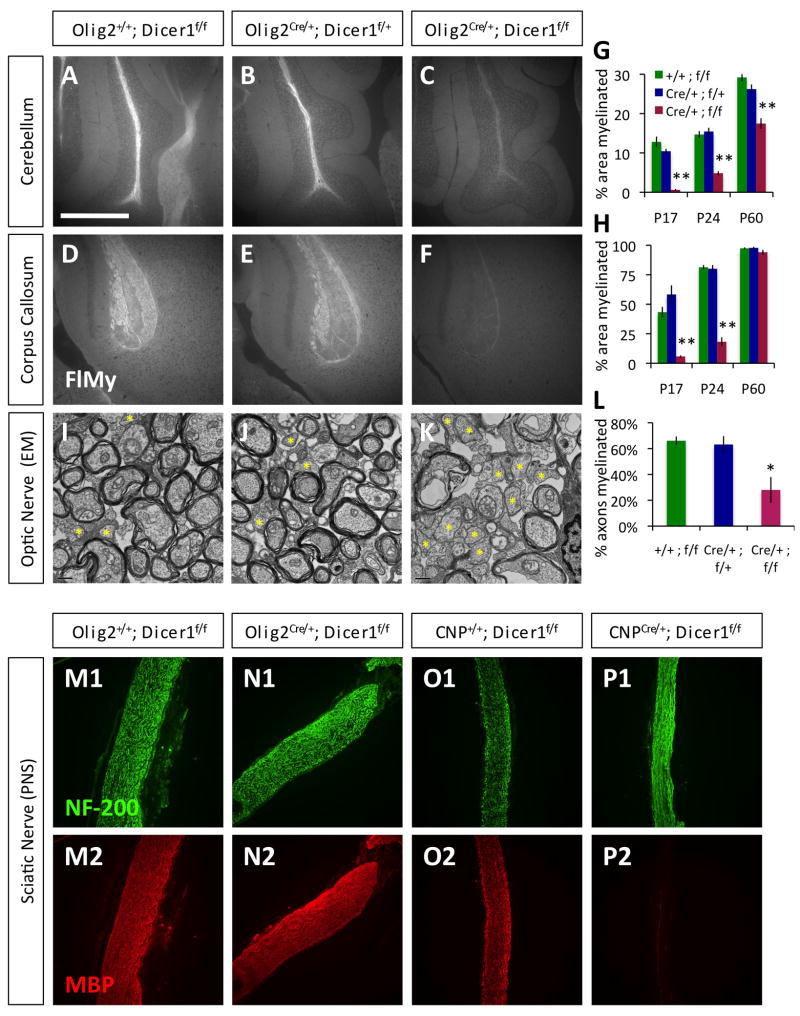
To investigate the ability of miRNAs to regulate CNS myelination, we used mice in which a floxed Dicer1 exon can be conditionally deleted by Cre recombinase to generate non-functional Dicer1 protein (Harfe et al., 2005). We crossed mice containing the floxed Dicer1 exon with mice expressing Cre recombinase from either the Olig2 or the CNP gene locus (Lappe-Siefke et al., 2003; Schuller et al., 2008); Olig2 is normally highly expressed by OPCs, whereas CNP is weakly expressed by OPCs but strongly induced very early during OL differentiation (Baumann and Pham-Dinh, 2001; Zhou et al., 2000). Both of these lines have been used previously to delete floxed gene expression in the OPC/OL lineage (Emery et al., 2009; Lappe-Siefke et al., 2003). We observed that mutant Olig2Cre/+ Dicer1flox/flox or CNPCre/+ Dicer1flox/flox mice developed a notable tremor from postnatal day 9–10 (P9–10) (Supp. movies M1–2), reminiscent of previously reported CNS myelin deficits (Baumann and Pham-Dinh, 2001). This phenotype was pronounced and reproducible when compared to littermate control genotype mice, yet appeared to lessen with age in older Olig2Cre/+ Dicer1flox/flox mice (Fig. S1), and by P60 these mutant mice were indistinguishable from littermate controls (data not shown). Interestingly, in addition to the shivering phenotype seen early in development, CNPCre/+ Dicer1flox/flox mice developed increasing functional deficits consistent with peripheral neuropathy at older ages, and generally did not live past 4 weeks. Knowing that CNP, but not Olig2, is expressed in PNS-myelinating Schwann cells, we stained peripheral nerves of mutant Olig2Cre/+ Dicer1flox/flox and CNPCre/+ Dicer1flox/flox mice for expression of myelin basic protein (MBP), a typical marker of mature myelin (Baumann and Pham-Dinh, 2001). We found that peripheral myelination was severely disrupted specifically in CNPCre/+ Dicer1flox/flox mice (Fig. 1M–P), and concluded that this likely accounted for the differences observed in motor function and lethality between older Olig2Cre/+ Dicer1flox/flox and CNPCre/+ Dicer1flox/flox mice.
Figure 1. Disruption of Dicer1 in myelinating cells disrupts normal myelin formation.
A–F. Visualization of compact CNS myelin by FluoroMyelin (FlMy) staining in the cerebellar arms (A–C) and splenium regions of the corpus callosum (D–F) from similar depth sagital brain sections from P24 littermate control Olig2+/+;Dicer1f/f (A,D), Olig2Cre/+;Dicer1f/+ (B,E), and mutant Olig2Cre/+;Dicer1f/f (C,F) mice. Scale bar = 500μm. G–H. Percentage area myelinated, as determined by FlMy staining, in the cerebellar arms (G) and corpora callosa (H) of mutant Olig2Cre/+;Dicer1f/f and littermate control mice at P17–18, P23–24, and P60. I–K. EM of optic nerve cross sections collected from P24 littermate control Olig2+/+;Dicer1f/f (I), Olig2Cre/+;Dicer1f/+ (J), and mutant Olig2Cre/+;Dicer1f/f (K) mice. Yellow asterisks = example unmyelinated axons. Scale bar = 0.5μm. L. Percentages of axons myelinated in the optic nerves of P23–24 mutant Olig2Cre/+;Dicer1f/f and littermate control mice. M–P. Sciatic nerve longitudinal sections from P23 littermate control Olig2+/+;Dicer1f/f (M) and mutant Olig2Cre/+;Dicer1f/f (N) mice, P22 littermate control CNP+/+;Dicer1f/f (O) and mutant CNPCre/+;Dicer1f/f (P) mice stained for axons (NF-200, M1–P1) and myelin (MBP, M2–P2). Error bars ±S.E.M., n=3–5 animals/condition. * p < 0.05, ** p < 0.001 post-hoc all pairwise SNK tests, mutant compared to either control condition. See also Fig. S1–S2, Movie S1–S2.
We next investigated whether the shivering phenotype observed in both Olig2Cre/+ Dicer1flox/flox and CNPCre/+ Dicer1flox/flox mice resulted from a disruption of normal CNS myelination. When we stained sagital brain sections with FluoroMyelin, a stain for compact myelin, we found that mature myelin formation in both the cerebellum and corpus callosum was significantly reduced through P24 in mutant Olig2Cre/+ Dicer1flox/flox and CNPCre/+ Dicer1flox/flox mice relative to littermate controls (Fig. 1A–H, S2A–R). Consistent with observed behavioral recovery in Olig2Cre/+ Dicer1flox/flox mice, at older ages (P60) these mutant mice had levels of compact CNS myelination that approached control levels (due to strain lethality, we were unable to assess myelination levels in older CNPCre/+ Dicer1flox/flox mice). We also observed reduced, patchy myelination early in development in both Olig2Cre/+ Dicer1flox/flox and CNPCre/+ Dicer1flox/flox mutant mice when measured by MBP expression (Fig. S2S-D′). Interestingly, we found that MBP expression levels in mutant mice appeared closer to control littermate levels by P24 (Fig. S2A′-D′), despite the fact that compact myelin levels were still low (Fig. 1, S2A–R). Therefore, to further investigate the extent of CNS myelination in older mutant mice, we examined the optic nerves of mutant P23–24 Olig2Cre/+ Dicer1flox/flox mice by electron microscopy (EM). We found that optic nerves from Olig2Cre/+ Dicer1flox/flox mice contained approximately 50% less myelinated axons than littermate controls (Fig. 1I–L). However, we did not observe any gross morphological defects in the myelin or paranodes present in the Olig2Cre/+ Dicer1flox/flox mice relative to controls (data not shown). By P60, mutant Olig2Cre/+ Dicer1flox/flox mice and littermate controls had similar numbers of myelinated axons (data not shown). Cumulatively, these data suggest that myelin generation is significantly delayed in Olig2Cre/+ Dicer1flox/flox mice, but that, eventually, normal myelination is achieved in these animals.
Do OLs that lack functional Dicer1, though delayed, eventually form normal myelin sheaths, or is the myelin observed in older Olig2Cre/+ Dicer1flox/flox and CNPCre/+ Dicer1flox/flox mice formed by OLs that have failed to correctly excise the floxed Dicer1 allele? To address this question, we compared Dicer1 expression levels between mutant P23 Olig2Cre/+ Dicer1flox/flox/CNPCre/+ Dicer1flox/flox and control genotype littermate OLs. By qRT-PCR, we found that Dicer1 expression was approximately 10-fold reduced in acutely purified mutant Olig2Cre/+ Dicer1flox/flox OLs relative to littermate control OLs, despite the fact that MBP expression levels were only slightly reduced in the mutant sample (Fig. S4A); similar results were observed with CNPCre/+ Dicer1flox/flox OLs (data not shown). Interestingly, when we repeated these experiments with P45 mice, we found that the levels of functional Dicer1 transcripts were significantly increased in the pool of mutant Olig2Cre/+ Dicer1flox/flox OLs, up to 50% of control levels, indicating that at this later age as many as half of the Olig2Cre/+ Dicer1flox/flox OLs may be expressing functional Dicer1. These data indicate that Olig2Cre/+ Dicer1flox/flox OLs that fail to disrupt functional Dicer1 expression may significantly contribute to the recovery of compact myelin formation in older mutant animals, and that the Olig2Cre/+ Dicer1flox/flox OLs lacking functional Dicer1 expression, which account for the vast majority of OLs in younger mutant animals, are likely severely delayed or disrupted in their ability to form mature myelin sheaths.
Finally, to determine whether axons were disrupted in the mutant mice, we stained Olig2Cre/+ Dicer1flox/flox and CNPCre/+ Dicer1flox/flox brains for neurofilament expression (NF-200), and found that axons appeared normal in areas of reduced myelination (Fig. S2E′-G′). These data indicate that the reductions in myelination we have observed in postnatal mice are likely not secondary to axonal loss in the CNS.
Loss of Dicer1 function in OPCs delays OL differentiation in vivo
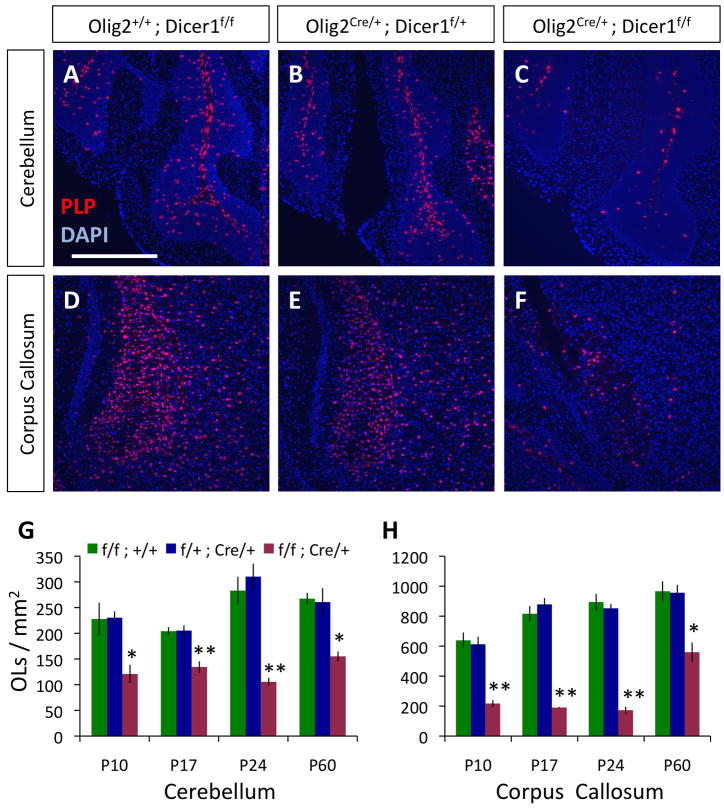
Because both Olig2Cre/+ Dicer1flox/flox and CNPCre/+ Dicer1flox/flox mice exhibited behavioral defects and delayed CNS myelin formation, we hypothesized that these deficits resulted directly from a disruption of normal OL differentiation. To test this hypothesis, we began by quantifying mature OLs in mutant Olig2Cre/+ Dicer1flox/flox and littermate control mice. We performed fluorescent in situs against proteolipid protein (PLP), a gene expressed in mature OLs, and found that OL numbers in mutant Olig2Cre/+ Dicer1flox/flox mice were significantly reduced at various postmitotic ages, and were still lower than control levels at P60 (Fig. 2, S3A–L). Reduced OL numbers were not the result of a failure to generate OL precursors, as OPC numbers in mutant Olig2Cre/+ Dicer1flox/flox mice were similar to or greater than numbers observed in control animals (Fig. S2M). These data likely indicate that loss of Dicer1 function in the OPCs of these mutant mice delays or disrupts the normal program of OL differentiation.
Figure 2. Mature OL numbers are reduced in mice lacking Dicer1 function in OPCs and OLs.
A–F. In situs for PLP expression (red) to visualize mature OLs in the cerebellar arms (A–C) and corpus callosum (D–F) in P24 littermate control Olig2+/+;Dicer1f/f (A,D), Olig2Cre/+;Dicer1f/+ (B,E), and mutant Olig2Cre/+;Dicer1f/f (C,F) mice. Scale bar = 500μm. G–H. Quantification of mature OLs/mm2 in the cerebellar arms (G) and corpora callosa (H) of mutant Olig2Cre/+;Dicer1f/f and littermate control mice at P10, 17, 24, and 60. Error bars ±S.E.M., n=3 animals/condition. * p < 0.01, ** p < 0.001 post-hoc Holm-Sidak/SNK tests, mutant compared to either control condition. See also Fig. S3.
Surprisingly, when we performed similar experiments with mutant CNPCre/+ Dicer1flox/flox mice, we detected no significant reductions in mature OL numbers, even at P9, when OL maturation is just commencing in the brain (Fig. S3N–U). As CNP expression is induced just as OPCs begin to differentiate into OLs (Baumann and Pham-Dinh, 2001), it is possible that Dicer1 is still functional at the earliest stages of OL differentiation in the CNPCre/+ Dicer1flox/flox OPCs, but that Dicer1 is lost soon after the initiation of differentiation, and this is sufficient to disrupt subsequent maturation and/or myelin sheath formation in these mutant OLs.
OPCs lacking functional Dicer1 fail to differentiate normally in vitro
Is disruption of normal CNS myelination and OL differentiation a cell autonomous effect resulting from the loss of Dicer1 function within OPCs and OLs in mutant Olig2Cre/+ Dicer1flox/flox and CNPCre/+ Dicer1flox/flox mice? To address this question, we purified immature OPCs from the brains of P7 mutant CNPCre/+ Dicer1flox/flox and control littermate mice, and analyzed their ability to completely differentiate in vitro. Purified OPCs can carry out the normal program of OL differentiation in vitro when cultured in differentiation-promoting conditions, such as withdrawal of proliferation-inducing mitogens platelet-derived growth factor alpha (PDGF) and neurotrophin 3 (NTF3), and presentation of thyroid hormone (T3) (Dugas et al., 2006). In vitro studies therefore allowed us to directly determine the effects of intrinsic Dicer1 disruption on OL differentiation, removed from the influence of interacting heterologous cell types in the CNS.
We first confirmed that OPCs purified from CNPCre/+ Dicer1flox/flox mice had indeed lost their expression of the functional Dicer1 gene. When purified OPCs were induced to differentiate in media lacking mitogens for 4 days in vitro (DIV), we found that Dicer1 expression was almost completely eliminated in mutant CNPCre/+ Dicer1flox/flox OLs relative to control genotype OLs (Fig. S4B). Similar results were obtained with mutant Olig2Cre/+ Dicer1flox/flox OLs (data not shown).
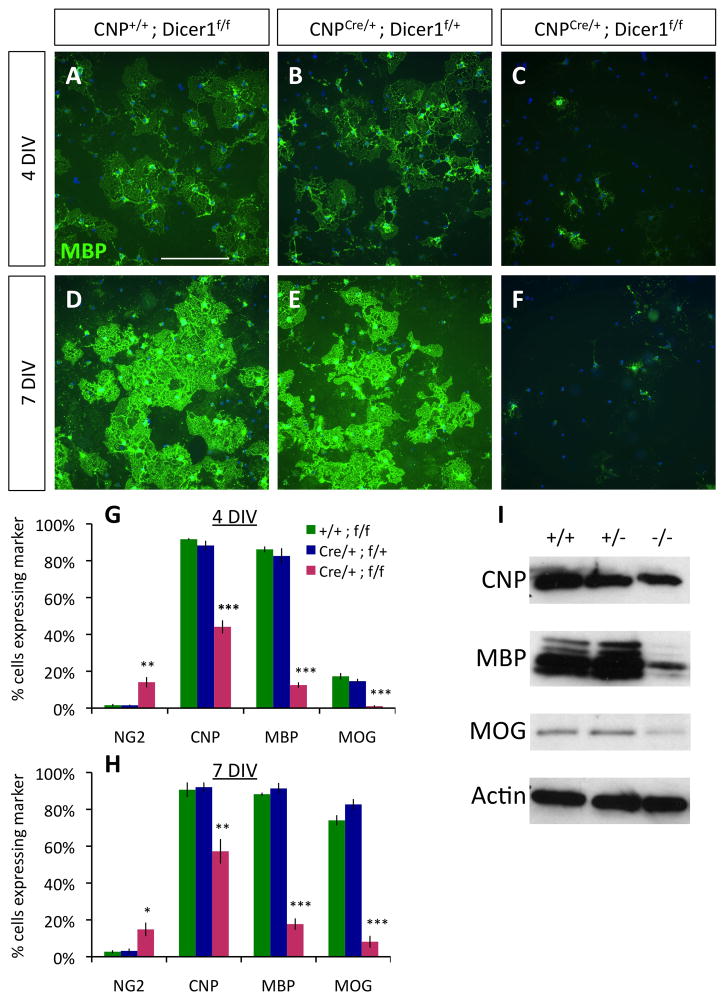
Next, we examined whether OPCs lacking Dicer1 function could differentiate normally in vitro. When mutant CNPCre/+ Dicer1flox/flox OPCs were cultured for 4 or 7 DIV in mitogen-free media, the proportion of OPCs able to differentiate into normal, MBP-expressing OLs was strongly decreased (Fig. 3A–F). When we quantified the proportion of healthy cells expressing markers of early (CNP), intermediate (MBP), or late (myelin oligodendrocyte glycoprotein: MOG) stage OL differentiation (Baumann and Pham-Dinh, 2001; Dugas et al., 2006), we found that a significant portion of mutant OPCs initiated differentiation (being CNP+), but were severely delayed in expressing markers of later OL differentiation (MBP, MOG) (Fig. 3G–H). Conversely, a small but significant proportion of mutant OPCs continued to express chondroitin sulfate proteoglycan 4 (NG2), a marker of undifferentiated OPCs, whereas expression of NG2 was nearly completely abolished by 4 DIV in control genotype OPCs. Comparable reductions in OL protein expression were observed at 4 DIV in mutant CNPCre/+ Dicer1flox/flox OPC cultures when assayed by western blotting (Fig. 3I). Similar reductions in OL differentiation were noted when comparing cultured Olig2Cre/+ Dicer1flox/flox OPCs to control genotype OPCs (data not shown). These data show that impairment of mature miRNA production strongly retards complete OL differentiation in a cell autonomous manner.
Figure 3. Purified OPCs lacking Dicer1 fail to differentiate normally.
A–F. OPCs purified from P7 control CNP+/+;Dicer1f/f (A,D), CNPCre/+;Dicer1f/+ (B,E), and mutant CNPCre/+;Dicer1f/f mice (C,F), cultured for 4 DIV (A–C) or 7 DIV (D–F) in −PDGF−T3 media, then stained for MBP expression (green); nuclei stained by DAPI (blue). Scale bar = 200μm. G–H. Percentages of cultured cells expressing the indicated markers (NG2, CNP, MBP, or MOG) at 4 DIV (G) or 7 DIV (H). Error bars ±S.E.M., n=3 samples/condition. * p < 0.05, ** p < 0.005, *** p < 0.001 post-hoc all pairwise SNK tests, mutant compared to either control condition. I. Western blots showing reduced expression of CNP, MBP, and MOG in mutant CNPCre/+;Dicer1f/f (−/−) OLs relative to control CNP+/+;Dicer1f/f (+/+) and CNPCre/+;Dicer1f/+ (+/−) OLs; protein from purified OPCs cultured 4 DIV in −PDGF−T3 media. Actin blot shown is stripped and re-probed MBP blot. See also Fig. S4.
Analysis of miRNAs induced during OL differentiation
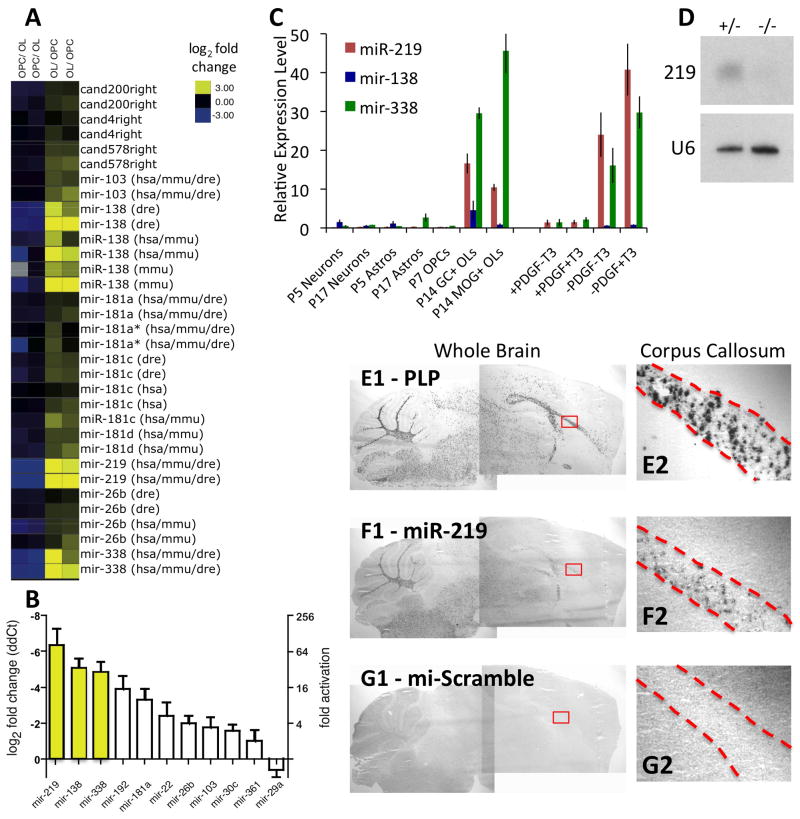
The defects observed in vivo and in vitro as a result of the loss of Dicer1 function in OPCs and OLs indicate that mature miRNA production is crucial to normal OL differentiation and CNS myelination. Which specific miRNAs regulate these processes? To investigate this question, we used miRNA microarrays identify miRNAs regulated during OL differentiation. Several miRNAs found to be differentially expressed are depicted in Fig. 4A, and the top statistically ranked candidates are listed in Table S1. Interestingly, most miRNAs that showed a significant change were expressed at a higher level in OLs; very few miRNAs were strongly repressed as OPCs differentiated into OLs. qRT-PCR performed on independently-obtained OPC and OL samples confirmed these results (Fig. 4B), and indicated that the top three ranked candidate miRNAs, miR-219, miR-138, and miR-338, were induced 100-, 30-, and 30-fold, respectively, in differentiating OLs. As further independent confirmation, the strong induction of these miRNAs during OL differentiation has also recently been reported by Lau et al. (2008).
Figure 4. miR-219, -138, and -338 are most highly expressed by OLs in the CNS and are induced specifically by mitogen withdrawal.
A. miRNA expression was compared between OPCs and OLs. Averaged results from 4x two-color microarrays are shown. “Cand” = candidate miRNAs at the time of microarray printing. Hsa, mmu, and dre = human, mouse, and zebrafish annotated miRNAs. B. qRT-PCR to determine relative OL/OPC expression level of individual miRNAs. Error bars ±S.E.M.; n=3 samples, 4x independent qRT-PCR runs per sample. C. qRT-PCR to determine expression levels of miR-219, -138, and -338 in acutely purified P5 or P17 neurons and astrocytes, P7 OPCs, P14 GC+ MOG− immature OLs, and P14 MOG+ mature OLs (left), or in OPCs cultured for 7 DIV in indicated media: ±PDGF±T3 (right). Error bars ±S.E.M., n=2–8 samples. D. Northern blot showing mature miR-219 expression in acutely purified P23 Olig2Cre/+;Dicer1f/+ OLs (+/−), which is strongly reduced in Olig2Cre/+;Dicer1f/f OLs (−/−). Blots were stripped and re-probed for U6 snRNA as loading control. E–G. In situ hybridizations showing expression of PLP (E) and miR-219 (F) in sagital sections of a wt P17 mouse brain, compared to short LNA negative control probe (G). Boxed regions in E1–G1 shown at higher magnification in E2–G2, corpus callosum outlined by dashed lines. See also Fig. S5, Table S1.
Having identified three miRNAs that are strongly induced during OL differentiation, we next wanted to investigate whether expression of these candidate miRNAs is specific to cells of the OL lineage within the CNS. Previous reports had already indicated that expression of these three miRNAs in studied vertebrates and invertebrates is highest in the CNS (Kim et al., 2004; Landgraf et al., 2007; Ruby et al., 2007; Wienholds et al., 2005). To investigate the identity of the neural cell types that express these miRNAs, we utilized previously established protocols in our lab (Cahoy et al., 2008) to acutely isolate pure populations of OPCs, immature GC+ MOG− OLs, mature MOG+ OLs, and also young (P5) and older (P17) astrocytes and neurons. qRT-PCR analyses of miRNA expression in these samples indicated that expression of all three candidate miRNAs is highest in acutely purified OLs relative to other CNS cell types (although miR-138 expression is weaker than miR-219 and -338), and also confirmed that all three miRNAs are induced in OLs relative to OPCs in vivo as well as in vitro (Fig. 4C). We also investigated the most highly expressed miRNA, miR-219, by northern blotting. We found that purified OLs expressed detectable levels of fully processed miR-219, and that this expression was strongly reduced in mutant Olig2Cre/+ Dicer1flox/flox OLs (Fig. 4D). To further investigate the in vivo expression of these miRNAs, we performed in situ hybridizations on wt P17 brain sections with LNA probes for all three miRNAs. We found that miR-219 is expressed specifically and robustly in the white matter areas of the brain, in a pattern similar to PLP expression (Fig. 4E–G). miR-138 was also expressed in the white matter, but expression was weaker than miR-219, and was also detected in other areas of the brain (Fig. S5A–F). Expression of these miRNAs persists in mature OLs, as consistent expression patterns were observed at P24 (data not shown) and P60 (Fig. S5G–O). In contrast, we detected miR-338 expression only at later developmental ages (Fig. S5H,K,N). In vivo expression of miR-219 and miR-338 in OLs of the murine CNS was also independently confirmed by X. Zhao and Q. Lu (personal communication).
Several groups have previously established that OL differentiation can be stimulated by two distinct pathways. OPCs cultured in the absence of proliferation-promoting mitogens immediately leave the cell cycle and initiate differentiation (Barres et al., 1993; Raff et al., 1983). Alternatively, in the presence of mitogens T3 can trigger OPCs to differentiate (Barres et al., 1994a; Temple and Raff, 1986). To investigate whether miR-219, miR-138, and miR-338 expression is induced equally by these two distinct differentiation-promoting pathways, we analyzed miRNA expression in OPCs cultured either in the presence of T3 (plus saturating amounts of mitogens) for 7 DIV, or in the absence of mitogens for 4 DIV. All three miRNAs were reproducibly induced by mitogen withdrawal, but not by T3 (Fig. 4C). These data indicate that miR-219, miR-138, and miR-338 are primarily induced in response to mitogen-withdrawal in OPCs.
miR-219 and miR-138 promote OL differentiation
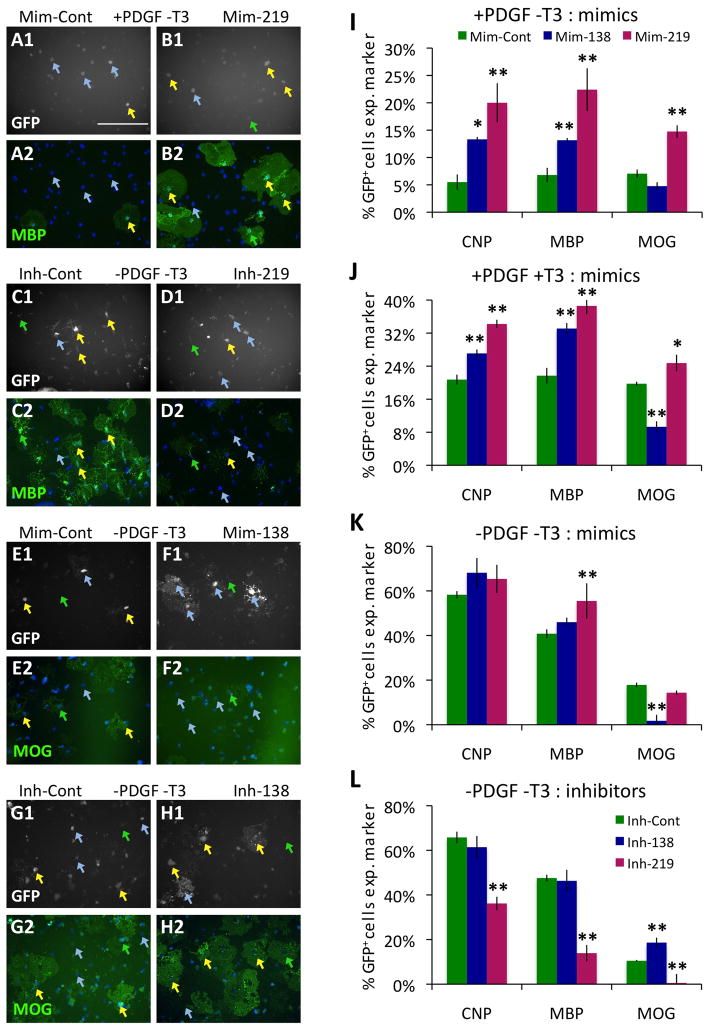
Do these three highly OL-induced miRNAs play a functional role in regulating OL differentiation? To find out, we transfected purified OPCs with nucleotide reagents homologous to the mature miRNAs (mimics), and cultured transfected cells in media that should maintain OPCs in an immature, proliferating state (+PDGF−T3) for 7 DIV. Introduction of miR-219, the most strongly induced miRNA, into OPCs doubled the number of cells that differentiated in proliferative conditions (Fig. 5A–B, S6A–D). OPCs induced to differentiate by miR-219 mimic expressed markers of both early and late OL differentiation (Fig. 5I), whereas OPCs induced to differentiate by miR-138 mimic expressed markers of the earlier stages of OL differentiation (CNP, MBP), but not later differentiation (MOG). When OPCs were induced to differentiate by mitogen withdrawal, transfecting cells with miR-219 and miR-138 mimics had very little ability to further increase OL differentiation (Fig. 5K). This result is consistent with the previous finding that expression of these miRNAs is induced endogenously in OPCs by mitogen withdrawal. In contrast, addition of miR-219 or miR-138 mimics to OPCs cultured in mitogens and induced to differentiate by T3 greatly increases the rate of OL differentiation (Fig. 5J). Consistent with the findings in +PDGF−T3 media, miR-219 is the stronger inducer of OL differentiation in the combined presence of mitogens and T3 (+PDGF+T3), and only miR-219 increases expression of the late differentiation marker MOG. Interestingly, we found that miR-138 significantly repressed expression of the late differentiation marker MOG during differentiation promoted by either mitogen withdrawal (−PDGF−T3) or T3 (+PDGF+T3) (Fig. 5E–F, J–K). These data indicate that both miR-219 and miR-138 are sufficient to induce OL differentiation, but only miR-219 induced both the early and late stages of OL differentiation. In contrast, miR-138 appears to have the interesting role of promoting the early phase of OL differentiation (CNP+, MBP+) while delaying the later phase of OL differentiation (MOG+).
Figure 5. miR-219 and -138 regulate normal OL differentiation.
A–H. Transfected OPCs identified by GFP expression (white, A1–H1) that express indicated markers (green, MBP: A2–D2; MOG: E2–H2) are indicated by yellow arrows; transfected GFP+ cells negative for marker expression are indicated by blue arrows; untransfected GFP− cells positive for marker expression are indicated by green arrows. Nuclei marked by DAPI (blue, A2–H2). Scale bar = 200 μm. A–B. OPCs transfected with control mimic (A) or miR-219 mimic (B) cultured for 7 DIV in +PDGF−T3 media. C–D. OPCs transfected with control inhibitor (C) or miR-219 inhibitor (D) cultured for 3 DIV in −PDGF−T3 media. E–F. OPCs transfected with control mimic (E) or miR-138 mimic (F) cultured for 4 DIV in −PDGF−T3 media. G–H. OPCs transfected with control inhibitor (G) or miR-138 inhibitor (H) cultured for 4 DIV in −PDGF−T3 media. I–L. Percentages of transfected, GFP+ cells expressing the indicated markers (CNP, MBP, or MOG). OPCs transfected with control, miR-138, and miR-219 mimics (I–K) or control, miR-138, and miR-219 inhibitors (L). Transfected cells cultured for 7 DIV in +PDGF−T3 (I) or +PDGF+T3 (J) media, or 3–4 DIV in −PDGF−T3 (K–L) media. Error bars ±S.E.M., n=3 (I–J); n=9 (K), n=6 (L). * p < 0.05, ** p < 0.01 post-hoc Holm-Sidak test vs. control. See also Fig. S6.
To determine whether these miRNAs are necessary for mitogen-withdrawal mediated OL differentiation, we transfected OPCs with competitive inhibitors of miR-219 and miR-138 and cultured the cells for 4 DIV in −PDGF−T3 media. Inhibition of miR-219 strongly repressed OL differentiation (Fig. 5C–D, L, S6E–H). In contrast, inhibition of miR-138 did not repress the early stages OL differentiation, but did promote expression of the late-phase gene MOG (Fig. 5G–H,L). Consistent with the finding that miR-219 and miR-138 are not induced by T3 presentation, T3-promoted OL differentiation is not significantly affected by inhibition of either of these miRNAs (Fig. S6I). In contrast to the findings for miR-219 and miR-138, neither mimics nor inhibitors of the third OL-induced miRNA, miR-338, had any significant effect on OL myelin gene expression tested in any of these assays (data not shown). However, miR-338 may still play an important role in regulating CNS myelination, as disruption of miR-338 function has been shown to promote OL differentiation in other vertebrate model system assays (X. Zhao and Q. Lu, personal communication).
Cumulatively, these data indicate that miR-219 is both necessary and sufficient to promote rapid OL differentiation promoted by mitogen-withdrawal, whereas miR-138 is necessary and sufficient to delay the later stage of OL differentiation. We have previously noted that OL differentiation appears to progress in at least two distinct stages of gene expression (Dugas et al., 2006). As only immature, MOG− OLs are able to robustly initiate the process of axon wrapping that leads to compact myelin formation (Watkins et al., 2008), it could be that miR-138 plays a role in elongating the immature phase of OL differentiation, thereby extending the window of time in which a terminally differentiating OL can select and correctly myelinate nearby axons.
miR-219 partially rescues OL differentiation in Dicer1− OLs
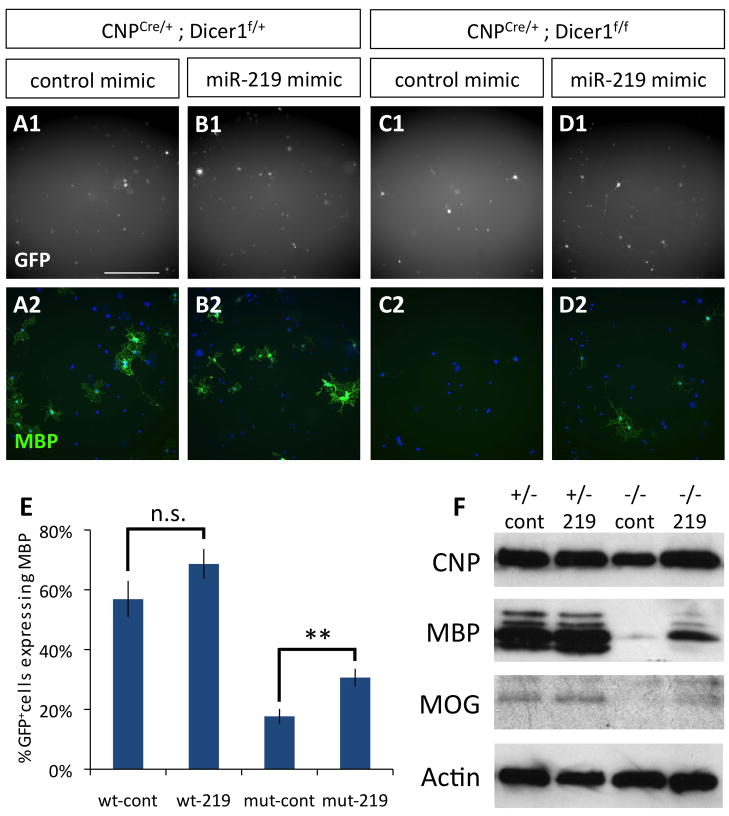
Based on our finding that miR-219 strongly induces OL differentiation, we next investigated whether miR-219, on its own, could rescue differentiation disrupted in OLs lacking functional Dicer1 expression. To address this question, we purified both mutant CNPCre/+ Dicer1flox/flox OPCs and control genotype CNPCre/+ Dicer1flox/+ OPCs, and then transfected them with either miR-219 or negative control mimic. These cells were then cultured for 4 DIV in mitogen withdrawal media to determine the effect of adding miR-219 back to OPCs lacking mature miRNA expression. As expected, loss of Dicer1 function strongly repressed myelin gene expression; however, when miR-219 mimic was transfected into these mutant OPCs, the number of transfected cells expressing MBP (Fig. 6A–E), as well as the overall expression of CNP, MBP, and MOG (Fig. 6F), robustly increased relative to control transfected mutant OPCs. These data implicate miR-219 as a key miRNA regulator of mitogen-withdrawal mediated OL differentiation.
Figure 6. miR-219 partially rescues OL differentiation in Dicer− OPCs.
A–D. Transfected OPCs (GFP+ = white, A1–D1), stained for MBP expression (green) and co-stained with DAPI (blue, A2–D2). OPCs purified from P7 control littermate CNPCre/+;Dicer1f/+ (A–B) and mutant CNPCre/+;Dicer1f/f mice (C–D) were transfected with control mimic (A,C) or miR-219 mimic (B,D) and cultured 4 DIV in −PDGF−T3 media. Scale bar = 200 μm. E. Percentages of healthy control (cont) or miR-219 (219) mimic transfected CNPCre/+;Dicer1f/+ (wt) and mutant CNPCre/+;Dicer1f/f (mut) OLs expressing MBP. Error bars ±S.E.M., n=9. ** p < 0.005, n.s. = not significant T-test control vs. miR-219 transfections. F. Western blots to determine CNP, MBP, and MOG expression levels in control CNPCre/+;Dicer1f/+ (+/−) and mutant CNPCre/+;Dicer1f/f (−/−) OPCs transfected with miR-219 or control mimic miRNA and cultured 4 DIV in −PDGF−T3 media. Actin blot shown is stripped and re-probed MBP blot. See also Fig. S7, Table S2–S4.
OL-expressed miRNAs preferentially target genes repressed during OL differentiation
miRNAs generally function by altering the expression or translation of “targeted” mRNAs, which are recognized by miRNAs via complementary sequences in their 3′ UTRs. Therefore, identification of miRNA targets often reveals the mechanisms by which miRNAs exert their influence. To analyze the genes targeted by miR-219, miR-138, and miR-338, we obtained the lists of their predicted targets from TargetScan 5.1 (http://www.targetscan.org/). We then ascertained the expression patterns of these targeted genes by consulting genomic expression data obtained from acutely purified OPCs, immature GC+ OLs, and mature MOG+ OLs (Cahoy et al., 2008). In this way, we were able to determine the percentages of miRNA-targeted genes that were either induced or repressed during OL differentiation (Fig. S7A). Interestingly, when compared to the percentage of total characterized, unique genes that are induced or repressed during OL differentiation, we found that all three OL-expressed miRNAs preferentially targeted genes that are repressed as OLs differentiate. To determine whether the enriched targeting of genes repressed during OL differentiation is restricted to the miRNAs that are highly expressed in OLs, we also analyzed two unrelated miRNAs, miR-142 and miR-196, which are most strongly expressed outside of the nervous system (Landgraf et al., 2007), and found that they do not similarly target such a high percentage of genes repressed during the full course of OL differentiation (miR-142 targets a higher percentage of genes repressed in GC+ OLs, but not in the more mature MOG+ OLs). Cumulatively, these data indicate that miR-219, miR-138, and miR-338 may function by targeting OPC-expressed genes whose repression is necessary to promote the rapid cessation of proliferation coupled to OL differentiation.
miR-219 directly represses the expression of several genes that inhibit OL differentiation
To further investigate the mechanisms by which miR-219 promotes OL differentiation, we next wanted to determine whether miR-219 directly targets any genes that functionally regulate OL differentiation. We looked for predicted miR-219 targets that were repressed > 2-fold in OLs relative to OPCs that were also either (a) computationally determined to be very likely targets of miR-219 (Table S2), (b) very highly repressed during OL differentiation (Table S3), or (c) very highly expressed in OPCs (Table S4). From these analyses, we selected four candidate genes for further study: PDGFRα, the receptor for the OPC mitogen PDGF (Besnard et al., 1987), Sox6, a transcription factor previously reported to repress terminal OL differentiation (Stolt et al., 2006), and also FoxJ3 and ZFP238 (a.k.a. RP58), two transcription factors with no previously reported functions in OPCs. ZFP238 is a transcriptional repressor expressed in neuronal precursors and distinct subsets of mature neurons that has been implicated in regulating neuronal differentiation (Aoki et al., 1998; Okado et al., 2009), and FoxJ3 expression has been detected various developing tissues, including the embryonic neural tube (Landgren and Carlsson, 2004). These facts, coupled with the observed expression of these transcription factors in OPCs and subsequent repression in OLs (Table S2, Fig. S7B), implicated them as potential novel regulators of the OPC-OL transition.
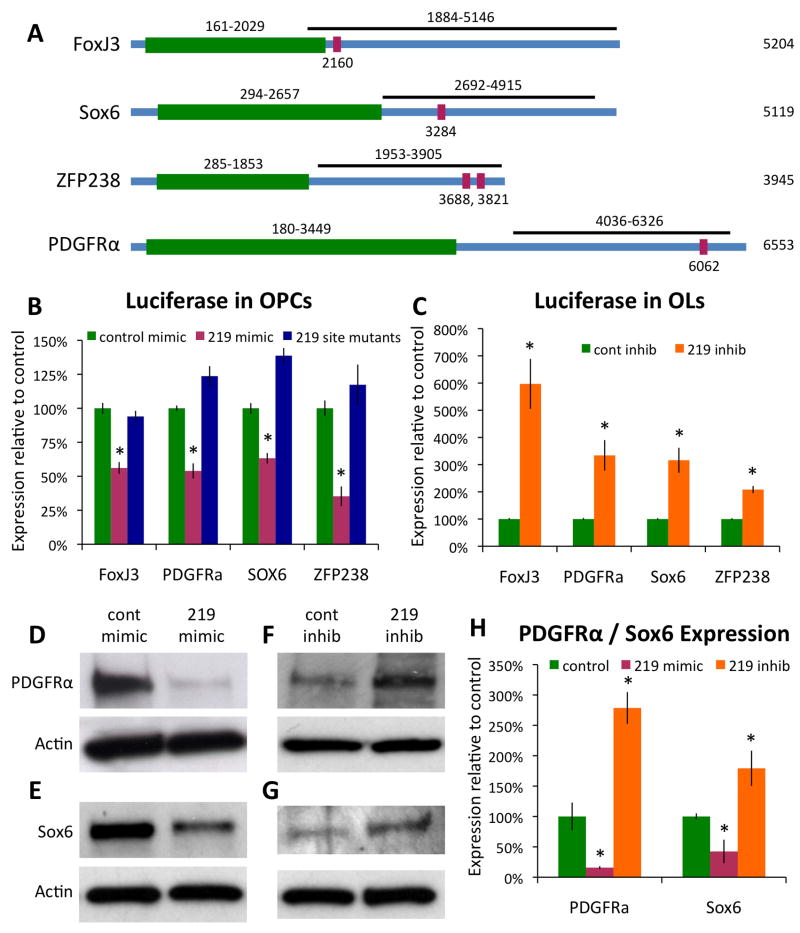
We first confirmed that these genes were in fact targets of miR-219. We subcloned the 3′ UTRs of these candidate genes, which contain the predicted miR-219 target sites, into a reporter construct that fused these 3′ UTRs to the end of a constitutively expressed luciferase gene (Fig. 7A). These constructs were then co-transfected into HEK-293 cells along with miR-219 mimic to determine the extent to which miR-219 could repress expression of mRNAs physically linked to these candidate 3′ UTRs. When compared to the negative control, non-targeting mimic, miR-219 reproducibly repressed, by about 50%, the translation of luciferase linked to the 3′ UTRs of FoxJ3, PDGFRα, and ZFP238 (Fig. S8A). To further investigate targeting of these candidate 3′ UTRs by miR-219, we performed similar transfection experiments in purified OPCs. We observed robust miR-219 mediated translational repression of luciferase linked to all four candidate 3′ UTRs when expressed in OPCs (Fig. 7B). This level of repression is similar to or greater than previously reported repression of protein expression by miRNA (Cheng et al., 2007; Kocerha et al., 2009; Wu and Belasco, 2008), and therefore we believe that PDGFRα, Sox6, ZFP238, and FoxJ3 all represent true functional targets of miR-219 repression in vivo. To further confirm the repression of these genes by direct interaction with miR-219, we utilized site-directed mutagenesis to specifically disrupt each of the predicted miR-219 targeted sequences in our reporter constructs. This mutagenesis completely abolished the ability of miR-219 to repress translation from each of our candidate gene 3′UTRs in both HEK-293 cells and OPCs (Fig. 7B, S8A). To test the role of endogenously expressed miR-219 in regulating PDGFRα, Sox6, ZFP238, and FoxJ3, we co-transfected our reporter constructs into OPCs along with miR-219 inhibitor and immediately induced OL differentiation. We found that expression of all four reporter constructs was strongly de-repressed (2–6 fold increases) relative to control transfected cells (Fig. 7C), indicating that sufficient miR-219 is produced in newly differentiating OLs to repress translation of PDGFRα, Sox6, ZFP238, and FoxJ3.
Figure 7. miR-219 represses the expression of PDGFRα, Sox6, FoxJ3, and ZFP238.
A. Regions of the FoxJ3, Sox6, ZFP238, and PDGFRα 3′ UTRs cloned into the luciferase reporter construct are depicted (black bars). Green = coding regions of depicted genes, red = locations of predicted miR-219 targeted sites. B. Luciferase activity at 1–2 DIV in OPCs co-transfected with indicated luciferase reporter constructs and either control (green) or miR-219 (red) mimic, blue = co-transfections of miR-219 mimic with reporter constructs containing mutated 219 binding sites; data normalized to control mimic co-transfection luciferase activity. C. Luciferase activity at 4 DIV in OLs co-transfected with indicated luciferase reporter constructs and either control (green) or miR-219 (orange) inhibitor. D–G. Western blot showing the levels of PDGFRα (D,F) or Sox6 (E,G) expression in OPCs transfected with control or miR-219 mimic, 2 DIV +PDGF−T3 (D,E), or transfected with control or miR-219 inhibitor, 1–3 DIV −PDGF+T3 (F,G). All blots stripped and re-probed for actin as loading control. H. Actin-normalized PDGFRα and Sox6 expression, relative to control transfection levels, in OPCs transfected with miR-219 mimic (red) or inhibitor (orange). Error bars ±S.E.M., n = 8–12 (B–C) or 3–4 (H). * p < 0.001 (B–C) or < 0.05 (H) T-test control vs. miR-219 mimic/inhibitor transfections (T-tests 219 site mutants vs. control not done). See also Fig. S8.
To further confirm the targeting of endogenous PDGFRα and Sox6 by miR-219, we compared the expression of PDGFRα and Sox6 protein in OPCs transfected with either miR-219 or control mimic. PDGFRα and Sox6 protein expression, normally repressed in OLs, was high in OPCs transfected with control mimic and then cultured for 2 DIV in +PDGF−T3 media. In contrast, in OPCs transfected with miR-219 mimic both PDGFRα and Sox6 expression was strongly repressed, consistent with our luciferase assay data (Fig. 7D–E, H). Also consistent with our luciferase assay data was the finding that both PDGFRα and Sox6 expression was increased in OLs transfected with miR-219 inhibitor (Fig. 7F–G, H). These data indicate that miR-219 directly represses endogenous PDGFRα and Sox6 protein expression, and likely does so in response to mitogen withdrawal in OPCs. Interestingly, the repression of PDGFRα would set up a positive feedback loop, wherein reduced levels of mitogen stimulation would induce miR-219 expression, leading to a repression of PDGFRα expression, and a subsequent further reduction of mitogen stimulation and a more rapid initiation of OL differentiation. This effect would be further enhanced by the loss of repressors of OL differentiation, such as Sox6.
ZFP238 and FoxJ3 repress OL differentiation
After confirming that miR-219 targets and represses the 3′ UTRs of FoxJ3 and ZFP238, we next wanted to determine whether these OPC-enriched transcription factors functionally regulate OL differentiation. To determine whether the repression of these genes was required for normal OL differentiation, we drove the constitutive expression of FoxJ3 or ZFP238 in transfected OPCs, and then induced differentiation by culturing these cells for 3 DIV in mitogen withdrawal (−PDGF−T3) media (Fig. S8B–D). We found that overexpression of either of these OPC-expressed transcription factors significantly inhibited normal OL differentiation, and also increased expression of the OPC marker NG2 (Fig. S8E). We also found that overexpression of either of these genes did not strongly affect the survival of immature OPCs cultured in PDGF, but did reduce survival when mitogens were withdrawn (Fig. S8F). These data indicate that ZFP238 and FoxJ3, both previously uncharacterized in OPCs, are each sufficient to repress OL differentiation, and that failure to inhibit either of these transcription factors leads to decreased differentiation and survival in the absence of proliferation-supporting mitogens. It is interesting to note that one of the unexpected results we obtained from characterizing OL-induced miRNAs was the identification of novel proteins involved in regulating OL differentiation. Similar analyses aimed at correlating expressed miRNAs with expressed miRNA targets may prove a fruitful means of identifying previously undiscovered gene functions in future studies.
DISCUSSION
Dicer1 is required for normal OL differentiation and myelination
Our findings show that miRNA processing is required to promote normal OL differentiation and myelination in vitro and in vivo. The role of miRNAs specifically in promoting myelin formation is further confirmed in the CNPCre/+ Dicer1flox/flox mice, in which the initial stages of OL differentiation commence normally, but complete compact myelin formation is still significantly delayed. The myelin that does eventually form in the absence of OL-expressed miRNAs appears to be functionally normal, as evidenced by both the formation of normal paranodal structures in the optic nerve and by alleviation of behavioral symptoms in older Olig2Cre/+ Dicer1flox/flox mice. However, at older ages, when compact myelin levels are recovering, a significant portion (up to 50%) of Olig2Cre/+ Dicer1flox/flox OLs do apparently express functional Dicer1, raising the possibility that mature OLs that fail to express functional Dicer1 are less healthy or are otherwise at a competitive disadvantage compared to OLs that continue to express Dicer1, and that these mutant OLs that fail to excise Dicer1 eventually generate the majority of healthy compact myelin sheaths in Olig2Cre/+ Dicer1flox/flox mice. Future experiments with strains of mice that more completely disrupt miRNA processing in the OL lineage will be required to clarify this issue.
Several factors lead us to believe that the myelination deficits we have observed result directly from a loss of Dicer1 function in the OPCs and OLs, rather than from an alternative defect in a secondary cell type. First, we observe an identical CNS dysmyelination phenotype when either Olig2 or CNP drives Cre recombinase mediated disruption of Dicer1. In particular, CNP expression is restricted to cells of the OL lineage in the CNS (Yuan et al., 2002), and we therefore would not predict that Dicer1 disruption would extend to other CNS cell types in the CNPCre/+ Dicer1flox/flox mice. Second, OLs purified from either Olig2Cre/+ Dicer1flox/flox or CNPCre/+ Dicer1flox/flox mice both fail to fully differentiate in vitro, indicating that complete OL differentiation is directly impaired by the disruption of miRNA processing. Third, axons present in dysmyelinated areas appear normal and comparable to littermate control genotype mice, implying that observed myelination deficits are not the result of disrupted axon tracts. All of these data implicate OL-expressed miRNAs as playing a crucial role in promoting normal CNS myelin development.
One distinction between the Olig2Cre/+ Dicer1flox/flox and CNPCre/+ Dicer1flox/flox mice is the loss of PNS myelin. In the CNPCre/+ Dicer1flox/flox mice, but not the Olig2Cre/+ Dicer1flox/flox mice, we observe a near complete ablation of sciatic nerve myelination at P22–23. This phenotype likely indicates a requirement for Dicer1 function in the myelinating Schwann cells of the PNS, as CNP, but not Olig2, is expressed by Schwann cells (Matthieu et al., 1980; Zhou et al., 2000). In addition, the axons of the sciatic nerve are still present in the CNPCre/+ Dicer1flox/flox mice, implying that ablation of PNS myelin precedes loss of PNS axons. Also, PNS myelin and axons both appears normal in Olig2Cre/+ Dicer1flox/flox mice, despite the fact that Olig2 (and therefore Cre recombinase) is transiently expressed in the myelinated motor neurons of the sciatic nerve (Takebayashi et al., 2000; Zhou and Anderson, 2002). Loss of mature miRNAs appears to severely disrupt the ability of Schwann cells to produce myelin, and therefore the role of miRNAs in PNS myelination will be an important area of future study.
OL-induced miRNAs link initiation of OL differentiation to cessation of OPC proliferation
In several vertebrate cell types, particularly in the nervous system, cessation of proliferation and initiation of differentiation are temporally linked (Buttitta and Edgar, 2007; Kitzmann and Fernandez, 2001; Politis et al., 2008; Truong and Khavari, 2007). How is the timing of the induction of one genetic program specifying a post-mitotic cell fate so tightly associated with the repression of a distinct genetic program that drives the proliferation of the immature precursor cell? In OLs, miRNAs induced by the withdrawal of OPC mitogens, and miR-219 in particular, appear to be crucial to linking the cessation of proliferation to the initiation of OL differentiation. Indeed, all three of the miRNAs highly induced by mitogen withdrawal in OPCs preferentially target genes that are expressed by OPCs and repressed as OLs differentiate. A similar role for miRNAs in repressing proliferation has already been postulated in other cell types (Carleton et al., 2007). In fact, one of the additional miRNAs induced during OL differentiation, miR-192, has been directly implicated in arresting the cell cycle by coordinately repressing several G1-S and G2-M cell cycle checkpoint control genes (Georges et al., 2008), and another OL-induced miRNA, miR-181a, functions as a tumor suppressor in gliomas (Shi et al., 2008). Of equal importance may be the repression of inhibitors of OL differentiation, as several immature cell types, including OPCs, have been shown to express not only genes that promote the cell cycle, but also genes (e.g. transcription factors) that actively inhibit adoption of a differentiated cell fate (Dugas et al., 2006; Lang et al., 2007; Stolt et al., 2006; Wang et al., 2001).
Several labs have demonstrated that OL differentiation can be induced by distinct pathways. When OPC mitogens become limiting, proliferation ceases and OL differentiation rapidly commences (Barres et al., 1993; Besnard et al., 1987). In contrast, exposure to T3 promotes a more gradual program of OL differentiation: OPCs exposed to T3 in the presence of saturating amounts of mitogens continue to proliferate and only differentiate once they have become sufficiently “mature” enough to respond to T3 (Barres et al., 1994a; Durand and Raff, 2000; Temple and Raff, 1986). T3 exposure does not simply recapitulate the program of mitogen-withdrawal triggered differentiation, as distinct sets of genes are induced by mitogen withdrawal and T3 exposure (Tokumoto et al., 2001). The fact that several miRNAs appear to be specifically induced by mitogen withdrawal, and not T3, in OPCs is consistent with their proposed role in inhibiting proliferation, as exposure to T3 alone in the presence of mitogens is not sufficient to trigger the immediate arrest of the cell cycle. In addition, the finding that the loss of OL-expressed miRNAs, which are specifically induced by mitogen withdrawal, leads to a dysmyelination phenotype in vivo implies that, consistent with previous reports (Calver et al., 1998), limiting mitogen levels in vivo play an important role in regulating the timing of OL differentiation.
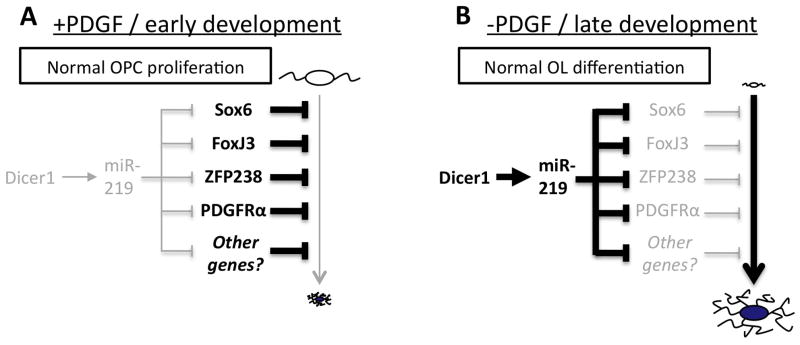
Model of miR-219 function in OL differentiation
Our data indicate that miR-219 in particular plays an important role in promoting OL differentiation. Not only does inhibition of miR-219 strongly impair OL differentiation, but miR-219 alone can both instigate OL differentiation in OPCs immersed in mitogens, and can also partially rescue the differentiation deficit caused by the loss of mature miRNA production in OLs. We have identified several genes directly targeted by miR-219 whose repression accelerates the program of OL differentiation. Cumulatively, these data lead us to the following model (Fig. 8). Early in development, or in the presence of saturating amounts of mitogens, OL-induced miRNAs, and Dicer1 expression in general, are low (Fig. S4C). In contrast, targets of miR-219, such as the mitogen receptor PDGFRα and several transcription factors that repress OL differentiation, are highly expressed (Fig. 8A). Later in development or when mitogens are withdrawn, genes that inhibit OL differentiation are downregulated. The reduced expression/residual transcripts of these genes are then further repressed by miR-219, whose expression is induced, at least in part, by Dicer1 expression promoted by mitogen withdrawal (Fig. 8B). The importance of this compounded repression is demonstrated in instances where miR-219 function is blocked either by introduction of miR-219 inhibitor or disruption of Dicer1, as both of these manipulations lead to increased repression of OL differentiation (Fig. 1–3, 5). In contrast, introduction of miR-219 into proliferating OPCs reduces the expressed levels of miR-219-targeted inhibitors of differentiation, leading to a de-repression of OL differentiation (Fig. 5, 7).
Figure 8. Model of Dicer1 and miR-219 promotion of OL differentiation.
Arrows indicate a promotion of pathway or expression, and bars indicate inhibition of pathway or expression. Stronger activity/expression is denoted by bold lines/text, and weaker activity/expression is denoted by grey lines/text. A. OL differentiation is inhibited by several OPC-expressed genes in the presence of large amounts of PDGF/early development under normal conditions. B. When mitogens become limiting, Dicer1 and miR-219 expression is induced, leading to a direct, accelerated repression of several OPC-expressed genes and a de-repression of OL differentiation. See text for additional details.
We hypothesize that the OL-induced miRNAs, and miR-219 in particular, help to coordinate proliferation and differentiation. When mitogens are withdrawn, these miRNAs are induced as part of the program of OL differentiation, at which point they rapidly repress several genes that normally promote the immature OPC phenotype. The importance of rapidly repressing all of these inhibitors of OL differentiation is demonstrated in a number of experiments. If the normal repression of a single inhibitor of OL differentiation is blocked by transfection with an overexpression plasmid, OL differentiation is impaired (Fig. S8C–G). Conversely, loss of a single inhibitory transcription factor in proliferating OPCs does not instantly promote differentiation, likely because of the persistent high expression of several additional inhibitors of OL differentiation (Stolt et al., 2006).
miR-219 in the CNS and glioblastoma
Our data indicate that miR-219 is expressed at the highest level in the OLs of the developing CNS, and the strong degree of miR-219 induction we observe is consistent with previous findings (Lau et al., 2008). However, other labs have reported biological roles for miR-219 in mature neurons of the suprachiasmatic nuclei (Cheng et al., 2007) and prefrontal cortex (Kocerha et al., 2009). Potentially, miR-219 expressed at a lower level in these neurons may still be able to affect neuronal function. Alternatively, miR-219 may be expressed at a high level in a few distinct populations of neurons. As we had assayed the expression level of miR-219 in the pooled sample of all telencephalic neurons, a high level of expression in less than 5% of the total population of neurons would be masked by the remaining >95% of the non-expressing cells. Nevertheless, OLs are the first mammalian cells in which a developmental role for miR-219 has been demonstrated.
The potential role of miR-219 as an inhibitor of cellular proliferation has been indicated in previous oncogenetic research. Wong et al. (2008) identified miR-219 and miR-138 as being downregulated in squamous cell carcinomas, and Izzotti et al. (2009) identified both miR-219 and miR-192 as being downregulated in potentially cancerous lung tissue. In CNS tumors, miR-219, miR-138, and miR-192 are all downregulated in medulloblastomas relative to normal cerebellar tissue (Ferretti et al., 2009). These data, in combination with our current findings, raise the intriguing possibility that loss of miR-219 expression, along with other OL-enriched miRNAs, may contribute to the aberrant proliferation observed in some types of glioblastomas, particularly those derived from or expressing genes of the OL lineage. In fact, overexpression of PDGF in glial progenitor cells is sufficient to induce glioblastoma in mice (Assanah et al., 2006; Assanah et al., 2009; Uhrbom et al., 2000). Therefore, reintroduction of miRNAs identified here, such as miR-219, into glioblastomas could be investigated as a potential means of slowing tumor progression.
EXPERIMENTAL PROCEDURES
Complete protocols available upon request from jcdugas@alum.mit.edu.
Purification and culturing of cells
Unless otherwise indicated, all in vitro culturing and transfection experiments were performed with OPCs purified from P7–P8 Sprague-Dawley (Charles Rivers) rat brains as described previously (Dugas et al., 2006). For acutely purified samples, OPCs, OLs, astrocytes, and neurons were isolated from wild type C57Bl6 (Charles Rivers) or transgenic S100β-eGFP mice as described previously (Cahoy et al., 2008). See supplemental methods for additional details.
Immunohistochemistry and histology
Immunostaining of OPC and OL cultures for CNP, MBP, MOG, and GFP expression was performed as described previously (Dugas et al., 2006). Immunostaining for NG2 expression was performed with 1/400 rabbit-anti-NG2 antibody (Millipore AB5320).
For tissue sections, mice were perfused with DPBS followed by 4% paraformaldehyde (PFA), then immersion fixed in 4% PFA, equilibrated in 30% sucrose, and mounted in 2:1 30% sucrose:O.C.T. (Tissue-Tek) to generate 8 μm longitudinal (nerves) or 10 μm sagital (brains) sections. Sections were blocked in 50% goat serum with 0.4% Triton X-100 in PBS, then stained with 1/100 rat-anti-MBP (Abcam ab7349) and 1/1000 rabbit-anti-NF200 (Sigma N4142) antibodies. In all cases, Alexa dye conjugated antibodies (Invitrogen) were used to visualize staining, and sections/cells were mounted in Vectashield (Vector Labs) + DAPI to allow identification of healthy nuclei.
To stain tissue sections for compact myelin, brain sections were rehydrated in PBS, then incubated in a 1/300 dilution FluoroMyelin (Invitrogen F-34652) in PBS for 20 minutes, rinsed extensively in PBS, then mounted in Vectashield + DAPI.
To quantify cells expressing OPC/OL markers, slides were scored blind, and only cells with healthy nuclei were counted. To quantify CNS myelination cerebellar arms and corpora callosa were outlined, and the percentage area myelinated was determined as number of pixels +10 arbitrary units (0–255 scale in 8-bit images) brighter than background/total number of pixels. For each animal, two sagital sections at different depths (~200–300 μm apart) were analyzed; the two depths analyzed were the same across all littermates.
miRNA microarrays and RT-PCR
For miRNA microarrays, OPC samples were collected from purified rat OPCs cultured for 4 DIV in +PDGF−T3 media, OL samples were collected from purified rat OPCs cultured for 3 DIV in +PDGF−T3 media, then 4 DIV in −PDGF+T3 media. Total RNA samples (including small RNA molecules) were isolated using Qiagen miRNeasy Mini Kit, and were labeled and hybridized to mircroarrays as described in supplemental materials.
For qRT-PCR to analyze miRNA expression, RNA samples were obtained as described above from purified rat OPCs cultured for 4 DIV in ±PDGF±T3, or from acutely purified neural cell samples as described in Cahoy et al. (2008). Taqman miRNA qRT-PCR analysis was performed on 10ng of RNA for indicated miRNAs, following the manufacturer’s protocol (Applied Biosystems) and normalized to the U6 snRNA. Sybr green incorporation qRT-PCR to detect rat/mouse mRNA expression was performed on an ABI StepOnePlus Real Time PCR system following manufacturer’s instructions, normalized to GAPDH mRNA levels. Data were analyzed using the ddCt method (Livak and Schmittgen, 2001).
For RT-PCR to analyze gene expression, RNA was purified from OPC/OL samples using the Qiagen RNeasy Mini Kit, then 400 μg total RNA was reverse transcribed using SuperScriptIII (Invitrogen). Equivalent volumes from RT samples to be compared were then amplified using Platinum Taq (Invitrogen). All primers in supplemental materials.
In situ hybridization
In situ hybridizations were performed as described previously (Cahoy et al., 2008). 5′-end DIG-labeled LNA probes were obtained from Exiqon for miR-138, miR-219, miR-338, and scramble-miR (negative control). As a positive control, a 747bp DIG-labeled antisense fragment encompassing the coding region of the DM-20 isoform of the PLP gene and recognizing both isoforms (gift from Prof. William Richardson) was transcribed as described in the manufacturer’s protocol (Roche T7/SP6 In Vitro Transcription Kit). All LNA hybridizations/initial washes were performed at 50 °C; all PLP at 72 °C. For fluorescent in situs, the same DIG-labeled PLP probe was used with the TSA-Plus Cyanine 3 labeling system (PerkinElmer) according to manufacturer’s instructions, and slides were mounted in Vectashield + DAPI.
Western blots
Protein samples were collected from cultured cells in RIPA buffer + Complete Protease Inhibitor (Roche) at 4 °C. Equivalent total protein amounts were loaded onto 4–15% polyacrylamide gels (Bio-Rad) and then transferred to Immobilon-P (Millipore) membranes. Blots were probed with 1/500 rat-anti-MBP (Millipore MAB386), 1/500 mouse-anti-CNP, 1/100 mouse-anti-MOG, 1/1000 mouse-anti-PDGFRα (Abnova H00005156-M02), 1/10000 guinea pig-anti-Sox6 (gift M. Wegner), 1/5000 mouse-anti-β-actin (Sigma A5441), then HRP-linked goat-anti-mouse, –rabbit, or -guinea pig (Millipore) or -rat (Jackson Immunology). Blots were developed with ECL Western blotting detection reagent (Amersham) and exposed to film. Blots were quantified with NIH ImageJ 10.2 Gel Analyzer, and normalized to in-lane actin levels as loading control.
Cell transfections
OPCs were transfected as described previously (Dugas et al., 2006), using Lonza/Amaxa nucleofector kit; 2–3×106 rat OPCs or 4–5×106 mouse OPCs per transfection. For luciferase assays, 6 μg test firefly luciferase plasmid, 0.6 μg pRL-TK Renilla luciferase control plasmid (Promega), and 0.4 nmol miRNA mimic reagent were combined in transfections. OPCs were then cultured 24–48 hours in +PDGF−T3 media. For other miRNA transfection experiments, 0.4 nmol miRNA mimic or inhibitor was used and OPCs were cultured ±PDFF±T3 as indicated. In immunostaining experiments, 3.5 ug pC1-eGFP was included to allow identification of transfected cells. In FoxJ3/ZPF238 experiments, 2.5 ug pSPORT6-FoxJ3 or -ZFP238 was co-transfected with 1.5 ug pC1-eGFP, and OPCs were cultured ±PDFF±T3 as indicated.
In all immunostained transfection experiments, slides were scored blind, and data was quantified as the percentage of GFP+ transfected cells with healthy nuclei that co-expressed the indicated marker protein (NG2, CNP, MBP, or MOG); > 150 cells/coverslip were quantified.
Supplementary Material
Acknowledgments
We thank Prof. Klaus-Armin Nave for kindly providing the CNP-Cre mouse line, Prof. David Rowitch for providing the Olig2-Cre mouse line, and Prof. Chang-Zheng Chen for providing the floxed-Dicer mouse line. We would also like to thank Prof. Michael Wegner for generously providing the Sox6 antibody. This work was funded by the Myelin Repair Foundation and by NIH grant 5R03DA022201.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aoki K, Meng G, Suzuki K, Takashi T, Kameoka Y, Nakahara K, Ishida R, Kasai M. RP58 associates with condensed chromatin and mediates a sequence-specific transcriptional repression. J Biol Chem. 1998;273:26698–26704. doi: 10.1074/jbc.273.41.26698. [DOI] [PubMed] [Google Scholar]
- Assanah M, Lochhead R, Ogden A, Bruce J, Goldman J, Canoll P. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J Neurosci. 2006;26:6781–6790. doi: 10.1523/JNEUROSCI.0514-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assanah MC, Bruce JN, Suzuki SO, Chen A, Goldman JE, Canoll P. PDGF stimulates the massive expansion of glial progenitors in the neonatal forebrain. Glia. 2009 doi: 10.1002/glia.20895. [DOI] [PubMed] [Google Scholar]
- Barres BA, Lazar MA, Raff MC. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development. 1994a;120:1097–1108. doi: 10.1242/dev.120.5.1097. [DOI] [PubMed] [Google Scholar]
- Barres BA, Raff MC. Control of oligodendrocyte number in the developing rat optic nerve. Neuron. 1994;12:935–942. doi: 10.1016/0896-6273(94)90305-0. [DOI] [PubMed] [Google Scholar]
- Barres BA, Schmid R, Sendnter M, Raff MC. Multiple extracellular signals are required for long-term oligodendrocyte survival. Development. 1993;118:283–295. doi: 10.1242/dev.118.1.283. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev. 2001;81:871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Besnard F, Perraud F, Sensenbrenner M, Labourdette G. Platelet-derived growth factor is a mitogen for glial but not for neuronal rat brain cells in vitro. Neurosci Lett. 1987;73:287–292. doi: 10.1016/0304-3940(87)90260-6. [DOI] [PubMed] [Google Scholar]
- Buttitta LA, Edgar BA. Mechanisms controlling cell cycle exit upon terminal differentiation. Curr Opin Cell Biol. 2007;19:697–704. doi: 10.1016/j.ceb.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calver AR, Hall AC, Yu WP, Walsh FS, Heath JK, Betsholtz C, Richardson WD. Oligodendrocyte population dynamics and the role of PDGF in vivo. Neuron. 1998;20:869–882. doi: 10.1016/s0896-6273(00)80469-9. [DOI] [PubMed] [Google Scholar]
- Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell cycle (Georgetown, Tex. 2007;6:2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Liu A. Relationship between cell cycle molecules and onset of oligodendrocyte differentiation. Journal of neuroscience research. 2003;72:1–11. doi: 10.1002/jnr.10565. [DOI] [PubMed] [Google Scholar]
- Chen Y, Stevens B, Chang J, Milbrandt J, Barres BA, Hell JW. NS21: re-defined and modified supplement B27 for neuronal cultures. J Neurosci Methods. 2008;171:239–247. doi: 10.1016/j.jneumeth.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E, McManus MT. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci U S A. 2008;105:5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional Genomic Analysis of Oligodendrocyte Differentiation. J Neurosci. 2006;26:10967–10983. doi: 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B, Raff M. A cell-intrinsic timer that operates during oligodendrocyte development. Bioessays. 2000;22:64–71. doi: 10.1002/(SICI)1521-1878(200001)22:1<64::AID-BIES11>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, Ibrahim A, Ligon KL, Rowitch DH, Barres BA. Identification of Myelin-gene Regulatory Factor as a Critical Transcriptional Regulator Required for CNS Myelination. Cell. 2009 doi: 10.1016/j.cell.2009.04.031. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, De Smaele E, Po A, Di Marcotullio L, Tosi E, Espinola MS, Di Rocco C, Riccardi R, Giangaspero F, Farcomeni A, et al. MicroRNA profiling in human medulloblastoma. Int J Cancer. 2009;124:568–577. doi: 10.1002/ijc.23948. [DOI] [PubMed] [Google Scholar]
- Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA, Chau BN. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer research. 2008;68:10105–10112. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806–812. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci U S A. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzmann M, Fernandez A. Crosstalk between cell cycle regulators and the myogenic factor MyoD in skeletal myoblasts. Cell Mol Life Sci. 2001;58:571–579. doi: 10.1007/PL00000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocerha J, Faghihi MA, Lopez-Toledano MA, Huang J, Ramsey AJ, Caron MG, Sales N, Willoughby D, Elmen J, Hansen HF, et al. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc Natl Acad Sci U S A. 2009;106:3507–3512. doi: 10.1073/pnas.0805854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgren H, Carlsson P. FoxJ3, a novel mammalian forkhead gene expressed in neuroectoderm, neural crest, and myotome. Dev Dyn. 2004;231:396–401. doi: 10.1002/dvdy.20131. [DOI] [PubMed] [Google Scholar]
- Lang D, Powell SK, Plummer RS, Young KP, Ruggeri BA. PAX genes: roles in development, pathophysiology, and cancer. Biochem Pharmacol. 2007;73:1–14. doi: 10.1016/j.bcp.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, Griffiths IR, Nave KA. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28:11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- Matthieu JM, Costantino-Ceccarini E, Beny M, Reigner J. Evidence for the association of 2′,3′-cyclic-nucleotide 3′-phosphodiesterase with myelin-related membranes in peripheral nervous system. J Neurochem. 1980;35:1345–1350. doi: 10.1111/j.1471-4159.1980.tb09008.x. [DOI] [PubMed] [Google Scholar]
- Okado H, Ohtaka-Maruyama C, Sugitani Y, Fukuda Y, Ishida R, Hirai S, Miwa A, Takahashi A, Aoki K, Mochida K, et al. The transcriptional repressor RP58 is crucial for cell-division patterning and neuronal survival in the developing cortex. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.04.030. [DOI] [PubMed] [Google Scholar]
- Politis PK, Thomaidou D, Matsas R. Coordination of cell cycle exit and differentiation of neuronal progenitors. Cell Cycle. 2008;7:691–697. doi: 10.4161/cc.7.6.5550. [DOI] [PubMed] [Google Scholar]
- Raff MC, Durand B, Gao FB. Cell number control and timing in animal development: the oligodendrocyte cell lineage. Int J Dev Biol. 1998;42:263–267. [PubMed] [Google Scholar]
- Raff MC, Miller RH, Noble M. A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on culture medium. Nature. 1983;303:390–396. doi: 10.1038/303390a0. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, Huillard E, Sun T, Ligon AH, Qian Y, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Cheng Z, Zhang J, Li R, Zhao P, Fu Z, You Y. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–193. doi: 10.1016/j.brainres.2008.07.085. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Schlierf A, Lommes P, Hillgartner S, Werner T, Kosian T, Sock E, Kessaris N, Richardson WD, Lefebvre V, Wegner M. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev Cell. 2006;11:697–709. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Yoshida S, Sugimori M, Kosako H, Kominami R, Nakafuku M, Nabeshima Y. Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech Dev. 2000;99:143–148. doi: 10.1016/s0925-4773(00)00466-4. [DOI] [PubMed] [Google Scholar]
- Temple S, Raff MC. Differentiation of a bipotential glial progenitor cell in a single cell microculture. Nature. 1985;313:223–225. doi: 10.1038/313223a0. [DOI] [PubMed] [Google Scholar]
- Temple S, Raff MC. Clonal analysis of oligodendrocyte development in culture: evidence for a developmental clock that counts cell divisions. Cell. 1986;44:773–779. doi: 10.1016/0092-8674(86)90843-3. [DOI] [PubMed] [Google Scholar]
- Tokumoto YM, Tang DG, Raff MC. Two molecularly distinct intracellular pathways to oligodendrocyte differentiation: role of a p53 family protein. Embo J. 2001;20:5261–5268. doi: 10.1093/emboj/20.18.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Khavari PA. Control of keratinocyte proliferation and differentiation by p63. Cell Cycle. 2007;6:295–299. doi: 10.4161/cc.6.3.3753. [DOI] [PubMed] [Google Scholar]
- Uhrbom L, Hesselager G, Ostman A, Nister M, Westermark B. Dependence of autocrine growth factor stimulation in platelet-derived growth factor-B-induced mouse brain tumor cells. Int J Cancer. 2000;85:398–406. doi: 10.1002/(sici)1097-0215(20000201)85:3<398::aid-ijc17>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001;29:603–614. doi: 10.1016/s0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Watkins TA, Emery B, Mulinyawe S, Barres BA. Distinct stages of myelination regulated by gamma-secretase and astrocytes in a rapidly myelinating CNS coculture system. Neuron. 2008;60:555–569. doi: 10.1016/j.neuron.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Yuan X, Chittajallu R, Belachew S, Anderson S, McBain CJ, Gallo V. Expression of the green fluorescent protein in the oligodendrocyte lineage: a transgenic mouse for developmental and physiological studies. J Neurosci Res. 2002;70:529–545. doi: 10.1002/jnr.10368. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–343. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.