Abstract
Serum prostate-specific antigen screening has led to earlier detection and surgical treatment of prostate cancer, favoring an increasing incidence-to-mortality ratio. However, about one third of tumors that are diagnosed when still confined to the prostate can relapse within 10 years from the first treatment. The challenge is therefore to identify prognostic markers of aggressive versus indolent tumors. Although several preclinical models of advanced prostate tumors are available, a model that recapitulates the genetic and growth behavior of primary tumors is still lacking. Here, we report a complete histopathological and genomic characterization of xenografts derived from primary localized low- and high-grade human prostate tumors that were implanted under the renal capsule of immunodeficient mice. We obtained a tumor take of 56% and show that these xenografts maintained the histological as well as most genomic features of the parental tumors. Serum prostate-specific antigen levels were measurable only in tumor xenograft-bearing mice, but not in those implanted with either normal prostate tissue or in tumors that likely regressed. Finally, we show that a high proliferation rate, but not the pathological stage or the Gleason grade of the original tumor, was a fundamental prerequisite for tumor take in mice. This mouse xenograft model represents a useful preclinical model of primary prostate tumors for their biological characterization, biomarker discovery, and drug testing.
Screening based on serum prostate-specific antigen (PSA) has led to earlier detection of prostate cancer (PCa) and to the possibility of excision of organ confined tumors by surgical treatment. However, about one third of tumors that are diagnosed when still confined to the prostate relapse within 10 years. The parameters used for risk classification and clinical decision-making include purely clinical and pathological features and are not sufficiently discriminatory. The challenge, therefore, is to identify those patients at risk for relapse after surgery as well as those with indolent tumors not requiring further intervention, especially in patients with intermediate risk of recurrence.1,2,3 If primary tumors have embedded the indolent or the aggressive signature, molecular profiling should in theory identify these and help guide therapy. Unfortunately, tumors are often too small to perform thorough and comprehensive molecular analyses to arrive at these signatures, begging for a model that will allow expansion of the primary tumor while retaining its genetic characteristics.
As a result, considerable efforts have been devoted to generate clinically relevant models of prostate tumors. Although a number of cell lines and in vivo models have been developed, to date, most are based on metastatic disease, and no model exists that is able to recapitulate human primary tumors.4,5
Available mouse models of PCa include both genetically engineered mice (GEM), in which one gene at a time is either overexpressed or knocked-out, and mouse xenografts, where usually human tumor cells are injected into the mouse by different techniques (ie, subcutaneous, orthotopic, intravenous, intracardiac injections).6,7 Although a substantial number of preclinical studies are available, most of these do not show a good predictivity of drug response. In addition, prostate tumors developed in a genetically conditioned background (GEM) show histopathological features that differ somewhat from that of human cancers. These tumors are driven by single specific alterations, at least in their first phase of development, reflecting the complexity of human tumor biology only to a limited extent.8,9 GEMs may therefore be ideal to screen compounds that target a particular molecular pathway, which is activated in this case by genetic manipulations. On the other hand, mouse models such as xenografts are able to recapitulate the histopathological and molecular heterogeneity of human cancer. These models are commonly generated by injecting established cell lines, because of the fact that freshly isolated primary cells rarely form tumors in immunodeficient mice.10 Unfortunately, genetic variability ensues in these established cancer cells over several passages while the risk of contamination from other commonly used cell lines is frequent. Importantly, established prostatic cancer cell lines are derived mostly from metastases, thus not recapitulating human primary prostate tumors.4,5
The ideal alternative would be to generate mouse xenografts of human primary tumor tissues. A debated theme is whether growth efficiency of human prostate tumors in the murine host can be improved. In fact, subcutaneous implantations of human localized prostate tumors in immunodeficient mice have been performed for many years with scarce results in terms of tumor take.11,12
Here we report our experience in generating xenografts by implanting a large cohort of fresh tumor tissue samples from radical prostatectomy specimens in mice with different immunodeficient status. We show that 56% of a cohort of human organ-confined prostate cancers with Gleason score (GS) comprised between 7 and 9 was able to grow in mice in the subrenal capsule site, whereas scarce or no success was achieved by both orthotopic and subcutaneous implantations.
To our knowledge, this is the first in-depth characterization of a cohort of human prostate tumors and paired mouse xenografts. These results will be relevant when planning future in vivo studies on the use of human prostate tissues either for biological approaches, drug testing, or biomarker discovery.
Materials and Methods
Patients and Mouse Xenografts
A total of 143 immunodeficient mice were implanted with 53 human primary localized prostate tumors (see Supplemental Figure S1 at http://ajp.amjpathol.org) obtained from patients undergoing radical prostatectomy at the Brigham and Women’s Hospital (Boston, MA). The research protocol was approved by our institutional review board, and all human subjects provided appropriate informed consent. None of these patients had received any neoadjuvant hormone or radiation therapy. Fresh tissue samples were dissected by an expert pathologist, and fragments of average size 6 × 6 × 3 mm were immediately transferred on ice from the surgical room to the laboratory. These tissues were cut in half and used within 24 hours after surgery. Half of the sample was used to confirm the presence of at least 50% tumor by histology and to perform molecular analyses, whereas the other half was implanted in mice or mechanically disaggregated for primary cell isolation.
Both Nu/Nu and NOD/SCID mice (Charles River Laboratories International, Inc., Wilmington, MA) were used for the implantations. The mice were maintained under specific pathogen-free conditions at constant temperature and humidity. The research protocol was approved by the Institutional Animal Care and Use Committee (IACUC), and regulations concerning the use of animals in research were followed.
All mice were castrated at least two weeks before surgery and implanted with 90-day release subcutaneous testosterone pellets (Innovative Research of America, Sarasota, FL) to maintain the hormone concentration constant in the blood.
Subrenal Capsule Implantation of Human Prostate Tissue
Thirty prostate tumors and ten normal prostate tissue specimens were implanted under the renal capsule of 70 and 20 immunodeficient mice, respectively. Tumors were staged in accordance with the American Joint Committee on Cancer (AJCC Cancer Staging Manual, Sixth Edition, 2002). Thirteen of these 30 prostate cancers (44%) were diagnosed as GS 7 (3 + 4), 7 (23%) were GS 7 (4 + 3), 5 (16.6%) were GS 8, and 5 (16.6%) were GS 9. Also, 16 (53%) prostate adenocarcinomas presented with pathological stage T2 (pT2, which includes tumors confined to the organ and not invading the capsule), and the remaining 14 (47%) were pT3 (the prostate capsule and/or the seminal vesicles were invaded).
Mice were anesthetized by i.p. injection of Avertin (2,2,2-Tribromoethanol and amylene hydrate, Sigma-Aldrich, St. Louis, MO) at the dose of 250 mg/kg of body weight. A skin incision of approximately 2 cm was made along the dorsal midline and then in the body wall slightly shorter than the long axis of the kidney. The kidney was slipped out of the body cavity by applying pressure on the other side of the organ using forefinger and thumb. Renal capsule was then gently pinched, lifted from the renal parenchyma, and cut to create a pocket, in which the graft was inserted. Two or three grafts of size 2 × 2 × 1 mm were placed under the renal capsule of each mouse. One to four mice were implanted with each tumor.
Primary Cell Cultures of Human PCa Cells
Eight primary cell cultures were derived as previously described.13 Briefly, tissues were chopped in small fragments and left in a T25 flask with one ml of Iscove’s modified Dulbecco’s medium (IMDM, Invitrogen Corporation) until outgrowth of cells was visible (1 to 4 days). Cells were trypsinized and passaged when 80% confluence was reached.
Orthotopic Implantation of Human PCa Cells and Tissues
Primary cells and cell suspensions obtained from 11 tissue samples by one-hour enzymatic digestion (collagenase, Sigma-Aldrich) at 37°C were injected orthotopically in 24 and 11 mice, respectively. Mice were anesthetized using Avertin at the dose of 250 mg/kg of body weight. An incision was made through skin and body wall at the ventral lower-midline and the bladder together with the seminal vesicles were exteriorized. Human cells (at least 2 × 105) resuspended in 10 microliters of Phosphate Buffer Solution (PBS, Invitrogen Corporation) were then injected in one of the anterior prostate lobes using a 27-gauge needle. Four tissue samples were directly grafted in 12 mice. To this end, a small pocket was created in the anterior prostate to implant 1 × 2 × 1 mm tissue fragments. Both the peritoneum and skin were then sutured with absorbable stitches.
Subcutaneous Implantation of Human Tissues
Three tumors for which more tissue was available were implanted subcutaneously in six mice. An incision was made through the skin at both flanks of the anesthetized mouse, and tissue fragments (3 × 6 × 3 mm) were inserted in a pocket far from the incision hole.
Histology and Immunohistochemistry
Hematoxylin and eosin as well as immunohistochemical stainings were performed on five-micron sections of formalin-fixed and paraffin-embedded (FFPE) samples (xenografts and parental human tumors). The following primary antibodies were used: mouse anti–androgen receptor (AR) clone AR441 (Dako North America, Inc., Carpinteria, CA) at 1:100 dilution, mouse anti-PSA (Dako) at 1:200 dilution, mouse anti–α-methylacyl-CoA racemase (AMACR; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at 1:300 dilution, mouse anti-p63 (Santa Cruz Biotechnology) at 1:50 dilution, rabbit anti–phospho-AKT clone D9E (Cell Signaling Technology, Inc. Danvers, MA) at 1:50 dilution, rabbit anti–phospho-S6 (Cell Signaling Technology) at 1:100 dilution, and mouse anti–Ki-67 (Dako) at 1:100 dilution.
Tissue sections were deparaffinized, rehydrated, and subjected to microwaving in 10 mmol/L citrate buffer (pH 6.0) in a 750-W microwave oven for 15 minutes. The primary antibody was applied at room temperature in an automated tissue stainer (i6000, Biogenex, San Ramon, CA). Standardized 3,3-diaminobenzidine or methyl green development times allowed accurate comparison of all samples. Antibody isotype-specific IgG served as negative control.
Apoptosis was analyzed in FFPE tissue sections using the Apoptag peroxidase in situ assay (Chemicon International, Billerica, MA) according to the manufacturer’s instructions. The apoptotic rate was evaluated as the number of stained apoptotic cells divided by the total number of prostate epithelial cells. Four high-power microscopic fields were analyzed for each sample counting at least 200 cells as previously described.14 The proliferation rate was calculated similarly, counting the Ki-67–positive nuclei.
Hoechst staining was performed on FFPE sections for all of the xenografts. Tissue sections were deparaffinized, rehydrated, and incubated with the fluorescent dye for 30 seconds. Slides were then mounted with Vectashield aqueous mounting media (Vector Laboratories, Burlingame, CA).
PSA Dosage
Mice were subjected to monthly facial vein bleeding according to the IACUC guidelines. Fifty microliters of serum were used to measure human total PSA with an enzyme immunoassay kit (Alpco Diagnostics Ltd, Salem, NH) following the manufacturer’s instructions. Duplicates were assayed for all of the samples.
Fluorescence in Situ Hybridization
Five-micron sections of FFPE tissues (xenografts and original tumors) were mounted on standard glass slides and baked at 60°C for at least two hours, then deparaffinized and digested using methods described previously.15 The following bacterial artificial chromosome (BAC) clones were obtained from CHORI (Oakland, CA) and were used as probes: (1) RP11-476D17 (including the 3′ portion of ERG), labeled in aqua (PerkinElmer Inc., Waltham, MA); (2) RP11-95I21 (including the 5′portion of ERG), labeled with Spectrum Orange (Abbott Molecular/Vysis Inc., Des Plaines, IL); and (3) RP11-35C4 (maps just distal to TMPRSS2), labeled in Spectrum Green (Abbott Molecular/Vysis Inc., Des Plaines, IL). Approximately 1 microgram of each BAC DNA was direct-labeled using nick translation and precipitated using standard protocols. Final probe concentration was approximately 50 to 100 ng/ul. All three BAC clones were hybridized simultaneously in the ERG break-apart assay. Tissue sections and probes were codenatured, hybridized at least 16 hours at 37°C in a darkened humid chamber, washed in 2× saline-sodium citrate buffer (SSC) at 70°C for 10 minutes, rinsed in room temperature 2× SSC, and counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Abbott Molecular/Vysis, Inc.). Slides were imaged using an Olympus BX51 fluorescence microscope. Individual images were captured using an Applied Imaging system running CytoVision Genus version 3.9.
DNA Extraction, Whole Genome Amplification, and Array-Based Comparative Genomic Hybridization
Tumor and normal epithelium were independently isolated from seven pairs of xenografts and parental human tumors. FFPE tissues were processed by laser capture microdissection (LCM) or manual needle-dissection followed by DNA isolation (QIAamp DNA Micro Kit, QIAGEN, Valencia, CA) and whole genome amplification (WGA2, Sigma-Aldrich, St. Louis, MO). Eight-micron sections of FFPE samples from xenografts and parental human tumors were cut on membrane slides (Molecular Devices, Sunnyvale, CA), stained with HistoGene LCM Paradise Staining kit (Molecular Devices), and then microdissected with a Veritas LCM Instrument (Arcturus Bioscience Inc., Molecular Devices). Amplified DNA was directly labeled and hybridized onto 244K agilent comparative genomic hybridization (CGH) 60-mer oligoarrays as performed previously.16 Data were analyzed with DNA analytics Version 4.0.81 (Agilent Technologies, Inc., Santa Clara CA) software.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism version 4.00 software. Results were compared using Mann–Whitney, Wilcoxon, and Fisher exact tests. P values less than 0.05 were considered significant (*P < 0.05; **P < 0.01; ***P < 0.001).
Results
Generation and Histopathological Characterization of Subrenal Capsule Xenografts of Human Prostate Tissues in Immunodeficient Mice
Tumor and Normal Prostate Xenografts
Thirty human primary prostate tumors were implanted in the subrenal capsule site of 70 immunodeficient mice, and 23 (55 mice) of them were suitable for analysis (seven transplanted tumor samples could not be recovered because the mice died of non–tumor-related causes).
All murine hosts were sacrificed at 3 months to obtain a complete morphological, immunophenotypic, and molecular characterization at a preset time point. Thirteen of these 23 (56%) prostate adenocarcinomas grew in at least one of the murine hosts, whereas three (13%) probably underwent regression, as suggested by the calcifications present in the recovered tissue at the site of implantation. In addition, in seven cases (30%) we found only benign tissue in the xenografts.
Three tumors were implanted in both the subcutaneous and subrenal capsule sites. Take of subcutaneous grafts was observed in one of six mice, whereas all of these tumors were able to grow under the renal capsule. Unfortunately, only a few cases could be implanted subcutaneously because of the limited tissue availability from most of the tumor specimens. These three tumors were exceptionally large and high grade, and the fact that all of them were able to grow under the renal capsule but not under the skin (except one) suggests that the subrenal capsule microenvironment favors tumor take more efficiently than the subcutaneous location.
To compare the tumor take under the renal capsule with that in the orthotopic site, eight early-passage primary cell cultures and 15 freshly dissected prostate tissues were implanted in the prostate of 24 and 23 immunodeficient mice, respectively. Eleven of these tissues were first digested with collagenase, then resuspended in PBS and injected in the anterior prostate lobe. The remaining four prostate tumor samples were instead reduced in 1 × 2 × 1 mm fragments and implanted orthotopically. Three months after surgery, mice were sacrificed and prostate dissected. None of the orthotopic injections/implantations gave a successful outcome in terms of tumor take in mice. In fact, digested tissues injected as cell suspensions produced in most cases an inflammatory reaction in the prostate stroma at the injection site, whereas only normal human prostate tissue was recovered from the xenografts when implantation of entire fragments of tissue was performed. This may be attributable to the histological heterogeneity of prostate tissue or explained as the tumor failed to grow and regressed.
Ten normal tissue samples, dissected from the peripheral zone of the prostate, were implanted under the renal capsule of 20 mice, two per case. Surprisingly, in four of ten normal tissues (eight mice), three months after implantation we still found presence of florid glands mixed with stroma (Figure 1A, upper right panel). Only stroma was found in the other six normal xenografts.
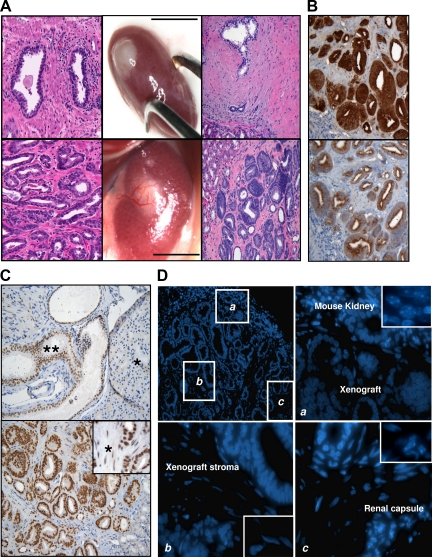
Figure 1.
Generation of mouse xenografts from primary localized human prostate tumors. Subrenal capsule xenografts of prostate tumor tissues (A, lower middle panel) often showed a robust vascularization that was not present in those derived from normal samples (A, upper middle panel). Hematoxylin and eosin staining of 5-micron FFPE sections of normal (A, upper right panel, ×200 magnification) and tumor (A, lower right panel, ×200 magnification) xenografts revealed that both maintain the histopathological features of the parental tissues (A, upper left and lower left panels, respectively). Tumor xenografts showed immunohistochemical markers of human PCa (PSA in B, upper panel, AMACR in B, lower panel, ×200 magnification; androgen receptor in C, lower panel, ×200 magnification, inset ×400). An example of Gleason 3 xenograft is shown. Scale bar = 10 mm. The xenografts did not show infiltration of mouse fibroblasts in the stroma, which was confirmed of human origin by both immunohistochemistry against the human androgen receptor (C, lower panel [asterisk represents human stromal cells]; C, upper panel, shows an orthotopic xenograft of human normal prostate as control [asterisk represents mouse prostate gland, double asterisk represents human prostate tissue]) and Hoechst staining (D; magnifications ×100 in the upper left panel, ×400 in the upper right and bottom panels, ×600 in the insets).
To assess whether the type of immunodeficiency in the murine host affected the tumor take,17,18 we placed 9 of 23 (39%) prostate tumors in the subrenal capsule site of Nu/Nu, 11 of 23 (48%) in NOD/SCID mice, and 3 of 23 (13%) in mice of both strains. The two groups of human tumors were similar in Gleason grade and pathological stage. Nu/Nu and NOD/SCID mice differ mainly because of their immunodeficient status: whereas athymic Nu/Nu mice lack T cells, NOD/SCID mice lack both T and B cells. The tumor take was 7 of 9 (77%) in Nu/Nu mice and 5 of 11 (45%) in NOD/SCID mice, whereas one of the three tumors implanted in both strains grew in all of them. Our experiments showed no significant difference in tumor take between the two mouse strains (P = 0.26).
Morphological and Immunophenotypic Characterization of Tumors
Macroscopically, subrenal capsule implants of normal tissues showed rare or no surrounding blood vessels (Figure 1A, upper middle panel), whereas tumors were usually overlayed by a robust vascularization (Figure 1A, lower middle panel). Despite their ability to recruit vessels, tumors did not show a significant increase in size over 3 months, suggesting that they display the slow growing characteristics of human primary prostate tumors.
All tumor xenografts maintained the histopathological features of the parental tumors (Figure 1A, lower left and right panels). In fact, they not only showed the same histotype and grade of differentiation (assessed by GS), but also resulted in a mix of tumor, stroma, and sometimes benign glands. To further confirm that human tumors implanted in murine hosts were able to maintain the same phenotypic profile over time, paired original prostate cancers and xenografts were screened for canonical PCa markers by immunohistochemistry. Stainings for PSA, AMACR,19 and AR were positive in all of the xenografts (Figures 1B, upper and lower panel, and 1C, lower panel), whereas p63 was negative (image not shown), consistent with human prostate tumors.
Murine Xenografts Maintain the Original Human Stroma
Next, we wanted to determine whether the stroma in the xenografts maintained an exclusive human origin or infiltrating mouse fibroblasts were present mixed with or replacing the human component. To this end, we used two approaches. First, taking advantage of the fact that prostate stroma expresses the androgen receptor, we used a human-specific anti-AR antibody to stain the xenografts. All of the tumor xenografts showed strong nuclear positivity in both epithelium and stroma, thus confirming the presence of human stromal cells. The human specificity of the antibody was tested on an orthotopic mouse xenograft of normal human prostate, in which the antibody exclusively stained the human nuclei, whereas the surrounding mouse tissue remained negative (Figure 1C, upper panel).
The second approach was based on a previous observation20 that human and mouse cell nuclei show a different distribution pattern of the chromatin by a fluorescent dye (Hoechst). The mouse kidney and the renal capsule were clearly different from the nuclei of the human implanted tissue (Figure 1D).
Serum PSA Levels in Mouse Xenografts
To monitor tumor take in vivo, we tested the serum PSA levels in all of the recipient mice. Mice do not produce PSA, and therefore this protein, when detected in the serum, must be produced and secreted into the blood by the grafted human tissue. Sera from mice implanted either with tumor or normal tissues were assayed for total PSA by immunoassay. Only those cases in which at least a duplicate measurement was made were considered.
As expected, serum PSA values in mice implanted with normal tissues were low, in some cases undetectable (mean 1.02 ng/ml ± 1), whereas in the tumor cohort PSA levels displayed a wide range (mean 13 ng/ml ± 10.7). A significant difference between the two groups was found (P < 0.001; Figure 2A). The 3-month PSA was not detectable in any mouse in which tumor propagation failed or tumor presumably regressed.
Figure 2.
A: Serum total PSA was significantly increased (***P < 0.001) in tumor xenograft-bearing mice compared with those implanted with human normal tissues. The box plots show PSA values in the two groups of tumor and normal xenografts. Lower and higher whiskers indicate 10th and 90th percentile, respectively; lower and higher edges of box indicate 25th and 75th percentile, respectively; the inner line in the box indicates 50th percentile. The PSA levels increased over time as the tumor proliferated (B, graph and inset) and dropped to undetectable levels when the tumor regressed (C, graph and inset; asterisk represents calcifications).
Serum PSA progressively increased when the tumor was growing (Figure 2B), whereas it did not change over time in the cases of normal tissue implantation and dropped to undetectable levels in the cases of tissue regression (Figure 2C).
Effect of Clinical-Pathological Parameters, Proliferation, and Apoptotic Rates of Parental Human Tissues on Tumor Take in Mice
We asked whether clinical-pathological parameters such as GS and pathological stage would affect the tumor take in mice. In our cohort of implanted human prostate tumors, 5/10 (50%) of low GS (7 [3 + 4]) cancers grew in mice, whereas, among high GS (7 [4 + 3], 8, and 9) tumors, 8/13 (61%) did (P = 0.68) (Figure 3B). In terms of stage, 8/12 (67%) of pT3 tumors successfully developed xenografts versus 5/11 (45%) of pT2 tumors (P = 0.41; Figure 3C). No association was found between tumor take rate in mice and differentiation grade or pT even when we combined the two pathological parameters to compare tumors with both low GS and pT2 stage (3/6, 50% tumor take) versus tumors with either high GS or pT3 stage (10/17, 59% tumor take; P = 1).
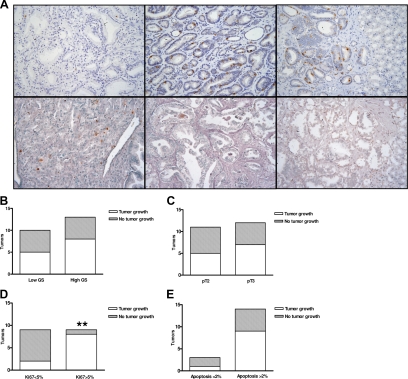
Figure 3.
Proliferation rate (percentage of Ki-67–positive nuclei) and apoptosis (percentage of Apoptag-positive nuclei) in parental prostate tumors and mouse xenografts. Tumors with higher proliferation rate (Ki-67 > 5%; A, upper middle panel, and D; **P < 0.01) grew preferentially in mice compared with those with low proliferation (Ki-67 < 5%; A, upper left panel). Representative examples of tumors with low and high apoptotic rates are reported in panel A, lower middle and lower left, respectively. Apoptotic rate, pathological stage, and GS did not affect the tumor take in mice (P > 0.05; B, C, and E). A, upper right and lower right, shows immunostaining of the xenografts derived from the tumors reported in A, upper middle and lower middle, which maintains the same proliferation rate and apoptosis of the parental tissues (×200 magnification).
To assess the proliferative and apoptotic rates in the original tumors as well as in the corresponding xenografts, we used FFPE tissue samples. Paraffin sections with residual tumor were available for 10 of the 13 pairs of human tumors and xenografts as well as for 8 of the 10 human tumors that did not grow in mice. These sections were immunostained for Ki-67 (proliferation) and Apoptag TUNEL assay (apoptosis).
First, we compared percentages of Ki-67–positive and apoptotic nuclei in each pair of human tumor/xenograft, and found that there was no significant difference between them (P = 0.50 and P = 0.62, respectively; Figure 3A, middle and right panels). Interestingly, when the parental tumor showed areas with high heterogeneity in proliferation, the xenograft displayed the highest proliferation rate found in the original tumor, suggesting that proliferative capacity might represent a major requirement for tumor growth in mice. In fact, tumors with a Ki-67 positivity of ≥5% cut-off (range, 5–16; median 7) had a tumor take of 8/9 (89%) versus those with a Ki-67 posivity lower than 5% (range, 1 to 4%; median 3) that had a tumor take of 2/9 (22%; P < 0.01; Figure 3D). As expected, normal cells showed only rare Ki-67–positive nuclei (see Supplemental Figure S2 at http://ajp.amjpathol.org). Also, a moderately significant correlation (R2 = 0.62) between serum PSA levels and Ki-67% in the xenografts was found (data not shown). Conversely, the apoptotic rate (cut-off 2%) in the human primary tumors did not affect the xenograft take (P = 0.53; Figure 3E).
Genome-Wide Analysis, TMPRSS2-ERG Gene Fusion, and AKT Pathway Activation Signature in Human Primary Tumors and Mouse Xenografts
To investigate whether the xenografts growing in mice recapitulate the genetic background of the original human tumors or acquire genomic alterations that might confer growth advantage, we performed an array-based CGH (aCGH) analysis on micro- (Figure 4A) and needle-dissected FFPE tissues. Fluorescence in situ hybridization (FISH) analysis was also performed to investigate the presence of the TMPRSS2-ERG rearrangement at the 21q22 locus.21 Because of the multiple challenges (scant amount of tissue, highly fragmented genomic DNA), respectively 7 and 9 of the 13 available pairs of human primary tumors and subrenal capsule xenografts were suitable for the two analyses.
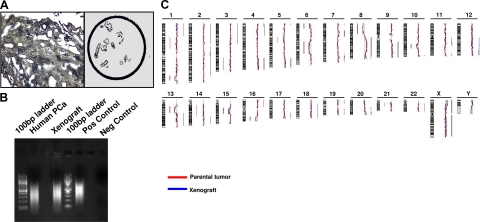
Figure 4.
Genome-wide analysis (aCGH) was performed after laser capture microdissection (A) and whole genome amplification (WGA; B). Genome view of a representative pair human tumor/xenograft shows that the most significant genomic aberrations (losses and gains as reported on the left and on the right of each bar, respectively) are consistent between the two profiles (C).
Importantly, significant genomic gains and losses that have been described as hallmarks of PCa22 were consistent between the human primary tumors and the paired xenografts (Figure 4, B and C). When we looked at the penetrance of these alterations in our tumor cohort, we found that 57% of human tumors and xenografts harbored losses at the PTEN gene locus (10q23) and at 8p, either limited to the 8p21 locus (NKX3A gene) or extended to the entire short arm of the chromosome. Also, 42% of the pairs exhibited loss at 6q15 and at the 16q23 locus, 29% showed loss at 21q22 (responsible for the TMPRSS2-ERG gene fusion) and 13q, and 22% showed loss at 17p (where TP53 gene is located). Gain at 8q, which included Myc oncogene, was present in only one pair of samples (Figure 5, A and B). Of note, all of the tumors and derived xenografts showed at least two of these genetic aberrations.
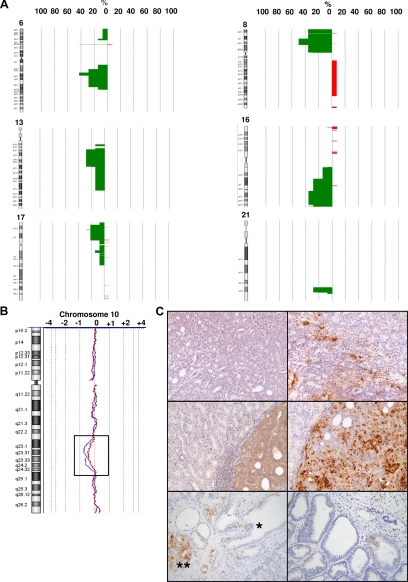
Figure 5.
Penetrance plots of the most frequent genomic alterations occurring in the human parental tumors and in the xenografts. Chromosomes 6, 8, 13, 16, 17, and 21 are shown (A). PTEN loss is shown in a pair of human tumor and xenograft (B), where it is associated with AKT pathway activation (C). Phospho-AKT and phospho-S6 staining is heterogeneous in the parental tumor (C, upper left and right, respectively), whereas the xenograft shows a more intense and diffuse positivity for both proteins (C, middle left and right, respectively; ×400 magnification). Conversely, normal prostate epithelium and PTEN-positive tumors in three-month xenografts maintain low or absent staining for phospho-S6 (C, lower left, asterisk represents normal glands and double asterisk represents tumor, and C, lower right, respectively).
In the attempt to correlate genomic and functional data, we investigated the AKT pathway status in our samples. Phospho-AKT and phospho-S6 were chosen as key proteins in the pathway and detected by immunohistochemical analysis. As expected, the human parental tumors with PTEN loss by aCGH were positive for both phosphoproteins (Figure 5C, upper left and right panels) and, more importantly, AKT pathway activation was maintained and in some cases enhanced in the xenografts (Figure 5C, middle left and right panels). Low or absent phospho-AKT pathway activation was found in normal prostate glands and PTEN-positive tumor xenografts (Figure 5C, lower left and right panels), respectively.
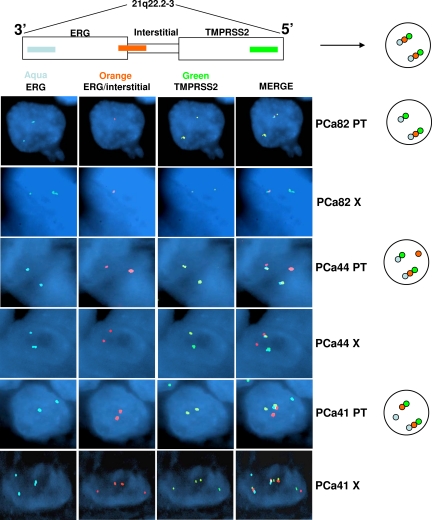
To investigate the presence of the TMPRSS2-ERG gene fusion23 in our human tumors and corresponding xenografts, we also performed FISH using a tricolor assay. In this experiment, three probes were cohybridized as follows: (1) a probe against the 3′ (or, centromeric) sequence of ERG (aqua), (2) a probe including the 5′ (or, telomeric) sequence of ERG (orange), and (3) a probe mapping distal to TMPRSS2 (green; Figure 6, scheme). In the absence of ERG rearrangement the two ERG probes give a fused signal (aqua + orange). In contrast, when interstitial deletion leads to TMPRSS2-ERG fusion, the aqua and green probes colocalize but the orange signal is absent. The assay is designed to identify any ERG rearrangement, which can manifest as physical separation of the red and aqua signals. Fusion of the aqua (3′ ERG) and green (TMPRSS2) signals occurs when TMPRSS2 is the rearrangement partner for ERG. However, in the absence of colocalized aqua and green signals, the resulting pattern is consistent with ERG rearrangement in the absence of TMPRSS2 involvement.
Figure 6.
FISH break-apart assay showed abnormalities of the 21q22.2-3 locus in 77% of the paired human tumors and xenografts. PCa82 (PT indicates parental tumor; X, xenograft) and PCa44 are examples of TMPRSS2-ERG gene fusion by interstitial deletion and translocation, respectively. PCa41, instead, shows a rearrangement of ERG, which could have a 5′ partner different from TMPRSS2. The corresponding xenograft (PCa41 X) presents the same alteration in association with polisomy for the TMPRSS2 and ERG regions (×1000 magnification, oil immersion). Schematic representations of the rearrangements are reported on the right.
Five of the nine (55%) human tumor/xenograft pairs suitable for FISH showed TMPRSS2-ERG fusion, whereas two of them presented polysomy for the respective chromosome region (three to four copies in the original tumors and up to six copies in the xenografts) without TMPRSS2-ERG rearrangement. Interstitial deletion was the mechanism for two of the five fusions (Figure 6, PCa82). ERG rearrangement leading to fusion between the 5′ TMPRSS2 probe and the 3′ ERG probe was identified in the other three pairs (Figure 6, PCa44). In one case, a pattern suggestive ERG rearrangement in the absence of TMPRSS2 involvement was also identified (Figure 6, PCa41). In these three tumors, polysomy (three to four copies) for the region was observed concomitantly with rearrangement of a single copy of ERG in ≈25% of cells (Figure 6, PCa41 X). Taken together, the results from both genetic analyses endorse the hypothesis that human prostate tumors maintain a similar genomic profile until analysis at 3 months.
Further, we asked whether the molecular heterogeneity of the original human tissue is maintained and might contribute to the outcome of the xenograft. In particular, while we observed by FISH analysis that the primary tumors do not present an homogenous TMPRSS2-ERG fusion status, with 35% to 45% of the cells being positive for the rearrangement, the xenograft’s cell population showed the genetic alteration in 60% to 80% of the cells (P < 0.05). This supports the hypothesis that the xenografts tend to enrich in tumor cells bearing driving genetic alterations that could facilitate cell survival and proliferation.
Discussion
The size of prostate tumors at the time of diagnosis is diminishing as a result of clinical screening. Despite this, some cancers continue to behave aggressively. The biomarkers currently available, such as clinical-pathological stage and differentiation grade, have therefore lost their discriminatory power. There is a dire need for novel molecular markers to distinguish aggressive from indolent tumors (eg, 19 prostatectomies are performed seemingly unnecessarily to do a curative procedure on the 20th24).
The search for such predictive markers has, however, become challenging as a result of insufficient tissue available. Our approach to this problem has been to expand primary tumors in the murine host. Provided that in this setting the genomic analysis of the propagated tissue is stable through passaging and reflects the tumor whence it originated, this model allows the identification of biologically aggressive tumors based on their ability to grow in mice and provides a tool for “personalized” preclinical studies. In this article, we describe our attempts at propagating primary tumors of the prostate in the murine host with analysis of xenografts at three months after implantation.
Propagation of human primary prostate cancers in immunodeficient mice is challenging, and successful growth has previously been achieved only with high grade and advanced tumors.25 This study presents a complete histopathological and genomic characterization of a model of prostate cancer xenografts derived from primary localized human tumors.
The reasons for the limited success in the propagation of primary tumors thus far are multiple. These include interspecies incompatibilities (receptor-ligand interactions) between human xenograft and surrounding mouse tissue,26 limited availability of tissue from primary tumor specimens, technical difficulties in the manipulation of the tissue and isolation of cells (enzymatic digestion, mechanical disruption), and characteristics of the mouse anatomical site chosen for implantation (vascularization, surgical accessibility, etc).
Here, we have attempted to address some of these issues. A series of conditions were tested to grow human primary localized prostate tumors in immunodeficient mice (Nu/Nu and NOD/SCID), including orthotopic and subcutaneous implantations to compare the tumor take in these sites with that in the subrenal capsule.
We have found that, regardless of the mouse strain, subrenal capsule implantation of fresh surgical specimens allows tumor take in 56% of localized low (with prevalent Gleason 3 component) and high-grade primary prostate adenocarcinomas. In contrast, orthotopic and subcutaneous implantations were largely unsuccessful. Importantly, the histopathological characterization of these subrenal capsule xenografts has revealed that they maintain both grading and expression of phenotypic markers (AR, PSA, AMACR, p63 loss) of the parental prostate tumors.
Many factors may affect the growth in size of the grafts and the take rate, including biological and histological characteristics of PCa and the ability of the tumor to attract blood vessels. Because of the small size and multifocality of most human primary prostate adenocarcinomas, only two to three 2-mm fragments of histologically confirmed primary tumor were implanted under the renal capsule of each mouse. In addition, consistent with the original adenocarcinomas, all xenografts showed heterogeneity in terms of presence of stroma, which was confirmed to be of human origin, and sometimes of benign glands admixed with tumor cells. Because of such heterogeneity, the percentage of tumor, stroma, and normal tissue components differed in the various implantations. This and the evidence that prostate tumors are usually slow growing in humans may in part be responsible for the limited size of the xenografts at three months after implantation. Neverthless, the initial phase after grafting is critical for tumor take and growth, mainly because of the need to vascularize. The ability to recruit blood vessels from the surroundings or to induce angiogenesis depends on both the angiogenic potential of the tumor27 and the mouse microenvironment. Because the kidney is one of the most vascularized organs, we believe this is one of the reasons for the success in tumor take under the renal capsule as opposed to the subcutaneous or even the orthotopic location. In this context, the balance between the number of proliferating and apoptotic cells may also play a role. In fact, the tumor take in our xenografts was affected primarily by the proliferation rate as measured in the original tumors, but not by their Gleason grade, pathological stage, and apoptotic rate. In addition, when the primary tumor showed heterogeneous areas of Ki-67 positivity, the xenograft always homogeneously displayed the highest proliferation rate recorded in the original tumor. These results strongly suggest that relatively brisk proliferation rate may be a prerequisite for the growth of localized prostate tumors in mice, regardless of their histopathological features.
We were able to follow the tumor take and growth by measuring serum PSA levels in the mice. Only mice in which the tumors grew had in fact measurable levels of PSA. This result is of great impact for future applications of our model. First, it represents a reliable screening method when carrying out preclinical studies on the mouse xenograft population. Second, it suggests that these tumors were vascularized and that other tumor-specific proteins secreted into the blood may be detectable as well. This in vivo model could therefore represent a good tool for biomarker discovery and/or validation. Also, PSA secretion by the epithelium, together with evidence that both the histological architecture and the grade of differentiation of the tumor are conserved, serve as proof of the preservation of the microenvironment in our model. In fact, these properties ensure that physiological hormone concentrations are able to reach and stimulate the epithelial cells, allowing us to conclude that the subrenal capsule site is ideal for prostate cancer grafting experiments.
Third, as PSA is used in the clinic as a marker of PCa recurrence after primary treatment and progression,28 its monthly increment in mice may be considered a surrogate of xenograft’s growth. Indeed, its decline in the presence of putative tumor regression confirms this assumption. Consistent with these data, only tumor cells showed expression of the proliferative marker Ki-67, whereas normal cells did not.
Wang et al29 reported a higher take of both low-grade prostate tumor and normal tissues for subrenal capsule and orthotopic grafts in SCID mice, defining the success as recovery of viable tissues. Here, we wanted to use more stringent parameters to define growth of tumor populations by biological and genetic means, to investigate whether first passage xenografts are consistent with their parental tumors and feasible for further biological and genomic studies.
When we looked at the genome scan of the parental human tumors that grew in mice and their paired xenografts, we observed that all of them harbored an association of two to eight genetic alterations (gains or losses) that have been defined as hallmarks of PCa.22,30 Of relevance, the tumors showed a similar genomic profile over a three-month life span in the mouse subrenal capsule site. Significant genetic alterations included losses at 6q15, 8p, 10q23, 13q, 16q23, and 17p as well as 8q gain. Also, 77% of these pairs of samples harbored the TMPRSS2-ERG fusion, one of the most common genetic alterations in prostate tumors,31 and/or a polisomy of the same chromosomal region involved in the fusion, 21q22.2-3.
We found that in our cohort of tumors, 60% of the fusions occurred by translocation of telomeric ERG and adjacent interstitial DNA, and in one of these cases the hybridization pattern was suggestive also for a rearrangement of centromeric ERG that did not lead to the TMPRSS2-ERG fusion. Forty percent of the fusions occurred by interstitial deletion, independently confirmed by the aCGH results.
Interestingly, the majority (60% to 80%) of tumor cells in the xenograft showed such aberrations, versus 35% to 45% of cells in the parental human tumors. It is tempting to speculate that population of cells in the original tumor bearing either the TMPRSS2-ERG rearrangement or other defined genetic alterations such as PTEN loss, which are associated with tumor aggressiveness,31,32,33 would preferentially grow in the xenografts, under selective pressure. In fact, activation of the AKT pathway, which was associated with PTEN deletion in our cases, was diffusely present in the xenografts whereas heterogeneous areas of activation were seen in the original tumors.
Overall, we observed that the subrenal capsule xenografts recapitulate the complexity of the genomic background of the primary tumors, and no new significant genomic aberrations occurred. In addition, although enrichment in cells bearing the TMPRSS2-ERG fusion or showing activation of the AKT pathway in association with PTEN loss was found in the xenografts, there was no case in which these aberrations were present in the xenograft but absent in the primary tumor.
A limitation of our xenograft model is that the tumors do not appear to grow in size over a time frame of three months, thus making drug testing studies challenging. However, these experiments could be performed either after serial transplantation or using first generation xenografts, particularly when large tumor specimens are available. This would require the implantation of many mice and an imaging system able to detect biological changes in the tumor (molecular imaging), eg, apoptosis labeling in vivo, instead of size measurement, together with the circulating PSA levels. In fact, we showed that the apoptosis rate does not increase in the xenografts compared with the parental tumors, whereas serum PSA may be used to monitor prostate cancer progression or response to therapy.
In conclusion, the mouse xenograft model that we have established through direct implantation of human primary prostate tumors represents a useful preclinical model for future applications in biological studies aimed at the identification of the most aggressive and possibly recurrent human localized prostate tumors as well as in biomarker discovery.
Acknowledgments
We thank Levi Garraway for critical reading of the manuscript, Lingxian Wu for technical help, and Alexei Protopopov for array CGH.
Footnotes
Address reprint requests to Massimo Loda, M.D., Dana Farber Cancer Institute, D1536, 44 Binney Street, Boston, MA 02115. E-mail: massimo_loda@dfci.harvard.edu.
Supported by Department of Defense grant W81XWH-06-1-0053 and National Cancer Institute RO1CA131945, PO1CA89021 and P50CA90381 (to M.L.). C.P. was funded by a Ph.D. program from the Catholic University of Rome (Italy).
Supplemental material for this article can be found at http://ajp.amjpathol.org.
References
- D'Amico AV, Whittington R, Malkowicz SB, Weinstein M, Tomaszewski JE, Schultz D, Rhude M, Rocha S, Wein A, Richie JP. Predicting prostate specific antigen outcome preoperatively in the prostate specific antigen era. J Urol. 2001;166:2185–2188. [PubMed] [Google Scholar]
- Thompson I, Thrasher JB, Aus G, Burnett AL, Canby-Hagino ED, Cookson MS, D'Amico AV, Dmochowski RR, Eton DT, Forman JD, Goldenberg SL, Hernandez J, Higano CS, Kraus SR, Moul JW, Tangen CM. Guideline for the management of clinically localized prostate cancer: 2007 update. J Urol. 2007;177:2106–2131. doi: 10.1016/j.juro.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Stark JR, Perner S, Stampfer MJ, Sinnott JA, Finn S, Eisenstein AS, Ma J, Fiorentino M, Kurth T, Loda M, Giovannucci EL, Rubin MA, Mucci LA. Gleason score and lethal prostate cancer: does 3 + 4 = 4 + 3? J Clin Oncol. 2009;27:3459–3464. doi: 10.1200/JCO.2008.20.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel RE, Sadar MD. Cell lines used in prostate cancer research: a compendium of old and new lines–part 2. J Urol. 2005;173:360–372. doi: 10.1097/01.ju.0000149989.01263.dc. [DOI] [PubMed] [Google Scholar]
- Sobel RE, Sadar MD. Cell lines used in prostate cancer research: a compendium of old and new lines–part 1. J Urol. 2005;173:342–359. doi: 10.1097/01.ju.0000141580.30910.57. [DOI] [PubMed] [Google Scholar]
- Cespedes MV, Casanova I, Parreno M, Mangues R. Mouse models in oncogenesis and cancer therapy. Clin Transl Oncol. 2006;8:318–329. doi: 10.1007/s12094-006-0177-7. [DOI] [PubMed] [Google Scholar]
- Van Dyke T, Jacks T. Cancer modeling in the modern era: progress and challenges. Cell. 2002;108:135–144. doi: 10.1016/s0092-8674(02)00621-9. [DOI] [PubMed] [Google Scholar]
- Roy-Burman P, Wu H, Powell WC, Hagenkord J, Cohen MB. Genetically defined mouse models that mimic natural aspects of human prostate cancer development. Endocr Relat Cancer. 2004;11:225–254. doi: 10.1677/erc.0.0110225. [DOI] [PubMed] [Google Scholar]
- Garber K. Realistic rodents? Debate grows over new mouse models of cancer. J Natl Cancer Inst. 2006;98:1176–1178. doi: 10.1093/jnci/djj381. [DOI] [PubMed] [Google Scholar]
- Peehl DM. Are primary cultures realistic models of prostate cancer? J Cell Biochem. 2004;91:185–195. doi: 10.1002/jcb.10691. [DOI] [PubMed] [Google Scholar]
- Pretlow TG, Wolman SR, Micale MA, Pelley RJ, Kursh ED, Resnick MI, Bodner DR, Jacobberger JW, Delmoro CM, Giaconia JM, Pretlow TP. Xenografts of primary human prostatic carcinoma. J Natl Cancer Inst. 1993;85:394–398. doi: 10.1093/jnci/85.5.394. [DOI] [PubMed] [Google Scholar]
- van Weerden WM, de Ridder CM, Verdaasdonk CL, Romijn JC, van der Kwast TH, Schroder FH, van Steenbrugge GJ. Development of seven new human prostate tumor xenograft models and their histopathological characterization. Am J Pathol. 1996;149:1055–1062. [PMC free article] [PubMed] [Google Scholar]
- Nanni S, Priolo C, Grasselli A, D'Eletto M, Merola R, Moretti F, Gallucci M, De Carli P, Sentinelli S, Cianciulli AM, Mottolese M, Carlini P, Arcelli D, Helmer-Citterich M, Gaetano C, Loda M, Pontecorvi A, Bacchetti S, Sacchi A, Farsetti A. Epithelial-restricted gene profile of primary cultures from human prostate tumors: a molecular approach to predict clinical behavior of prostate cancer. Mol Cancer Res. 2006;4:79–92. doi: 10.1158/1541-7786.MCR-05-0098. [DOI] [PubMed] [Google Scholar]
- Migita T, Ruiz S, Fornari A, Fiorentino M, Priolo C, Zadra G, Inazuka F, Grisanzio C, Palescandolo E, Shin E, Fiore C, Xie W, Kung AL, Febbo PG, Subramanian A, Mucci L, Ma J, Signoretti S, Stampfer M, Hahn WC, Finn S, Loda M. Fatty acid synthase: a metabolic enzyme and candidate oncogene in prostate cancer. J Natl Cancer Inst. 2009;101:519–532. doi: 10.1093/jnci/djp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, Chheda MG, Tamayo P, Finn S, Shrestha Y, Boehm JS, Jain S, Bojarski E, Mermel C, Barretina J, Chan JA, Baselga J, Tabernero J, Root DE, Fuchs CS, Loda M, Shivdasani RA, Meyerson M, Hahn WC. c17 CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protopopov A, Feng B, Chin L. Full complexity genomic hybridization on 60-mer oligonucleotide microarrays for array comparative genomic hybridization (aCGH). Methods Mol Biol. 2008;439:87–100. doi: 10.1007/978-1-59745-188-8_6. [DOI] [PubMed] [Google Scholar]
- Visonneau S, Cesano A, Torosian MH, Miller EJ, Santoli D. Growth characteristics and metastatic properties of human breast cancer xenografts in immunodeficient mice. Am J Pathol. 1998;152:1299–1311. [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Yamaguchi H, Watanabe M, Yamamoto T, Takahara T, Takeuchi T, Furukawa T, Kase S, Kodaira S, Ishibiki K, Kitajima M. Growth of human tumor xenografts in nude mice and mice with severe combined immunodeficiency (SCID). Surg Today. 1993;23:375–377. doi: 10.1007/BF00309059. [DOI] [PubMed] [Google Scholar]
- Kumar-Sinha C, Shah RB, Laxman B, Tomlins SA, Harwood J, Schmitz W, Conzelmann E, Sanda MG, Wei JT, Rubin MA, Chinnaiyan AM. Elevated alpha-methylacyl-CoA racemase enzymatic activity in prostate cancer. Am J Pathol. 2004;164:787–793. doi: 10.1016/s0002-9440(10)63167-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser FG, Dorman BP, Ruddle FH. Mouse-human heterokaryon analysis with a 33258 Hoechst-Giemsa technique. J Cell Biol. 1975;66:676–680. doi: 10.1083/jcb.66.3.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Ishkanian AS, Mallof CA, Ho J, Meng A, Albert M, Syed A, van der Kwast T, Milosevic M, Yoshimoto M, Squire JA, Lam WL, Bristow RG. High-resolution array CGH identifies novel regions of genomic alteration in intermediate-risk prostate cancer. Prostate. 2009;69:1091–1100. doi: 10.1002/pros.20959. [DOI] [PubMed] [Google Scholar]
- Tu JJ, Rohan S, Kao J, Kitabayashi N, Mathew S, Chen YT. Gene fusions between TMPRSS2 and ETS family genes in prostate cancer: frequency and transcript variant analysis by RT-PCR and FISH on paraffin-embedded tissues. Mod Pathol. 2007;20:921–928. doi: 10.1038/modpathol.3800903. [DOI] [PubMed] [Google Scholar]
- Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, Denis LJ, Recker F, Berenguer A, Maattanen L, Bangma CH, Aus G, Villers A, Rebillard X, van der Kwast T, Blijenberg BG, Moss SM, de Koning HJ, Auvinen A. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- Pienta KJ, Abate-Shen C, Agus DB, Attar RM, Chung LW, Greenberg NM, Hahn WC, Isaacs JT, Navone NM, Peehl DM, Simons JW, Solit DB, Soule HR, VanDyke TA, Weber MJ, Wu L, Vessella RL. The current state of preclinical prostate cancer animal models. Prostate. 2008;68:629–639. doi: 10.1002/pros.20726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan A, Weinberg RA. Opinion: comparative biology of mouse versus human cells: modelling human cancer in mice. Nat Rev Cancer. 2003;3:952–959. doi: 10.1038/nrc1235. [DOI] [PubMed] [Google Scholar]
- Naumov GN, Akslen LA, Folkman J. Role of angiogenesis in human tumor dormancy: animal models of the angiogenic switch. Cell Cycle. 2006;5:1779–1787. doi: 10.4161/cc.5.16.3018. [DOI] [PubMed] [Google Scholar]
- Loeb S, Catalona WJ. Prostate-specific antigen in clinical practice. Cancer Lett. 2007;249:30–39. doi: 10.1016/j.canlet.2006.12.022. [DOI] [PubMed] [Google Scholar]
- Wang Y, Revelo MP, Sudilovsky D, Cao M, Chen WG, Goetz L, Xue H, Sadar M, Shappell SB, Cunha GR, Hayward SW. Development and characterization of efficient xenograft models for benign and malignant human prostate tissue. Prostate. 2005;64:149–159. doi: 10.1002/pros.20225. [DOI] [PubMed] [Google Scholar]
- Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P, Ferrari M, Hernandez-Boussard T, Brooks JD, Pollack JR. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–8510. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto M, Cunha IW, Coudry RA, Fonseca FP, Torres CH, Soares FA, Squire JA. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer. 2007;97:678–685. doi: 10.1038/sj.bjc.6603924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall P, Witton CJ, Grimsley S, Nielsen KV, Edwards J. Is PTEN loss associated with clinical outcome measures in human prostate cancer? Br J Cancer. 2008;99:1296–1301. doi: 10.1038/sj.bjc.6604680. [DOI] [PMC free article] [PubMed] [Google Scholar]