Abstract
We report the synthesis and characterization of polyvalent RNA-gold nanoparticle conjugates (RNA-Au NPs), nanoparticles that are densely functionalized with synthetic RNA oligonucleotides and designed to function in the RNAi pathway. The particles were rationally designed and synthesized to be free of degrading enzymes, have a high surface loading of siRNA duplexes, and contain an auxiliary passivating agent for increased stability in biological media. The resultant conjugates have a half-life six times longer than free dsRNA, readily enter cells without the use of transfection agents, and demonstrate a high gene knockdown capability in a cell model.
Over the past decade, researchers have designed, synthesized, studied, and applied polyvalent DNA-functionalized gold nanoparticles (DNA-Au NPs).1 These efforts have resulted in a new fundamental understanding of hybrid nanostructures,2 important and in certain cases commercially viable detection and diagnostic assays,3 and the ability to program materials assembly through the use of DNA synthons.1,4 Polyvalent DNA-Au NPs have several unique properties, such as sharp and elevated melting temperatures,2b enhanced binding properties2c (as compared with free strands of the same sequence), and distance-dependent optical properties.5 In agreement with research on polyvalent molecular systems,6 the high surface DNA density and the ability of the nanoparticles to engage in multidentate interactions are the proposed origin of these unique properties.
Recently, we demonstrated the utility of the polyvalent DNA-Au NP for antisense gene regulation, where the unique ensemble properties of the conjugate confer several important advantages in the context of intracellular target recognition and binding.7 These properties include resistance to nuclease degradation and high cellular uptake as a result of their oligonucleotide functionalization. Although antisense DNA is an important way of regulating genes, an even more promising route is through the use of siRNA.8 However, no methods have been developed for utilizing polyvalent particles and their unusual properties to load and transport RNA across cell membranes. Indeed, one must develop synthetic routes and materials that overcome one of the most challenging problems associated with RNA, most notably its chemical instability.
Based upon our observations with DNA-modified particles, we hypothesized that gold nanoparticles densely functionalized with synthetic RNA oligonucleotides would take advantage of the ensemble properties that result from the dense surface functionalization of oligonucleotides, increase the stability and efficacy of the bound RNA, while retaining the ability to act in the highly potent and catalytic RNA interference pathway. While others have prepared siRNAs with chemical modifications, peptide conjugation, or as nanomaterials coated or embedded with small or synthetic RNA for the purposes of enzymatic detection, siRNA delivery, or even control of nanoparticle shape,9 here, we describe a unique nanoparticle conjugate that has a dense, covalently attached RNA monolayer. This functionalization results in a conjugate with unique properties that make it a potent material for cellular uptake, serum stability, and genetic regulation without the use of auxillary transfection strategies or chemical modifications.
The synthesis of polyvalent RNA-nanoparticle conjugates (RNA-Au NPs) requires that all components are free of nucleases, such as RNase, which degrades RNA ligands and results in unstable Au NP interactions by exposing the Au surface (as evidenced by aggregation). While conditions for preparing RNase-free organic components and solutions are well established, conditions for creating RNase-free inorganic gold nanoparticles have not yet been described. We determined that treatment of citrate-capped Au NPs with diethylpyrocarbonate (DEPC) followed by autoclaving at 121°C for 60 minutes yields sterile, RNase-free gold colloids, as measured using an RNase activity kit (Supporting Information). Importantly and quite surprisingly, the optical and physical properties of the nanoparticles are unaffected by this relatively extreme treatment, as measured by UV spectroscopy and TEM analysis (Supporting Information). The resultant RNase-free nanoparticles were amenable to further modification by thiolated oligonucleotides using published procedures.2a Without this pretreatment, subsequent functionalization with RNA could not be accomplished, presumably due to rapid degradation of the RNA-based surface capping ligands. Duplexes composed of a 27-base RNA strand, and 25-base complement terminated with an ethylene glycol spacer and alkylthiol, were hybridized and added to the RNase-free Au NPs, where they were allowed to chemisorb via the thiol-gold bond. For this work, the sequences were designed to target the firefly luciferase gene (Supporting Information).
To prevent dehybridization of the RNA duplex, the salt concentration of the Au NP solution was adjusted to 0.1 M with NaCl prior to duplex addition. Subsequently, the salt concentration was increased from 0.1 to 0.3 M NaCl over 12 hours with brief ultrasonication following each addition to increase the coverage of oligonucleotides on the nanoparticle surface.10 To yield a more stable conjugate, RNA-functionalized particles were treated with 30 μmol/mL of oligoethylene glycol-thiol (OEG-Thiol) as an additional surface passivating ligand (Scheme 1). OEG-Thiol passivation was found to stabilize these nanomaterials under cell culture conditions for extended periods of time (Supporting Information).
Scheme 1.
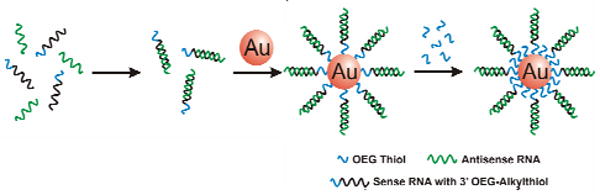
Preparation of Polyvalent RNA Gold Nanoparticle Conjugates. RNA duplexes (containing a double-stranded RNA sequence, an ethylene glycol spacer, and an alkylthiol group) are formed via hybridization and subsequently incubated with RNase free nanoparticles. Further passivation with oligoethylene glycol-thiol (OEG-Thiol) adds additional stability in cell culture.
Following synthesis, RNA-Au NPs were purified by three successive centrifugation, supernatant removal, and addition of phosphate buffered saline (PBS) steps at 4 °C. Refrigerated centrifugation was necessary to prevent duplex dehybridization due to heating caused by the centrifugation process. To determine dsRNA loading, antisense strands were labeled with Cyanine 3 (Cy3) fluorescent dye. The number of duplexes per particle was calculated by oxidizing the gold using 25 μM potassium cyanide, measuring the number of fluorescent antisense strands (indicative of formed duplexes), and dividing by the concentration of nanoparticles. Each RNA-gold nanoparticle conjugate contains 33 ± 4 RNA duplexes per 13 nm Au NP.
The ability of the conjugates to enter cells was investigated by confocal microscopy using fluorescent (Cyanine 5) conjugates prepared as above. RNA-Au NPs were added to cultures of HeLa cells. Imaging studies reveal fluorescence throughout the cytoplasm of HeLa cells after 6 hours (Figure 1a). It is interesting to note that, like DNA-Au NPs, RNA-Au-NPs require no transfection agent to enter cells.11 Indeed, analytical flow cytometry confirmed uptake of the RNA-Au NPs in >99% of the cell population (Figure 1b).
Figure 1.
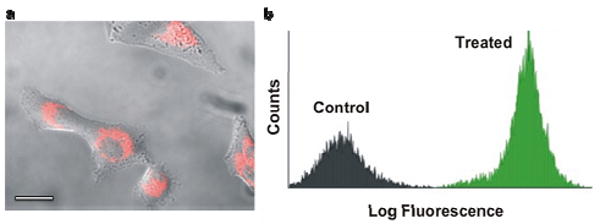
Cellular uptake of RNA-Au NPs. (a) Fluorescence microscopy images of HeLa cells incubated for 6 hours with RNA-Au NPs (Cy5 labeled RNA). Scale bar is 20 μm (b) Flow cytometry analysis comparing RNA –Au NP treated cells to untreated controls.
Having determined that RNA-Au NPs are internalized by cells, we next examined the intracellular activity of RNA-gold nanoparticle conjugates. We conducted protein knockdown studies in HeLa cells using a transfected luciferase plasmid as a target for this model system. Briefly, HeLa cells were seeded in 96 well plates and transfected with a plasmid containing both Renilla and firefly luciferase. The media was replaced 24 hours after plasmid introduction with media containing the functionalized RNA-Au NPs (3 nM nanoparticle concentration, ∼100 nM RNA duplex concentration) directed against firefly luciferase. For comparison, we transfected the same number of luciferase siRNA duplexes (100 nM) using the commercial agent Lipofectamine 2000. Quantification of luciferase expression reveals that the nanoparticle agents down- regulate firefly luciferase in a dose and time dependent manner. Control (Renilla) expression is not affected by RNA-Au NPs designed against firefly luciferase, indicating that the knockdown is also sequence specific. No changes in cell viability that would indicate off-target effects were observed, although this remains an active area of investigation. Interestingly, the results of three independent experiments with the RNA-Au NPs show knockdown that exceeds that of the free RNA 4 days after treatment (73 ± 7% RNA-Au NPs vs. 33 ± 2% free, Figure 2a).
Figure 2.
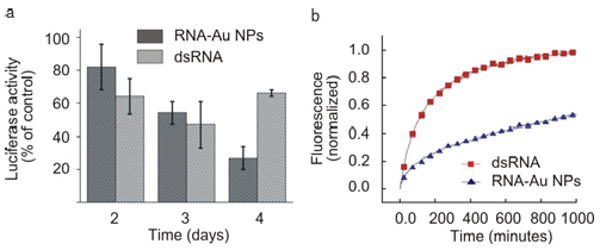
(a) Knockdown of luciferase expression over 4 days. (b) Stability of RNA- Au NPs. Comparison of the stability of dsRNA (red) and RNA- Au NPs (blue) in 10% serum.
We hypothesize that the persistent knockdown of luciferase may be the result of the stabilization of the RNA on the nanoparticle. In stability experiments, conjugates incubated in serum show greatly enhanced stability relative to their molecular RNA counterparts. For example, in the presence of 10% serum, RNA-Au NP conjugates have a 6 times greater half-life than molecular RNA duplexes (816 ± 59 mins vs. 133 ± 30 mins, Figure 2b). These data indicate that nanoparticle conjugation provides significant protection from degradation in an extracellular context. This stabilization is likely due to the high local salt concentration, which is known to deactivate enzymatic activity.12 Since the extracellular lifetime of RNA is of great importance for their storage, handling, and potential therapeutic application, nanoparticle conjugation may provide a significant advantage for the protection and delivery of functional RNA ligands. Importantly, this enhanced stability does not require chemical modifications to protect the integrity of the RNA.
We have developed the first polyvalent RNA-gold nanoparticle conjugates, which can be used to effectively regulate genes in the context of RNA interference. In contrast to other materials for introducing RNA to a cell, these conjugates are not simply a delivery tool for RNA, but rather single entity agents which take advantage of the cooperative properties that result from the arrangement and high density of the surface ligands.
The novel conjugate described here is a densely functionalized nanomaterial that is protected from degradation in a manner that enhances knockdown compared to conventional lipid carriers and enters cells without transfection agents. Note that in recent work we have shown that DNA density results in a decrease in nuclease activity as a result of high local salt concentration.13 The enhanced stability we observe here for the RNA-Au NPs may be due to the same phenomena. Given their straightforward synthesis, these RNA-Au NPs, in addition to useful intracellular gene regulation materials, may be promising candidates for therapeutic and diagnostic applications.
Supplementary Material
Acknowledgments
C.A.M. acknowledges a Cancer Center for Nanotechnology Excellence (NCI-CCNE) award for support of this research. C.A.M. is also grateful for an NIH Director's Pioneer Award. D.S.S. was supported by the LUNGevity Foundation – American Cancer Society Postdoctoral Fellowship in Lung Cancer. P.C.P. was supported by a Ryan Fellowship.
Footnotes
Supporting Information Available: Experimental conditions, sequences, and materials synthesized. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 2.(a) Demers LM, Mirkin CA, Mucic RC, Reynolds RA, III, Letsinger RL, Elghanian R, Viswanadham G. Anal Chem. 2000;72:5535. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]; (b) Jin RC, Wu GS, Li Z, Mirkin CA, Schatz GC. J Am Chem Soc. 2003;125:1643. doi: 10.1021/ja021096v. [DOI] [PubMed] [Google Scholar]; (c) Lytton-Jean AK, Mirkin CA. J Am Chem Soc. 2005;127:12754–12754. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]; (d) Storhoff JJ, Lazarides AA, Mucic RC, Mirkin CA, Letsinger RL, Schatz GC. J Am Chem Soc. 2000;122:4640. [Google Scholar]; (e) You C, Verma A, Rotello VM. Soft Matter. 2006;2:190. doi: 10.1039/b517354j. [DOI] [PubMed] [Google Scholar]; (f) Wang L, O'Donoghue MB, Tan W. Nanomed. 2006;1:413. doi: 10.2217/17435889.1.4.413. [DOI] [PubMed] [Google Scholar]
- 3.(a) Nam JM, Thaxton CS, Mirkin CA. Science. 2003;301:1884. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]; (b) Stoeva SI, Lee JS, Smith JE, Rosen ST, Mirkin CA. J Am Chem Soc. 2006;128:8378. doi: 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]; (c) Liu JW, Lu Y. J Am Chem Soc. 2004;126:12298. doi: 10.1021/ja046628h. [DOI] [PubMed] [Google Scholar]; (d) Faulds K, Smith WE, Graham D. Anal Chem. 2004;76:412. doi: 10.1021/ac035060c. [DOI] [PubMed] [Google Scholar]
- 4.(a) Park SY, Lytton-Jean AK, Lee B, Weigand S, Schatz GC, Mirkin CA. Nature. 2008;451:553. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]; (b) Nykypanchuk D, Maye MM, van der Lelie D, Gang O. Nature. 2008;451:549. doi: 10.1038/nature06560. [DOI] [PubMed] [Google Scholar]
- 5.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 6.Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. J Am Chem Soc. 2002;124:14922. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]
- 7.(a) Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Science. 2006;312:1027. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]; (b) Seferos DS, Giljohann DA, Rosi NL, Mirkin CA. Chembiochem. 2007;8:1230. doi: 10.1002/cbic.200700262. [DOI] [PubMed] [Google Scholar]; (c) Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. J Am Chem Soc. 2007;129:15477. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.lbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 9.(a) Kim JH, Estabrook RA, Braun G, Lee BR, Reich NO. Chem Comm. 2007:4342. doi: 10.1039/b710306a. [DOI] [PubMed] [Google Scholar]; (b) Motoi Oishi JN, Ishii T, Nagasaki Y. Chemistry Letters. 2006;35:1046. [Google Scholar]; (c) Derfus AM, Chen AA, Min DH, Ruoslahti E, Bhatia SN. Bioconj Chem. 2007;18:1391. doi: 10.1021/bc060367e. [DOI] [PubMed] [Google Scholar]; (d) Khaled A, Guo SC, Li F, Guo PX. Nano Lett. 2005;5:1797. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Gugliotti LA, Feldheim DL, Eaton BE. J Am Chem Soc. 2005;127:17814. doi: 10.1021/ja055039o. [DOI] [PubMed] [Google Scholar]; (f) Baigude H, McCarroll J, Yang CS, Swain PM, Rana TM. ACS Chem Biol. 2004;2:2317. doi: 10.1021/cb7000582. [DOI] [PubMed] [Google Scholar]; (g) Watts JK, Deleavey GF, Damha MJ. Drug Discovery Today. 2008;13:842. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Hurst SJ, Lytton-Jean AK, Mirkin CA. Anal Chem. 2006;78:8313. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Nano Lett. 2007;7:3818. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shack J. J Biol Chem. 1959;234:3003. [PubMed] [Google Scholar]
- 13.Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Nano Lett. 2009;9:308. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


