Abstract
Autosomal dominant facioscapulohumeral muscular dystrophy (FSHD) is likely caused by epigenetic alterations in chromatin involving contraction of the D4Z4 repeat array near the telomere of chromosome 4q. The precise mechanism by which deletions of D4Z4 influence gene expression in FSHD is not yet resolved. Regulatory models include a cis effect on proximal gene transcription (position effect), DNA looping, non-coding RNA, nuclear localization and trans-effects. To directly test whether deletions of D4Z4 affect gene expression in cis, nascent RNA was examined in single myonuclei so that transcription from each allele could be measured independently. FSHD and control myotubes (differentiated myoblasts) were subjected to sequential RNA–DNA FISH. A total of 16 genes in the FSHD region (FRG2, TUBB4Q, FRG1, FAT1, F11, KLKB1, CYP4V2, TLR3, SORBS2, PDLIM3 (ALP), LRP2BP, ING2, SNX25, SLC25A4 (ANT1), HELT and IRF2) were examined for interallelic variation in RNA expression within individual myonuclei. Sequential DNA hybridization with a unique 4q35 chromosome probe was then applied to confirm the localization of nascent RNA to 4q. A D4Z4 probe, labeled with a third fluorochrome, distinguished between the deleted and normal allele in FSHD nuclei. Our data do not support an FSHD model in which contracted D4Z4 arrays induce altered transcription in cis from 4q35 genes, even for those genes (FRG1, FRG2 and SLC25A4 (ANT1)) for which such an effect has been proposed.
Keywords: facioscapulohumeral muscular dystrophy, FSHD, allelic expression, RNA FISH, FRG1l, ANT1
Introduction
Facioscapulohumeral muscular dystrophy (FSHD) is an autosomal dominant neuromuscular disorder.1, 2 With a frequency of 1 in 20 000, FSHD is the third most common hereditary myopathy.3 Clinically, FSHD manifests as facial muscle weakness during adolescence with progression to the upper and then lower extremities later in adulthood. The primary tissue affected is the skeletal muscle, although other symptoms may include sensorineural hearing loss, retinal telangiectasias, epilepsy and, in the most severely affected cases, mental retardation.4, 5 The disease generally presents in the second decade of life. Disease onset, progression and symptoms present with considerable variability between patients even within the same family, ranging from mild muscle disease to wheelchair dependence.
A vast majority of patients with FSHD (97%) present with a contraction of the macrosatellite repeat D4Z4 on the subtelomere of chromosome 4q (4q35).6, 7 D4Z4 is a polymorphic tandem repeat array consisting of 3.3 kb units. In the general population, the D4Z4 array spans 11 to 150 units, whereas the repeat array is contracted to only 1 to 10 elements on one allele in patients with FSHD. As a result of an ancient duplication, the D4Z4 repeat array is also present on the subtelomere of chromosome 10.8 However, contractions of the D4Z4 repeat on 10q26 are not associated with FSHD despite extensive homology proximal and distal to the array on 4q35 and 10q26.
The presence of a contracted D4Z4 repeat array on 4qter alone is not sufficient to cause FSHD. Distal to the D4Z4 repeat, a genetic variation has been identified and is distinguished as alleles 4qA and 4qB. Although the two alleles are equally distributed in the general population, all reported patients with FSHD carry the 4qA variant in parallel to the contracted D4Z4 repeat.9 The predominant difference between the two allelic variants is the presence of a 6.2 kb β-satellite repeat on the 4qA allele. Furthermore, only certain haplotypes of 4qA are associated with the disease, indicating that specific sequence polymorphisms contribute to the disease mechanism.10 Just how these polymorphic variations are involved in FSHD pathogenesis is unclear.
Over the past two decades, several models have been proposed to explain the disease mechanism underlying FSHD. These modes of gene regulation include a cis effect on proximal gene transcription (position effect),11, 12 DNA looping,13 non-coding RNA,14 nuclear localization15, 16 and trans-effect.17 Perhaps the most generally accepted model is that of a cis effect in which the intact D4Z4 repeat array is involved in controlling the expression of genes located proximally on 4q35, and possibly at large distances from the array. Such position effects are known to influence human disease genes at distances >1 Mb.18 Contraction of D4Z4 array to <11 units is thought to cause an inappropriate expression of 4q35 genes, resulting in FSHD pathology. Upregulation of several genes: SLC25A4 (ANT1), FRG1 and FRG2, was detected in one study of FSHD muscle tissue using chromosome-4-specific reverse transcriptase PCR.19 However, this upregulation has not been confirmed by several other studies using techniques varying from microarray analysis to quantitative real-time PCR.20, 21, 22, 23, 24 In addition, chromatin studies have also not supported a cis-spreading effect.13, 20
This study aims to resolve the controversy as to whether 4q35 genes are transcriptionally upregulated by a cis effect in FSHD. To this end, we investigated nascent RNA transcription in single myonuclei using quantitative fluorescent hybridization.25, 26, 27 Earlier expression studies have assayed pooled mRNA levels without unambiguously distinguishing transcription from the contracted versus the unaffected allele. Our approach uses sequential RNA–DNA FISH for quantitation of variations in interallelic RNA transcription within individual nuclei. Thus, any potential cis effect can be directly assayed within individual nuclei. In addition, by examining pre-mRNA (nascent) transcription, variation in RNA quantities attributed to posttranscriptional processing of transcripts (that is, unrelated to nascent cis effects) is avoided. Our approach has the further advantage in that, by examining individual nuclei, the chromosome-4-specific expression of multicopy genes, such as FRG1 and FRG2, can be unambiguously determined.
In this study, transcription was measured specifically in differentiated myotubes derived from normal and mildly affected muscle cells (quadriceps and deltoid). As FSHD is a progressive neuromuscular disorder, cultures were derived from muscle before the onset of severe dystrophy, so that transcriptional activity leading to the muscle phenotype could be observed as a primary effect and not secondary to the dystrophic process. Previous studies have used pooled mRNA isolated from muscle tissue containing a variety of cell types or cultured cells at different stages of differentiation, leading to variability in expression patterns that might not be correlated with the disease. By restricting our analysis to one myogenic cell type at a specific stage of differentiation, such variation in transcription unrelated to disease was minimized. Here, we present an overview of nascent RNA transcription from 16 genes in the FSHD region of chromosome 4q35.
Materials and methods
Cell lines and tissue culture
The FSHD primary myoblast cell lines, FM41j and FM1243j, and the control myoblast cell lines, Cl-02 and CM020399, were propagated on gelatin-coated flasks in F10 medium supplemented with 20% FBS, 100 U/ml penicillin/streptomycin, 10 ng/ml bFGF (Invitrogen, Carlsbad, CA, USA) and 1 μ dexamethasone (Sigma-Aldrich, St Louis, MO, USA). Myoblast cultures were derived from quadriceps or deltoid biopsies; FSHD samples were from mildly affected muscle based on histopathology. To allow for myotube differentiation, cells were grown on laminin-coated coverslips in DMEM medium supplemented with 2% horse serum, 2 m-glutamine and 100 U/ml penicillin/streptomycin (Invitrogen) for 5 days. Before fixation in 4% paraformaldehyde, cells were treated with a cytoskeleton extraction buffer containing 0.5% Triton X-100 and 10 m VRC (vanadyl ribonucleoside complex).26 Subsequently, coverslips were stored in 70% ethanol at 4°C. Myoblast lineage was verified by standard immunostaining with the polyclonal desmin antibody Y-20 (1:200) and a rhodamine-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Differentiation was confirmed by the appearance of fused, multinucleated myotubes and the expression of myosin heavy chain by immunofluorescence (Sigma MY-32 anti-MHC; data not shown).
Probes
A total of 16 genes in the FSHD region were analyzed using cosmid probes. The relative map location of each of these genes with respect to the D4Z4 repeat array is shown in Table 1. Cosmids were isolated in-house from the Los Alamos human chromosome 4 library.28 Isolated cosmids were verified by EcoRI digestion and sequencing of insert ends with T3 and T7 primers. Cosmids were labeled with Cy3-dCTP (Amersham, Louisville, CO, USA) using a nick translation mix (Roche, Palo Alto, CA, USA), with the exception of the chromosome 4-specific c88F8 (D4S139) localization cosmid, which was labeled with fluorescein-12-dUTP (Roche). The SNRPN probe was labeled with red fluorochrome in the Texas Red spectrum (Rainbow Scientific Inc, Windsor, CT, USA). A Bluescript KSII+ plasmid containing a single unit of the 3.3 kb D4Z4 repeat was labeled with Cy5-dCTP. Labeled probes were subsequently filtered through Microspin G-50 columns (Amersham) and ethanol precipitated. The localization of all probes to chromosome 4q35 was verified by hybridization to metaphase cells.
Table 1. FSHD region genes analyzed by RNA–DNA FISH.
| Gene | Alternate | Name | Kb to D4Z4 | Cosmid | Copy number |
|---|---|---|---|---|---|
| FRG2 | FSHD region gene 2 | 55 | — | Multi | |
| TUBB4Q | Tubulin 4q pseudogene | 97 | — | Multi | |
| FRG1* | FSHD region gene 1 | 141 | — | Multi | |
| FAT1* | FAT tumor suppressor homolog | 3508 | 67F9 | Single | |
| F11 | Coagulation factor XI | 3816 | 10C7 | Single | |
| KLKB1 | Kallikrein B | 3855 | 10C7 | Single | |
| CYP4V2 | Cytochrome P450, family 4 | 3890 | 190B1 | Single | |
| TLR3 | Toll-like receptor 3 | 4012 | 249D4 | Single | |
| SORBS2 | ARGBP2 | Sorbin and SH3 domain containing 2 | 4540 | 144B4 | Single |
| PDLIM3* | ALP | PDZ and LIM domain 3 | 4581 | 49D9 | Single |
| LRP2BP* | LRP2-binding protein | 4717 | 79A10 | Single | |
| SNX25 | Sorting nexin 25 | 4888 | 164D7 | Single | |
| SLC25A4* | ANT1 | Solute carrier family 25 | 4936 | 168D11 | Single |
| HELT | Hey-like transcriptional repressor | 5060 | 234E12 | Single | |
| IRF2* | Interferon regulatory factor 2 | 5702 | 66B6 | Single | |
| ING2 | Inhibitor of growth family, member 2 | 6574 | 191A10 | Single |
—, Null or low expression.
Gene not shown in bold exhibit null or expression levels too low to be analyzed by RNA–FISH.
Genes names given in bold represent those that are analyzed by two-color RNA–DNA FISH only.
Genes names given in bold along with asterisk symbol represent those that are analyzed by two- and three-color RNA–DNA FISH.
Fluorescence in situ hybridization
Cosmid DNA together with 100-fold human Cot1 DNA (Invitrogen) was lyophilized and resuspended in 50% formamide, 20% dextran sulfate, 4 × SSC, 0.1 DTT, 0.5 × Denhardts solution, and ssDNA, tRNA and PolyA (250 μg/ml each). The Cy3-labeled RNA-FISH probes were denatured for 10 min at 80°C and immediately put on ice. Non-denatured coverslips were dehydrated and hybridized overnight with the RNA-FISH probes in a humidified environment at 37°C. Coverslips were washed in 50% formamide/2 × SSC and 2 × SSC at 45°C with a final wash in 1 × SSC at room temperature. Cells were then fixed in 4% paraformaldehyde and treated with 0.15 NaOH/70% ethanol (alkaline hydrolysis) for 10 min each at room temperature before dehydration with increasing concentrations of ethanol. As negative controls, coverslips were either treated with 0.1 mg/ml RNase A (Applied Biosystems, Foster City, CA) at 37°C for 1 h before RNA hybridization or 0.5 U/μl RNase H (Epicentre Biotechnologies, Madison, WI, USA) at 37°C for 1 h after hybridization, but before paraformaldehyde fixation. For chromosomal localization and allele identification, the fluorescein isothiocyanate (FITC)-labeled c88F8 and Cy5-labeled D4Z4 DNA-FISH probes were prepared identically to the RNA-FISH probes. However, for nuclei hybridized with c88F8 and D4Z4 DNA-FISH probes, the chromosomal DNA and probes were simultaneously denatured by placing the slide on a 70°C hot plate for 2 min. After overnight hybridization at 37°C, the coverslips were washed as described above and counterstained with DAPI.
Fluorescence microscopy and deconvolution
Fluorescence signals were captured with a Photometrics Coolsnap HQ CCD camera (Photometrics, Tucson, AZ, USA) using an inverted widefield microscope (Nikon Eclipse TE2000-U, Nikon, Melville, NY, USA) with a × 63 Nikon Plan-Apo objective (N.A. 1.4) and dual motorized 10-position filterwheels with separate excitation and emission filters. The focal plane of the microscope passed through the whole nucleus with z-steps of 0.2 μm creating individual image series for each excitation filter. For each hybridization, at least 60 nuclei were captured. Images were deconvoluted with the program Autoquant version 9.3 (Media Cybernetics, Bethesda, MD, USA) and combined into single RGB composite images using the program ImageJ (NIH, Bethesda, MD, USA) as previously described.15
Quantification and scoring
RNA–DNA RGB composite images were presented to an operator who used the FITC-labeled c88F8 DNA signal to identify and mark the location of each 4q allele. Occasional nuclei were excluded because the two 4q loci were in such close proximity that distinct allelic RNA signals could not be measured, or rarely because two 4q signals were not clearly visualized. Census, an ImageJ plugin, and an automated algorithm were used to quantify the Cy3 fluorescence signal adjacent to the DNA signal as a measure of pre-mRNA transcription. The algorithm opened the corresponding deconvoluted 3D image stacks, using them to quantify the signal intensities by measuring the total probe intensity (TPI) within a fixed 500 nm diameter sphere. The sum of the intensity of the voxels within the sphere was calculated and normalized by subtracting the median intensity of signals within a cylindrical shell around the signal. The algorithm assessed the entire image region near the operator's mark and located the coordinates that provided the brightest signal within the sphere. Those coordinates were recorded as the signal center and the corresponding TPI was used for data analysis. For nuclei studied using a Cy5-labeled D4Z4 probe, the Cy5 image was included in the RGB composites as the blue signal. Using these RGB composites, the operator visually compared the signals on each allele and designated one of the telomeres as ‘affected' and the other as ‘normal' based on hybridization intensity with the D4Z4 probe. In over 95% of nuclei with measured RNA and DNA signals, one of the two 4q D4Z4 signals was absent or very faint, identifying the affected (deleted) allele. The rare nuclei with signals similar in intensity, or those confounded by the near presence of another signal from a D4Z4 repeat elsewhere in the genome, were excluded from the analysis. It is noted that for SNRPN analysis, only one allele would produce a visible nascent RNA signal in virtually all nuclei (as expected). For these, the brightest background spot near the non-transcribing 15q allele was used for fold comparison.
Statistical analysis
All data were analyzed using the R Project version 2.5.1 (http://www.r-project.org). When comparing allele ratios between normal and FSHD cell lines in the two-color experiments, the Mann–Whitney U-test analysis was applied. When comparing allele signals within FSHD nuclei in the three-color experiments, a single sample Wilcoxon signed-rank test was used to evaluate for symmetry of expression between the alleles. Interallelic ratios for the positive control gene, SNRPN, were analyzed by comparing the fold distribution of SNRPN FSHD nuclei with the FSHD fold distribution for all the other genes studied combined in one pool (Supplementary Figure 2), and significance was established by using the Mann–Whitney U-test. It is noted that FRG1 and IRF2 were only studied using three-color RNA–DNA FISH. However, control cell lines were also analyzed for the sake of completeness. Note, however, that the same FSHD data sets were used for the two- and three-color significance testing shown.
Results
Validation of methodology
The sensitivity and specificity of nascent RNA transcript detection was determined using several approaches. First, as a positive control, we detected monoallelic RNA transcription from SNRPN, an imprinted gene from the Prader-Willi/Angelman locus.29 The SNRPN probe was hybridized independently to coverslips either after denaturation (DNA-FISH conditions) or without denaturation (RNA-FISH conditions). As expected, the SNRPN probe detected allele-specific transcription, with a single 15q allele signal under RNA hybridization conditions and both 15q alleles under DNA hybridization conditions (Figure 1). The percent myonuclei displaying monoallelic expression of SNRPN (83%) is similar to that reported by Kohda et al29 for HL60 and WI38 cells, with only 4% displaying biallelic expression and 13% null expression. A second positive control used to test for sensitivity was the correlation of D4Z4 FISH signal intensity with the size of the D4Z4 target array. Extrapolation of DNA-FISH results to RNA-FISH results can be made here to address the correlation between fold change of nucleic acid quantity and that of signal intensity, as both DNA-FISH and RNA-FISH were performed within the same nuclei, fixed with identical conditions and washed using the same stringent conditions. From our previous FISH studies, we have shown that we can discriminate between the normal and disease allele on the basis of signal intensity of the D4Z4 repeat.15 The two chromosome 4 in the cell line FM1243j have 4 D4Z4 repeats (2 complete, 1 degenerate and 1 inverted) on the affected (contracted) allele and 23 D4Z4 repeats (21 complete, 1 degenerate and 1 inverted) on the normal allele. D4Z4 probe hybridization to the 2 alleles in 46 nuclei had a signal intensity ratio with a mean of 6.2, median of 6.4 and median confidence interval of 5.9–6.8. As the ratio of nucleic acid target is 5.75 (23/4 D4Z4 repeats), this correlates extremely well with the ratio of signal intensity (6.2). As a negative control, FITC-labeled 4q35 probe c88F8 (which does not contain coding sequences) was mixed with the Cy3-labeled HELT probe (a 4q35 gene), and hybridized independently to denatured and non-denatured nuclei. Two green and two red signals were detected on denatured nuclei (DNA conditions), corresponding to the FITC-labeled c88F8 DNA and the Cy3-labeled HELT DNA and RNA (single-stranded DNA and RNA), whereas only red signals were detected on non-denatured nuclei, corresponding to single-stranded HELT RNA only (Figure 2a, b). A second negative control was provided by the lack of RNA transcript detection at the 4q locus when cells were pretreated with RNase A before hybridization or when treated post-hybridization with RNase H (Figure 2c, d).
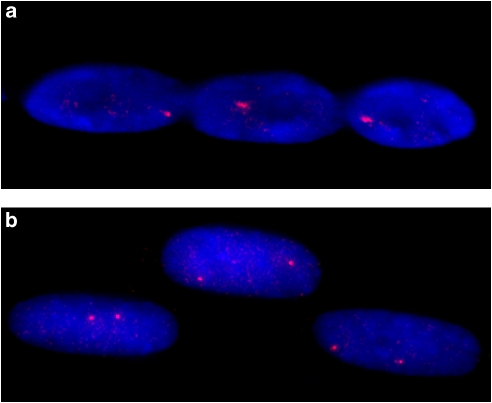
Figure 1.
RNA- and DNA-FISH of the imprinted gene SNRPN in myonuclei. (a) Monoallelic expression of RNA from one 15q allele is detected when non-denatured nuclei are probed with Cy3-labeled SNRPN, corresponding to the actively transcribed gene. (b) Two loci are detected when denatured nuclei are probed with Cy3-labeled SNPRN, corresponding to the DNA at both chromosome 15 alleles.
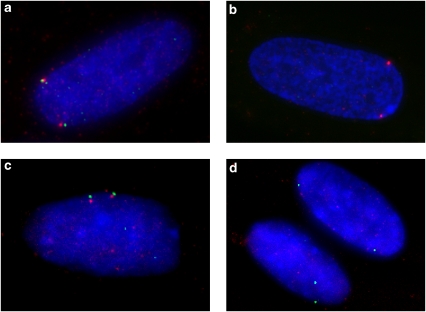
Figure 2.
Controls for RNA expression detection. (a and b) Influence of denaturation on RNA and DNA detection. (a) DNA signals from both HELT (Cy3, red fluorescence) and c88F8 FITC (green fluorescence) are detected on denatured nuclei. (b) RNA signals are detected only from the HELT locus on non-denatured nuclei, as c88F8 does not contain coding sequence. (c and d) Influence of RNase H on RNA and DNA detection. SLC25A4 (red) and c88F8 (green) were labeled with Cy3 and FITC, respectively. (c) Nuclei not treated with Rnase H show signals from both ANT1 and c88F8 on sequential RNA–DNA hybridization. (d) Nuclei treated with RNase H after RNA hybridization reveal the loss of the signals from ANT1, confirming that these are RNA signals.
Several 4q35 genes are not expressed in differentiated myotubes
Of the 16 genes studied, 6 were expressed at too low a level to be detected or adequately quantified. These six are FRG2, TUBB4Q, F11, KLKB1, SNX25 and ING2. For the multicopy loci FRG2 and TUBB4Q, >85% of nuclei show no pre-mRNA signals at the chromosome 4 alleles in both control and FSHD nuclei, making statistical analysis impossible. This high number of ‘null expression' nuclei suggests that these genes are not expressed (or expressed at a very low level) from chromosome 4 in FSHD and normal myotubes. For the genes ING2 and SNX25, no RNA expression was detected in >55% of the captured nuclei, both for the control and FSHD myotubes. Statistical analysis over the remainder of values was deemed unreliable and transcription of these genes in myotubes appears too low to measure. For F11 and KLKB1, no RNA transcription was detected in any of the nuclei examined.
Relative allele ratios as determined by two-color RNA–DNA FISH
All genes were first analyzed using two-color RNA–DNA FISH to identify those genes with either a monoallelic or skewed biallelic ratio suggestive of an altered cis effect on expression. Figure 2c depicts typical nuclei of differentiated myoblasts in which the green signal corresponds to the 4q localization probe D4S139, and the red signal corresponds to the RNA (gene) of interest. For two-color FISH, pre-mRNA transcription signals at each 4q allele were compared with those from the other 4q allele for each individual nucleus. Intranuclear transcription ratios between alleles for each gene were quantified, and the distribution of ratio values was compared between normal and FSHD myonuclei. If a cis effect on transcription attributed to short D4Z4 array exists in FSHD myonuclei, then one would expect higher interallelic signal ratios in FSHD than in normal nuclei. Figure 3 displays results for two-color analysis of six genes present in 3.9–5.1 Mb proximal to D4Z4: SORBS2, HELT, CYP4V2, TLR3, PDLIM3 and ANT1. Data for four other genes expressed in myotubes (FRG1, FAT1, IRF2 and LRP2BP) are presented in Supplementary Figure 1.
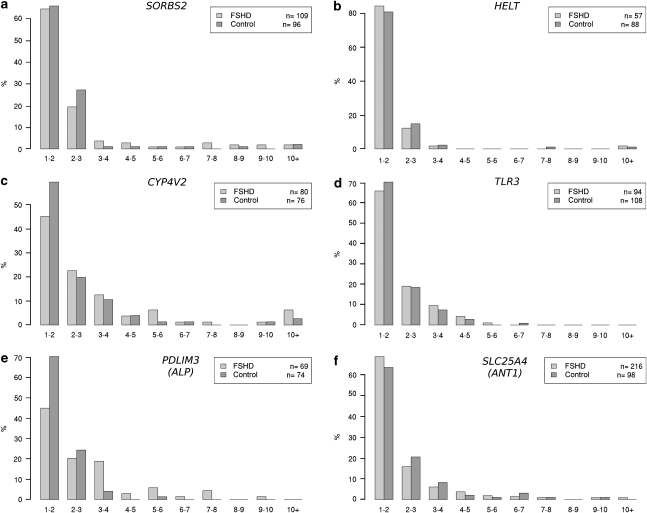
Figure 3.
Interallelic ratios of RNA signal intensity. Two-color RNA–DNA FISH analysis results for (a) SORBS2, (b) HELT, (c) CYP4V2, (d) TLR3, (e) PDLIM3 (ALP) and (f) SLC25A4 (ANT1). For each nucleus measured, the total probe intensity of the brighter RNA signal was divided by the intensity of the weaker signal. The fold difference in signal intensity between the two alleles is plotted on histograms (x axis) with the percent of nuclei displaying each ratio on the y axis. Typically, most nuclei have signals with similar intensities, with fold differences between one and three. Some nuclei have much brighter signals at one chromosome compared with the other, with the majority of transcription occurring only at one allele. Intranuclear (interallelic) ratios for control myonuclei (light gray) are plotted alongside FSHD myonuclei (dark gray). If a cis effect was modifying RNA transcription at the FSHD-affected allele, one would expect to see a greater number of nuclei with larger fold changes in the FSHD myonuclei compared with the control myonuclei. Instead, with the exception of PDLIM3, the histograms are nearly identical.
For 9 out of the 10 expressed genes analyzed by two-color RNA–DNA FISH, the distribution of fold difference between interallelic signal intensities is not significantly different (Table 2). An FSHD-specific cis effect was not shown for these nine genes, as there was no conserved asymmetry of interallelic expression in either of the two FSHD samples or two control samples. One gene, PDLIM3, was suggestive of a possible cis effect (Figure 3 and Table 2). To study the expression of this gene further, as well as of several others implicated previously in FSHD,19 we performed allele-specific (three-color) RNA–DNA FISH.
Table 2. Comparison of RNA signal intensities.
| Gene | Two-color FISH: FSHD cell line versus control cell line (median of ratios, FSHD/normal; P-values by Wilcoxon test) | Three-color FISH: mean of ratios of affected allele versus normal allele (95% CI of median) |
|---|---|---|
| SORBS2 | 0.986 (P=0.47) | — |
| CYP4V2 | 0.504 (P=0.11) | — |
| HELT | 0.952 (P=0.82) | — |
| TLR3 | 0.924 (P=0.95) | — |
| PDLIM3 | 0.326 (P=0.0000098) | 1.035 (0.92–1.26) |
| SLC25A4 | 1.106 (P=0.39) | 1.096 (0.94–1.19) |
| FAT1 | 1.026 (P=0.96) | 0.982 (0.78–1.27) |
| LRP2BP | 0.834 (P=0.38) | 0.902 (0.66–1.23) |
| FRG1 | 0.688 (P=0.52) | 0.918 (0.72–1.33) |
| IRF2 | 0.917 (P=0.26) | 0.945 (0.76–1.14) |
| SNRPN | 9.35 (P=9.8e−26) | — |
Allele-specific detection of RNA transcription using three-color RNA–DNA FISH
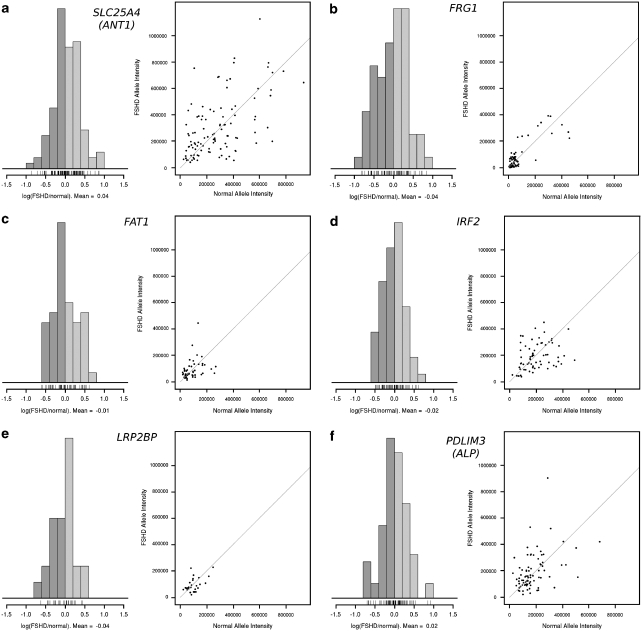
The more definitive three-color analysis on FSHD nuclei allows unequivocal identification of transcription from the contracted allele versus the normal allele within a given nucleus. A Cy5-labeled D4Z4 probe was used to identify the affected (contracted) allele in each FSHD myotube nucleus (as exemplified in Figure 4). In the analysis of a single nucleus, the TPI of the FSHD-affected allele (with a D4Z4 repeat deletion) was compared with the unaffected allele (with normal D4Z4 repeat count). Importantly, the strength of this methodology is that comparisons are made within a given nucleus. As such, experimental variability is greatly reduced, and a direct comparison of expression between affected and non-affected alleles can be carried out. The candidate genes SLC25A4 (ANT1) and FRG1, as well as PDLIM3 (ALP), FAT1, LRP2BP and IRF2, were further analyzed in this three-color RNA–DNA FISH. Often, the two signals measured were either both high, indicating biallelic active transcription, or both low, indicating little transcription at either allele. For some nuclei, only one of the two alleles had a high intensity, implying active transcription at only that allele. If there was a cis effect modifying the level of transcription at the affected FSHD allele, one would expect a bias such that either the FSHD or normal allele preferentially showed transcription. Conversely, if there was no cis effect, the FSHD and the normal allele would be equally represented among the single bright transcription signals. For all the six genes studied, RNA transcription occurred without bias as to normal/affected allele status (Figure 5, Table 2). We further tested for a possible overall trend of increased expression at one of the transcription sites by dividing the intensity of the FSHD allele by the intensity of the normal allele for each nucleus. For all six genes studied, the average ratio was close to one, indicating no bias toward expression at either allele.
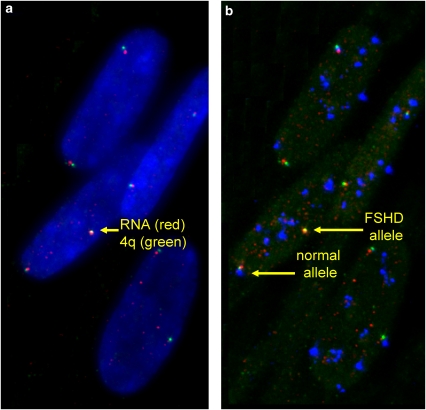
Figure 4.
Identification of affected versus normal allele status: representative myonuclei analyzed by three-color sequential RNA–DNA FISH. SLC25A4, c88F8 and D4Z4 were labeled with Cy3, FITC and Cy5, respectively. (a) SLC25A4 RNA from both 4q alleles is detected adjacent to the DNA signal from the 4q localization probe c88F8 in DAPI-stained nuclei. (b) Hybridization with a Cy5-labeled D4Z4 probe enables identification of the affected (deleted) allele and the normal allele. Although D4Z4 is represented at many chromosomal loci (as seen in blue), the 4q D4Z4 locus is revealed on co-hybridization with the 4q35-specific probe c88F8. The deleted (affected allele) does not hybridize adequately with the D4Z4 probe because of truncations of the target site.15 Thus, the absence of the Cy5 signal adjacent to the 4q-specific probe c88F8 identifies the affected allele. The normal 4q allele harbors a sufficient number of D4Z4 repeat units to allow for hybridization detection with the D4Z4 probe.
Figure 5.
Ratios of RNA signal intensity between FSHD normal and contracted alleles. Three-color RNA–DNA FISH analysis for the genes (a) SLC25A4 (ANT1), (b) FRG1, (c) FAT1, (d) IRF2, (e) LRP2BP and (f) PDLIM3 (ALP) on FSHD myotube nuclei. Hybridization to D4Z4 was used to identify normal and affected alleles. The intensity of the RNA signal at the FSHD allele was divided by the intensity of the RNA signal at the normal allele and plotted in the histogram on the left. The log of the ratio was used. As such, nuclei with signals of similar intensity (with a ratio near 1) would have a log(ratio) near 0. The greater the fold difference between the two signals was, the greater the distance the log(ratio) would be from 0. The average of the log(ratio) for each gene is listed below the histogram and is very near to 0 for all of the genes. Log(ratios) above 0 (light gray) represent myonuclei in which the affected allele shows greater transcription than the normal allele, whereas ratios below 0 (dark gray) represent myonuclei in which the normal allele shows greater transcription than the affected allele The data depict a balance of transcription between the normal and the affected alleles, and the absence of a cis effect bias to favor either of the alleles. The same procedure was performed using control cell lines with statistically identical results (data not shown). On the scatterplots given on the right, the horizontal axis represents the total probe intensity of the RNA signal at the normal allele. The vertical axis represents the intensity of the signal at the affected allele (with the D4Z4 repeat deletion). Points near the diagonal line represent nuclei in which the two alleles had even signal intensities. Points that stray away from the diagonal line represent nuclei in which one signal was brighter than the other. For all the genes tested, the points remained symmetric across the diagonal line, showing no bias for brighter signals on the affected or unaffected allele.
Data are shown pooled for the two FSHD samples, leaving open the question of whether the data are consistent across patients. Supplementary Table 3 further displays an analysis of the samples individually for the two genes of primary interest in the FSHD field: FRG1 and ANT1. The ratio of interallelic signal intensity is shown (FSHD allele versus normal allele, three-color RNA–DNA FISH using D4Z4 probe to identify the alleles) for each FSHD sample independently, with both the mean signal intensity as well as upper and lower confidence intervals of the median. As can be seen, both FSHD samples behave similarly, with no cis effect for either gene.
Conclusion
This study provides a comprehensive overview of allele-specific, nascent 4q35 gene expression in single primary myotube nuclei. Using this approach, we were able to directly and unequivocally test whether or not a cis effect on gene expression underlies the pathology in FSHD myotubes. RNA–DNA FISH is well suited for this analysis, as it does not compare total transcription levels between cell lines or tissues, but is able instead to measure transcription independently at each allele within single nuclei. A total of 16 genes from the FSHD region at 4q35 were studied with both two-color and three-color (allele specific) RNA–DNA FISH, with none of them showing a cis effect on gene expression in FSHD. The power of the three-color RNA–DNA FISH approach is evidenced by the fact that the PDLIM3 data in the two-color experiments suggested a possible cis effect, but when studied with three-color FISH, the absence of allele-specific differences in expression was clearly revealed.
Data analysis was based on fold change (ratio) between the signal intensities at the two alleles. The precise linear relationship of this signal ratio to that of nascent RNA (that is, sensitivity of the technique) cannot as yet be defined, as to date there is no ‘gold standard' for intranuclear, interallelic nascent RNA expression. That is, there is no gene for which it is known that one allele consistently transcribes a precise quantity of nascent RNA relative to the other allele. However, it is not unreasonable to expect a direct relationship between the amount of nascent RNA present, the number of probe molecules bound and the intensity of the fluorescent signal within a single nucleus. Indeed, others have validated this relationship with mRNA FISH.30 As such, when we find rather narrow confidence intervals for the median fold difference between affected and unaffected allele probe intensities (Table 2, ∼0.75–1.25), this likely reflects a similarly narrow range for the ratio of the biological amount of nascent RNA present.
Our data show a direct correlation between the D4Z4 allele DNA target size ratio (5.75) and the DNA-FISH signal intensity (mean of 6.2), validating the sensitivity of this approach to measure relative differences in nucleic acid between the two alleles. One argument against this line of reasoning would be that the residual folding of the RNA molecule might prevent comparison of RNA-FISH with DNA-FISH. However, considering the fact that the normal D4Z4 repeat is heterochromatic and thus less accessible to our probes than the contracted repeat, which is more euchromatic, the fact that we can easily detect these fold changes strongly argues that our hybridization conditions are adequate and that the folding of the RNA molecules is not likely to have a major role in our studies. Therefore, although the precise quantitative relationship of our measured intensity differences to the RNA present is not known, it is reasonable to assume that a persistent twofold difference in expression between the alleles would be detected by this system. The previously reported transcriptional upregulation of 10- to 30-fold for the genes ANT1 and FRG119 would certainly have been identified in our system analyzing interallelic fold changes had they been present.
Altered expression of three candidate genes, FRG1, FRG2 and SLC25A4 (ANT1), in FSHD was reported by Gabellini et al,19 although several others have reported no such effect.20, 21, 22, 23, 24 In particular, FRG1 has been the subject of controversy concerning an increased level of expression in FSHD as FRG1 overexpressing mice show a muscle phenotype.31 This study finds no evidence of a cis effect for FRG1, and suggests an explanation for the confusion over FRG1 expression. In both control and FSHD myotubes, the RNA-hybridized FRG1 probe detects strong red fluorescence signals dispersed throughout the nucleus (data not shown). These signals are not comparable with the minimal background signals obtained when using the probes for all other investigated genes. As FRG1 is a multicopy gene, which is present not only on chromosome 4 but also on chromosomes 8, 9, 12, 20 and all of the acrocentric chromosomes, these signals likely represent transcription from these homologs. BLAST searches in dbEST also support active FRG1 transcription from multiple loci (data not shown). This widespread and abundant transcription of FRG1 from non-4q loci has obscured the definitive analysis of nascent 4q FRG1. Using RNA–DNA FISH, which unequivocally detects chromosome-4-specific transcription, we conclusively exclude cis effects on FRG1 transcription at 4q35 as underlying the pathology in FSHD myotubes. A trans-effect on FRG1 expression remains a possibility, as this study examines intranuclear allelic ratios and not internuclear ratios, that is, overall ratios of FSHD myotube transcription relative to control myotubes.
This study was designed to analyze interallelic expression in myotube nuclei, as this stage of differentiation in culture most closely mimics that of mature myofibrils. We specifically analyzed myonuclei within myotubes (that is, not unfused myoblasts) to minimize variability in expression data because of differing stages of development. This gave us the added opportunity to examine the behavior of individual myonuclei within the same myotube. We would not expect, a priori, to find that all myonuclei show consistent interallelic expression ratios. Indeed, this is what we found – myonuclei not only vary in interallelic signal intensity ratios but they also indicate which allele (deleted versus normal) shows higher transcription from a particular gene (data not shown). Such variance in expression between individual myonuclei has been noted in other published work32, 33 and between alleles in other studies of non-muscle cells.34
The current expression data using RNA-FISH corroborates earlier studies using microarrays.21, 22 Transcriptional upregulation of 4q genes was not detected in three independent studies using microarray expression profiling on myoblasts, myotubes and skeletal muscles. The accompanying paper by Klooster et al,24 using Q-PCR and western blot analysis of myoblasts, myotubes and skeletal muscle, also reaches largely similar conclusions. Importantly, the data presented here expand FSHD expression analysis to rule out a cis effect in FSHD. It must be stressed that this technique is not suitable for the identification of genes that are transcriptionally upregulated in FSHD on both chromosomes, as RNA-FISH compares interallelic expression levels within single nuclei and is therefore the only technique capable of identifying a cis effect.
The RNA-FISH approach confirms the difficulty of detecting low levels of transcript from specific genes in the FSHD region. Klooster et al24 show that very low levels of FRG2 could be detected by RT-PCR in FSHD myotubes only. We found expression of FRG2 by RNA-FISH in <15% of myonuclei. In a recent study by Snider et al,14 very low levels of DUX4 expression were reported in myotubes on the basis of nested RT-PCR. It is likely that such low levels of expression cannot be reliably detected or quantified by RNA-FISH. Several other genes for which we did not detect expression by RNA-FISH, such as KLKB1, F11, ING2 and SNX25, were also not detected by microrray analysis.21, 22
Several characteristics of FSHD implicate a developmental origin for the disease and therefore warrant investigation of transcription at earlier stages, that is, before the differentiation of muscle. Even though the typical age of phenotypic onset in FSHD is during adolescence, there are many indications that the disease is influenced by earlier developmental processes: D4Z4 contains the putative gene DUX4 (encoded within the D4Z4 repeat), which is homologous to the homeobox genes regulating development;35 DUX4 regulates PITX1,36 a developmental gene involved in left–right asymmetry (common in FSHD); and developmentally controlled features such as pectus excavatum, and a complete lack of specific muscles such as the pectoralis, often present with the disease.1, 2, 3
Epigenetic chromatin modifications such as DNA methylation, altered binding of chromatin proteins and histone modifications have a central role in regulating developmental gene expression. The D4Z4 repeat itself shows many of these epigenetic modifications in FSHD-affected tissues, including hypomethylation37, 38 and alterations in heterochromatin protein association.39 As the inability to repress developmental genes is known to interfere with the proper differentiation of many tissues, these epigenetic modifications of D4Z4 may underlie the pathophysiology of FSHD. Although this study finds no evidence for a cis effect on FSHD region gene expression in myonuclei, developmental stages before the differentiation of the muscle lineage have not been studied. Of particular interest is the pluripotent stem cell gene ZFP42 (REX1) that lies proximal to the D4Z4 repeat and is downregulated on differentiation.40, 41 Continued expression of this gene in early FSHD progenitor cells may lead to the defects in differentiation seen in FSHD.21, 23, 42, 43 Therefore, analysis of FSHD developmental genes in pluripotent and early mesodermal stem cells, before the program of muscle differentiation is initiated, may be especially crucial to our understanding of this disease.
Acknowledgments
We are grateful to the Muscular Dystrophy Association (STW), the FSH Society (STW), the FisherShaw Foundation (STW), the Fields Center for FSHD and Neuromuscular Research (RT and SvdM), the NIH (ME), and the Dutch FSHD Foundation (OYC) for support of this study and their ongoing commitment to FSHD research.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Tawil R, van der Maarel SM. Facioscapulohumeral muscular dystrophy. Muscle Nerve. 2006;34:1–15. doi: 10.1002/mus.20522. [DOI] [PubMed] [Google Scholar]
- van der Maarel SM, Frants RR. The D4Z4 repeat-mediated pathogenesis of facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2005;76:375–386. doi: 10.1086/428361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padberg GW, Adams C.Facioscapulohumeral muscular dystrophyin: Pulst S-M (ed.): Neurogenetics Oxford, UK: Oxford University Press; 2000105–116. [Google Scholar]
- Padberg GW, Brouwer OF, de Keizer RJ, et al. Retinal vascular disease and sensorineural deafness are part of facioscapulohumeral muscular dystrophy. Am J Hum Genet. 1992;51 S:A104. [Google Scholar]
- Funakoshi M, Goto K, Arahata K. Epilepsy and mental retardation in a subset of early onset 4q35-facioscapulohumeral muscular dystrophy. Neurology. 1998;50:1791–1794. doi: 10.1212/wnl.50.6.1791. [DOI] [PubMed] [Google Scholar]
- Wijmenga C, Hewitt JE, Sandkuijl LA, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Wijmenga C, van Tienhoven EA, et al. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- Clapp J, Mitchell LM, Bolland DJ. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81:264–279. doi: 10.1086/519311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, de Kievit P, Sandkuijl L, et al. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet. 2002;32:235–236. doi: 10.1038/ng999. [DOI] [PubMed] [Google Scholar]
- Lemmers RJ, Wohlgemuth M, van der Gaag KJ, et al. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81:884–894. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winokur ST, Bengtsson U, Feddersen J, et al. The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Res. 1994;2:225–234. doi: 10.1007/BF01553323. [DOI] [PubMed] [Google Scholar]
- Hewitt JE, Lyle R, Clark LN, et al. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Mol Genet. 1994;3:1287–1295. doi: 10.1093/hmg/3.8.1287. [DOI] [PubMed] [Google Scholar]
- Yang F, Shao C, Vedanarayanan V, Ehrlich M. Cytogenetic and immuno-FISH analysis of the 4q subtelomeric region, which is associated with facioscapulohumeral muscular dystrophy. Chromosoma. 2004;112:350–359. doi: 10.1007/s00412-004-0280-x. [DOI] [PubMed] [Google Scholar]
- Snider L, Asawachaicharn A, Tyler AE, et al. RNA transcripts, miRNA-sized fragments, and proteins produced from D4Z4 units. New candidates for the pathophysiology of facioscapulohumoral dystrophy. Hum Mol Genet. 2009;18:2414–2430. doi: 10.1093/hmg/ddp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masny PS, Bengtsson U, Chung SA, et al. Localization of 4q35.2 to the nuclear periphery: is FSHD a nuclear envelope disease. Hum Mol Genet. 2004;13:1857–1871. doi: 10.1093/hmg/ddh205. [DOI] [PubMed] [Google Scholar]
- Tam R, Smith KP, Lawrence JB. The 4q subtelomere harboring the FSHD locus is specifically anchored with peripheral heterochromatin unlike most human telomeres. J Cell Biol. 2004;167:269–279. doi: 10.1083/jcb.200403128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers T, Deidda G, van Koningsbruggen S, et al. FRG2, an FSHD candidate gene, is transcriptionally upregulated in differentiating primary myoblast cultures of FSHD. J Med Genet. 41:826–836. doi: 10.1136/jmg.2004.019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L, Marshall CR, Wintle RF, Scherer SW. Structural variants: changing the landscape of chromosomes and design of disease studies. Hum Mol Genet. 2006;15:R57–R66. doi: 10.1093/hmg/ddl057. [DOI] [PubMed] [Google Scholar]
- Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- Jiang G, Yang F, van Overveld PG, et al. Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum Mol Genet. 2003;12:2909–2921. doi: 10.1093/hmg/ddg323. [DOI] [PubMed] [Google Scholar]
- Winokur ST, Chen YW, Masny PS, et al. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum Mol Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- Osborne RJ, Welle S, Venance SL, Thornton CA, Tawil R. Expression profile of FSHD supports a link between retinal vasculopathy and muscular dystrophy. Neurology. 2007;68:569–577. doi: 10.1212/01.wnl.0000251269.31442.d9. [DOI] [PubMed] [Google Scholar]
- Celegato B, Capitanio D, Pescatori M, et al. Parallel protein and transcript profiles of FSHD patient muscles correlate to the D4Z4 arrangement and reveal a common impairment of slow to fast fibre differentiation and a general deregulation of MyoD-dependent genes. Proteomics. 2006;6:5303–5321. doi: 10.1002/pmic.200600056. [DOI] [PubMed] [Google Scholar]
- Klooster R, Straasheijm K, Shah B, et al. Comprehensive expression analysis of FSHD candidate genes on mRNA and protein levels Eur J Hum Genet 2009. This issue. [DOI] [PMC free article] [PubMed]
- Herzing LB, Cook EH, Jr, Ledbetter DH. Allele-specific expression analysis by RNA-FISH demonstrates preferential maternal expression of UBE3A and imprint maintenance within 15q11- q13 duplications. Hum Mol Genet. 2002;11:1707–1718. doi: 10.1093/hmg/11.15.1707. [DOI] [PubMed] [Google Scholar]
- Tam R, Shopland LS, Johnson CV, et al. Applications of RNA FISH for visualizing gene expression and nuclear architectureIn: Beatty BMSSJ (ed).: FISH Practical ApproachVolume 260New York: Oxford University Press; 200293–118. [Google Scholar]
- van Raamsdonk CD, Tilghman SM. Optimizing the detection of nascent transcripts by RNA fluorescence in situ hybridization. Nucleic Acids Res. 2001;29:E42–E42. doi: 10.1093/nar/29.8.e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riess O, Siedlaczck I, Kredtke S, Melmer G, Epplen JT, Deaven LL. Characterization of a human chromosome 4 flow-sorted cosmid library. Cytogenet Cell Genet. 1994;65:238–242. doi: 10.1159/000133638. [DOI] [PubMed] [Google Scholar]
- Kohda A, Taguchi H, Okumura K. Visualization of biallelic expression of the imprinted SNRPN gene induced by inhibitors of DNA methylation and histone deacetylation. Biosci Biotechnol Biochem. 2001;5:1236–1239. doi: 10.1271/bbb.65.1236. [DOI] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Gabellini D, D'Antona G, Moggio M, et al. Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature. 2006;439:973–977. doi: 10.1038/nature04422. [DOI] [PubMed] [Google Scholar]
- Myers TK, Andreuzza SE, Franklin DS. p18INK4c and p27KIP1 are required for cell cycle arrest of differentiated myotubes. Exp Cell Res. 2004;300:365–378. doi: 10.1016/j.yexcr.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Smith KP, Moen PT, Wydner KL, Coleman JR, Lawrence JB. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol. 1999;144:617–629. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Vambrie S, Steinlein P, et al. Independent regulation of the two Pax5 alleles during B-cell development. Nat Genet. 1999;21:390–395. doi: 10.1038/7720. [DOI] [PubMed] [Google Scholar]
- Beckers M, Gabriëls J, van der Maarel S, et al. Active genes in junk DNA? Characterization of DUX genes embedded within 3.3 kb repeated elements. Gene. 2001;264:51–57. doi: 10.1016/s0378-1119(00)00602-8. [DOI] [PubMed] [Google Scholar]
- Dixit M, Ansseau E, Tassin A, et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci USA. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Sandkuijl LA, et al. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- de Greef JC, Wohlgemuth M, Chan OA, et al. Hypomethylation is restricted to the D4Z4 repeat array in phenotypic FSHD. Neurology. 2007;69:1018–1026. doi: 10.1212/01.wnl.0000271391.44352.fe. [DOI] [PubMed] [Google Scholar]
- Zeng W, de Greef JC, Chen YY, et al. Specific loss of histone H3 lysine 9 trimethylation and HP1g/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS 20095e1000559in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongan NP, Martin KM, Gudas LJ. The putative human stem cell marker, Rex-1 (Zfp42): structural classification and expression in normal human epithelial and carcinoma cell cultures. Mol Carcinog. 2006;45:887–900. doi: 10.1002/mc.20186. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morosetti R, Mirabella M, Gliubizzi C, et al. Isolation and characterization of mesoangioblasts from facioscapulohumeral muscular dystrophy muscle biopsies. Stem Cells. 2007;25:3173–3182. doi: 10.1634/stemcells.2007-0465. [DOI] [PubMed] [Google Scholar]
- Barro M, Carnac G, Flavier S, Mercier J, Vassetzky Y, Laoudj-Chenivesse D.Myoblasts from affected and non affected FSHD muscles exhibit morphological differentiation defects J Cell Mol Med 2008. published online 24 May 2009, DOI 10.1111/j.1582-4934.2008.00368.x. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.