Summary
Two-component systems (TCS) are universal among bacteria and play critical roles in gene regulation. Our understanding of the contributions of TCS in the biology of the Borrelia is just now beginning to develop. Borrelia burgdorferi, a causative agent of Lyme disease, harbours a TCS comprised of open reading frames (ORFs) BB0419 and BB0420. BB0419 encodes a response regulator designated Rrp1, and BB0420 encodes a hybrid histidine kinase–response regulator designated Hpk1. Rrp1, which contains a conserved GGDEF domain, undergoes phosphorylation and produces the secondary messenger, cyclic diguanylate (c-di-GMP), a critical signaling molecule in numerous organisms. However, the regulatory role of the Rrp1–Hpk1 TCS and c-di-GMP signaling in Borrelia biology are unexplored. In this study, the distribution, conservation, expression and potential global regulatory capability of Rrp1 were assessed. rrp1 was found to be universal and highly conserved among isolates, co-transcribed with hpk1, constitutively expressed during in vitro cultivation, and significantly upregulated upon tick feeding. Allelic exchange replacement and microarray analyses revealed that the Rrp1 regulon consists of a large number of genes encoded by the core Borrelia genome (linear chromosome, linear plasmid 54 and circular plasmid 26) that encode for proteins involved in central metabolic processes and virulence mechanisms including immune evasion.
Introduction
Borrelia burgdorferi, B. garinii and B. afzelii, causative agents of Lyme disease (Burgdorfer et al., 1982; Benach et al., 1983; Steere et al., 1983), are maintained in an enzootic cycle involving Ixodes ticks and diverse mammalian reservoirs (Anderson and Magnarelli, 2008). During the transmission cycle, the spirochetes must adapt to changing environmental parameters including temperature, pH, nutrient availability, host-derived factors and immune pressures. Differential expression of Borrelia genes in response to environmental conditions has been demonstrated (Revel et al., 2002; Roberts et al., 2002; Brooks et al., 2003; Ojaimi et al., 2003; Caimano et al., 2007; Lybecker and Samuels, 2007); however, many aspects of the molecular basis of differential regulation of Borrelia gene expression are unknown.
Two-component systems (TCS), typically consisting of a histidine kinase and response regulator, play important roles in the adaptive responses of bacteria (Stock et al., 2000; West and Stock, 2001). Signal transduction begins when the histidine kinase senses an environmental signal, leading to autophosphorylation at a conserved histidine residue. The phosphoryl group is then transferred to an aspartate in the receiver domain of the response regulator, allowing for activation of the output domain which may have DNA-binding, RNA-binding, protein-binding or enzymatic functions (Galperin et al., 2001; Shu and Zhulin, 2002; Galperin, 2006). Almost all bacteria possess TCS, with most species containing 20–30 response regulator–histidine kinase pairs (Galperin et al., 2001; Skerker et al., 2008). In contrast, the genome of B. burgdorferi has only two gene pairs that encode for TCS that are likely to have global regulatory capabilities. Of these, the Hpk2–Rrp2 (histidine protein kinase 2–response regulatory protein 2) TCS was the first to be studied (Yang et al., 2003a,b; Burtnick et al., 2007; He et al., 2007; Boardman et al., 2008; Ouyang et al., 2008). Initial analyses demonstrated that Rrp2 influences the expression of several B. burgdorferi virulence factors. The DNA-binding output domain of Rrp2 binds to the −24/−12 region upstream of rpoS (σS) to enhance transcription with the help of RpoN (σ54). The Rrp2–RpoN–RpoS regulatory pathway positively regulates mainly plasmid-carried genes including ospC (outer surface protein C), dbpA (decorin-binding protein A) and the mlp proteins (multi-copy lipoproteins) (Brown et al., 1999; Hubner et al., 2001; Pikas et al., 2003; Yang et al., 2003b). It has recently been demonstrated that Rrp2 regulates genes independent of RpoS, the majority of which are plasmid-encoded proteins that have no known function (Boardman et al., 2008).
In this study, we investigate the regulatory potential of the B. burgdorferi Rrp1 (BB0419, a response regulator) and Hpk1 (BB0420, a hybrid sensory transduction histidine kinase-response regulator) TCS. Rrp1 differs from Rrp2 in that it lacks a DNA-binding output domain, but carries an enzymatic GGDEF domain with diguanylate cyclase activity. Diguanylate cyclase enzymes convert two molecules of GTP, via the linear intermediate diguanosine tetraphosphate (pppG3′p5′G) to cyclic dimeric guanosine monophosphate (cyclic diguanylate or c-di-GMP) (Ausmees et al., 2001; Galperin et al., 2001; Simm et al., 2004; Jenal and Malone, 2006). The ability of recombinant Rrp1 to produce c-di-GMP has been demonstrated to be dependent on phosphorylation (Ryjenkov et al., 2005). Rrp1 is the only B. burgdorferi protein that harbours a GGDEF domain and thus appears to be the only B. burgdorferi protein capable of c-di-GMP production. C-di-GMP is a secondary messenger molecule that influences cellular processes including motility, cell differentiation, phage and heavy metal resistance, expression of virulence genes, cell–cell signaling, fimbrial synthesis, exopolysaccharide production, and the transition between biofilm and planktonic modes of growth (Garcia et al., 2004; Simm et al., 2004; Tischler and Camilli, 2004; Karaolis et al., 2005; Romling et al., 2005; Beyhan et al., 2006; Galperin, 2006; Mendez-Ortiz et al., 2006; Romling and Amikam, 2006; Bobrov et al., 2007; Cotter and Stibitz, 2007; Kim and McCarter, 2007; Lee et al., 2007; Tamayo et al., 2007; Wolfe and Visick, 2008). The intracellular pool of c-di-GMP is controlled by the opposing activities of diguanylate cyclase and phosphodiesterase enzymes (EAL or HD-GYP domain-containing proteins). The B. burgdorferi genome harbours one EAL (BB0363) and one HD-GYP (BB0374) domain-containing protein. The identification of GGDEF (in Rrp1/BB0419), EAL (in BB0363) and HD-GYP (in BB0374) domains indicates that B. burgdorferi has the full set of proteins required for regulating c-di-GMP levels within the cell. The mechanisms by which c-di-GMP exerts its regulatory effects are largely unknown; however, c-di-GMP has been shown to bind to proteins harbouring the PilZ domain (Amikam and Galperin, 2006; Ryjenkov et al., 2006; Benach et al., 2007). B. burgdorferi BB0733 harbours this domain, although its ability to bind c-di-GMP has not been demonstrated. Additionally, c-di-GMP riboswitches with GEMM domains have been implicated as a possible mechanism for translational control (Sudarsan et al., 2008).
In this report, we demonstrate that rrp1 and hpk1 are conserved and universal among Lyme disease isolates. Deletion of rrp1 through allelic exchange replacement and subsequent microarray analyses revealed that Rrp1 influences the expression of a large and functionally diverse set of genes. The majority of these genes are present on the chromosome and code for proteins with basic cellular functions. This study is the first to explore the regulatory capability of a protein with diguanylate cyclase activity in any spirochete and the first to identify a central regulator of the core genome. The data presented shed significant new light on the global regulatory mechanisms of B. burgdorferi and indirectly suggest that c-di-GMP may be a key secondary messenger molecule in the Borrelia.
Results
Polymerase chain reaction and Southern hybridization analyses of rrp1 and hpk1
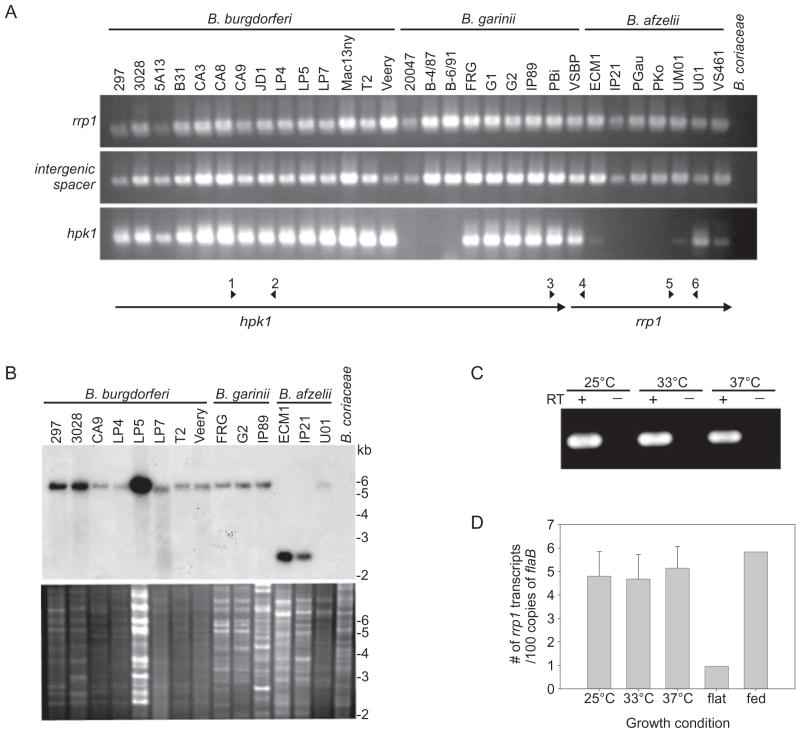
To assess the distribution of rrp1–hpk1 in a diverse panel of Borrelia species and strains, polymerase chain reaction (PCR) analyses were performed on 30 Lyme disease isolates (B. burgdorferi, B. garinii and B. afzelii), 4 relapsing fever isolates (Borrelia hermsii and B. turicatae) and 1 Borrelia coriaceae isolate (a causative agent of epizootic bovine abortion). Representative data are presented in Fig. 1A. All Lyme disease isolates were PCR positive with an internal rrp1 primer set (primers 5 and 6) and with a primer set designed to amplify from the 3′ end of hpk1 into the 5′ end of rrp1 (primers 3 and 4). Amplification with internal hpk1 primers yielded more variable results (primers 1 and 2) suggesting sequence variation within this gene. rrp1–hpk1-derived amplification products were also obtained from the relapsing fever spirochetes B. hermsii and B. turicatae (data not shown). Hybridization analyses were also performed to screen for rrp1. The probe was generated by radiolabeling a PCR product of rrp1 from B. burgdorferi B31MI (Fig. 1B). All Lyme disease spirochete isolates screened (n = 14) were hybridization positive while B. coriaceae was hybridization negative (relapsing fever spirochetes were not assessed by this approach). Collectively, these data indicate that the rrp1–hpk1 operon is carried by Borrelia species that are pathogenic in humans.
Fig. 1.
rrp1 and hpk1 are universally distributed, highly conserved, co-transcribed, and constitutively expressed in vitro. (A) presents PCR analyses conducted with primers designed to amplify an internal region of rrp1 (primers 5 + 6), across the rrp1–hpk1 intergenic spacer (primers 3 + 4), and an internal region of hpk1 (primers 1 + 2). The isolates analysed are indicated above each lane with the species names above the isolates. The PCR results for a select group of isolates were confirmed through Southern hybridization using a radiolabeled rrp1 probe and EcoRI-restricted genomic DNA from the isolates listed above each lane (B). The top and bottom portions of (B) present the hybridization results and ethidium bromide-stained gel of the restricted DNA respectively. Molecular weight markers are shown to the right in kilobases (kb). (C) presents the results of RT-PCR analyses conducted using RNA isolated from B. burgdorferi B31 grown at 25°C, 33°C or 37°C. The plus (+) and minus (−) signs indicate reactions that were run with or without reverse transcriptase (RT) enzyme, respectively. Using the same RNA, transcripts of rrp1 were quantified using qRT-PCR and are graphed as number of rrp1 transcripts per 100 copies of the flaB gene (D). (E) presents an amino acid sequence alignment of deduced Rrp1 sequences. Dashes indicate gaps in the sequence and dots indicate an identical amino acid to the reference sequence (BbB31). Species and isolate names are shown on the left. The sequence shown for B. afzelii represents three isolates with identical amino acid sequences (PKo, ECM1, and VS461), B. turicatae represents three isolates with identical sequences (91E135, PE1926, and OZ1), and B. hermsii represents two isolates with identical sequences (DAH and MAN). The six critical residues associated with receiver domain function of response regulators are indicated by shaded boxes with the phosphorylated aspartate (D) residue indicated with a star. The box indicates the location of the GGDEF domain and the filled circles above each amino acid indicate the residues that are most conserved among proteins with GGDEF domains. The active site in this domain is underlined in the reference sequence.
Analysis of the transcriptional expression patterns of rrp1 and hpk1
Most histidine kinase–response regulator pairs are co-transcribed as a polycistronic operon. The separation of rrp1 and hpk1 by a 3 bp spacer is indicative of co-transcription. To assess co-transcription, reverse transcription PCR (RT-PCR) was performed using primers that span the intergenic spacer (3 and 4). Co-transcription of these genes was detected in B. burgdorferi grown at 25°C, 33°C and 37°C (Fig. 1C). Transcript levels were determined using real-time reverse transcription PCR (qRT-PCR) with an internal rrp1 primer set (primers 9 and 10). In these specific qRT-PCR experiments, transcript levels were normalized against flaB transcript. rrp1 was expressed at equivalent levels in spirochetes grown at the different temperatures tested (Fig. 1D).
To determine if rrp1 is expressed in infected ticks, transcript levels were determined in flat (unfed) and fed Ixodes scapularis nymphs. rrp1 expression was sixfold higher in fed nymphs as compared with unfed nymphs. The transcriptional analyses demonstrate that rrp1 and hpk1 are co-transcribed, expressed in vitro in a temperature-independent manner, and induced upon tick feeding.
Rrp1 sequence analyses
To determine if residues of Rrp1 thought to be involved in diguanylate cyclase activity (i.e. c-di-GMP production) and response regulator function are conserved across species lines, rrp1 amplicons from Lyme disease and relapsing fever isolates were cloned and sequenced (Fig. 1E). Residues within the receiver and GGDEF domains that have been identified as functionally important in other bacteria were conserved in all sequences (Ausmees et al., 2001; Romling et al., 2005; Simm et al., 2005; Gjermansen et al., 2006; Cotter and Stibitz, 2007). Within the Lyme spirochetes, at the intraspecies level, the amino acid identity values were 97.7–100%. The relapsing fever spirochetes Borrelia hermsii, B. turicatae, B. duttonii, and B. recurrentis also carry rrp1 and the residues associated with diguanylate cyclase activity are conserved (Lescot et al., 2008). In summary, rrp1 and hpk1 are widely distributed among the Borrelia and have conserved genetic organization and sequence. It is likely that the Rrp1 regulons in the Lyme disease and relapsing fever spirochetes have significant overlap and play important roles in the pathophysiology of the Borrelia.
Analysis of Rrp1 cellular location and production during in vitro cultivation
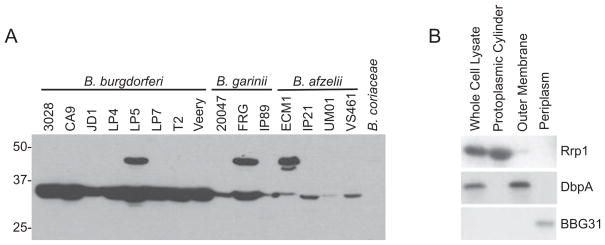
To determine if Rrp1 is produced during cultivation, antiserum was generated against r-Rrp1 derived from B. burgdorferi B31MI. The antiserum was used to screen immunoblots of cell lysates from 17 Borrelia strains (Fig. 2A). Rrp1 (35 kDa) was detected in all Lyme disease strains tested. The antiserum also detected a protein of ~48 kDa in some isolates; however, the identity of this protein is unknown. B. coriaceae, which was PCR and Southern hybridization negative for rrp1, did not produce a protein that was detected with the Rrp1 antiserum.
Fig. 2.
Demonstration of the universal production of Rrp1 by Lyme disease isolates and analysis of its Triton X-114 extraction and phase partitioning properties. A panel of Lyme disease isolates was cultivated under standard conditions and then cell lysates were fractionated, immunblotted and screened with Rrp1 antiserum (A). Species and isolate names are indicated above each lane and molecular weight markers are indicated to the left. (B) presents the results of immunoblot analyses of B. burgdorferi B31 fractions obtained from Triton X-114 extraction and phase partitioning. Identical blots of the fractionated proteins were screened with antiserum to Rrp1, DbpA (an established lipidated outer membrane protein) and BBG31 (a member of the inner membrane-localized Bdr protein family).
To assess the basic properties of Rrp1, Triton X-114 extraction and phase partitioning was performed as described in Experimental procedures. The resulting fractions and the whole-cell lysate were screened by immunoblotting (Fig. 2B). The anti-Rrp1 antiserum detected Rrp1 in the whole-cell lysate and in the fraction containing the protoplasmic cylinder which consists of the inner membrane and cytoplasmic proteins. As controls, additional blots were screened with antiserum against DbpA (a known outer membrane protein) and BBG31 (a periplasmic protein). The control proteins partitioned as expected. These analyses, coupled with the lack of export signals or predicted transmembrane-spanning domains, indicate that Rrp1 is most likely a cytoplasmic protein. This putative cellular location is consistent with that of other response regulators (Galperin et al., 2001; Guvener and Harwood, 2007).
Construction and characterization of an rrp1 deletion mutant
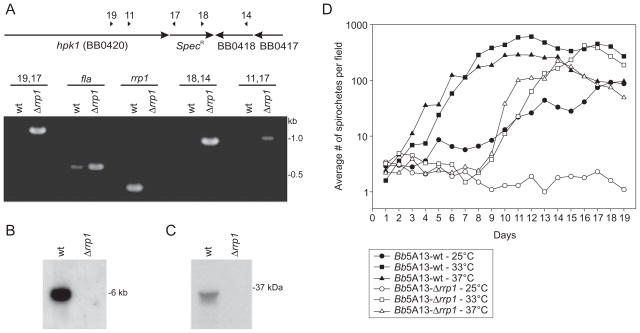
To determine the physiological role of Rrp1, the gene was deleted by allelic exchange replacement from the B. burgdorferi 5A13 background. Numerous clonal populations were obtained, and deletion of rrp1 was confirmed using PCR (Fig. 3A), Southern hybridization (Fig. 3B) and immunoblotting (Fig. 3C). Attempts at deleting rrp1 from the infectious B. burgdorferi 5A4 strain, which has been reported to have an inherently low transformation efficiency (Purser and Norris, 2000), were not successful. However, with the recovery of stable 5A13 transformants, it was possible to assess the potential global regulatory capability of Rrp1 in vitro.
Fig. 3.
Verification of rrp1 deletion by allelic exchange replacement and analysis of growth rate of Bb5A13-wt and Bb5A13-Δrrp1. To confirm allelic exchange replacement was successful, PCR (A), Southern hybridization (B) and immunoblot analyses (C) were performed. (A) shows a schematic of the chromosome after allelic exchange, in which rrp1 (BB0419) has been replaced by the spectinomycin/streptomycin resistance cassette (SpecR). Primers are indicated by numbers and listed in Table S2. Numbers above each lane in the ethidium bromide-stained gel indicate primer combinations. In (B), EcoRI-restricted genomic DNA was fractionated, blotted and screened with a radiolabeled rrp1 probe. In (C), cell lysates were screened with Rrp1 antiserum. In (D), Bb5A13-wt and Bb5A13-Δrrp1 cells were cultivated at 25°C, 33°C and 37°C in BSK-H complete media. All methods were as described in the text.
Electroporation commonly results in the loss of some of plasmids. To determine the plasmid content of clonal populations of the rrp1 knockout (Bb5A13-Δrrp1), a PCR-based approach using plasmid-specific primer sets was employed. Clonal population 9, which lost the fewest plasmids (cp9, lp28-1 and lp21; plasmids C, F and U respectively) relative to 5A13, was selected for all future analyses. cp9, lp21 and lp28-1 are relatively unstable plasmids that can be lost upon cultivation and/or passage through mice and are not essential for in vitro growth (Xu et al., 1996; 2003; Palmer et al., 2000; McDowell et al., 2001; Lawrenz et al., 2002; Iyer et al., 2003; Stewart et al., 2005; Embers et al., 2008). It is noteworthy that lp21 is comprised largely of repeat elements and pseudogenes (Casjens et al., 2000; Palmer et al., 2000). In addition, earlier microarray analyses and the microarray data presented below demonstrate that the overwhelming majority of genes encoded by these plasmids are either poorly expressed or not expressed at all in vitro (Table S1) (Brooks et al., 2003; Ojaimi et al., 2003; 2005; Tokarz et al., 2004; Caimano et al., 2007; Boardman et al., 2008; Ouyang et al., 2008). Furthermore, none of the open reading frames (ORFs) present on these plasmids encodes proteins that are putative transcriptional regulators or intermediaries in metabolic pathways. Consistent with this, these plasmids do not influence infective potential or in vitro growth characteristics, and hence their absence in clone 9 does not compromise this initial and preliminary analysis of the potential global regulatory influence of Rrp1 during in vitro cultivation.
To examine potential growth defects due to the inactivation of Rrp1, cultures of Bb5A13-wt and Bb5A13-Δrrp1 were diluted to equal cell densities in fresh media and incubated at 25°C, 33°C and 37°C. The average number of spirochetes in 10 high-powered fields was determined every day for 19 days (Fig. 3D). All Bb5A13-Δrrp1 cultures displayed a growth defect compared with Bb5A13-wt. Bb5A13-Δrrp1 cultures grown at 33°C and 37°C were found to undergo an extended (1 week) lag phase. This is in contrast to Bb5A13-wt, which entered exponential phase within 2 days. Growth of Bb5A13-Δrrp1 at 25°C was severely impaired, and while the spirochetes remained viable, the population never entered exponential phase over the 19-day period of analysis. These data demonstrate that while Rrp1 is not essential for survival in vitro, it does influence growth characteristics. The inability of the Bb5A13-Δrrp1 mutant to grow at lower temperatures, coupled with its upregulation in fed ticks (Fig. 1D), suggests that Rrp1 may be important in environmentally induced adaptive responses in the tick.
Deletion of rrp1 results in wide-scale changes in the B. burgdorferi transcriptome
To identify genes regulated by Rrp1, gene-specific mRNA levels were compared between Bb5A13-wt and Bb5A13-Δrrp1 by microarray analyses. RNA was isolated from three independent cultures of each strain, converted to cDNA, labeled, and applied to oligonucleotide microarrays that represent the entire genome of B. burgdorferi B31MI. The arrays have been previously described (Terekhova et al., 2006). Hybridization to oligonucleotides representing 1140 ORFs (65.5% of the total ORFs) was above background in at least one of the strains. ORFs for which hybridization was below background in both strains are listed in Table S1.
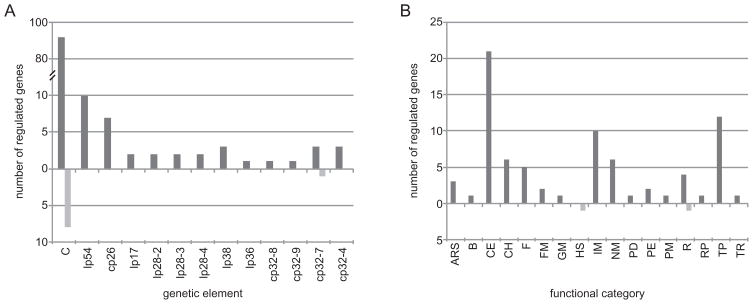
The expression of 140 genes (8% of the genome) was influenced by Rrp1 (Table 1). The majority of these genes are encoded by the chromosome (102 genes), lp54 (plasmid A; 10 genes) and cp26 (plasmid B; 7 genes) (Fig. 4A). These genetic elements are the most conserved components of the Lyme disease spirochete genome and comprise the core Borrelia genome (Terekhova et al., 2006). Of the genes regulated by Rrp1, 131 had lower expression in Bb5A13-Δrrp1 while only 9 genes had elevated expression (Table 1). Most genes with lower mRNA levels in Bb5A13-Δrrp1 (80 out of 131) have predicted functions and span almost all functional categories except cell division, translation factors, heat shock and haemolysis (Fig. 4B). The functional categories most affected were: cell envelope (21 genes), transport (12 genes), intermediary metabolism (10 genes), nucleotide metabolism (6 genes), chemotaxis (6 genes) and flagellar biosynthesis (5 genes). Of the nine genes negatively regulated by Rrp1 (i.e. with elevated expression in the rrp1 knockout), only two have predicted functions: ruvB helicase (BB0022) and groES (BB0741). Since the deletion of rrp1 resulted predominantly in lower expression of the affected genes, it can be concluded that Rrp1 exerts its regulatory effects primarily through gene activation (directly or indirectly).
Table 1.
Borrelia burgdorferi genes (a) activated and (b) repressed by Rrp1.
| Gene number and annotationa,b | Functional categorya | WT/Mc | P-value | Relevant citationd |
|---|---|---|---|---|
| (a) Borrelia burgdorferi genes activated by Rrp1 | ||||
| BB0014 primosomal protein N′ (priA) | R | 9.55 | 0.0117 | |
| BB0021 S-adenosylmethionine: tRNA ribosyltransferase-isomerase | ARS | 8.36 | 0.00924 | Caimano et al. (2007) |
| BB0047 conserved hypothetical protein | HX | 11.0 | 0.0275 | Ojaimi et al. (2003); Hyde et al. (2006) |
| BB0048 hypothetical protein | U | 122 | 0.0350 | Ojaimi et al. (2003) |
| BB0084 nifS protein (nifS) | ARS | 5.79 | 0.0269 | |
| BB0100 glutamate racemase (murI) | CE | 94.0 | 0.0232 | |
| BB0107 N-utilization substance protein B (nusB) | TR | 4.02 | 0.00995 | |
| BB0110 hypothetical protein | U | 6.13 | 0.0209 | |
| BB0114 single-stranded DNA-binding protein (ssb) | R | 4.15 | 0.00559 | |
| BB0129 conserved hypothetical protein | HX | 37.1 | 0.0203 | Narasimhan et al. (2002) |
| BB0140 acriflavine resistance protein (acrB) | X | 11.6 | 0.0136 | |
| BB0146 glycine betaine, L-proline ABC transporter, ATP-binding protein (proV) | TP | 80.1 | 0.0132 | Narasimhan et al. (2002) |
| BB0149 flagellar hook-associated protein 2 (fliD) | F | 7.87 | 0.0300 | |
| BB0151 N-acetylglucosamine-6-phosphate deacetylase (nagA) | IM | 12.3 | 0.0163 | Narasimhan et al. (2002) |
| BB0152 glucosamine-6-phosphate isomerase (nagB) | IM | 66.7 | 0.0260 | Brooks et al. (2003) |
| BB0153 superoxide dismutase (sodA) | X | 129 | 0.0484 | Hyde et al. (2006) |
| BB0156 hypothetical protein | U | 9.78 | 0.0183 | |
| BB0160 alanine racemase (alr) | CE | 10.8 | 0.0277 | |
| BB0162 hypothetical protein | U | 8.86 | 0.00235 | Ojaimi et al. (2003); Tokarz et al. (2004) |
| BB0167 outer membrane protein (tpn50)b | CE | 29.8 | 0.00587 | |
| BB0170 hypothetical protein | U | 136 | 0.0207 | |
| BB0173 hypothetical protein | U | 131 | 0.0142 | |
| BB0174 hypothetical protein | U | 27.0 | 0.0112 | |
| BB0175 conserved hypothetical protein | HX | 4.49 | 0.00218 | Ojaimi et al. (2003); Tokarz et al. (2004) |
| BB0178 glucose inhibited division protein A (gidA) | R | 4.15 | 0.00934 | |
| BB0179 thiophene and furan oxidation protein (thdF) | X | 30.5 | 0.0193 | |
| BB0180 flagellar protein, putative | F | 30.9 | 0.00809 | |
| BB0182 flagellar hook-associated protein 3 (flgL) | F | 10.8 | 0.00882 | |
| BB0185 conserved hypothetical protein | HX | 5.65 | 0.00311 | |
| BB0186 conserved hypothetical protein | HX | 4.98 | 0.0338 | |
| BB0221 flagellar motor switch protein (fliG-1) | F | 9.67 | 0.0130 | |
| BB0241 glycerol kinase (glpK)b | IM | 5.43 | 0.000391 | Narasimhan et al. (2002); Ojaimi et al. (2003); Hyde et al. (2006); Caimano et al. (2007) |
| BB0242 hypothetical protein | U | 367 | 0.00274 | Ojaimi et al. (2003); Tokarz et al. (2004); Hyde et al. (2006); Caimano et al. (2007) |
| BB0243 glycerol-3-phosphate dehydrogenase, anaerobic (glpA)b | IM | 21.9 | 0.00201 | Ojaimi et al. (2003); Hyde et al. (2006); Caimano et al. (2007) |
| BB0250 dedA protein (dedA) | X | 14.9 | 0.0363 | |
| BB0261 hypothetical protein | U | 14.9 | 0.0310 | Tokarz et al. (2004) |
| BB0287 flagellar protein (flbA) | F | 11.6 | 0.0233 | |
| BB0319 exported protein (tpn38b) | CE | 53.9 | 0.0114 | |
| BB0349 hypothetical protein | U | 57.0 | 0.00831 | |
| BB0358 conserved hypothetical protein | HX | 29.6 | 0.0376 | |
| BB0367 hypothetical proteinb | U | 18.8 | 0.00123 | Tokarz et al. (2004) |
| BB0407 mannose-6-phosphate isomerase (manA) | IM | 59.0 | 0.0397 | |
| BB0420 sensory transduction histidine kinase/response regulator (hpk1) | GM | 236 | 0.0343 | Narasimhan et al. (2002) |
| BB0451 chromate transport protein, putative | TP | 63.0 | 0.0475 | |
| BB0466 ABC transporter, ATP-binding protein | TP | 12.4 | 0.0404 | |
| BB0475 lipoprotein, putative | CE | 4.66 | 0.0231 | |
| BB0530 hypothetical protein | U | 7.66 | 0.0172 | |
| BB0531 hypothetical protein | U | 11.0 | 0.0387 | |
| BB0553 hypothetical protein | U | 6.61 | 0.0438 | |
| BB0567 chemotaxis histidine kinase (cheA-1) | CH | 45.1 | 0.0348 | Tokarz et al. (2004); Fisher et al. (2005) |
| BB0568 protein-glutamate methylesterase (cheB-2) | CH | 6.50 | 0.0198 | |
| BB0570 chemotaxis response regulator (cheY-2) | CH | 4.79 | 0.0322 | |
| BB0571 uridylate kinase (smbA) | NM | 99.7 | 0.0177 | |
| BB0572 glycosyl transferase (lgtD) | CE | 67.5 | 0.0367 | |
| BB0575 CTP synthase (pyrG) | NM | 15.5 | 0.0130 | |
| BB0578 methyl-accepting chemotaxis protein (mcp-1) | CH | 31.8 | 0.0163 | Caimano et al. (2007) |
| BB0583 MATE efflux family protein | TP | 264 | 0.0382 | |
| BB0584 conserved hypothetical integral membrane protein | CE | 7.78 | 0.0173 | |
| BB0586 femA protein (femA) | X | 6.43 | 0.0153 | |
| BB0587 methionyl-tRNA synthetase (metG) | ARS | 19.5 | 0.0125 | |
| BB0588 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase, putative (pfs-2) | NM | 14.2 | 0.0104 | Caimano et al. (2007) |
| BB0589 phosphate acetyltransferase (pta) | IM | 26.6 | 0.0149 | |
| BB0590 dimethyladenosine transferase (ksgA) | NM | 198 | 0.0166 | |
| BB0596 methyl-accepting chemotaxis protein (mcp-2) | CH | 240 | 0.0114 | |
| BB0598 UDP-N-acetylmuramate dehydrogenase (murB) | CE | 104 | 0.0475 | |
| BB0600 hypothetical protein | U | 13.6 | 0.0181 | Narasimhan et al. (2002) |
| BB0604 L-lactate permease (lctP) | TP | 4.63 | 0.0297 | |
| BB0617 hypothetical protein | U | 33.9 | 0.0146 | |
| BB0620 beta-glucosidase, putative | IM | 17.8 | 0.0142 | |
| BB0625 N-acetylmuramoyl-L-alanine amidase, putative | CE | 6.25 | 0.00591 | |
| BB0629 PTS system, fructose-specific IIABC component (fruA-2)b | TP | 20.1 | 0.0128 | |
| BB0630 1-phosphofructokinase (fruK) | IM | 15.1 | 0.0326 | |
| BB0635 conserved hypothetical protein | HX | 5.02 | 0.0102 | Narasimhan et al. (2002) |
| BB0636 glucose-6-phosphate 1-dehydrogenase (zwf) | IM | 60.9 | 0.0230 | |
| BB0637 Na+/H+ antiporter (nhaC-1) | TP | 33. 7 | 0.0154 | |
| BB0638 Na+/H+ antiporter (nhaC-2) | TP | 6.14 | 0.00977 | |
| BB0642 spermidine/putrescine ABC transporter, ATP-binding protein (potA) | TP | 17.4 | 0.0158 | |
| BB0645 PTS system, glucose-specific IIBC component (ptsG) | TP | 30.5 | 0.0481 | |
| BB0648 serine/threonine kinase, putative | PM | 4.11 | 0.0253 | |
| BB0650 hypothetical protein | U | 22.0 | 0.0229 | |
| BB0652 protein-export membrane protein (secD) | PE | 37.7 | 0.0136 | Brooks et al. (2003) |
| BB0653 protein-export membrane protein (secF) | PE | 55.9 | 0.00618 | |
| BB0656 oxygen-independent coproporphyrinogen III oxidase, putative | B | 147 | 0.00662 | |
| BB0664 hypothetical protein | U | 108 | 0.0394 | |
| BB0672 chemotaxis response regulator (cheY-3) | CH | 7.63 | 0.0141 | |
| BB0675 hypothetical protein | U | 4.45 | 0.0137 | |
| BB0683 3-hydroxy-3-methylglutaryl-CoA synthaseb | FM | 14.7 | 0.00139 | |
| BB0687 phosphomevalonate kinase, putative | FM | 5.34 | 0.0126 | |
| BB0702 lipopolysaccharide biosynthesis-related protein (kdtB) | X | 17.5 | 0.0497 | |
| BB0769 sialoglycoprotease (gcp) | PD | 84.2 | 0.0117 | |
| BB0785 stage V sporulation protein G | X | 11.2 | 0.0176 | Tokarz et al. (2004); Caimano et al. (2007) |
| BB0804 ribosomal protein S15 (rpsO) | RP | 8.54 | 0.0263 | |
| BB0837 excinuclease ABC, subunit A (uvrA) | R | 11.8 | 0.0212 | |
| BB0842 ornithine carbamoyltransferase, catabolic (arcB) | IM | 72.1 | 0.0359 | |
| BBA03 outer membrane protein | CE | 247 | 0.00704 | Carroll et al. (2000); Tokarz et al. (2004); Hyde et al. (2006) |
| BBA19 conserved hypothetical protein | HX | 26.9 | 0.00915 | |
| BBA30 hypothetical proteinb | U | 157 | 0.00375 | |
| BBA41 conserved hypothetical protein | HX | 3.42 | 0.0218 | Ojaimi et al. 2003) |
| BBA52 outer membrane protein | CE | 575 | 0.0216 | Brooks et al. (2003); Tokarz et al. (2004) |
| BBA59 lipoprotein | CE | 741 | 0.00397 | Brooks et al. (2003); Tokarz et al. 2004) |
| BBA61 conserved hypothetical protein | HX | 5.32 | 0.0331 | Brooks et al. (2003) |
| BBA69 hypothetical protein | U | 5.77 | 0.0460 | Brooks et al. (2003); Tokarz et al. (2004); Caimano et al. (2007) |
| BBA74 outer membrane-associated periplasmic protein | CE | 159 | 7.76E-05 | Brooks et al. (2003); Ojaimi et al. (2003); Tokarz et al. (2004); Caimano et al. (2007) |
| BBA76 thymidylate synthase-complementing protein (thy1) | X | 324 | 0.0335 | |
| BBB14 hypothetical protein | U | 206 | 0.0261 | Ojaimi et al. (2003) |
| BBB16 oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (oppAIV)b | TP | 17.0 | 7.26E-05 | |
| BBB17 IMP dehydrogenase (guaB)b | NM | 6.91 | 0.0307 | |
| BBB18 GMP synthase (guaA)b | NM | 8.30 | 0.000847 | |
| BBB19 outer surface protein C (ospC)b | CE | 33.9 | 0.0476 | Brooks et al. (2003); Ojaimi et al. (2003); Yang et al. (2003a); Tokarz et al. (2004); Fisher et al. (2005); Caimano et al. (2007) |
| BBB22 conserved hypothetical protein | HX | 366 | 0.0182 | |
| BBB29 PTS system, maltose and glucose-specific IIABC component (malX) | TP | 25.7 | 0.0154 | |
| BBD09 hypothetical protein | U | 10.6 | 0.0130 | Hyde et al. (2006) |
| BBD10 hypothetical protein | U | 47.2 | 0.00975 | |
| BBG06 conserved hypothetical protein | HX | 6.09 | 0.0103 | Tokarz et al. (2004) |
| BBG33 Borrelia direct repeat protein (bdrF2) | CE | 27.5 | 0.0151 | |
| BBH06 factor H-binding protein (cpsZ)b | U | 159 | 0.0140 | |
| BBH27 conserved hypothetical protein | HX | 34.9 | 0.00127 | |
| BBI29 hypothetical protein | U | 223 | 0.0241 | Ojaimi et al. (2003); Caimano et al. (2007) |
| BBI38 hypothetical protein | U | 224 | 0.0302 | Brooks et al. (2003); Caimano et al. (2007) |
| BBJ08 hypothetical protein | U | 11.6 | 0.0340 | Brooks et al. (2003); Ojaimi et al. (2003); Tokarz et al. (2004) |
| BBJ09 outer surface protein D (ospD) | CE | 7.13 | 0.0146 | Brooks et al. (2003); Ojaimi et al. (2003); Tokarz et al. (2004) |
| BBJ34 hypothetical protein | U | 4.68 | 0.0432 | Ojaimi et al. (2003) |
| BBK19 hypothetical protein | U | 15.4 | 0.0131 | Caimano et al. (2007) |
| BBL28 lipoprotein | CE | 111 | 0.047 | Brooks et al. (2003); Ojaimi et al. (2003); Tokarz et al. (2004) |
| BBN38 factor H-binding protein (ospE)b | CE | 5.36 | 0.0126 | |
| BBO21 conserved hypothetical protein | HX | 45.0 | 0.0102 | |
| BBO27 Borrelia direct repeat protein (bdrE3) | HX | 484 | 0.00972 | Brooks et al. (2003) |
| BBO28 lipoprotein (lp) | CE | 292 | 0.0131 | Tokarz et al. (2004) |
| BBR28 lipoprotein (lp) | CE | 9.63 | 0.00599 | Brooks et al. (2003); Tokarz et al. (2004) |
| BBR36 conserved hypothetical protein | HX | 11.1 | 0.0475 | Tokarz et al. (2004) |
| BBR42 outer surface protein F (ospF)b | CE | 151 | 0.0336 | Ojaimi et al., 2003; Yang et al., 2003a; Caimano et al. (2007) |
| (b) Borrelia burgdorferi genes repressed by Rrp1 | ||||
| BB0022 Holliday junction DNA helicase (ruvB) | R | 15.0 | 0.0108 | |
| BB0340 hypothetical protein | U | 16.0 | 0.0192 | |
| BB0398 lipoprotein, putative | U | 12.3 | 0.0239 | |
| BB0470 hypothetical protein | U | 14.4 | 0.0124 | |
| BB0509 hypothetical protein | U | 4.98 | 3.73E-05 | |
| BB0510 hypothetical protein | U | 126 | 0.0129 | |
| BB0741 chaperonin (groES) | HS | 4.96 | 0.0440 | |
| BB0794 hypothetical protein | U | 82.1 | 0.0410 | |
| BBO05 hypothetical protein | U | 4.06 | 0.0254 | |
As published in TIGR B. burgdorferi B31MI genome database and in Fraser et al. (1997).
Genes differentially expressed that were validated by qRT-PCR and/or western blotting are bolded.
M, mutant mRNA hybridization intensity values; WT, wild-type mRNA hybridization intensity values.
Only genes that pass the statistical criteria of log2(M/WT) < −2 or > 2 and P < 0.05 are listed.
Fig. 4.
Rrp1 is a central regulator of the core Borrelia genome. Genes differentially expressed in Bb5A13-Δrrp1 relative to Bb5A13-wt were identified by microarray analysis as described in Experimental procedures. Numbers of genes with lower or higher mRNA levels (dark grey and light grey bars respectively) in Bb5A13-Δrrp1 relative to Bb5A13-wt are organized by genetic location (A) and by functional category (B). Abbreviations: C, chromosome; lp, linear plasmid; cp, circular plasmid; ARS, amino acid biosynthesis; B, biosynthesis; CE, cell envelope; CH, chemotaxis; F, flagellar biosynthesis; FM, fatty acid metabolism; GM, general metabolism; HS, heat-shock proteins; IM, intermediary metabolism; NM, nucleotide metabolism; PD, protein degradation; PE, protein export; PM, protein metabolism; R, replication; RP, ribosomal proteins; TP, transporter proteins; TR, transcription.
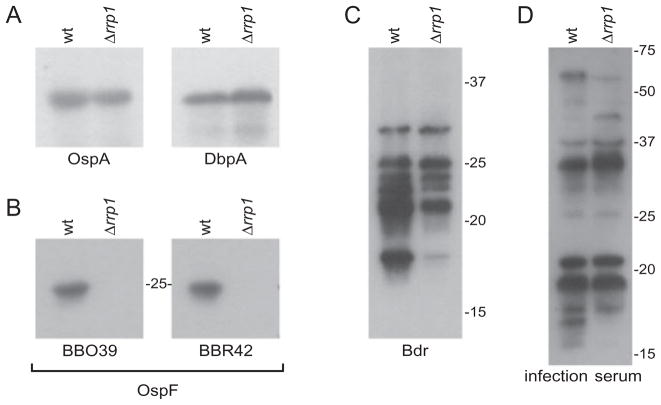
Cell envelope protein genes with lower mRNA levels in Bb5A13-Δrrp1 belong to several highly studied paralogous protein families including the OspE (PF162), OspF (PF164), Mlp (PF113) and Bdr (PF80) families. As noted above, the majority of Rrp1-regulated genes are chromosomal; however, all of the cell envelope-encoding genes listed above are carried on the cp32 plasmids. The OspE proteins have been implicated as important virulence factors that facilitate complement evasion by binding negative regulators of the complement system such as factor H (FH) (Hellwage et al., 2001; McDowell et al., 2003; Alitalo et al., 2004; Kraiczy and Würzner, 2005; Hovis et al., 2006a). There is also evidence that members of this protein family bind to additional serum proteins and that this may influence the host–pathogen interaction (Hovis et al., 2006b; Rogers and Marconi, 2007). The microarray analyses revealed that the FH-binding OspE paralogue, BBN38, had significantly lower expression in Bb5A13-Δrrp1. In addition, the two other members of this gene family, BBP38 and BBL39 (identical paralogues), also had lower transcript levels in Bb5A13-Δrrp1, but the difference was below the cut-off value set for significance. However, it is important to note that the dependence of this gene family on Rrp1 as inferred from the arrays is strongly supported by western blot and immunofluorescence assay (IFA) analysis (see below Fig. 6A and B). The expression of several members of the OspF protein family was also lower in Bb5A13-Δrrp1. Several of the ospF genes are co-transcribed with ospE gene family members or are located just downstream of ospE genes (Lam et al., 1994). The OspF proteins have been postulated to serve as adhesins (Antonara et al., 2007), and earlier work demonstrated that they might carry out stage-specific functions during infection in mice (McDowell et al., 2004). The microarrays demonstrated that the OspF paralogue, BBR42, has significantly lower expression in Bb5A13-Δrrp1. This observation is supported by immunoblot analyses (Fig. 5).
Fig. 6.
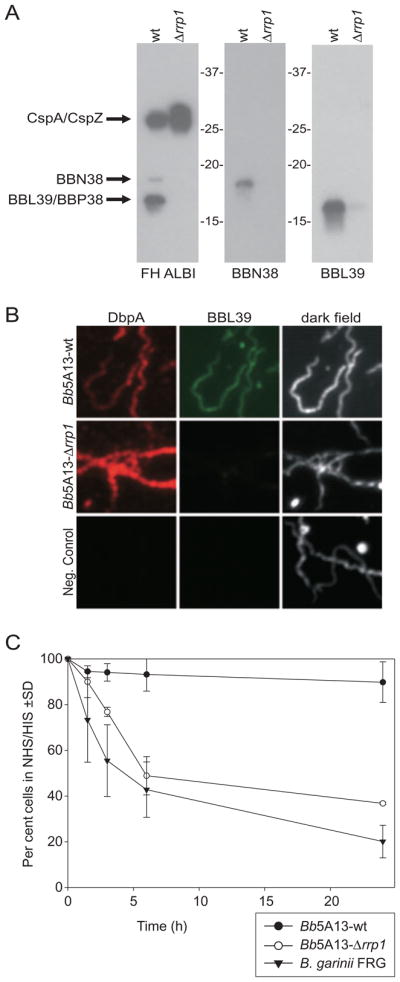
Rrp1 regulation of the OspE factor H binding proteins and analysis of Bb5A13-wt and Bb5A13-Δrrp1 cells for serum sensitivity. Western blots of the Bb5A13-wt and Bb5A13-Δrrp1 cell lysates were screened for factor H binding ability and with anti-BBN38 and anti-BBL39 (A). For the IFA analysis in (B), Bb5A13-wt and Bb5A13-Δrrp1 cells were washed, dried onto slides, and screened simultaneously with anti-DbpA and anti-BBL39. The negative control slide was screened with secondary antibodies only. For (C), Bb5A13-wt, Bb5A13-Δrrp1 and B. garinii FRG cells were incubated in 50% normal human serum (NHS) or in 50% heat-inactivated serum (HIS) for 1.5, 3, 6, and 24 h. At each time, the number of cells in 10 high-powered fields was counted and averaged. The graph shows the number of cells in NHS divided by the number of cells in HIS, expressed as a percentage. The error bars represent standard deviation (SD) from three independent experiments.
Fig. 5.
Comparative immunoblot analysis of Bb5A13-wt and Bb5A13-Δrrp1. The results of the microarray analyses were confirmed by western blot analysis of a select group of proteins. Cell lysates of the Bb5A13-wt and Bb5A13-Δrrp1 were separated by SDS-PAGE, immunoblotted, and screened with the antiserum indicated below each blot. (A) represents proteins not affected in the Rrp1 knockout, (B) represents an example of two members of a protein family that are coordinately regulated by Rrp1, and (C) represents family members that are differentially regulated by Rrp1. In (D), cell lysates were screened with serum collected from mice infected with B. burgdorferi B31MI.
The expression of the eight-member mlp gene family (PF113) (Yang et al., 1999; Porcella et al., 2000) was also influenced by Rrp1. The function(s) of this cp32-carried gene family have not yet been defined. Three mlp paralogues (BBL28, BBO28 and BBR28) had lower expression in Bb5A13-Δrrp1. The mlp paralogues BBN28 and BBP28 were also repressed in the mutant but missed the stringent statistical criteria set for significance. The data suggest that this family as a whole is activated by Rrp1.
The transcription of select members of the bdr gene family (PF80) (Zuckert et al., 1999; Carlyon et al., 2000a,b) was also influenced by deletion of rrp1. Expression of BdrF2 (BBG33) and BdrE3 (BBO27) was significantly reduced in Bb5A13-Δrrp1. The expression of BdrE4 (BBR27), Bdr frameshift (BBR35), and BdrE5 (BBS29) were similarly affected in Bb5A13-Δrrp1, but missed the significance cut-off. The Rrp1 dependence of some Bdr proteins was also demonstrated by western blot (Fig. 5C). The varying influence of the rrp1 deletion on the expression of individual Bdr paralogues is consistent with earlier analyses demonstrating that the individual bdr paralogues respond differently to environmental variables and thus may be regulated through different mechanisms (Roberts et al., 2002).
Genes encoding proteins involved in the transport of ions, carbohydrates, amino acids, and oligopeptides were affected by the deletion of Rrp1. The Na+/H+ antiporters nhaC-1 (BB0637) and nhaC-2 (BB0638) had lower mRNA levels in Bb5A13-Δrrp1. The Na+/Ca+ exchange protein, BB0164, also appears to be regulated by Rrp1. Carbohydrate transporter genes with lower mRNA levels in Bb5A13-Δrrp1 included lctP, a L-lactate permease (BB0604), fruA-2, a fructose-specific IIABC component (BB0629), and proteins of the glucose, maltose, glucosamine, and N-acetylglucosamine transporter systems [PTS system, maltose- and glucose-specific IIABC component malX (BBB29), and glucose-specific IIBC component ptsG (BB0645)]. Thus, expression of almost all carbohydrate transport genes, with the exception of those specifically involved in chitobiose, ribose and galactose transport, was affected in Bb5A13-Δrrp1. Transcript levels for numerous carbohydrate metabolism genes were also significantly reduced in Bb5A13-Δrrp1. These include N-acetylglucosamine-6-phosphate deacetylase (nagA; BB0151), glucosamine-6-phosphate isomerase (nagB; BB0152), glycerol kinase (glpK; BB0241), glycerol-3-phosphate dehydrogenase (glpA; BB0243), mannose-6-phosphate isomerase (manA; BB0407), phosphate acetyltransferase (pta; BB0589), beta-glucosidase (BB0620), 1-phosphofructokinase (fruK; BB0630), glucose-6-phosphate 1-dehydrogenase (zwf; BB0636), and ornithine carbamoyltransferase (arcB; BB0842). Strikingly, most of the affected carbohydrate metabolism genes code for enzymes that generate substrates for glycolysis. Beta-glucosidase (BB0620) results in generation of glucose-6-phosphate; manA, nagA and nagB result in production of fructose-6-phosphate; fruK converts fructose-1-phosphate into fructose-1,6-bisphosphate and glpK and glpA produce dihydroxyacetone phosphate.
The expression of almost all genes involved in amino acid and oligopeptide transport was affected in Bb5A13-Δrrp1. Lower mRNA levels were observed in Bb5A13-Δrrp1 for the glycine/proline transporter [glycine betaine, L-proline ABC transporter, ATP-binding protein proV (BB0146)], the spermidine/putrescine transporter (potA, BB0642) and the oligopeptide transporter [oligopeptide ABC transporter, periplasmic oligopeptide-binding protein oppAIV (BBB16)].
Some genes involved in nucleotide biosynthesis were found to have reduced expression in Bb5A13-Δrrp1. These include uridylate kinase (smbA; BB0571), CTP synthase (pyrG; BB0575), 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase (pfs-2; BB0588), IMP dehydrogenase (guaB; BBB17) and GMP synthase (guaA; BBB18). Only pfs-2 had been previously shown to be regulated by RpoS (Caimano et al., 2007). Thus, the data suggest that Rrp1 regulates a unique set of nucleotide metabolism genes.
In several bacterial systems, intracellular c-di-GMP levels influence chemotaxis and motility (Wolfe and Visick, 2008). In that Rrp1 is the sole protein of B. burgdorferi likely to have diguanylate cyclase activity, it was predicted that deletion of rrp1 would influence the expression of some chemotaxis and motility genes. Consistent with this, numerous genes involved in chemotaxis and motility had reduced expression in Bb5A13-Δrrp1. Examples include cheA-1 (BB0567), cheB-2 (BB0568), cheY-2 (BB0570), cheY-3 (BB0672), mcp-1 (BB0578), and mcp-2 (BB0596). Expression of mcp-3 (BB0597) and mcp-4 (BB0680) was also reduced in the Bb5A13-Δrrp1, but missed the cut-off for statistical significance. The flagellar hook-associated proteins fliD (BB0149) and flgL (BB0182), the flagellar motor switch protein fliG-1 (BB0221), and flaB (BB0285) had lower expression levels in Bb5A13-Δrrp1. Thus, Rrp1 appears to be involved in the positive regulation of chemotaxis and motility in B. burgdorferi.
Validation of microarray analyses by qRT-PCR, immunoblot, IFA, and FH binding assays
To validate the microarray data, several approaches were applied including qRT-PCR, immunoblotting, and IFA. The RNA used for the microarray and qRT-PCR analyses and the cell lysates used for the immunoblot studies were derived from the same log-phase cultures. For the qRT-PCR analyses, relative transcripts levels of 20 of the differentially expressed genes were assessed (Table 2). In general, the trends in differential expression for these genes (i.e. either elevated or reduced expression in wild-type and mutant spirochetes) are similar. It should be noted that for Lyme disease spirochetes, qRT-PCR data are typically normalized against the levels of flaB, which is considered to be constitutively expressed under most conditions. However, the array data indicate that flaB, as well as several other genes involved in motility and chemotaxis, are regulated to varying degrees either directly or indirectly by Rrp1. This may have a modest effect on the fold changes determined by qRT-PCR. As an additional means of validating the microarray analyses, a series of identical immunoblots were generated and screened with antiserum to a number of B. burgdorferi proteins including OspA, DbpA, OspE (BBL39 and BBN38), OspF (BBO39 and BBR42) and the Bdr protein family in order to assess protein expression (Figs 5 and 6A). Consistent with the array analysis, production of OspA and DbpA were not affected by the absence of Rrp1, whereas production of certain Bdr, OspF, and OspE proteins was abrogated. Furthermore, the OspE protein, BBL39, was not detected at the cell surface by IFA analysis in Bb5A13-Δrrp1 (Fig. 6B). Since OspE proteins function as FH-binding proteins, the ability of Bb5A13-Δrrp1 to bind FH was also assessed (Fig. 6A). Consistent with the expression data, FH binding to proteins in the OspE size range (15–20 kDa) was not observed; FH binding to the CspZ and CspA proteins (25–27 kDa) was detected.
Table 2.
Quantitative RT-PCR analysis of select genes.
| Gene | Annotation | Array | qRT-PCR |
|---|---|---|---|
| BB0167 | Outer membrane protein (tnp50) | 29.8 | 6.2 |
| BB0241 | Glycerol kinase (glpK) | 5.4 | 2.9 |
| BB0243 | Glycerol-3-phosphate dehydrogenase (glpA) | 21.9 | 11.3 |
| BB0367 | Hypothetical protein | 18.8 | 10.0 |
| BB0420 | Sensory transduction histidine kinase (hpk1) | 236.0 | 1.3 |
| BB0596 | Methyl-accepting chemotaxis protein (mcp-2) | 240.0 | 1.3 |
| BB0629 | Fructose-specific IIABC component (fruA-2) | 20.1 | 2.9 |
| BB0683 | 3-Hydroxyl-3-methylglutaryl-CoA synthase | 14.7 | 2.7 |
| BB0687 | Phosphomevalonate kinase | 5.3 | 1.1 |
| BB0769 | Sialoglycoprotease (gcp) | 84.2 | 1.5 |
| BB0785 | Stage V sporulation protein G | 11.2 | 3.5 |
| BBA30 | Hypothetical protein | 157.0 | 4.9 |
| BBA74 | Periplasmic protein | 159.0 | 4.5 |
| BBB16 | Oligopeptide ABC transporter (oppAIV) | 17.0 | 6.0 |
| BBB17 | IMP dehydrogenase (guaB) | 6.9 | 2.1 |
| BBB18 | GMP synthase (guaA) | 8.3 | 2.1 |
| BBB19 | Outer surface protein C (ospC) | 33.9 | 3.7 |
| BBB29 | Maltose-glucose-specific IIABC component | 25.7 | 3.2 |
| BBH06 | Factor H-binding protein (cspZ) | 159.0 | 3.4 |
| BBR28 | Lipoprotein | 9.6 | 2.1 |
Values expressed as a ratio of expression for the given gene in the Bb5A13-wt relative to Bb5A13-Δrrp1.
To determine if the Rrp1 regulon includes proteins that are antigenic during infection, whole-cell lysates of Bb5A13-wt and Bb5A13-Δrrp1 strains were screened with serum recovered from mice infected with B. burgdorferi B31 (Fig. 5D). While the identity of individual antigens was not determined in this particular experiment, the data demonstrate that inactivation of Rrp1 eliminated or decreased production of several proteins that elicit an antibody response during murine infection with B. burgdorferi B31.
Increase in serum sensitivity of the rrp1 knockout
The FH binding capabilities of the Lyme spirochetes have been correlated with serum resistance (McDowell et al., 2003; Kraiczy et al., 2004; Brooks et al., 2005). While the Rrp1 regulon appears to be comprised primarily of housekeeping genes, the regulation of several FH-binding proteins by Rrp1 indicates a connection between this specific immune evasion/virulence mechanism and Rrp1-mediated transcriptional regulation. To determine if the inactivation of Rrp1 influences serum resistance, a serum sensitivity assay was performed on the Bb5A13-wt and Bb5A13-Δrrp1 strains (Fig. 6C). Significant differences in serum sensitivity were evident by 6 h. After a 24 h incubation with normal human serum, less than 40% of the Bb5A13-Δrrp1 cells were alive compared with cells incubated with heat-inactivated serum. This is in contrast to the Bb5A13-wt cells, in which 90% of cells survived in normal human serum after 24 h. The serum sensitivity of the Bb5A13-Δrrp1 cells proved to be equivalent to that of B. garinii, which has been demonstrated to be highly sensitive to complement-mediated killing (Brooks et al., 2005). It is noteworthy that the highly serum-sensitive Bb5A13-Δrrp1 strain still produces the FH-binding proteins, CspA and CspZ. While the dramatic increase in serum sensitivity of the Bb5A13-Δrrp1 strain suggests that OspE proteins are key players in serum resistance, it is possible that this effect could be indirect. Nonetheless, it can be concluded that the significant changes in overall protein production in the Rrp1 mutant results in a highly serum-sensitive phenotype.
Discussion
Borrelia burgdorferi encodes only two TCS with global regulatory capability. The Rrp2–Hpk2 system controls a regulon that consists primarily of plasmid-carried genes (94% of its regulon) (Boardman et al., 2008; Ouyang et al., 2008). The majority of genes regulated by Rrp2 encode proteins of unknown function and many have been demonstrated in earlier analyses to be environmentally regulated (Revel et al., 2002; Brooks et al., 2003; Ojaimi et al., 2003; Yang et al., 2003a; Burtnick et al., 2007; Caimano et al., 2007; He et al., 2007; Lybecker and Samuels, 2007; Boardman et al., 2008). This study is the first to report on the regulatory capability of Rrp1, a unique response regulator with diguanylate cyclase activity (Ryjenkov et al., 2005). Rrp1 is the only Borrelia protein that carries a GGDEF domain, which is associated with c-di-GMP production. C-di-GMP is an important secondary messenger molecule that influences the expression of proteins involved in a wide range of core biological functions (Cotter and Stibitz, 2007). Recombinant Rrp1 converts GTP to c-di-GMP and this diguanylate cyclase activity is dependent on phosphorylation of Rrp1 (Ryjenkov et al., 2005). It is important to note that the B. burgdorferi genome possesses the full set of proteins that are required for the control of c-di-GMP levels including GGDEF, EAL, HD-GYP, and PilZ domain-containing proteins. To date, only the enzymatic activity of the GGDEF-containing protein, Rrp1, has been assessed. It remains to be determined if B. burgdorferi also harbours a GEMM riboswitch motif, an RNA domain that interacts with c-di-GMP (Weinberg et al., 2007; Sudarsan et al., 2008). Although such a domain has not been detected in B. burgdorferi, this could simply be a reflection of sequence divergence in GEMM domains consistent with the deep phylogenetic divergence of the Borrelia from other eubacteria.
As a first step in investigating the potential biological importance of Rrp1, and by extension of c-di-GMP, we assessed the distribution of Rrp1 and conservation of its critical functional residues. Using PCR and Southern hybridization, rrp1 was shown to be widely distributed among Borrelia as it was detected in all analysed species that are associated with human disease (Lyme disease and relapsing fever spirochetes). Although not analysed as part of this report, recent genome sequence analyses of B. duttonii and B. recurrentis have revealed that these species also possess genes that encode for the Rrp1–Hpk1 TCS (Lescot et al., 2008). The residues thought to be essential for diguanylate cyclase and response regulator activity were also found to be absolutely conserved in all Borrelia species. While this study is focused primarily on Rrp1, the functional link between rrp1 and hpk1 is supported by the conservation of their gene organization and co-transcription. Using RT-PCR and immunoblot analyses, we further demonstrate that all Lyme disease spirochetes produce Rrp1 and Hpk1 during in vitro cultivation under all temperatures assessed. Importantly, qRT-PCR demonstrated that rrp1 expression is detectable in unfed nymphal ticks, but is upregulated sixfold upon tick feeding, indicating that Rrp1 may play a role in regulation of genes required for the transition from the tick to mammalian environment.
To identify the Rrp1 regulon, rrp1 was deleted from the genome of B. burgdorferi by allelic exchange replacement and comparative microarray analyses were conducted. Bb5A13, which is non-infectious, is widely used in B. burgdorferi studies that involve genetic manipulation because it is highly transformable (Purser et al., 2003). Attempts to inactivate rrp1 in the infectious Bb5A4 background were not successful. Nonetheless, the successful deletion of rrp1 from the Bb5A13 background allowed us to assess the overall global regulatory capability of this uncharacterized TCS. The microarray analyses revealed that Rrp1 is a key regulator of the core genome of B. burgdorferi which consists of the linear chromosome, lp54, and cp26 (Terekhova et al., 2006). Of the 140 genes differentially regulated by Rrp1, 85% are encoded on these three genetic elements (102 genes on the chromosome, 10 on lp54 and 7 on cp26). In contrast only 5.6% of the genes regulated by Rrp2 map to the chromosome (Boardman et al., 2008). Hence it is evident that there are fundamental differences in the Rrp1 and Rrp2 regulons.
As a first step in assessing the influence of Rrp1 on Borrelia biology, the growth rates of Bb5A13-wt and Bb5A13-Δrrp1 strains maintained at different temperatures were compared. The rrp1 knockout achieved equivalent cell densities during in vitro cultivation in standard BSK-H media when the spirochetes were grown at 33°C and 37°C. However, there was a significantly longer lag phase observed in Bb5A13-Δrrp1 at these temperatures than in the wild type and overall growth was significantly impaired at 25°C. While the Bb5A13-Δrrp1 spirochetes remained motile over the course of the growth curve analyses at 25°C, the culture densities remained low, suggesting that Rrp1 is required for spirochete replication at the lower temperature. These analyses suggest that Rrp1 either directly or indirectly controls genes involved in the transition to exponential growth and for overall growth at lower temperatures (such as that which might be encountered in the tick environment).
The microarray analyses revealed that the genes that are regulated by Rrp1 span a wide range of functional categories. Rrp1-activated genes encode for proteins that are involved in transport pathways, carbohydrate and nucleotide metabolism, chemotaxis, and flagellar biosynthesis. Additionally, the loss of Rrp1 resulted in pronounced effects on the expression of genes involved in carbohydrate and amino acid transport. These processes may be particularly important in B. burgdorferi in its naturally encountered environments because B. burgdorferi cannot synthesize nucleosides and amino acids de novo (Fraser et al., 1997). The ability of Bb5A13-Δrrp1 to grow, at least in the nutrient-rich BSK-H media under optimal laboratory conditions, possibly indicates that other, not yet identified, transporters may complement the functions of transporters affected in Bb5A13-Δrrp1. It is important to note that the spirochetes, as a whole, represent a deep phylogenetic branch and may have evolved many unique biological strategies for their core metabolic or physiological functions.
Chemotaxis and motility are likely key components of successful completion of the B. burgdorferi transmission cycle. As an example, it has been demonstrated that tick salivary gland extracts are a potent chemoattractant that may facilitate spirochete acquisition (Shih et al., 2002). The microarray analyses revealed that expression of a number of flagellar and chemotaxis genes including fliD, flgL, fliG-1, cheA-1, and mcp-1 are activated by Rrp1. Some of these genes are also regulated, at least in part, by RpoS and have been shown to be induced by exposure to blood (Tokarz et al., 2004; Fisher et al., 2005; Caimano et al., 2007). In other bacteria, c-di-GMP levels have been demonstrated to be a key regulator of chemotaxis and motility (Simm et al., 2004; Wolfe and Visick, 2008). The general paradigm for c-di-GMP-mediated signaling is that increases in c-di-GMP lead to decreases in motility and a transition to biofilm formation (Wolfe and Visick, 2008). In B. burgdorferi, inactivation of Rrp1, which presumably results in decreased intracellular c-di-GMP, resulted in repression of some motility and chemotaxis genes. However, comparative dark-field microscopic analyses of the Bb5A13-wt and Bb5A13-Δrrp1 strains did not reveal an obvious motility defect in the mutant. It is worth noting that c-di-GMP signaling has only been studied in bacteria that have distinct motile and non-motile stages during the infection process (including species of Salmonella, Yersinia, Pseudomonas, Escherichia and Vibrio) (Simm et al., 2004; Tischler and Camilli, 2004; Meissner et al., 2007; Tamayo et al., 2007; Wolfe and Visick, 2008). In contrast, there is no evidence that the life cycle of Borrelia includes a non-motile stage. Thus, B. burgdorferi may have developed a novel use for c-di-GMP signaling in the infection process. The number of key chemotaxis and motility genes that are regulated by Rrp1 serves as an additional example of the central regulatory contributions of Rrp1 to the core biology of B. burgdorferi.
Numerous genes encoding cell envelope-associated proteins and in vivo expressed antigens are also regulated by Rrp1. While the majority of the genes encoding these proteins reside on the chromosome, those located on extrachromosomal elements reside primarily on the cp32 plasmids. Based on the homology of many cp32-encoded proteins to phage proteins and on the transcriptional properties of the cp32s, it has been postulated that these genetic elements are prophage (Eggers and Samuels, 1999; Eggers et al., 2000; 2001; Casjens, 2003; Canchaya et al., 2004; Zhang and Marconi, 2005). Interestingly, all of the cp32-carried genes that are regulated by Rrp1 (ospE, bdr, ospF, and mlp gene families, for example) reside outside of the phage operon and appear to be of bacterial origin (Zhang and Marconi, 2005). Hence the genes of phage origin on the cp32 plasmids appear to be regulated by different mechanisms that are Rrp1-independent. The functional roles of the OspE and OspF proteins and their expression during infection suggest that the genes encoding these proteins represent ‘morons’ which have been defined as phage-carried genes of bacterial origin that enhance the fitness of the lysogen (Brussow et al., 2004). It is important to note that consistent with the upregulation of Rrp1 upon tick feeding reported here, the ospE and ospF proteins are also upregulated upon tick feeding (Hefty et al., 2001; 2002; El-Hage and Stevenson, 2002; Eggers et al., 2004; 2006). The functions of other cp32-carried, Rrp1-regulated genes, such as the mlp and bdr families, have not yet been determined. The Bdr protein family is large, with individual isolates possessing as many as 18 different Bdr paralogues (Carlyon et al., 2000a). This gene family is universal among the Borrelia, and these proteins possess putative serine/threonine phosphorylation motifs (Carlyon et al., 2000b; Roberts et al., 2000; Lescot et al., 2008). At least some members of the Bdr family have been demonstrated to be upregulated in spirochetes cultivated in implanted dialysis membrane chambers suggesting an important role in the pathogenic process (Roberts et al., 2002). Consistent with the potential importance of the OspE, OspF, and Bdr proteins in the host environment, a recent analysis of the B. burgdorferi proteome revealed that members of these protein families are among the dominant antigens recognized during murine infection (Barbour et al., 2008).
mlp expression has previously been demonstrated to be activated by the Rrp2–RpoN–RpoS regulatory pathway (Yang et al., 2003b; Boardman et al., 2008). However, it has been postulated that at least one additional layer of gene regulation contributes to mlp gene expression (Yang et al., 2003b). Consistent with this, we demonstrate that Rrp1 also serves as an activator of certain members of the mlp gene family as their expression was repressed in Bb5A13-Δrrp1. Other genes encoding cell membrane-associated proteins whose expression was affected in Bb5A13-Δrrp1 include BBA03, BBA52, BBA59 and BBA74. Several of the plasmid-encoded genes that we demonstrate in this report to be regulated by Rrp1 have previously been shown to be regulated by environmental conditions (Carroll et al., 2000; Revel et al., 2002; Roberts et al., 2002; Brooks et al., 2003; Ojaimi et al., 2003; Seshu et al., 2004; Tokarz et al., 2004; Hyde et al., 2007). Hence, for these specific genes, stimuli associated with their expression may be transduced by both the Rrp1 and Rrp2 regulatory networks (Yang et al., 2003a; Fisher et al., 2005; Caimano et al., 2007). It is now apparent that the regulation of the expression of cell membrane-associated protein genes in B. burgdorferi is complex with several contributing regulatory mechanisms.
In many bacteria, the regulons of individual TCS are often interconnected. However, in B. burgdorferi the two main response regulators do not appear to influence the expression of each other. The expression of rrp1 was not significantly affected by inactivation of rrp2 (Boardman et al., 2008), and as demonstrated here, inactivation of rrp1 did not influence rrp2 expression. Interestingly, rpoS expression does appear to be controlled at least in part by Rrp1 as inactivation of rrp1 led to reduced expression of rpoS (2.6-fold as measured by qRT-PCR). The converse regulation does not appear to occur (i.e. inactivation of rpoS do not influence rrp1 expression) (Fisher et al., 2005; Caimano et al., 2007; Boardman et al., 2008). Consistent with co-regulation of rpoS by Rrp1 and Rrp2, ospC expression is lower in both rrp1 and rrp2 mutants (Boardman et al., 2008). Other genes regulated by both Rrp1 and Rrp2 include ospE (BBL39/BBP38), ospF (BBO39 and BBR42), cspZ (BBH06), and a gene encoding a hypothetical protein (BBR36) (Table 3). However, the overall set of genes that require signals from both Rrp1 and Rrp2 for their activation appears to be small. When the Rrp1 and Rrp2 regulons are compared, one of the most striking differences observed lies in the genomic location of the genes that they regulate. Rrp1 primarily regulates genes from the core genome, while Rrp2 primarily regulates plasmid genes. Earlier studies suggest that the Rrp2 regulon may be particularly important in transmission and dissemination and that Rrp2-regulated genes may be repressed in ticks (Caimano et al., 2007; Boardman et al., 2008). The Rrp1 regulon may be important in controlling metabolic processes that are required for adapting to changes in nutrient availability, acquisition by and/or survival of spirochetes in ticks. The qRT-PCR analyses presented here indicate a sixfold induction of Rrp1 upon tick feeding. Consistent with this, the expression of hpk1, the gene that encodes for the sensory transduction histidine kinase, was previously shown to have elevated expression in fed ticks compared with flat ticks (Narasimhan et al., 2002). As the Rrp1–Hpk1 TCS is activated, genes that encode for proteins involved in the transport of ions, carbohydrates and amino acids are expressed. This may be followed by an increase in the expression of genes that encode proteins involved in carbohydrate and nucleotide metabolism, thus leading to activation of energy metabolism and nucleotide and protein synthesis. Additionally, Rrp1 would activate expression of motility and chemotaxis genes, as well as cell envelope genes and genes involved in immune evasion (examples include ospC, ospE, ospF and cspZ) to prepare the spirochetes for entry into the host. Hence, Rrp1 is likely a critical regulator during spirochete transmission and adaptation to the mammalian host. Further studies using an infectious Rrp1 knockout are required to examine this possibility.
Table 3.
Genes regulated by Rrp1, Rrp2 and RpoS.a
| Genes induced by Rrp1, Rrp2 and RpoS | |
| BBB19 | Outer surface protein C (ospC) |
| BBR36 | Conserved hypothetical protein |
| BBR42 | Outer surface protein F (ospF) |
| Genes induced by Rrp1 and Rrp2 | |
| BBH06 | Factor H-binding protein (cspZ) |
| BBL39/BBP38 | Factor H-binding protein (ospE)b |
| BBO39 | Outer surface protein F (ospF)b |
| Genes induced by Rrp1 and RpoS | |
| BB0021 | S-adenosylmethionine: tRNA ribosyltransferase-isomerase |
| BB0250 | dedA protein (dedA) |
| BB0567 | Chemotaxis histidine kinase (cheA-1)c |
| BB0578 | Methyl-accepting chemotaxis protein (mcp-1)c |
| BB0588 | 5′-Methylthioadenosine/S-adenosylhomocysteine nucleosidase |
| BB0648 | Serine/threonine kinase, putative |
| BBB14 | Hypothetical protein |
| BBN38 | Factor H-binding protein (ospE)c |
| BBO28 | Mlp lipoproteinc |
| Genes induced by Rrp1 but repressed by RpoS | |
| BB0241 | Glycerol kinase (glpK) |
| BB0242 | Hypothetical protein |
| BB0243 | Glycerol-3-phosphate dehydrogenase, anaerobic (glpA) |
| BB0785 | Stage V sporulation protein G |
| BBA69 | Hypothetical protein |
| BBA74 | Membrane-associated periplasmic protein |
| BBI29 | Hypothetical protein |
| BBI38 | Hypothetical protein |
| BBK19 | Hypothetical protein |
| Genes induced by Rrp1 but repressed by Rrp2 | |
| BB0048 | Hypothetical protein |
| BB0160 | Alanine racemase |
| BB0180 | Flagellar protein, putative |
Based on Caimano et al. (2007) and Boardman et al. (2008).
These genes did not appear in the Rrp1 arrays, but protein levels were downregulated in the rrp1 knockout as shown by western blot analysis.
Genes in these protein families are also regulated by Rrp2 [ex: OspE (BBL39), Mlp (BBM28 and BBP28), chemotaxis (mcp-4, mcp-5, cheR-1)].
In summary, the data presented within demonstrate that Rrp1 plays a central role in the global regulation of core metabolic functions in pathogenic Borrelia species. The Rrp1 regulon is unique and exhibits only minimal overlap with other regulatory networks. The data presented in this report provide important information that enhances our understanding of the global regulatory networks of the Borrelia and the ability of these bacteria to adapt to rapidly changing environmental conditions. While it remains to be determined if Rrp1 directly or indirectly influences the transcription of the Rrp1 regulon, the production c-di-GMP by Rrp1 (Ryjenkov et al., 2005) suggests that this secondary messenger is an important contributing factor. As detailed above, Borrelia possess the necessary machinery to regulate c-di-GMP levels. The presence of single proteins with the domains known to be associated with the formation and degradation of c-di-GMP in B. burgdorferi suggests that this important human pathogen may be an excellent model system to study the regulatory effects of c-di-GMP and c-di-GMP-producing TCS.
Experimental procedures
Bacterial isolates, cultivation, and ticks
Table 4 lists and describes the Borrelia isolates analysed in this report. All Borrelia isolates were cultivated in BSK-H complete media (Sigma-Aldrich) at 25°C, 33°C, or 37°C in sealed bottles under 5% CO2 and harvested by centrifugation (14 000 g; 4°C; 20 min). Infected I. scapularis nymphal stage ticks were kindly provided by Melissa Caimano and Justin Radolf (University Connecticut Health Sciences Center). All methods used to propagate and infect the ticks have been previously described (Caimano et al., 2007).
Table 4.
Borrelia isolates used in this study.
| Species/isolate | Geographic origin | Biological source |
|---|---|---|
| B. burgdorferi | ||
| 297 | Connecticut | Human CSF |
| 3028 | Texas | Human skin |
| 5A4 | New York | B31 derivative, infectious |
| 5A13 | New York | B31 derivative, non-infectious |
| B31 | New York | Ixodes scapularis |
| B348 | New York | Human skin |
| BL224 | New York | Human blood |
| CA3, CA8, CA9 | California | Ixodes pacificus |
| JD1 | Massachusetts | Ixodes scapularis |
| LP4, LP5, LP7 | Connecticut | Human EM |
| Mac13NY, N40 | New York | Human EM |
| T2 | USA | Ixodes scapularis |
| Veery | Connecticut | Veery bird |
| B. afzelii | ||
| ECM1 | Sweden | Human skin |
| IP21 | Russia | Ixodes persulcatus |
| PGau | Germany | Human ACA |
| PKo | Germany | Human skin |
| UM01, U01 | Sweden | Human skin |
| VS461 | Switzerland | Ixodes scapularis |
| B. garinii | ||
| 20047 | France | Ixodes ricinus |
| B-4/87, B-6/91 | Norway | Ixodes ricinus |
| FRG | Germany | Ixodes ricinus |
| G1, G2 | Germany | Human CSF |
| IP89 | Russia | Ixodes persulcatus |
| PBi | Germany | Ixodes ricinus |
| VSBP | Switzerland | Ixodes ricinus |
| B. turicatae | ||
| OZ1, PE-926 | Texas | Ornithodoros turicata |
| B. hermsii | ||
| MAN | California | Human blood |
| DAH | Washington | Human blood |
| B. coriaceae | California | Ornithodoros coriaceus |
CSF, cerebral spinal fluid; EM, erythema migrans; ACA, acrodermatitis chronica atrophicans.
PCR, cloning, and DNA sequencing
Polymerase chain reaction was performed using the Bio-Rad DNAEngine Peltier Thermal Cycler-200 and the following cycle conditions: one cycle at 95°C for 2 min followed by 30 cycles of 94°C for 30 s, 50°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 7 min. Each PCR reaction included 0.75 units of Taq polymerase (Promega), 1× polymerase buffer including 1.5 mM MgCl2, 0.75 pmol μl−1 of each primer, 200 μM each of dATP, dGTP, dCTP and dTTP, and 1–10 ng of genomic DNA for template. Primers are listed in Table S2 and were designed based on the genome sequence of B. burgdorferi B31MI. PCR amplicons were analysed by electrophoresis in 1% agarose gels with Tris-acetate-EDTA (TAE) buffer and visualized by staining with ethidium bromide. For sequence analyses, the PCR amplicons were cloned into the pCR2.1-TOPO vector (Invitrogen). DNA sequencing was performed on a fee-for-service basis (MWG Biotech). The sequence accession numbers for Rrp1 sequences determined or analysed as part of this report or in earlier genome sequence analyses are as follows: B. burgdorferi N40 –FJ643425, B348 – FJ643424, BL224 – FJ643423; B. garinii PBi – YP_072864, B. afzelii PKo – ABH01689, ECM1 –FJ345410, VS461 – FJ345411; B. spielmanii A14S –ZP_03099004; B. valaisiana VS116 – ZP_02368043; B. turicatae 91E135 – YP_945417, PE1926 – FJ263942, OZ1 – FJ263941; B. hermsii DAH – YP_001883847, MAN –FJ643426; B. duttonii Ly – ACH93363; and B. recurrentis A1 – ACH94659.
Southern blot hybridization analysis
For hybridization analyses, DNA was isolated using the MasturePure DNA purification kit (Epicentre), digested with EcoRI (New England Biolabs), and fractionated by agarose gel electrophoresis in 0.7% gels with TAE buffer. The DNA was transferred to Hybond-N membranes (Amersham Biosciences) by vacuum blotting, and the DNA was fixed to the membrane by UV irradiation. An rrp1 gene probe was generated by PCR (primers 7 + 8; Table S2) using purified B. burgdorferi B31 DNA as the template. Probe radiolabeling and all hybridization conditions were as previously described (Rogers and Marconi, 2007).
RNA isolation, RT-PCR, and qRT-PCR
RNA was isolated from ticks as previously described (Caimano et al., 2007). To isolate RNA from Borrelia cultures, cells were lysed with diethylpyrocarbonate-treated 1% SDS and RNA was recovered using the RNeasy Midi kit as instructed by the manufacturer (Qiagen) and with modifications that have been previously described (Zhang et al., 2005). Residual DNA was removed by treatment with DNase I (Invitrogen). RT-PCR analyses were performed as described (Zhang et al., 2005). qRT-PCR was performed using the DNA Engine Opticon 2 System (MJ Research) and SYBR Green PCR Master Mix (Applied Biosystems). Primer pairs (Table S2) were designed to amplify a 100–150 bp fragment of each target and a standard curve was generated using serial dilutions of a known amount of genomic DNA as template. qRT-PCR was performed in triplicate and error bars in Fig. 1D represent standard deviation. The limited quantity of RNA isolated from ticks prevented qRT-PCR analysis in triplicate and as a result there are no error bars. qRT-PCR was performed using the following cycling parameters: one cycle of 10 min at 95°C followed by 40 cycles of 10 s at 94°C, 20 s at 60°C and 20 s at 72°C. Melting curves were generated over a temperature range of 55–95°C. PCR product was quantified and the data analysed using software provided by the thermocycler manufacturer (MJ Research).
Generation of recombinant proteins, protein-specific antiserum, and infection serum
Recombinant (r-) proteins were generated by PCR amplification of the gene of interest with primers harbouring tail sequences that allow for cloning into the pET-46 Ek/LIC vector (Novagen). All methods were as previously described (Rogers and Marconi, 2007). Antiserum to r-proteins was generated in C3H/HeJ mice (Jackson Laboratories) by injection of 50 μg of each r-protein in Freund’s complete adjuvant (Pierce), followed by two injections of 50 μg of r-protein in Freund’s incomplete adjuvant at weeks 2 and 4. At week 6, the mice were euthanized and blood was collected. Serum was recovered from blood by centrifugation (5 min; 14 000 g) after a 3 h incubation at room temperature. To generate infection serum, C3H/HeJ mice were infected subcutaneously with 105B. burgdorferi B31, and infection was confirmed using ear punch biopsies.
Triton X-114 extraction and phase partitioning, SDS-PAGE, and immunoblotting
Triton X-114 extraction and phase partitioning was performed as described by Cunningham et al. (1988) with modifications (Zhang et al., 2005). A 1% Triton X-114 solution removes the outer membrane of the Borrelia, leaving the protoplasmic cylinder intact. The protoplasmic cylinder, consisting of the inner membrane and cytoplasmic proteins, is pelleted by centrifugation. The remaining outer membrane and periplasmic proteins are separated based on their hydrophobic/hydrophilic properties. For SDS-PAGE and immunoblotting, proteins or cell lysates were fractionated using 12.5% Criterion Precast Gels (Bio-Rad) and transferred to PVDF membranes (Millipore), blocked (5% non-fat dry milk, 0.2% Tween, 1× PBS), and screened with various antisera including mouse anti-BBO39 (1:500), mouse anti-BBR42 (1:500), mouse anti-BdrF2 (BBG31; 1:1000), mouse anti-Rrp1 (BB0419; 1:5000), mouse anti-BBN38 (1:5000), mouse anti-BBL39 (1:1000), rabbit anti-Bdr (1:40 000), rabbit anti-DbpA (BBA24; 1:400 000), rat anti-OspA (BBA15; 1:600 000) and mouse anti-B. burgdorferi B31 infection serum (1:1000). Detection was through the use of the appropriate horseradish peroxidase-conjugated secondary antibody (1:40 000; Pierce). All antibodies were diluted in blocking buffer, and in all cases, antibody binding was detected by chemiluminescence using the SuperSignal West Pico chemiluminescence substrate (Pierce) and exposure to X-ray film (Phenix).
Allelic exchange mutagenesis
Allelic exchange gene inactivation was accomplished using procedures described by Samuels et al. (1994). In separate PCR reactions, 1000 bp segments of DNA that reside immediately upstream and downstream of rrp1 (chromosomal co-ordinates 431 616–430 616 and 429 693–428 693 respectively) were PCR amplified and cloned into the pCR2.1-TOPO vector to yield pCR2.1-up and pCR2.1-dn respectively. Where appropriate, the primers were constructed with restriction sites to facilitate subsequent cloning steps. Both pCR2.1-up and pCR2.1-dn were digested with AatII and AgeI. The downstream fragment was excised from pCR2.1-dn and ligated into the linearized pCR2.1-up to yield pCR2.1-updn. The spectinomycin resistance cassette (1203 bp), derived from pKFSS1-AatII (Frank et al., 2003), was inserted between the upstream and downstream fragments at an AatII site to generate pΔBB0419. The plasmid was propagated, purified, linearized with BamHI, and electroporated into B. burgdorferi clones 5A13 and 5A4. Clonal populations were obtained by subsurface plating and plasmid profiles were assessed by PCR using plasmid-specific primers (Table S2).
Growth curve analysis
To compare the growth rates of the Bb5A13-wt and Bb5A13-Δrrp1, equal numbers of actively growing spirochetes were seeded into fresh media and incubated at 25°C, 33°C, and 37°C. Aliquots were removed once a day for 19 days and spirochete numbers determined by counting under dark-field microscopy. Ten fields were counted at each time point and the average number of spirochetes per field was determined.
Microarray hybridization, scanning, and data analysis
RNA isolated from three independent stationary-phase cultures of Bb5A13-wt and Bb5A13-Δrrp1 strains grown at 33°C was used for microarray hybridizations. RNA was labeled indirectly with cy3 and cy5 fluorescent dyes and co-hybridized onto the same microarray slide as previously described (Caimano et al., 2007). To avoid dye bias, two hybridizations were performed for each of the three biological replicates: in the first hybridization RNA from Bb5A13-wt was labeled with cy3 fluorescent dye and RNA from Bb5A13-Δrrp1 was labeled with cy5 fluorescent dye. In a second hybridization, RNA from Bb5A13-wt was labeled with cy5 and RNA from Bb5A13-Δrrp1 was labeled with cy3. The custom glass slide microarrays employed in this study consist of 1741 70-mer oligonucleotides representing all annotated ORFs and pseudogenes of B. burgdorferi B31MI. These arrays and all associated methods for hybridization and array scanning have been previously described (Terekhova et al., 2006; Caimano et al., 2007).
Data analysis was conducted as detailed previously. In brief, mean and standard deviation of fluorescence intensities were calculated for all blank control spots in cy3 and cy5 channels separately; the mean + 2SD was considered to be background and was subtracted from each intensity value in the appropriate channel. Background correction was performed separately for each slide. Spots yielding signals below background in both cy3 and cy5 channels were considered undetected. The background adjusted intensity values were normalized across all arrays. The resultant values were used to calculate log2(Bb5A13-Δrrp1/Bb5A13-wt) for each spot in each slide, and 18 log2(Bb5A13-Δrrp1/Bb5A13-wt) values for each ORF from two separate microarrays and three biological replicates were averaged yielding a final value. Two-tailed, two-sample unequal variance t-test between 18 background-subtracted normalized Bb5A13-Δrrp1 and 18 background-subtracted normalized Bb5A13-wt hybridization intensity values was used to determine which ORFs had statistically significant expression differences between wild-type and rrp1 knockout strains. ORFs with P < 0.05 and log2(Bb5A13-Δrrp1/Bb5A13-wt) > 2 or < −2 were considered induced or repressed in the Bb5A13-Δrrp1.
Factor H affinity ligand binding immunoblot (FH ALBI) assays
Factor H binding to membrane-immobilized proteins obtained from Bb5A13-Δrrp1 and Bb5A13-wt was assessed using the affinity ligand binding immunoblot (ALBI) assay approach (referred to as the FH ALBI assay) as previously described (Rogers and Marconi, 2007). In brief, the membranes were blocked, incubated with purified human FH (5 ng μl−1; Calbiochem), washed with PBS containing 0.2% Tween (PBS-T) and incubated with goat anti-human FH (1:1000; Calbiochem) in blocking buffer (1 h each). Detection was through HRP-conjugated rabbit anti-goat IgG (1:40 000; Pierce) and chemiluminescence.
Indirect IFA
To generate slides for IFA analysis, ~5–7 × 107B. burgdorferi cells (either Bb5A13-Δrrp1 or Bb5A13-wt) grown at 37°C were collected by centrifugation (7000 g; 10 min), washed three times with PBS and re-suspended in 1 ml of PBS. One hundred microlitres of this cell suspension was spread over a 2 cm2 area on Superfrost Plus charged slides (Fisher) and air dried. For screening, all steps were performed in a dark, humidified chamber. The slides were blocked (5% BSA in PBS-T) for 1 h and then screened simultaneously with a 1:100 dilution of mouse anti-BBL39 and rabbit anti-DbpA (1 h). The slides were washed and incubated with 1:200 dilutions of both Alexafluor 488-conjugated goat anti-mouse IgG and Alexafluor 555-conjugated goat anti-rabbit IgG (1 h). The slides were mounted with fluoromount-G (Electron Microscopy Sciences) and visualized by fluorescence and dark-field microscopy.
Serum sensitivity
To assess spirochetes for serum sensitivity, cells (Bb5A13-Δrrp1, Bb5A13-wt and B. garinii FRG) were incubated in 50% normal human serum (NHS) or heat-inactivated serum (HIS). Serum was heat-inactivated by incubation at 56°C for 20–30 min. Cell viability was assessed after 1.5, 3, 6, and 24 h using the BacLight LIVE/DEAD kit (Invitrogen) and viewed by fluorescent microscopy. At each time point, 10 fields were counted, averaged and graphed as a percentage of average number of cells in NHS divided by the average number of cells in HIS. The graph shows the average of three independent experiments. The error bars represent standard deviation.
Supplementary Material
Acknowledgments
The authors would like to thank members of the Marconi and Schwartz labs for their contributions to this study and Melissa Caimano and Justin Radolf for providing B. burgdorferi-infected ticks. This work was supported in part by funding from the National Institutes of Health (AI45801 to I. Schwartz) and by support from the Department of Microbiology and Immunology, Virginia Commonwealth University.
Footnotes
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alitalo A, Meri T, Chen T, Lankinen H, Cheng ZZ, Jokiranta TS, et al. Lysine-dependent multipoint binding of the Borrelia burgdorferi virulence factor outer surface protein E to the C terminus of factor H. J Immunol. 2004;172:6195–6201. doi: 10.4049/jimmunol.172.10.6195. [DOI] [PubMed] [Google Scholar]
- Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- Anderson JF, Magnarelli LA. Biology of ticks. Infect Dis Clin North Am. 2008;22:195–215. v. doi: 10.1016/j.idc.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Antonara S, Chafel RM, Lafrance M, Coburn J. Borrelia burgdorferi adhesins identified using in vivo phage display. Mol Microbiol. 2007;66:262–276. doi: 10.1111/j.1365-2958.2007.05924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausmees N, Mayer R, Weinhouse H, Volman G, Amikam D, Benziman M, Lindberg M. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol Lett. 2001;204:163–167. doi: 10.1111/j.1574-6968.2001.tb10880.x. [DOI] [PubMed] [Google Scholar]
- Barbour AG, Jasinskas A, Kayala MA, Davies DH, Steere AC, Baldi P, Felgner PL. A genome-wide proteome array reveals a limited set of immunogens in natural infections of humans and white-footed mice with Borrelia burgdorferi. Infect Immun. 2008;76:3374–3389. doi: 10.1128/IAI.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benach JL, Bosler EM, Hanrahan JP, Coleman JL, Bast TF, Habicht GS, et al. Spirochetes isolated from the blood of two patients with Lyme disease. N Engl J Med. 1983;308:740–742. doi: 10.1056/NEJM198303313081302. [DOI] [PubMed] [Google Scholar]
- Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, et al. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007;26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyhan S, Tischler AD, Camilli A, Yildiz FH. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J Bacteriol. 2006;188:3600–3613. doi: 10.1128/JB.188.10.3600-3613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman BK, He M, Ouyang Z, Xu H, Pang X, Yang XF. Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi. Infect Immun. 2008;76:3844–3853. doi: 10.1128/IAI.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov AG, Kirillina O, Perry RD. Regulation of biofilm formation in Yersinia pestis. Adv Exp Med Biol. 2007;603:201–210. doi: 10.1007/978-0-387-72124-8_17. [DOI] [PubMed] [Google Scholar]
- Brooks CS, Hefty PS, Joliff SE, Akins DR. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect Immun. 2003;71:3371–3383. doi: 10.1128/IAI.71.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CS, Vuppala SR, Jett AM, Alitalo A, Meri S, Akins DR. Complement regulator-acquiring surface protein 1 imparts resistance to human serum in Borrelia burgdorferi. J Immunol. 2005;175:3299–3308. doi: 10.4049/jimmunol.175.5.3299. [DOI] [PubMed] [Google Scholar]
- Brown EL, Guo BP, O’Neal P, Hook M. Adherence of Borrelia burgdorferi. Identification of critical lysine residues in DbpA required for decorin binding. J Biol Chem. 1999;274:26272–26278. doi: 10.1074/jbc.274.37.26272. [DOI] [PubMed] [Google Scholar]
- Brussow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. Lyme disease – a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- Burtnick MN, Downey JS, Brett PJ, Boylan JA, Frye JG, Hoover TR, Gherardini FC. Insights into the complex regulation of rpoS in Borrelia burgdorferi. Mol Microbiol. 2007;65:277–293. doi: 10.1111/j.1365-2958.2007.05813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, et al. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol. 2007;65:1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canchaya C, Fournous G, Brussow H. The impact of prophages on bacterial chromosomes. Mol Microbiol. 2004;53:9–18. doi: 10.1111/j.1365-2958.2004.04113.x. [DOI] [PubMed] [Google Scholar]
- Carlyon JA, Roberts DM, Marconi RT. Evolutionary and molecular analyses of the Borrelia bdr super gene family: delineation of distinct sub-families and demonstration of the genus wide conservation of putative functional domains, structural properties and repeat motifs. Microb Pathog. 2000a;28:89–105. doi: 10.1006/mpat.1999.0326. [DOI] [PubMed] [Google Scholar]
- Carlyon JA, Roberts DM, Theisen M, Sadler C, Marconi RT. Molecular and immunological analyses of the Borrelia turicatae Bdr protein family. Infect Immun. 2000b;68:2369–2373. doi: 10.1128/iai.68.4.2369-2373.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JA, Cordova RM, Garon CF. Identification of 11 pH-regulated genes in Borrelia burgdorferi localizing to linear plasmids. Infection Immunity. 2000;68:6677–6684. doi: 10.1128/iai.68.12.6677-6684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S. Prophage and bacterial genomics: what have we learned so far? Mol Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, et al. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- Cotter PA, Stibitz S. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Cunningham TM, Walker EM, Miller JN, Lovett MA. Selective release of the Treponema pallidum outer membrane and associated polypeptides with Triton X-114. J Bacteriol. 1988;170:387–395. doi: 10.1128/jb.170.12.5789-5796.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers C, Samuels DS. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J Bacteriol. 1999;181:7308–7313. doi: 10.1128/jb.181.23.7308-7313.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Casjens S, Hayes SF, Garon CF, Damman CJ, Oliver DB, Samuels DS. Bacteriophages of spirochetes. J Mol Microbiol Biotechnol. 2000;4:365–373. [PubMed] [Google Scholar]
- Eggers CH, Kimmel BJ, Bono JL, Elias AF, Rosa P, Samuels DS. Transduction by phiBB-1, a bacteriophage of Borrelia burgdorferi. J Bacteriol. 2001;183:4771–4778. doi: 10.1128/JB.183.16.4771-4778.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, Radolf JD. Analysis of promoter elements involved in the transcriptional initiation of RpoS-dependent Borrelia burgdorferi genes. J Bacteriol. 2004;186:7390–7402. doi: 10.1128/JB.186.21.7390-7402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, Radolf JD. Sigma factor selectivity in Borrelia burgdorferi: RpoS recognition of the ospE/ospF/elp promoters is dependent on the sequence of the −10 region. Mol Microbiol. 2006;59:1859–1875. doi: 10.1111/j.1365-2958.2006.05066.x. [DOI] [PubMed] [Google Scholar]
- El-Hage N, Stevenson B. Simultaneous coexpression of Borrelia burgdorferi Erp proteins occurs through a specific, erp locus-directed regulatory mechanism. J Bacteriol. 2002;184:4536–4543. doi: 10.1128/JB.184.16.4536-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embers ME, Alvarez X, Ooms T, Philipp MT. The failure of immune response evasion by linear plasmid 28-1-deficient Borrelia burgdorferi is attributable to persistent expression of an outer surface protein. Infect Immun. 2008;76:3984–3991. doi: 10.1128/IAI.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, Rosa PA, Gherardini FC. From the cover: Borrelia burgdorferi {sigma}54 is required for mammalian infection and vector transmission but not for tick colonization. Proc Natl Acad Sci USA. 2005;102:5162–5167. doi: 10.1073/pnas.0408536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank KL, Bundle SF, Kresge ME, Eggers CH, Samuels DS. aadA confers streptomycin resistance in Borrelia burgdorferi. J Bacteriol. 2003;185:6723–6727. doi: 10.1128/JB.185.22.6723-6727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- Garcia B, Latasa C, Solano C, Garcia-del Portillo F, Gamazo C, Lasa I. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol Microbiol. 2004;54:264–277. doi: 10.1111/j.1365-2958.2004.04269.x. [DOI] [PubMed] [Google Scholar]
- Gjermansen M, Ragas P, Tolker-Nielsen T. Proteins with GGDEF and EAL domains regulate Pseudomonas putida biofilm formation and dispersal. FEMS Microbiol Lett. 2006;265:215–224. doi: 10.1111/j.1574-6968.2006.00493.x. [DOI] [PubMed] [Google Scholar]
- Guvener ZT, Harwood CS. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol. 2007;66:1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Boardman BK, Yan D, Yang XF. Regulation of expression of the fibronectin-binding protein BBK32 in Borrelia burgdorferi. J Bacteriol. 2007;189:8377–8380. doi: 10.1128/JB.01199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefty PS, Jolliff SE, Caimano MJ, Wikel SK, Radolf JD, Akins DR. Regulation of the OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect Immun. 2001;69:3618–3627. doi: 10.1128/IAI.69.6.3618-3627.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefty PS, Brooks CS, Jett AM, White GL, Wikel SK, Kennedy RC, Akins DR. OspE-related, OspF-related and Elp lipoproteins are immunogenic in baboons experimentally infected with Borrelia burgdorferi and in human Lyme disease patients. J Clin Microbiol. 2002;40:4256–4265. doi: 10.1128/JCM.40.11.4256-4265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwage J, Meri T, Heikkila T, Alitalo A, Panelius J, Lahdenne P, et al. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi. J Biol Chem. 2001;276:8427–8435. doi: 10.1074/jbc.M007994200. [DOI] [PubMed] [Google Scholar]
- Hovis KM, Jones JP, Sadlon T, Raval G, Gordon DL, Marconi RT. Molecular analyses of the interaction of Borrelia hermsii FhbA with the complement regulatory proteins factor H and factor H-like protein 1. Infect Immun. 2006a;74:2007–2014. doi: 10.1128/IAI.74.4.2007-2014.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovis KM, Tran E, Sundy CM, Buckles E, McDowell JV, Marconi RT. Selective binding of Borrelia burgdorferi OspE paralogs to factor H and serum proteins from diverse animals: possible expansion of the role of OspE in Lyme disease pathogenesis. Infect Immun. 2006b;74:1967–1972. doi: 10.1128/IAI.74.3.1967-1972.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN–RpoS regulatory pathway. Proc Natl Acad Sci USA. 2001;98:12724–12729. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JA, Seshu J, Skare JT. Transcriptional profiling of Borrelia burgdorferi containing a unique bosR allele identifies a putative oxidative stress regulon. Microbiology. 2006;152:2599–2609. doi: 10.1099/mic.0.28996-0. [DOI] [PubMed] [Google Scholar]
- Hyde JA, Trzeciakowski JP, Skare JT. Borrelia burgdorferi alters its gene expression and antigenic profile in response to CO2 levels. J Bacteriol. 2007;189:437–445. doi: 10.1128/JB.01109-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R, Kalu O, Purser J, Norris S, Stevenson B, Schwartz I. Linear and circular plasmid content in Borrelia burgdorferi clinical isolates. Infect Immun. 2003;71:3699–3706. doi: 10.1128/IAI.71.7.3699-3706.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- Karaolis DK, Rashid MH, Chythanya R, Luo W, Hyodo M, Hayakawa Y. c-di-GMP (3′-5′-cyclic diguanylic acid) inhibits Staphylococcus aureus cell–cell interactions and biofilm formation. Antimicrob Agents Chemother. 2005;49:1029–1038. doi: 10.1128/AAC.49.3.1029-1038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, McCarter LL. ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J Bacteriol. 2007;189:4094–4107. doi: 10.1128/JB.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraiczy P, Würzner R. Complement escape of human pathogenic bacteria by acquisition of complement regulators. Mol Immunol. 2005;43:31–44. doi: 10.1016/j.molimm.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Kraiczy P, Hellwage J, Skerka C, Becker H, Kirschfink M, Simon MS, et al. Complement resistance of Borrelia burgdorferi correlates with the expression of BbCRASP-1, a novel linear plasmid-encoded surface protein that interacts with human factor H and FHL-1 and is unrelated to Erp proteins. J Biolog Chem. 2004;279:2421–2429. doi: 10.1074/jbc.M308343200. [DOI] [PubMed] [Google Scholar]
- Lam TT, Nguyen TP, Montgomery RR, Kantor FS, Fikrig E, Flavell RA. Outer surface proteins E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect Immun. 1994;62:290–298. doi: 10.1128/iai.62.1.290-298.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrenz MB, Kawabata H, Purser JE, Norris SJ. Decreased electroporation efficiency in Borrelia burgdorferi containing lineaer plasmids lp25 and lp56: Impact on transformation of infectious B. burgdorferi. Infect Immun. 2002;70:4798–4804. doi: 10.1128/IAI.70.9.4798-4804.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Audic S, Robert C, Nguyen TT, Blanc G, Cutler SJ, et al. The genome of Borrelia recurrentis, the agent of deadly louse-borne relapsing fever, is a degraded subset of tick-borne Borrelia duttonii. PLoS Genet. 2008;4:e1000185. doi: 10.1371/journal.pgen.1000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker MC, Samuels DS. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol Microbiol. 2007;64:1075–1089. doi: 10.1111/j.1365-2958.2007.05716.x. [DOI] [PubMed] [Google Scholar]
- McDowell JV, Sung SY, Labandeira-Rey M, Skare JT, Marconi RT. Analysis of mechanisms associated with loss of infectivity of clonal populations of Borrelia burgdorferi B31MI. Infect Immun. 2001;69:3670–3677. doi: 10.1128/IAI.69.6.3670-3677.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JV, Wolfgang J, Tran E, Metts MS, Hamilton D, Marconi RT. Comprehensive analysis of the factor H binding capabilities of Borrelia species associated with Lyme disease: delineation of two distinct classes of factor H binding proteins. Infect Immun. 2003;71:3597–3602. doi: 10.1128/IAI.71.6.3597-3602.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JV, Wolfgang J, Senty L, Sundy CM, Noto MJ, Marconi RT. Demonstration of the involvement of outer surface protein E coiled coil structural domains and higher order structural elements in the binding of infection-induced antibody and the complement-regulatory protein, factor H. J Immunol. 2004;173:7471–7480. doi: 10.4049/jimmunol.173.12.7471. [DOI] [PubMed] [Google Scholar]
- Meissner A, Wild V, Simm R, Rohde M, Erck C, Bredenbruch F, et al. Pseudomonas aeruginosa cupA-encoded fimbriae expression is regulated by a GGDEF and EAL domain-dependent modulation of the intracellular level of cyclic diguanylate. Environ Microbiol. 2007;9:2475–2485. doi: 10.1111/j.1462-2920.2007.01366.x. [DOI] [PubMed] [Google Scholar]
- Mendez-Ortiz MM, Hyodo M, Hayakawa Y, Membrillo-Hernandez J. Genome wide transcriptional profile of Escherichia coli in response to high levels of the second messenger c-di-GMP. J Biol Chem. 2006;281:8090–8099. doi: 10.1074/jbc.M510701200. [DOI] [PubMed] [Google Scholar]
- Narasimhan S, Santiago F, Koski RA, Brei B, Anderson JF, Fish D, Fikrig E. Examination of the Borrelia burgdorferi transcriptome in Ixodes scapularis during feeding. J Bacteriol. 2002;184:3122–3125. doi: 10.1128/JB.184.11.3122-3125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour AG, et al. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect Immun. 2003;71:1689–1705. doi: 10.1128/IAI.71.4.1689-1705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojaimi C, Mulay V, Liveris D, Iyer R, Schwartz I. Comparative transcriptional profiling of Borrelia burgdorferi clinical isolates differing in capacities for hematogenous dissemination. Infect Immun. 2005;73:6791–6802. doi: 10.1128/IAI.73.10.6791-6802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Blevins JS, Norgard MV. Transcriptional interplay among the regulators Rrp2, RpoN and RpoS in Borrelia burgdorferi. Microbiology. 2008;154:2641–2658. doi: 10.1099/mic.0.2008/019992-0. [DOI] [PubMed] [Google Scholar]
- Palmer N, Fraser C, Casjens S. Distribution of twelve linear extrachromosomal DNAs in natural isolates of Lyme disease spirochetes. J Bacteriol. 2000;182:2476–2480. doi: 10.1128/jb.182.9.2476-2480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikas DS, Brown EL, Gurusiddappa S, Lee LY, Xu Y, Hook M. Decorin-binding sites in the adhesin DbpA from Borrelia burgdorferi: a synthetic peptide approach. J Biol Chem. 2003;278:30920–30926. doi: 10.1074/jbc.M303979200. [DOI] [PubMed] [Google Scholar]
- Porcella SF, Fitzpatrick CA, Bono JL. Expression and immunological analysis of the plasmid-borne mlp genes of Borrelia burgdorferi strain B31. Infect Immun. 2000;68:4992–5001. doi: 10.1128/iai.68.9.4992-5001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purser JE, Norris SJ. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci USA. 2000;97:13865–13870. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purser JE, Lawrenz MB, Caimano MJ, Howell JK, Radolf JD, Norris SJ. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol Microbiol. 2003;48:753–764. doi: 10.1046/j.1365-2958.2003.03452.x. [DOI] [PubMed] [Google Scholar]
- Revel AT, Talaat AM, Norgard MV. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc Natl Acad Sci USA. 2002;99:1562–1567. doi: 10.1073/pnas.032667699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DM, Carlyon JA, Theisen M, Marconi RT. The bdr gene families of the Lyme disease and relapsing fever spirochetes: potential influence on biology, pathogenesis, and evolution. Emerg Infect Dis. 2000;6:110–122. doi: 10.3201/eid0602.000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DM, Caimano M, McDowell J, Theisen M, Holm A, Orff E, et al. Environmental regulation and differential production of members of the Bdr protein family of Borrelia burgdorferi. Infect Immun. 2002;70:7033–7041. doi: 10.1128/IAI.70.12.7033-7041.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EA, Marconi RT. Delineation of species-specific binding properties of the CspZ protein (BBH06) of Lyme disease spirochetes: evidence for new contributions to the pathogenesis of Borrelia spp. Infect Immun. 2007;75:5272–5281. doi: 10.1128/IAI.00850-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Amikam D. Cyclic di-GMP as a second messenger. Curr Opin Microbiol. 2006;9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Romling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryjenkov DA, Simm R, Romling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- Samuels DS, Mach K, Garon CF. Genetic transformation of the Lyme disease agent Borrelia burgdorferi with coumarin-resistant gyrB. J Bacteriol. 1994;176:6045–6049. doi: 10.1128/jb.176.19.6045-6049.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshu J, Boylan JA, Gherardini FC, Skare JT. Dissolved oxygen levels alter gene expression and antigen profiles in Borrelia burgdorferi. Infect Immun. 2004;72:1580–1586. doi: 10.1128/IAI.72.3.1580-1586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih CM, Chao LL, Yu CP. Chemotactic migration of the Lyme disease spirochete (Borrelia burgdorferi) to salivary gland extracts of vector ticks. Am J Trop Med Hyg. 2002;66:616–621. doi: 10.4269/ajtmh.2002.66.616. [DOI] [PubMed] [Google Scholar]
- Shu CJ, Zhulin IB. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem Sci. 2002;27:3–5. doi: 10.1016/s0968-0004(01)02036-9. [DOI] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Simm R, Fetherston JD, Kader A, Romling U, Perry RD. Phenotypic convergence mediated by GGDEF-domain-containing proteins. J Bacteriol. 2005;187:6816–6823. doi: 10.1128/JB.187.19.6816-6823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere AC, Grodzicki RL, Kornblatt AN, Craft JE, Barbour AG, Burgdorfer W, et al. The spirochetal etiology of Lyme disease. N Engl J Med. 1983;308:733–740. doi: 10.1056/NEJM198303313081301. [DOI] [PubMed] [Google Scholar]
- Stewart PE, Byram R, Grimm D, Tilly K, Rosa PA. The plasmids of Borrelia burgdorferi: essential genetic elements of a pathogen. Plasmid. 2005;53:1–13. doi: 10.1016/j.plasmid.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terekhova D, Iyer R, Wormser GP, Schwartz I. Comparative genome hybridization reveals substantial variation among clinical isolates of Borrelia burgdorferi sensu stricto with different pathogenic properties. J Bacteriol. 2006;188:6124–6134. doi: 10.1128/JB.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R, Anderton J, Katona L, Benach J. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect Immun. 2004;72:5419–5432. doi: 10.1128/IAI.72.9.5419-5432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- Wolfe AJ, Visick KL. Get the message out: cyclic-di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol. 2008;190:463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Kodner C, Coleman L, Johnson RC. Correlation of plasmids with infectivity of Borrelia burgdorferi sensu stricto type strain B31. Infect Immun. 1996;64:3870–3876. doi: 10.1128/iai.64.9.3870-3876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Bruno JF, Luft BJ. Detection of genetic diversity in linear plasmids 28-3 and 36 in Borrelia burgdorferi sensu stricto isolates by subtractive hybridization. Microb Pathog. 2003;35:269–278. doi: 10.1016/s0882-4010(03)00152-9. [DOI] [PubMed] [Google Scholar]
- Yang X, Popova TG, Hagman KE, Wikel SK, Schoeler GB, Caimano MJ, et al. Identification, characterization, and expression of three new members of the Borrelia burgdorferi Mlp (2.9) lipoprotein gene family. Infect Immun. 1999;67:6008–6018. doi: 10.1128/iai.67.11.6008-6018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Alani SM, Norgard MV. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc Natl Acad Sci USA. 2003a;100:11001–11006. doi: 10.1073/pnas.1834315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Hubner A, Popova TG, Hagman KE, Norgard MV. Regulation of expression of the paralogous Mlp family in Borrelia burgdorferi. Infect Immun. 2003b;71:5012–5020. doi: 10.1128/IAI.71.9.5012-5020.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Marconi RT. Demonstration of cotranscription and 1-methyl-3-nitroso-nitroguanidine induction of a 30-gene operon of Borrelia burgdorferi: evidence that the 32-kilobase circular plasmids are prophages. J Bacteriol. 2005;187:7985–7995. doi: 10.1128/JB.187.23.7985-7995.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Raji A, Theisen M, Hansen PR, Marconi RT. bdrF2 of Lyme disease spirochetes is coexpressed with a series of cytoplasmic proteins and is produced specifically during early infection. J Bacteriol. 2005;187:175–184. doi: 10.1128/JB.187.1.175-184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckert WR, Meyer J, Barbour AG. Comparative analysis and immunological characterization of the Borrelia Bdr protein family. Infect Immun. 1999;67:3257–3266. doi: 10.1128/iai.67.7.3257-3266.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.