Abstract
3-Methylindole (3MI) is a highly selective pneumotoxicant that is present in abundant amounts (as high as 1.4 µg/cigarette) present in cigarette smoke. Several human cytochrome P450 enzymes that are expressed in lung, such as CYP1A1, CYP2F1, CYP2A13, and CYP4B1 catalyze the dehydrogenation of 3MI to the reactive intermediate 3-methyleneindolenine, which alkylates DNA and induces cell death through apoptosis. In addition, 3MI potently damages DNA at low concentrations (observable at 0.1 µM). However, it seemed possible that 3MI could induce the levels of P450 enzymes, so transcription and translation of 1A1 and 2F1 genes were measured in primary normal human bronchial epithelial cells. In this study, 3MI-induced DNA damage at the 10µM concentration was ameliorated when P450 turnover was inactivated with the cytochrome P450 suicide substrate inhibitor 1-aminobenzotriazole. Thus, the observed DNA damage was cytochrome P450-dependent. Quantitative real-time PCR analysis revealed both concentration- and time-dependent increases in CYP1A1 and CYP2F1 transcription by the same 3MI concentrations that damaged DNA. Aryl hydrocarbon receptor (AhR) activation lead to CYP1A1 induction. Treatment with 3MI in combination with the AhR antagonist α-naphthoflavone prevented 3MI-mediated CYP1A1 induction, indicating that the induction was AhR-dependent. Conversely, CYP2F1 induction did not appear to require activation of AhR. These intriguing findings show that not only is induction of 1A1 and 2F1 caused by 3MI metabolites, rather than 3MI itself, but transcriptional activation of these pulmonary genes occurs through disparate mechanisms. Thus, the induction process, and subsequent increased bioactivation of 3MI to toxic intermediates, is a facile process that might enhance the acute toxicity and/or mutagenicity of this chemical.
Introduction
3-Methylindole (3MI) is formed by the decarboxylation of a fermentation product of tryptophan in the large intestine of humans (1). Upon formation, 3MI becomes systemically circulated, yet displays preferential pneumotoxicity, with high selectivity for Clara and epithelial cell damage (2). In addition to intestinal formation, cigarette smoke inhalation provides a significant source of 3MI exposure in humans, bringing the pneumotoxicant in direct contact with its target organ (3).
Cytochrome P450 enzymes catalyze the metabolism of both endogenous compounds and xenobiotics, including pharmaceutical agents and carcinogens (4). Although primarily expressed in the liver, cytochrome P450 enzymes that often are selectively expressed in the respiratory system and other organs, play a critical role in the tissue-selective formation of bioactivated metabolites and organ-specific toxicity of xenobiotic compounds (5). Respiratory bioactivation has been shown to be important in several disease processes, including lung cancer, where inhaled compounds such as polycyclic aromatic hydrocarbons (PAH), which are not themselves carcinogenic, can become bioactivated by pulmonary cytochrome P450s to reactive metabolites that alkylate DNA (6).
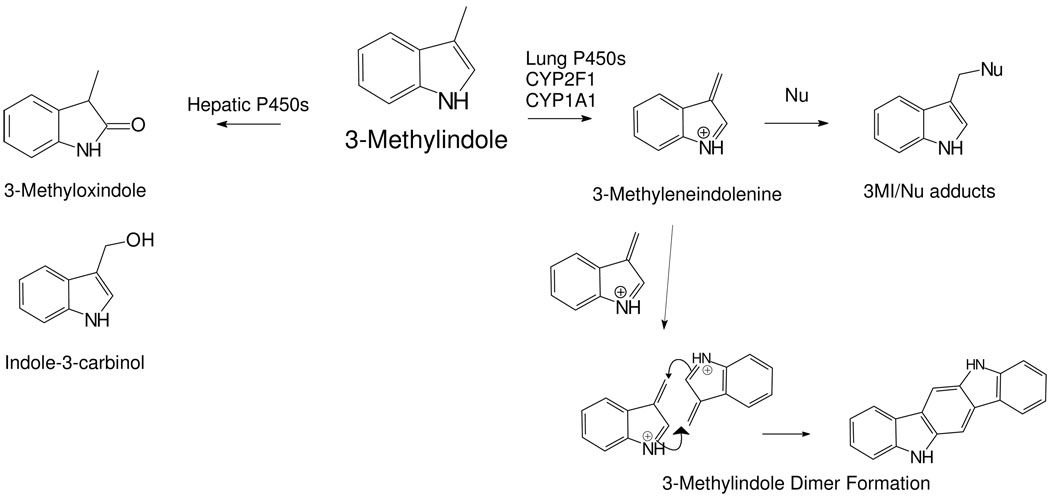
Several cytochrome P450 enzymes, including CYP1A1, CYP2F1, CYP2A13, and CYP4B1 are expressed in human lung (reviewed in (5) and have been shown to catalyze the dehydrogenation of 3MI (Scheme I) to the bioactivated intermediate 3-methyleneindolenine (3MEIN) (7). 3MEIN forms adducts with glutathione, proteins, and DNA (8, 9). Dehydrogenation of 3MI to 3MEIN by respiratory enzymes is, unfortunately, selectively and most efficiently catalyzed by lung P450s. The predominant hepatic enzymes like CYP3A4, CYP2E1, CYP2C9, and CYP2B4 oxygenate 3MI to 3-methyloxindole and indole-3-carbinol without appreciable formation of 3MEIN. The putative bifurcated pathways of dehydrogenation by respiratory enzymes, or oxygenation by hepatic enzymes, is illustrated in Scheme 1. Thus, selective production of 3MEIN by P450s selectively expressed in lung cells may lead to pneumotoxicity and lung cancer.
Scheme I.
Disparate Pathways of Hepatic and Respiratory Metabolism of 3MI. Formation of the 3-methyleneindolenine (3MEIN) reactive intermediate is primarily catalyzed by lung enzymes such as CYP1A1 and CYP2F1. Conversely, the major excreted metabolites such as 3-methoxindole and indole-3-carbinol are produced by several P450 enzymes that are predominately expressed in the liver. Metabolism of 3MI through 3MEIN could produce the dimer shown here, ICZ, that could be an inducer of CYP1A1 and CYP2F1. Nu = protein, DNA, glutathione
Concentrations of 3MI ranging from 0.1 to 10 µM did not initiate apoptosis, but did cause a concentration-dependent increase in DNA damage in primary normal human bronchial epithelial (NHBE) cells (10). This DNA damage was attenuated following a 30-minute pre-treatment with the suicide substrate inhibitor of cytochrome P450 activity 1-aminobenzotriazole (ABT), indicating that the observed DNA damage was cytochrome P450-dependent.
Xenobiotic-mediated induction of cytochrome P450 enzymes increases both the metabolic activation of carcinogens and the rate of formation of DNA adducts (11, 12). One of the most well established models of xenobiotic-mediated cytochrome P450 induction is 2,3,7,8-tetracholordibenzo-p-dioxin (TCDD)-mediated CYP1A1 induction. To initiate CYP1A1 induction, TCDD binds to the aryl hydrocarbon receptor (AhR), forming a complex which translocates into the nucleus and activates the xenobiotic response element (XRE), mediating the induction of CYP1A1 (13). This XRE-mediated transcriptional activation is inhibited with α-naphthoflavone (α-NF), which binds to the AhR to form an inactive complex (14) so (α-NF) can be used to evaluate AhR activation.
The use of tumorigenic or immortalized, non-neoplastic respiratory cell lines to mimic animal or human responses to xenobiotics can be highly controversial, and primary airway epithelial cells usually display the fundamental physiological and biochemical features of the intact respiratory system (15). Fortunately, commercial sources of primary human lung cells have recently become available. Primary NHBE and small airway epithelial cells (16) should provide more realistic in vitro models than neoplastic cells, so we (10) and others (17) have used them to study alterations in biochemical and cellular parameters. In fact, robust induction of CYP1A1 and CYP1B1 transcripts and proteins by TCDD have been documented in the small airway epithelial cells (16). Transcripts of AhR were also modestly increased (2.1-fold) by TCDD treatment.
3MI activates transcription of CYP2A6 and CYP2E1 in pig hepatocytes (18, 19). However, induction of human cytochrome P450 respiratory genes by 3MI in the target cells of lung cancer, airway epithelial cells, has not been reported. Thus, the aim of the present study was to examine the ability of 3MI to induce two important respiratory enzymes, CYP1A1 and CYP2F1, in primary human lung cells, and determine if the induction process required AhR activation.
Experimental Procedures
Cell Culture
Primary Normal Human Bronchial Epithelial (NHBE) cells, isolated in the presence of retinoic acid, were obtained from Lonza (Walkersville, MD) and cultured in Bronchial Epithelial Growth Medium with retinoic acid in a humidified incubator with 95% air and 5% CO2. For subculturing, cells were trypsin-dissociated and plated into fibronectin/collagen-coated culture plates. Cells were treated at passage three after achieving approximately 80% confluence. Drugs and toxins were dissolved in 0.5% DMSO and diluted in growth media prior to treating the cells. Control cells were treated with 0.5% DMSO (vehicle) alone.
Quantitative Real-Time PCR Analysis of Basal and 3MI-Induced CYP1A1 and CYP2F1 Transcript Levels
Total cellular RNA was isolated from control and treated NHBE cells using an RNeasy Plus Micro Kit (Qiagen, Valencia, CA) and QIAshredder microspin homogenizer according to the manufacturer’s recommended protocol. RNA yield was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Pittsburgh, PA) and RNA quality was assessed using the Experion RNA StdSens Analysis Kit (BioRad, Hercules, CA). First strand cDNA synthesis was performed with 1 µg total RNA using the SuperScript III First-Strand Synthesis System for rt-PCR (Invitrogen, Carlsbad, CA) and was random hexamer primed. Intron spanning primer pairs for both CYP1A1 and the internal reference housekeeping gene proteasome subunit β-type 6 (Psmb6) were designed using ProbeFinder Version 2.40 for Human (Roche, Nutley, NJ). Primers for CYP1A1 were: 5’- CCC AGC TCA GCT CAG TAC CT - 3’ (sense) and 5’-GGA GAT TGG GAA AAG CAT GA - 3’ (antisense). Primers for Psmb6 were: 5’- TAC CAG CTC GGT TTC CAC A – 3’ (sense) and 5’ - CCC GGT ATC GGT AAC ACA TC – 3’ (antisense). Primer pairs for CYP2F1 were previously published (Carr et al., 2003). Copy number standards spanning four orders of magnitude were generated for each gene using the StrataClone PCR Cloning Kit (Stratagene, La Jolla, CA) according to the manufacturer’s recommended protocol and were sequence verified. PCR reactions consisted of: 1 µL cDNA reaction mixture, 4 Units Platinum Taq DNA Polymerase (Invitrogen), 2 µL 10X PCR buffer, 0.25 µL 10mM dNTP Mix, 1 µL 50mM MgCl2, 0.5 µL 10X SYBR Green 1 (a cyanine dye used to detect double-stranded DNA), 0.2 µM of each primer, 1.3 M betaine, and water up to a final volume of 20 µL. Quantitative real-time PCR was performed using a Chromo4 PTC-200 cycler (BioRad) under the following reaction conditions: initial denaturing step at 95°C for 90 seconds, followed by 35 cycles of melting at 95°C for 5 seconds, annealing at 57°C for 5 seconds, and extending at 72°C for 20 seconds. After the final amplification cycle, a melting curve was run from 72°C to 90°C. All gene expression was normalized to the housekeeping gene Psmb6.
Western Blot Analysis of Basal and 3MI-Induced CYP1A1 and CYP2F1 Protein Levels
Cell culture media was aspirated from control and treated NHBE cells in 6-well plates. Cells were washed twice with chilled 1 X phosphate buffered saline (PBS) consisting of 137 mM NaCl, 10 mM phosphate, and 2.7 mM KCl at a pH of 7.4. PBS was aspirated and 300 µL chilled Radioimmunoprecipitation Assay (RIPA) buffer (Pierce, Rockford, IL) consisting of 50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, and 1% Triton-X 100, plus 3 µL protease inhibitor cocktail (Sigma), 3 µL 100 mM sodium orthovanadate, and 3 µL 57 mM phenylmethylsulphonyl fluoride (PMSF) was added to each well. Cells were placed on ice for 5 minutes. Lysates were gathered to one side of each well using a rubber policeman and transferred to microcentrifuge tubes. Lysates were centrifuged at 14,000 × g for 15 minutes at 4°C and supernatants were transferred to new tubes. Protein concentrations were determined via BCA Protein Assay Kit (Pierce) according to the manufacturer’s recommended protocol. Absorbance at 562 nm was measured using a SpectraMax 250 spectrophotometer (Molecular Devices, Sunnyvale, CA) and sample concentrations were extrapolated using a bovine serum albumin standard generated curve. 15 µg protein and 15 µL laemmli buffer containing 5% β-mercaptoethanol were heated for 5 minutes at 90°C. Samples were then fractionated by NuPAGE 4–12% Bis-Tris polyacrylamide gel (Invitrogen) electrophoresis at 200 V for 50 minutes. Following fractionation, proteins were transferred to a polyvinylidene fluoride (PVDF) membrane. The membrane was blocked with 5% nonfat milk in 1 X PBST (1 X phosphate buffered saline containing 0.1% Tween-20) for 1 hour at room temperature. The membrane was subsequently incubated overnight at 4°C with primary anti-CYP1A1 antibody (Santa Cruz Biotechnologies, Santa Cruz, CA) at a 1:200 dilution in the blocking buffer. After the overnight incubation, the membrane was rinsed twice with 1 X PBST and twice with 1 X PBS containing 1% NP-40 and was then incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (Sigma), at a 1:3000 dilution in 1 X PBST for 1 h at room temperature. After the secondary antibody was removed, the membrane was washed three times with 1 X PBST. For detection of CYP2F1 and the loading control glyceraldehyde 3-phosphate dehydrogenase (GAPDH), membranes were stripped with 0.2 M NaOH for 5 minutes at room temperature, rinsed once with 1 X PBS, blocked as indicated above, and incubated with either primary anti-CYP2F1 antibody (Santa Cruz Biotechnologies) at a 1:200 dilution in blocking buffer or primary anti-GAPDH antibody (ABCAM, Cambridge, MA) at a 1:5000 dilution in blocking buffer at 4°C overnight. Membranes were then washed and incubated with secondary antibody as indicated above. Protein was detected using Western Blot Luminol Reagent (Santa Cruz Biotechnologies) and visualized using a Kodak Image Station 400.
DNA Damage
DNA strand breaks were measured using alkaline single cell gel electrophoresis (alkaline Comet assay) according to the manufacturer’s recommended protocol (Trevigen Inc., Gaithersburg, MD). Briefly, control and treated primary NHBE cells were scraped and centrifuged to remove cell culture media. Cell pellets were then resuspended in 1 X PBS, combined with low melting point agarose, and plated on CometSlides. Slides were incubated at 4µ C in the dark for 30 minutes, followed by immersion in pre-chilled lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 mM Trizma base, 1% Triton X-100) for 1 hour, and subsequent incubation in alkaline (pH>13) electrophoresis buffer (10 N NaOH, 200 mM EDTA) for 30 minutes at room temperature. Following the post-lysis incubation, electrophoresis was applied at 300 mV for 30 minutes. Upon completion of electrophoresis, slides were immersed in 100% ethanol for 5 minutes and air dried. Slides were then flooded with 1 X ethidium bromide (EtBr), visualized using a fluorescent microscope equipped with an EtBr filter, and imaged. Images were scored using CASP freeware version 1.2.2 and DNA damage was computed as the average Olive Tail Moment (OTM) for 50 separate comets. OTM is defined as the fraction of tail DNA multiplied by the means of the head and tail DNA distributions.
Statistical analysis
All experiments were repeated in triplicate with cells from a minimum of two separate donors. All data were reported as mean ± standard deviation. The difference between control and treated cells was tested using one-way analysis of variance (ANOVA) and differences were considered significant with a probability of p ≤ 0.05. Post-hoc analysis of the differences between 3MI-treated cells and other treatment groups were calculated using student’s t-test and differences were considered significant with a probability of p ≤ 0.05.
Results
Induction of CYP1A1 and CYP2F1 in Response to 3MI Exposure
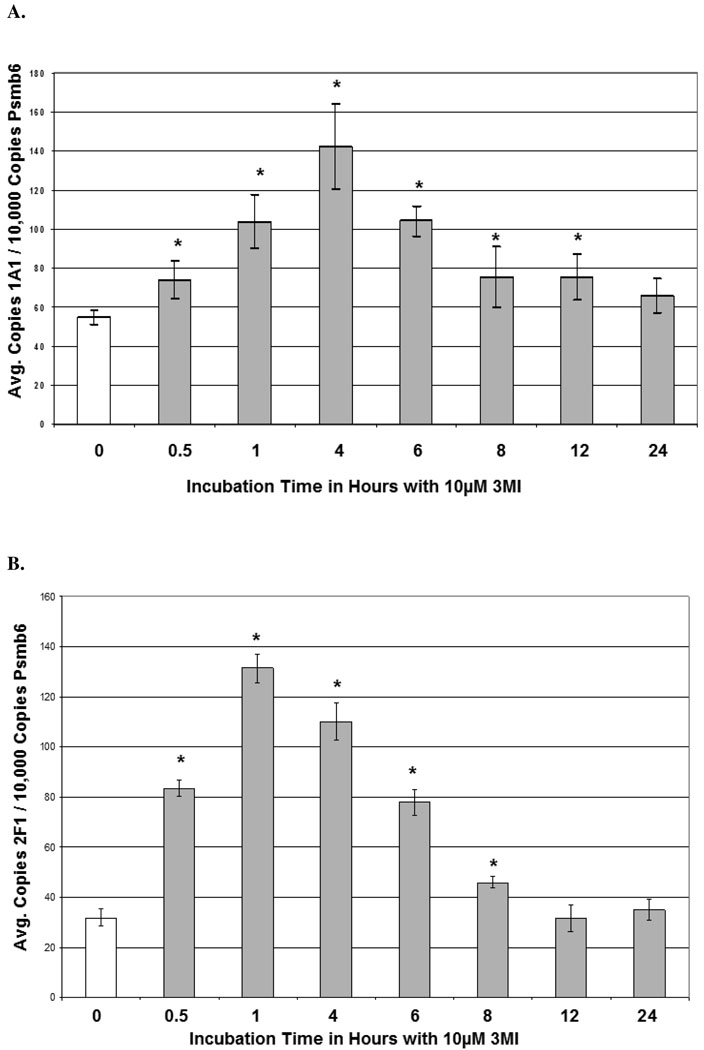
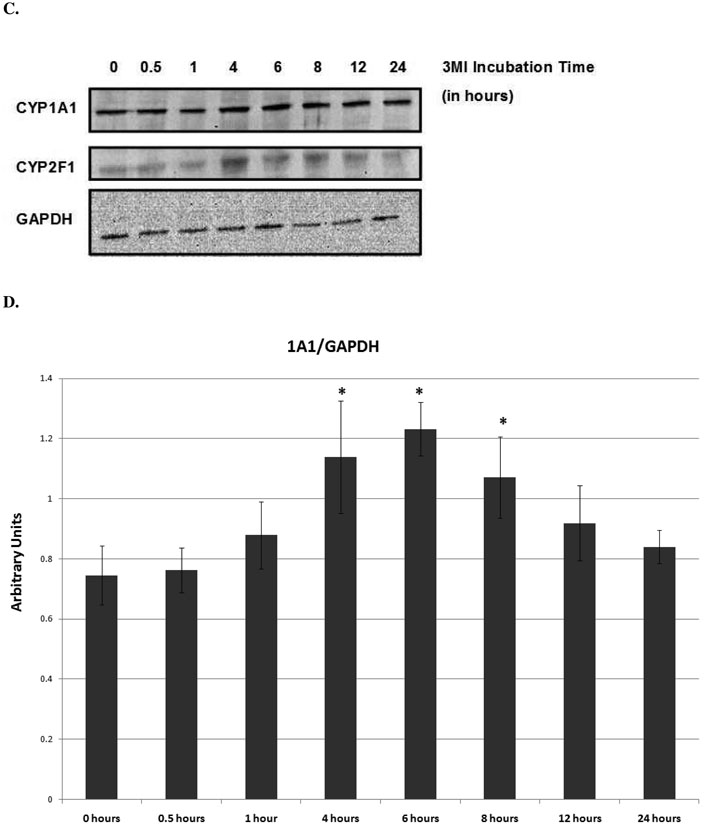
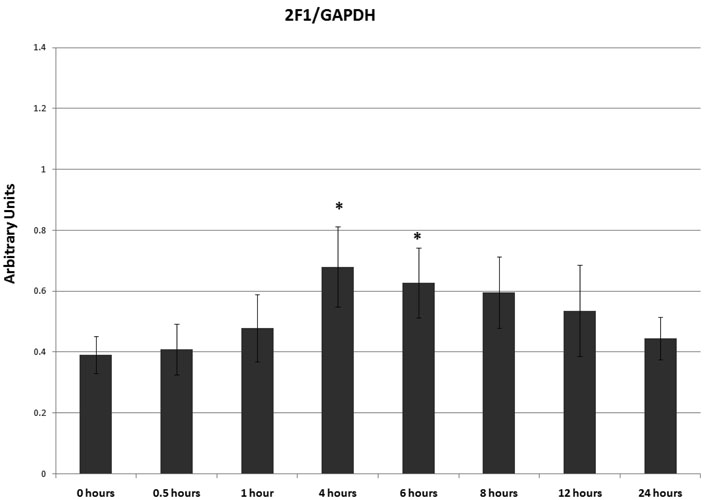
3MI-mediated CYP1A1 and CYP2F1 transcript and protein expression were assessed in NHBE cells in both a time- and concentration-dependent manner using quantitative real-time PCR and western blot analysis, respectively. Time course analysis consisted of 10 µM 3MI exposure for 0, 0.5, 1, 4, 6, 8, 12, and 24 hours. A statistically significant increase in transcription as compared to vehicle (0.5% DMSO)-treated control was observed within 30 minutes of exposure in both CYP1A1 and CYP2F1 (figure 1). Peak induction of CYP1A1 transcription was observed after 4 hours (figure 1A), whereas peak induction of CYP2F1 transcription occurred 1 hour after exposure to 3MI (figure 1B). CYP2F1 transcript levels remained elevated after 1 hour, but decreased in a time-dependent manner. Although initial transcript levels of CYP2F1 were roughly half that observed for CYP1A1, the peak induction of transcript levels for both enzymes was similar. Transcript levels were no longer significantly elevated by 12 (CYP2F1) and 24 (CYP1A1) hours following exposure. The time course of protein expression closely resembled transcript expression. CYP2F1 expression peaked at 4 hours, remained elevated to 6 hours, and decreased thereafter. CYP1A1 expression peaked at 6 hours and showed increased expression up to 8 hours, after which it returned to control levels (figure 1C). Densitometry analysis showed approximately 2-fold increases in protein expression for both enzymes (figure 1D).
Figure 1.
Time Course of 3MI-Mediated Cytochrome P450 Induction in NHBE Cells. Quantitative real-time PCR analysis revealed maximal induction of CYP1A1 (A) transcript expression 4 hours following 10 µM 3MI exposure, whereas maximal induction of CYP2F1 (B) transcript expression occurred 1 hour after exposure. Values were standardized to Psmb6 transcript expression and are the means of 3 separate experiments * p < 0.05 compared to vehicle (DMSO)-treated control (white bar). Western blot analysis of CYP1A1 and CYP2F1 protein expression (C) after exposure to 10 µM 3MI at various time points revealed peak induction of protein expression at 4 hours, after which point protein levels decreased to initial levels. GAPDH was included as a loading control. Protein expression levels were the means of 3 separate experiments and were verified using densitometric analysis (D). * p < 0.05 compared to the levels at 0 hours.
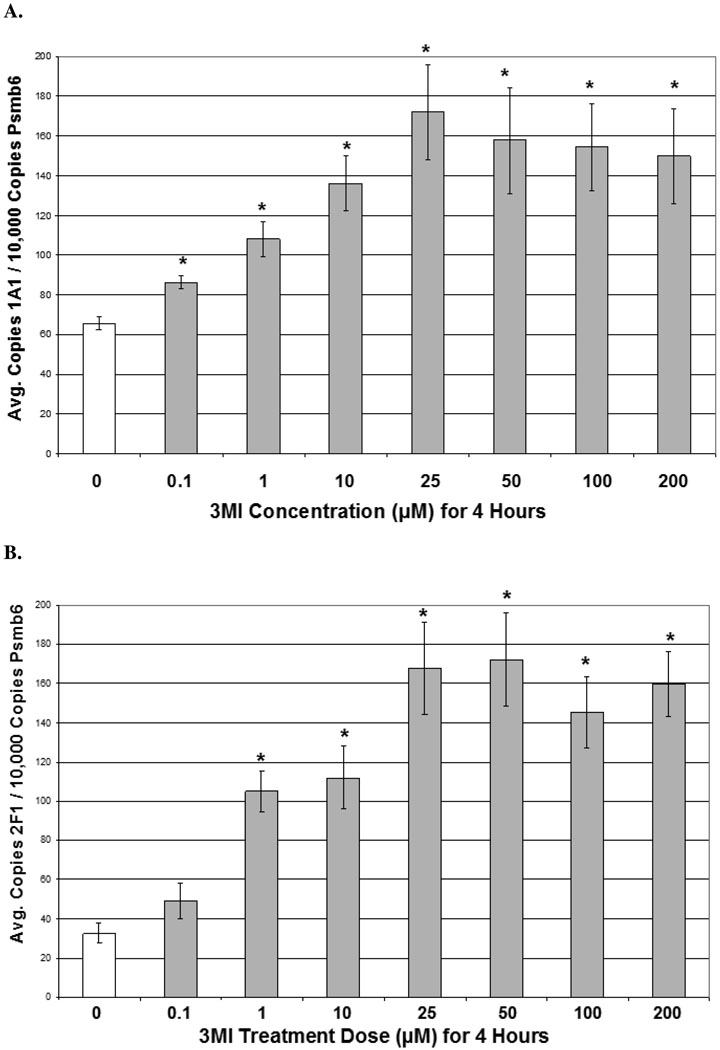
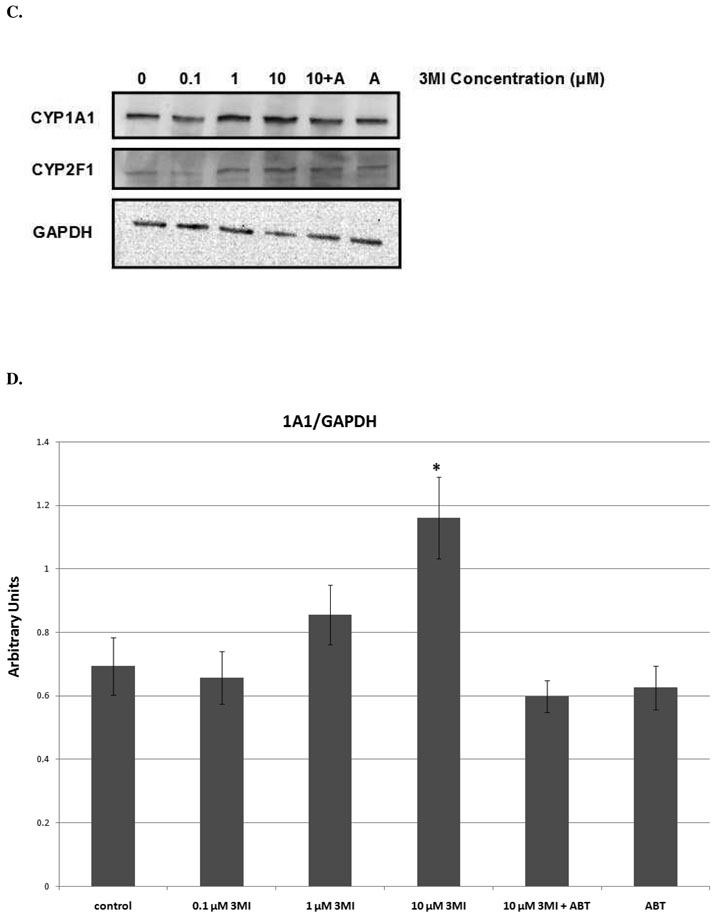
Dose response analysis consisted of 0, 0.1, 1, and 10 µM 3MI exposure at 1 (CYP2F1) and 4 (CYP1A1) hours (times observed to reach peak transcript induction of each gene). A concentration-dependent increase in transcript expression was observed for both genes (figure 2). As little as 0.1 µM 3MI was sufficient to induce a statistically significant increase in CYP1A1 transcript expression (figure 2A), while CYP2F1 required a concentration of 1 µM 3MI for significant induction (figure 2B). Both genes displayed a concentration-dependent increase in transcript expression up through exposure to 10 µM 3MI, after which the transcript expression leveled off. Protein expression of both enzymes (figures 2C & 2D) was significantly increased (p < 0.05) at the 10 µM concentration. Additionally, 30 minute pre-incubation with 5 mM ABT (the concentration previously determined to inhibit 3MEIN formation) (20) completely ameliorated 3MI-mediated protein induction for both enzymes. Thus, 3MI metabolites induced CYP1A1 and CYP2F1, not 3MI itself.
Figure 2.
Concentration Response of 3MI-Mediated Cytochrome P450 Induction in NHBE Cells. 4 hour incubation with increasing concentrations of 3MI (gray bars) produced a concentration-dependent induction of CYP1A1 (A) transcript expression. A similar effect on CYP2F1 (B) transcript expression was observed after 1 hour. Values were standardized to Psmb6 transcript expression and are the average of 3 separate experiments * p < 0.05 compared to vehicle (DMSO)-treated control (white bar). Western blot analysis of CYP1A1 and CYP2F1 protein expression (C) after 4 hours of exposure to increasing concentrations of 3MI revealed increased protein expression at 10 µM 3MI. Protein induction was attenuated by a 30-minute pre-treatment with ABT for both enzymes. GAPDH was included as a loading control. Protein expression levels were the means of 3 separate experiments and were verified using densitometric analysis (D). * p < 0.05 compared to vehicle (DMSO)-treated control
Elucidation of the Mechanisms of CYP1A1 and CYP2F1 Induction
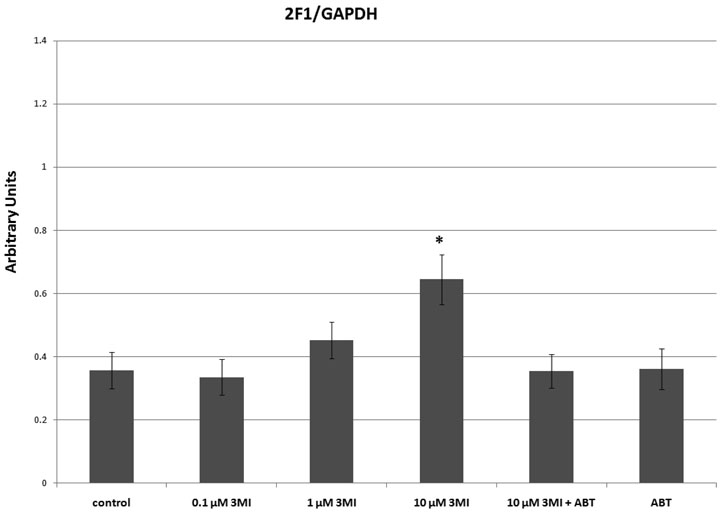
CYP1A1 transcript induction in NHBE cells was monitored using quantitative real-time PCR. One hour pre-treatment with the cytochrome P450 suicide substrate inhibitor 5 mM ABT significantly attenuated CYP1A1 transcript expression, compared to the 4-hour 10 µM 3MI treatment alone, indicating that 3MI-mediated induction was dependent upon the presence of active enzyme (figure 3A). One hour pre-treatment with the competitive inhibitor of AhR-mediated CYP1A1 induction, 1 µM α-NF, significantly attenuated induction mediated by both the positive control for AhR-mediated CYP1A1 transcript induction (14), 1 µM β-NF, and 3MI, indicating that the observed 3MI-mediated induction was AhR-dependent. One hour pre-treatment with the transcription inhibitor (21), 4 µM actinomycin D, was sufficient to completely block 3MI-mediated CYP1A1 transcription, indicating that 3MI-mediated CYP1A1 induction is dependent upon the formation of an active transcriptional complex.
Figure 3.
Mechanism of 3MI-Mediated CYP1A1 Induction in NHBE Cells. 1 hour pre-treatment with 5 mM ABT (gray and black diagonally striped bar), 4 µM actinomycin D (black bar), or 1 µM α-NF (horizontally striped gray and black bar) were sufficient to attenuate 10 µM 3MI-mediated (gray bar) CYP1A1 transcript induction (A). Pre-treatment with 1 µM α-NF also inhibited CYP1A1 induction mediated by 1 µM β-NF (white bar with open boxes). 1 hour pre-treatment with 35 µM cycloheximide (vertically striped gray and black bar) induced 3MI-mediated CYP1A1 expression more than 3-fold over that induced by 3MI (gray bar) treatment alone. ABT pre-treatment was sufficient to attenuate 3MI- and cycloheximide (gray and black checkered bar)-mediated superinduction of CYP1A1, while ABT co-treatment with cycloheximide (black and white checkered bar) did not attenuate induction compared to cycloheximide-mediated (vertically striped black and white bar) induction alone (A). CYP2F1 transcript expression (B) was successfully attenuated by 5 mM ABT, 35 µM cycloheximide, and 4 µM actinomycin D, whereas 1 µM β-NF did not significantly alter CYP2F1 transcript expression compared to vehicle treated control, indicating that the mechanisms of induction for CYP1A1 and CYP2F1 differ. Data are the average of 3 separate experiments and are standardized to Psmb6 transcript expression. * p < 0.05 compared to vehicle (DMSO) treated control (white bar), ‡ p < 0.05 compared to 10 µM 3MI (gray bar).
Surprisingly, a 1-hour pre-treatment with the inhibitor of translation, 35 µM cycloheximide, resulted in 3MI-mediated CYP1A1 transcript expression that was over 3-fold greater than that observed with 3MI treatment alone. This cycloheximide-mediated CYP1A1 superinduction effect has been previously observed in several cell lines, ranging from human MCF7 breast cancer cells (22) to Chinese hamster ovary (CHO) and human HepG2 liver cancer cells (23). The mechanism of the “superinduction” has not been established, but required certain intronic sequences of the CYP1A1 gene. Treatment with cycloheximide alone or in combination with ABT produced a much smaller level of CYP1A1 induction, which was significantly lower than that observed with both 3MI alone and 3MI in combination with cycloheximide. Additionally, pre-treatment with ABT was sufficient to attenuate 3MI and cycloheximide-induced superinduction.
CYP2F1 transcript induction (figure 3B) was also significantly attenuated following 5 mM ABT pre-treatment, indicating that induction of this enzyme was also dependent upon the presence of active enzyme. Additionally, pre-treatment with both the transcription inhibitor 4 µM actinomycin D and the translation inhibitor 35 µM cycloheximide completely blocked CYP2F1 transcript expression, indicating that 3MI-mediated CYP2F1 induction is dependent upon both transcription and translation. In contrast to 3MI-mediated CYP1A1 transcript expression, the positive control for AhR-mediated induction β-NF did not significantly alter transcription of CYP2F1. Interestingly, the inhibitor of AhR-mediated transcription, α-NF, did prevent 3MI-mediated CYP2F1 induction, indicating that crosstalk likely exists between the pathway governing CYP2F1 induction and the AhR pathway. However, although induction of CYP2F1 required metabolism of 3MI to presumably the same products that bound to AhR and induced CYP1A1, 3MI transcriptional activation of CYP2F1 did not require AhR binding.
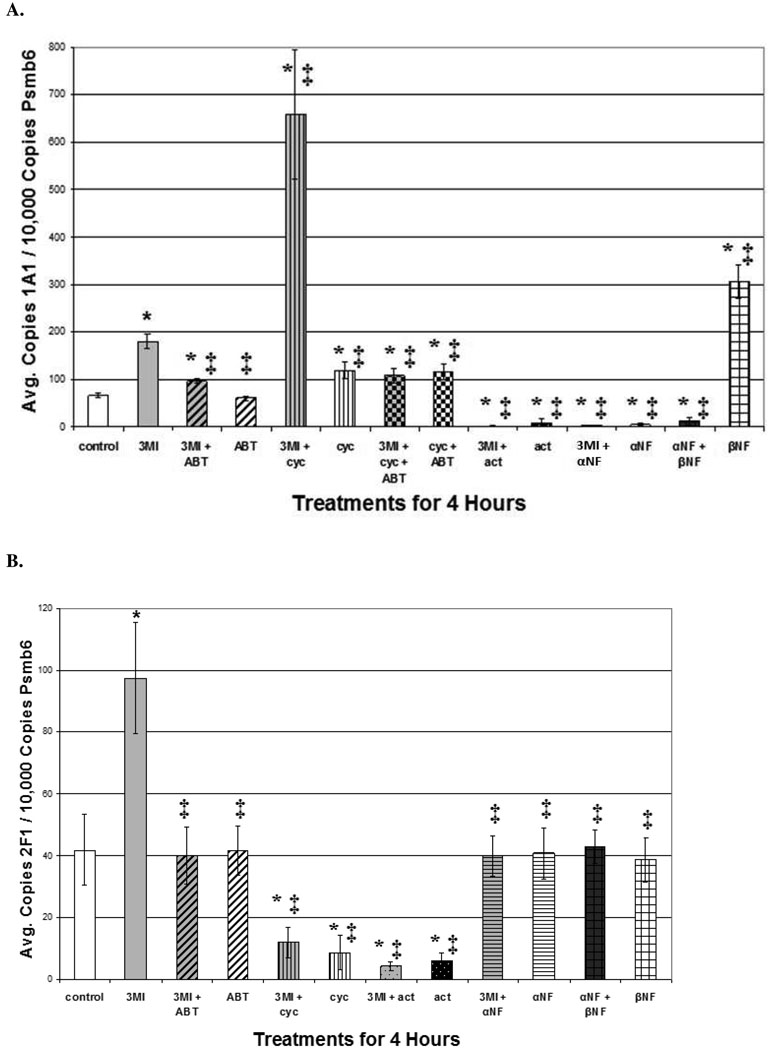
CYP1A1 Induction is Linked to DNA Damage
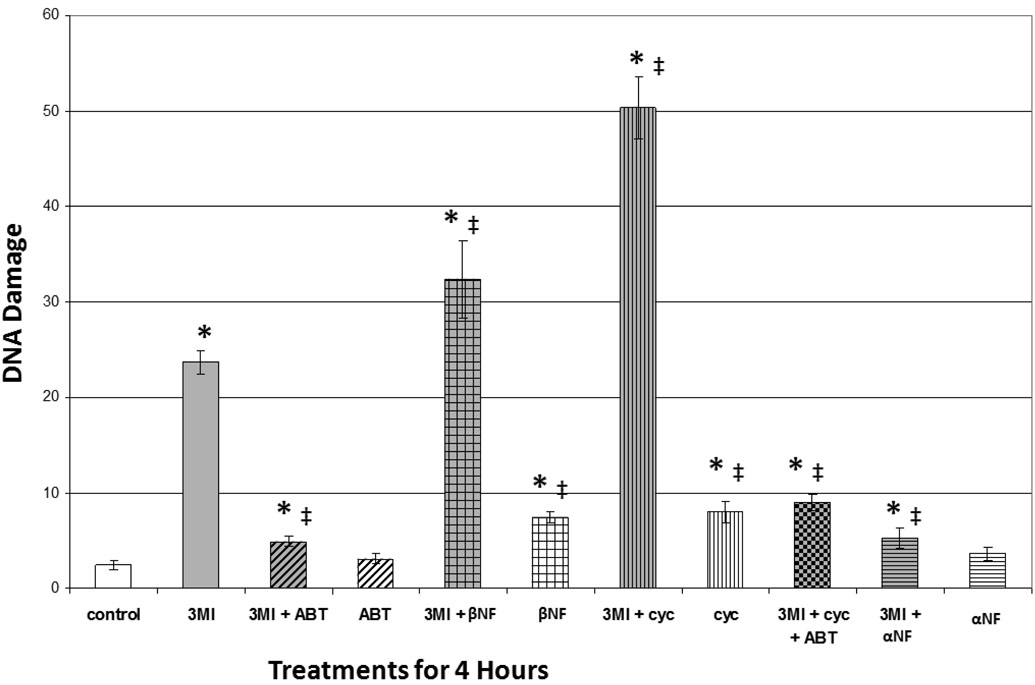
3MI-mediated DNA damage in NHBE cells was measured using the alkaline Comet assay. One-hour pre-treatment with 5 mM ABT significantly attenuated DNA damage, compared to the 4-hour 10 µM 3MI treatment alone (figure 4), which again showed requisite turnover of 3MI by cytochrome P450 enzymes. Although it was not sufficient to reduce DNA damage to control values, the 1-hour pre-treatment with α-NF significantly attenuated DNA damage, indicating that the majority of 3MI-mediated DNA damage required CYP1A1 induction. Both the 1-hour pre-treatment with either the positive control for CYP1A1 induction, β-NF, or the superinducer of CYP1A1, cycloheximide, increased the observed levels of 3MI-mediated DNA damage, which also confirmed the hypothesis that CYP1A1 induction was linked to DNA damage. Again, metabolites of 3MI caused DNA alterations, rather than 3MI itself, because ABT attenuated the process, even when cycloheximide was included as a “superinducer.”
Figure 4.
3MI-Mediated CYP1A1 Induction is Linked to DNA Damage in NHBE Cells. Comet assay analysis revealed that 1 hour pre-treatment with 5 mM ABT (gray and black diagonally striped bar) or 1 µM α-NF (gray and black horizontally striped bar) significantly attenuated 10 µM 3MI-mediated DNA damage, whereas treatment with ABT (white and black vertically striped bar) or α-NF alone (white and black horizontally striped bar) did not significantly alter DNA damage levels. 1 hour pre-treatment with the positive control 1 µM β-NF (gray and black bar with open boxes) or 35 µM cycloheximide (gray and black vertically striped bar) increased the levels of 3MI-mediated DNA damage to a greater extent than either 3MI (gray bar), β-NF (white and black bar with open boxes), or cycloheximide (white and black horizontally striped bar) treatment alone. DNA damage induced by 3MI co-treatment with cycloheximide was attenuated by pre-treatment with ABT (gray and black checkered bar). Data are the average OTM from 3 separate experiments. * p < 0.05 compared to vehicle (DMSO)-treated control (white bar), ‡ p < 0.05 compared to 10µM 3MI (gray bar).
Discussion
Cigarette smoke toxicants, such as PAH, have long been known to induce cytochrome P450 expression in the human lung (5). 3MI was previously reported to induce pig hepatic cytochrome P450 enzyme expression (18, 19). However, the present studies establish for the first time that 3MI induces human pulmonary cytochrome P450 enzymes. An interesting difference between hepatocytes and NHBE cells was that induction of CYP1A1 and CYP2F1 in NHBE cells was much faster than the induction of CYP2A6 and CYP2E1 in pig hepatocytes. In addition, significantly lower concentrations of 3MI (~ 10-fold) induced CYP1A1 and CYP2F1 in NHBE cells than concentrations required to induce CYP2E1 in pig hepatocytes (18). The concentrations examined in this study are well within the range of concentrations that could potentially exist at the site of deposition, the epithelial cells of airways, in people who smoke cigarettes. 3MI is formed by pyrolysis of tryptophan in burning tobacco and has been identified in cigarette smoke at a range of concentrations between 0.4 to 1.4 µg/cigarette (24). It seems reasonable to assume that concentrations that induce CYP1A1 and CYP2F1 transcription (0.1 to 1.0 µM) could be present in the airway cells if these amounts of 3MI were deposited directly on the cells from primary cigarette smoke. 3MI rapidly and potently induced cytochrome P450 expression in lung cells, which suggests that cells lining the human airway may be highly sensitive to 3MI-mediated CYP450 induction.
CYP1A1 and CYP2F1 have been previously reported to be expressed in human lung, with CYP2F1 displaying preferential expression in the lung (5). Unpublished data referred to in one report indicated that CYP1A1 expression in NHBE cells is lower than whole bronchial mucosa (25), whereas a recent study with bronchial cells from a single donor indicated that CYP1A1 is both present in NHBE cells, and is inducible by an average of 7.5-fold upon exposure to cigarette smoke condensate (17). Here, for the first time, we quantify transcript expression for CYP1A1 and CYP2F1 in NHBE cells from multiple donors, and report that both the mRNA and protein are readily detectable in NHBE cells (figure 1 and figure 2). These results are important because expression of CYP2F1 protein has been controversial, and it is possible that CYP2F1 may be an important catalyst of carcinogen bioactivation in human lung cells.
3MI-mediated induction of CYP1A1 transcript expression was blocked by actinomycin, but was superinduced by cycloheximide, indicating that the induction was due to increased transcription. Increased CYP1A1 protein expression followed roughly the same pattern as that observed for transcript expression (figure 1 and figure 2), so translation mimicked transcriptional increases. One of the most interesting differences in the mechanisms governing induction of the two genes was the observation that α-NF inhibited 3MI-mediated CYP2F1 induction, even though β-NF failed to induce expression, indicating that 3MI-mediated induction of CYP2F1 was probably not AhR-dependent. Several studies have indicated that crosstalk exists between the AhR receptor and other receptors that modulate cytochrome P450 expression, including the estrogen receptor (26), the nuclear pregnane X receptor (PXR) (27), and the constitutive androstane receptor (CAR) (27). CAR regulates several CYP2A and CYP2B family genes and has been shown to indirectly regulate AhR-controlled gene induction by crosstalk at the xenobiotic response element, making it a likely candidate for the observed effects in our study, although further investigation is required to confirm these results.
3MI-mediated induction of CYP1A1 and CYP2F1 protein expression (figure 2C), induction of CYP1A1 and CYP2F1 transcript expression (figure 3A), and superinduction by the combination of 3MI and cycloheximide, required metabolism of 3MI, indicating that a metabolite of 3MI was responsible for these events. Recent reports have identified the structures, biochemical pathways for production, and biochemical effects of several diindole tryptophan derivatives from plants. These tryptophan dimers are highly potent AhR agonists (28, 29). Another tryptophan dimer, 6-formylindolo[3,2-b]carbazole and its metabolites have been identified in human urine, and this dimer, one of the most potent activators of AhR, may be an important endogenous inducer of CYP1A1 (30).
Both CYP1A1 and CYP2F1 metabolize 3MI to a number of reactive intermediates (7), including the preferential metabolism of 3MI to the dehydrogenated product 3MEIN by CYP2F1. If 3MEIN dimerized, it seems possible that the dimer, indoldo[3,2-b]carbazole (ICZ), could be a metabolite that induces CYP1A1 and CYP2F1 (Scheme I). This chemical is produced in acidic aqueous solutions from indole-3-carbinol, and is a potent inducer of CYP1A1 (31). In fact, ICZ activates AhR 1.5 times greater than the prototypical AhR-mediated CYP1A1 inducer TCDD (30). Low amounts of ICZ have been detected in human feces (31), so it seems reasonable that it could be formed from 3MI, which is often called “skatole” in the literature, and is a normal component of human fecal matter. However, our preliminary studies showed that incubations of ICZ with NHBE cells did not appear to induce CYP2F1 (data not shown). It is possible, however, that ICZ or its metabolites, or other metabolites of 3MI, could induce CYP2F1, and more extensive experiments are required.
The effects of reactive metabolites of xenobiotic metabolism are not limited to cytochrome P450 induction. The reactive metabolite 3MEIN, for example, produces both protein and DNA adducts (8, 9). These findings lead us to examine the ability of 3MI to induce DNA damage in NHBE cells, using the comet assay to measure DNA fragmentation (10). We observed that 3MI-mediated DNA damage was cytochrome P450-dependent, indicating that a reactive metabolite of 3MI caused the damage (10). DNA fragmentation peaked at about 4 hours, and appeared to be at least partially repaired, because fragmentation returned to control levels by 24 hours. Concentrations of 3MI that fragmented DNA (0.1 µM) were not pro-apoptotic, so it seems reasonable that mutations produced from low concentrations of 3MI could be incorporated into proliferating epithelial cells, and enhance neoplastic processes in the lung.
In this study, we report for the first time that not only is 3MI-mediated DNA damage cytochrome P450-dependent in NHBE cells, but the majority of the damage is due to CYP1A1 activity (figure 4). In addition, when we treated the cells with 3MI in combination with known inducers of CYP1A1, including cycloheximide and β-NF, we observed increased levels of DNA damage. Inhibition of CYP1A1 did not attenuate all of the observed DNA damage, leading us to believe that other cytochrome P450 enzymes may also contribute to 3MI-mediated DNA damage. The current work indicates that CYP2F1, which preferentially catalyzes the formation of the reactive metabolite 3MEIN and is not induced in an AhR-dependent manner, is the most likely candidate.
The findings reported here parallel previous findings in our laboratory that low concentrations of 3MI damage DNA in NHBE cells, and demonstrate for the first time that similar concentrations can also induce cytochrome P450 expression. In addition, we have established a mechanistic link between cytochrome P450 induction and the DNA damage processes. These data indicate that 3MI is mechanistically similar to other well established cigarette smoke human lung carcinogens, such as PAH, and confirm that 3MI is a potential human lung carcinogen.
Acknowledgement
This work was supported by the National Heart, Lung, and Blood Institute [HL13645].
Contributor Information
Jessica M. Weems, Email: jessica.weems@pharm.utah.edu.
Garold S. Yost, Email: gyost@pharm.utah.edu.
References
- 1.Yost GS. Mechanisms of 3-methylindole pneumotoxicity. Chem Res Toxicol. 1989;2:273–279. doi: 10.1021/tx00011a001. [DOI] [PubMed] [Google Scholar]
- 2.Yost GS. Mechanisms of 3-methylindole pneumotoxicity. Chem Res Toxicol. 1989;2:273–279. doi: 10.1021/tx00011a001. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann D, Rathkamp G. Quantitative determination of 1-alkylindoles in cigarette smoke. Anal Chem. 1970;42:366–370. doi: 10.1021/ac60285a023. [DOI] [PubMed] [Google Scholar]
- 4.Nelson DR, K L, Kamataki T, Stegemann JJ, Feyereisen R, Wazman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, Gunsalus IC, Nebert DW. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Ding X, Kaminsky LS. Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol. 2003;43:149–173. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- 6.Guengerich FP. Bioactivation and detoxification of toxic and carcinogenic chemicals. Drug Metab Dispos. 1993;21:1–6. [PubMed] [Google Scholar]
- 7.Lanza DL, Yost GS. Selective dehydrogenation/oxygenation of 3-methylindole by cytochrome p450 enzymes. Drug Metab Dispos. 2001;29:950–953. [PubMed] [Google Scholar]
- 8.Ruangyuttikarn W, Appleton ML, Yost GS. Metabolism of 3-methylindole in human tissues. Drug Metab Dispos. 1991;19:977–984. [PubMed] [Google Scholar]
- 9.Regal KA, Laws GM, Yuan C, Yost GS, Skiles GL. Detection and characterization of DNA adducts of 3-methylindole. Chem Res Toxicol. 2001;14:1014–1024. doi: 10.1021/tx0100237. [DOI] [PubMed] [Google Scholar]
- 10.Weems JM, Cutler NS, Moore C, Nichols WK, Martin D, Makin E, Lamb JG, Yost GS. 3-Methylindole is mutagenic and a possible pulmonary carcinogen. Toxicol Sci. 2009;112:59–67. doi: 10.1093/toxsci/kfp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 12.Whitlock JP., Jr Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 13.Mimura J, Fujii-Kuriyama Y. Functional role of AhR in the expression of toxic effects by TCDD. Biochim Biophys Acta. 2003;1619:263–268. doi: 10.1016/s0304-4165(02)00485-3. [DOI] [PubMed] [Google Scholar]
- 14.Gasiewicz TA, Rucci G. Alpha-naphthoflavone acts as an antagonist of 2,3,7, 8-tetrachlorodibenzo-p-dioxin by forming an inactive complex with the Ah receptor. Mol Pharmacol. 1991;40:607–612. [PubMed] [Google Scholar]
- 15.Crystal RG, Randell SH, Engelhardt JF, Voynow J, Sunday ME. Airway epithelial cells: current concepts and challenges. Proc Am Thorac Soc. 2008;5:772–777. doi: 10.1513/pats.200805-041HR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang H, Chang LW, Cheng YH, Tsai WT, Tsai MX, Lin P. Preferential induction of CYP1A1 and CYP1B1 in CCSP-positive cells. Toxicol Sci. 2006;89:205–213. doi: 10.1093/toxsci/kfj025. [DOI] [PubMed] [Google Scholar]
- 17.Pickett G, Seagrave J, Boggs S, Polzin G, Richter P, Tesfaigzi Y. Effects of 10 Cigarette Smoke Condensates on Primary Human Airway Epithelial Cells by Comparative Gene and Cytokine Expression Studies. Toxicol Sci. 2009 doi: 10.1093/toxsci/kfp298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doran E, Whittington FW, Wood JD, McGivan JD. Cytochrome P450IIE1 (CYP2E1) is induced by skatole and this induction is blocked by androstenone in isolated pig hepatocytes. Chem Biol Interact. 2002;140:81–92. doi: 10.1016/s0009-2797(02)00015-7. [DOI] [PubMed] [Google Scholar]
- 19.Chen G, Cue RA, Lundstrom K, Wood JD, Doran O. Regulation of CYP2A6 protein expression by skatole, indole, and testicular steroids in primary cultured pig hepatocytes. Drug Metab Dispos. 2008;36:56–60. doi: 10.1124/dmd.107.017285. [DOI] [PubMed] [Google Scholar]
- 20.Nichols WK, Mehta R, Skordos K, Mace K, Pfeifer AM, Carr BA, Minko T, Burchiel SW, Yost GS. 3-methylindole-induced toxicity to human bronchial epithelial cell lines. Toxicol Sci. 2003;71:229–236. doi: 10.1093/toxsci/71.2.229. [DOI] [PubMed] [Google Scholar]
- 21.Sobell HM. Actinomycin and DNA transcription. Proc Natl Acad Sci U S A. 1985;82:5328–5331. doi: 10.1073/pnas.82.16.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joiakim A, Mathieu PA, Elliott AA, Reiners JJ., Jr Superinduction of CYP1A1 in MCF10A cultures by cycloheximide, anisomycin, and puromycin: a process independent of effects on protein translation and unrelated to suppression of aryl hydrocarbon receptor proteolysis by the proteasome. Mol Pharmacol. 2004;66:936–947. doi: 10.1124/mol.66.4.. [DOI] [PubMed] [Google Scholar]
- 23.Sakata Y, Yoshioka W, Tohyama C, Ohsako S. Internal genomic sequence of human CYP1A1 gene is involved in superinduction of dioxin-induced CYP1A1 transcription by cycloheximide. Biochem Biophys Res Commun. 2007;355:687–692. doi: 10.1016/j.bbrc.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 24.Wynder EL, Hoffmann D. Tobacco and tobacco smoke : studies in experimental carcinogenesis. New York: Academic Press; 1967. [Google Scholar]
- 25.Mace K, Bowman ED, Vautravers P, Shields PG, Harris CC, Pfeifer AM. Characterisation of xenobiotic-metabolising enzyme expression in human bronchial mucosa and peripheral lung tissues. Eur J Cancer. 1998;34:914–920. doi: 10.1016/s0959-8049(98)00034-3. [DOI] [PubMed] [Google Scholar]
- 26.Matthews J, Gustafsson JA. Estrogen receptor and aryl hydrocarbon receptor signaling pathways. Nucl Recept Signal. 2006;4:e016. doi: 10.1621/nrs.04016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- 28.Chowdhury G, Dostalek M, Hsu EL, Nguyen LP, Stec DF, Bradfield CA, Guengerich FP. Structural identification of Diindole agonists of the aryl hydrocarbon receptor derived from degradation of indole-3-pyruvic acid. Chem Res Toxicol. 2009;22:1905–1912. doi: 10.1021/tx9000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen LP, Hsu EL, Chowdhury G, Dostalek M, Guengerich FP, Bradfield CA. D-amino acid oxidase generates agonists of the aryl hydrocarbon receptor from D-tryptophan. Chem Res Toxicol. 2009;22:1897–1904. doi: 10.1021/tx900043s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wincent E, Amini N, Luecke S, Glatt H, Bergman J, Crescenzi C, Rannug A, Rannug U. The suggested physiologic aryl hydrocarbon receptor activator and cytochrome P4501 substrate 6-formylindolo[3,2-b]carbazole is present in humans. J Biol Chem. 2009;284:2690–2696. doi: 10.1074/jbc.M808321200. [DOI] [PubMed] [Google Scholar]
- 31.Kwon C-S, Grose KR, Riby J, Chen Y-H, Bjeldanes LF. In vivo production and enzyme-inducing activity of indolo[3,2-b]carbazole. J Agric Food Chem. 1994;42:2536–2540. [Google Scholar]