Abstract
Pancreatic cancer MIA PaCa cells were cultured in the presence and absence of 15N amino acids mixture for 72 hours. During protein synthesis, the incorporation of 15N amino acids results in a new mass isotopomer distribution in protein, which is approximated by the concatenation of two binomial distributions of 13C and 15N. Fraction of protein synthesis (FSR) can thus be determined from the relative intensities of the ‘labeled’ (new) and the ‘unlabeled” (old) spectra. Six prominent spots were picked from 2-D gels of proteins from lysates of cells cultured in 0% (control), and 50% and 33% 15N enriched media. These protein spots were digested and analyzed with MALDI-TOF/TOF. The isotopomer distribution of peptides after labeling can be fully accounted for by the labeled (new) and unlabeled (old) peptides. The ratio of the new and old peptide fractions was determined using multiple regression analysis of the observed spectrum as a linear combination of the expected new and the old spectra. The fractional protein synthesis rates calculated from such ratios of same peptide from cells grown in 50% and 33% 15N amino acid enrichments were comparable to each other. The FSR of these six identified proteins ranged between 44–76%.
The dynamics of protein turnover is key to the understanding of regulation of protein expression in cells.1;2 The level of expression of a protein depends on the rates of its synthesis and degradation. Thus the turnover of a protein is an important indicator of its functional significance in cells. Unlike other quantitative proteomics methods using stable isotope labeling of protein after its synthesis such as in ICAT3 and iTRAQ,4 measurement of protein synthesis requires labeling of protein in vitro or in vivo using pathways of protein synthesis and amino acid metabolism.1;5–9 In general, measurement of protein synthesis depends on the use of precursor/product relationship. For example, when a radioactive amino acid is used to trace protein synthesis, one has to first determine the specific activity of the radioactive amino acid in the cell and the specific activity of the same amino acid in the protein of interest.10 With the advent of mass spectrometry, many stable isotope methods have been developed for the estimation of protein turnover.2;5;7;8;11;12 In these methods precursor enrichment is either determined directly or indirectly. In the deuterated water methods of Busch et al.5 and Previs et al.,12 plasma alanine enrichment is used as the precursor enrichment, and fractional synthesis is determined from the ratio of alanine enrichment in protein to its enrichment in plasma. Precursor enrichment can also be determined indirectly by isotopomer ratio method such as those established for fatty acid synthesis using MIDA.13;14 Such an approach was employed in the recent method of Doherty et al.,1 where relative isotope abundance was estimated from peptides containing 2 or 3 molecules of leucine. Depending on how precursor enrichment is estimated, the approach by which enrichment in the labeled protein is determined also differs in these published methods for measurement of protein turnover.
Recently, we developed a general method for determination of protein synthesis using deuterated water and mass isotopomer distribution analysis.15 The approach is based on the concept that mass isotopomer distribution in the newly synthesized protein due to isotope incorporation is a concatenation of 13C isotopomers from 13C natural abundance with 2H isotopomersfootnote 1. Precursor and product enrichment can be determined by comparing the labeled and unlabeled spectra. Fraction of protein synthesis (FSR) is provided by the ratio of the old and new isotopomer distributions. Our method differs from the methods of Busch et al.5 and Previs et al.12 in that enrichment of 2H in amino acids as well as that in the peptide is determined from mass isotopomer distribution and average mass of the peptide. Once precursor enrichment is known, protein synthesis is determined from isotopomer distribution and average mass of the peptide similar to the approach of Vogt et al.16;17
In this study, we extend the application of this new technique of mass isotopomer distribution analysis for in vitro research using 15N labeling to illustrate the handling of spectral data when precursor enrichment is high resulting in sufficient mass shift in the new protein. Since the change in average mass (mass shift) of a new peptide is given by the product of the number of nitrogen atoms (NN) in the peptide and the average enrichment of 15N in the amino acids (p′), the 15N enrichment can be estimated from the mass shift by curve fitting and the expected isotopomer distribution of the new peptide can be generated by the concatenation function. FSR can be calculated by multiple linear regression analysis of the observed peptide spectrum on the expected new and the old (unlabeled) spectra. Since the method for determining precursor and product enrichment dictates the nature of the isotope and its infusion as well as the sensitivity and precision in estimating protein turnover of proteins with widely different half-lives, the approach described for use of low enrichment of deuterated water15 and 15N amino acids should be generally applicable in most in vivo and in vitro studies of quantitative proteomics.
EXPERIMENTAL SECTION
Reagents
Fetal bovine serum was purchased from Irvine Scientific (Santa Ana, CA). Dulbecco’s modified Eagles’s medium and antimycotic were from Gibco. U-15N enriched algal amino acid mixture was from Cambridge Isotope Lab Inc. (Andover, MA).
Cell culture
Mia PaCa cells were cultured in MEM supplemented with 10% fetal bovine serum, 1% antibiotic antimycotic, 5% CO2 at 37°C until 85–95% confluence when the experiment started. Experiments were set up in 3 groups: group A cells were cultured in MEM medium containing 1 mg/ml of natural amino acids; group B cells were cultured with 50% of 15N algal amino acid mixture (98% 15N); and group C cells were cultured with 33% of 15N algal amino acid mixture. Each treatment was repeated four times and had 4 flasks with 10 ml/flask. After incubation for 72 hours, the cell pellets were collected and stored at −80°C.
Protein extraction
Cell pellet was re-suspended in 300 μl lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, 100 mM DTT, 5% glycerol, protease inhibitor set III (100 mM AEBSF, 80 μM aprotinin, 5 mM bestatin, 1.5 mM E-64, 2 mM leupeptin, 1 mM pepstatin), phosphatase set II (200 mM imidazole, 100 mM sodium fluoride, 115 mM sodium Molybdate, 100 mM sodium orthovanadate, 400 mM sodium tartrate dihydrate) (Calbiochem, La Jolla, CA), 0.2% biolyte 3–10, 0.1% biolyte 4–6, and 0.1% biolyte 5–8) and sonicated for 3×5 sec. The mixture was spun down at maximum speed at 15 °C for 10 min. The protein concentration in the supernatant was measured by Bradford assay using bovine serum albumin as the standard.
2-D gel electrophoresis
After dilution with rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS, 50 mM DTT, 0.2% biolyte 3–10, 0.1% biolyte 4–6, and 0.1% biolyte 5–8), 540 μg (300 μl) sample was added to each pre-made 17cm immobilized pH gradient (IPG) strip (Bio-Rad, Hercules, CA). Prefocusing and focusing were performed on a Protean IEF cell (500 V hold 2.5 h, linear 500–1000V increase 1 h, 1000 V hold 1 h, linear 1000–8000 V increase 1.5 h, and 8000 V hold 60000 KVh). The equilibration was done sequentially by washing with equilibration buffer I and II (37.5 mM Tris-Cl, pH 8.8, 20% glycerol, 2% SDS, 6 M urea, with 2% DTT in buffer I and 2.5% iodoacetamide in buffer II, respectively). The second dimension electrophoresis was run on 8–16% Tris-HCl gel in a Protean Plus Dodeca cell (Bio-Rad, Hercules, CA). After running for 6 h, the gels were stained with Sypro-Ruby (Molecular Probes, Eugene, OR) overnight in darkness, and imaged with a Pharox FX™ molecular imager (Bio-Rad, Hercules, CA) under ultraviolet light.
Protein identification
PDQuest software (version 8.0.1, Bio-Rad, Hercules, CA) was used to select protein spots, which were excised by a spot-excision robot (EXQuest™ cutter, Bio-Rad, Hercules, CA). Gel slices were washed at 37°C in 100 mM NH4HCO3/50% ACN for 45 min twice. After dehydration in 100% ACN at room temperature for 5 min, the gel slices were dried in Speedvac and digested with trypsin (Promega, Madison, WI) in 40 mM NH4HCO3/10% ACN at 37°C overnight. The tryptic peptides were extracted once with water and twice with 50% ACN/5% TFA. After drying in Speedvac, the peptides were taken up in 10 μl of 0.1% TFA and purified with C18 ZipTip (Millippore, MA). Two microliter of CHCA matrix in 50% ACN/0.1% TFA was used to elute peptide onto ground steel plate (Bruker, Germany). MALDI-TOF/TOF (Ultraflex III, Bruker, Germany) was used for peptide fingerprint in reflector mode and sequence analysis in “lift” mode. Four fragments of each protein were used for identification and data analysis. The MS/MS data were searched against Homo sapiens proteins in the SWISS-PROT database, using the Mascot search program (Matrix science, London, United Kingdom) (www.matrixscience.com). The tolerance for parent ion and daughter ion is 100 ppm and 0.3 Da. A score of >72 was regarded as significant.
Spectral Data Analysis
Analysis of isotopomer distribution from spectral data is a well established mathematical method.18;19 A variation of the established technique to correct for 13C natural abundance was published by Jennings and Matthews for the study of kinetics of specifically labeled compounds such as [1-13C]Leucine and [1, 2-13C2]Leucine.20 We recently published an algorithm of mass isotopomer distribution analysis that allows us to separate natural and deuterium-labeled peptide spectrum using the inverse concatenation function 15. The fraction of labeled isotopomers represents the newly synthesized protein. In the current experiments, the 15N isotope enrichment is in the range of 30–50%. The mass shift due to isotope incorporation is substantial making the need for the application of inverse concatenation operation unnecessary. Therefore, a different strategy for data processing is devised.
The isotopomer distribution of a peptide is the concatenation of isotopomers from stable isotope species. For example, isotopomer distribution of a protein synthesized in the presence of 15N amino acids is the result of concatenation of the isotopomers contributed by natural abundance of 13C and the isotopomers contributed by the 15N in amino acids. When the natural abundance of the stable isotope species is low (e.g. 15N and deuterium), the mass shift of the isotope envelope formed by the isotopomers can be shown to be a function of N, the number of positions that can be substituted, and p, the enrichment of the isotope in question. The observed spectrum of a peptide (both preexisting and newly synthesized) is then linear combination of isotopomers of natural (unlabeled) and the expected labeled peptides, and the ratio of labeled to the total isotopomers provides a molar fraction of the newly synthesized protein.
Since the presence of 13C and 15N in an amino acid are independent of each other and are not mutually exclusive, the spectrum of the labeled peptide can be constructed using the concatenation operation on the 13C and 15N isotopomers. The 13C isotopomer distribution of a hypothetical natural peptide and 15N isotopomer distribution of a labeled peptide are typical binomial distribution (Figure S-1). The mean and variance of the 13C isotopomer distribution around the monoisotopic peak (m0) are NCp and NCp(1-p) respectively, where NC is the number of carbon atoms in the peptide, and p is the natural abundance of 13C. Similarly, the mean and variance of the 15N isotopomer distribution are NNp′ and NNp′(1-p′) respectively and p′ is the average 15N enrichment. The isotopomer distribution after concatenation is also shown in Figure S-1. The mean and variance of the new distribution is given by (NCp + NNp′) and [NCp(1-p) + NNp′(1-p′)] respectively. The mass shift of the new distribution is [(NCp + NNp′) − NCp] = NNp′. Therefore, the mass shift is a function of the 15N isotopomer distribution. Because NN can be determined from the known peptide sequence, the 15N enrichment (p′) can be determined by curve fitting (mass shift/NN). Once the natural isotopomers and the 15N isotopomers are known, the isotopomers of the 13C and 15N labeled peptide can be constructed using concatenation operation. The resultant distribution is the expected distribution of the new peptide and can be used to determine newly synthesized fraction.
The synthesis rate of each protein is the average of three to four fragments. One-way ANOVA with Tukey’s adjustment was used for multiple comparisons in SPSS 13.0 (SPSS Inc., Chicago, IL).
RESULTS
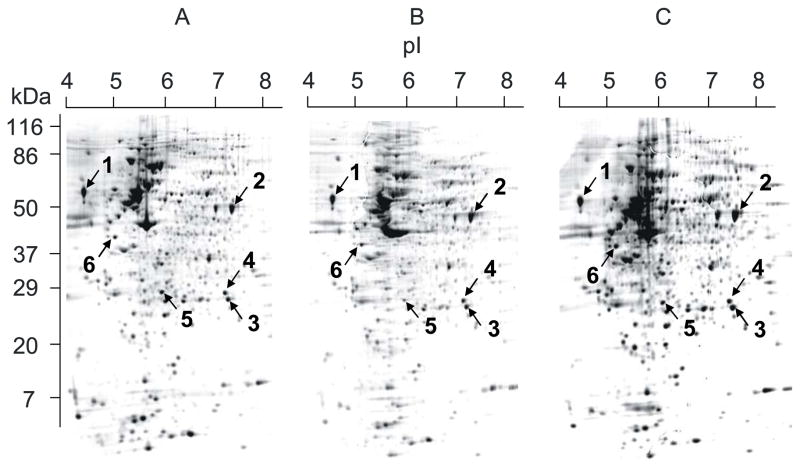
The MIA cells were cultured for 3 days with daily change of medium containing normal amino acids (group A), 50% 15N labeled amino acids (group B), and 33% 15N labeled amino acids (group C). The cell pellets were lysed and applied to 2-D electrophoresis. A total of greater than 1300 protein spots were detected in each gel. Among groups A, B, and C, we did not detect significant difference in protein expression (Figure 1), which indicates that stable isotope labeling does not affect normal protein metabolism. The same six strongly stained spots from each of the gels (Figure 1) were selected, digested with trypsin, and analyzed with MALDI-TOF/TOF in reflector mode. Three to four typical fragments from each protein spot were further identified in “lift” mode. The MALDI-TOF/TOF spectra were further analyzed for their mass isotopomer distribution. Protein identification and peptide sequences are listed in Table 1.
Figure 1.
Gel image after 2-D electrophoresis. MIA PaCa cells were cultured in MEM medium enriched with natural amino acids (A), 50% 15N labeled algal amino acids (B), and 33% 15N labeled algal amino acids (C) for 3 days. The cell pellets were lysed in 2-D lysis buffer and applied to 2-D electrophoresis. The marked spots were selected, cut out, digested, and analyzed. See Material and Methods for details.
Table 1.
The identified proteins by MALDI-TOF/TOFa
| spot | protein | fragment m/z | peptide sequence |
|---|---|---|---|
| 1 | Calreticulin precursor | 1410.6 | EQFLDGDGWTSR |
| 1607.8 | FYALSASFEPFSNK | ||
| 1800.8 | IKDPDASKPEDWDER | ||
| 2519.2 | IDNSQVESGSLEDDWDFLPPKK | ||
| 2 | Alpha-enolase | 1425.7 | YISPDQLADLYK |
| 1556.8 | VVIGMDVAASEFFRb | ||
| 1804.9 | AAVPSGASTGIYEALELR | ||
| 2510.1 | DYPVVSIEDPFDQDDWGAWQK | ||
| 3 | Triosephosphate isomerase | 1234.6 | SNVSDAVAQSTR |
| 1586.8 | DCGATWVVLGHSERc | ||
| 1807.9 | VAHALAEGLGVIACIGEKc | ||
| 2192.1 | VPADTEVVCAPPTAYIDFARc | ||
| 4 | Phosphoglycerate mutase I | 1150.7 | VLIAAHGNSLR |
| 1868.9 | YADLTEDQLPSCESLKc | ||
| 2131.1 | NLKPIKPMQFLGDEETVRb | ||
| 5 | Prohibitin | 1149.6 | FDAGELITQR |
| 1444.7 | IFTSIGEDYDER | ||
| 1606.8 | KLEAAEDIAYQLSR | ||
| 1998.1 | AAELIANSLATAGDGLIELR | ||
| 6 | 40S ribosomal protein SA | 1698.9 | FTPGTFTNQIQAAFR |
| 1740.9 | AIVAIENPADVSVISSR | ||
| 2633.3 | FLAAGTHLGGTNLDFQMEQYIYKb | ||
| 2996.5 | ADHQPLTEASYVNLPTIALCNTDSPLRc | ||
Each protein was identified from sequence analysis of four peptide fragments. The tolerance for parent ion and daughter ion is 100 ppm and 0.3 Da, respectively. A score of >72 was regarded as significant.
Oxidation on methionine.
Carbamidomethyl modification on cysteine.
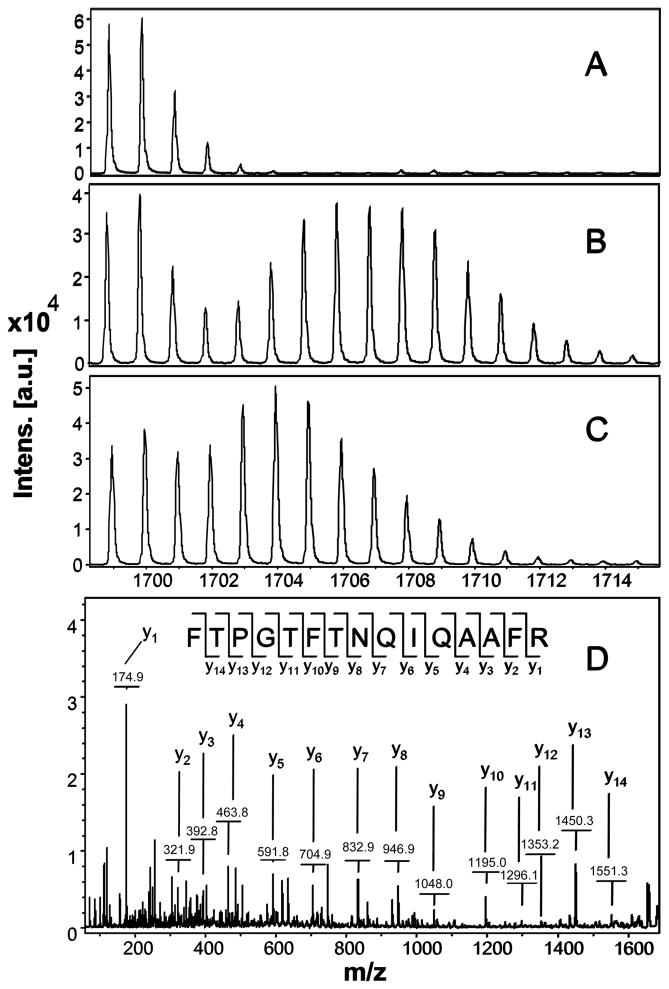
The effect of 15N incorporation on the isotopomer distribution of a peptide is illustrated in Figure 2. The isotopomer distribution of fragment 1699 m/z in spot 6, which is 40S ribosomal protein SA, from groups A, B, and C is shown (Figure 2A, B, C). The MS/MS spectrum of this fragment is shown in Figure 2D. Figure 2A shows the distribution of unlabeled (natural) fragment from control medium. Figure 2B spectrum is from the same peptide obtained from cells grown in 50% 15N enriched medium, showing the obvious spectrum shift in mass. Figure 2C spectrum is from the same peptide obtained from cells grown in 33% 15N enriched medium, which shows smaller mass shift than that of 50% 15N enrichment. Since the number of nitrogen atoms in the peptides is the same, the difference in mass shift reflects the difference in 15N enrichment in the medium. The 50% 15N labeling caused an almost complete separation of the isotopomers of the natural peptide from the distribution of the peptide of newly synthesized fraction. But there is still quite some overlapping of these two parts in the treatment of 33% 15N labeling (Figure 2C).
Figure 2.
Mass spectra of 1699 m/z fragment from spot 6 of lysates of cells grown in the presence of natural amino acids (Panel A), 50% enriched (Panel B) and 33% enriched (Panel C) 15N algal amino acid mixtures. MS spectrum of control in Panel A shows the binomial distribution of isotopomic peaks largely due to natural existence of 13C. Incorporation of 15N resulted in obvious mass shift in isotopomic distribution in Panel B and C. The degree of mass shift is a function of 15N enrichment. Panel D shows MS/MS spectrum of this fragment in ‘lift” mode. The peptide sequence is easily identified as almost every y ion is observed.
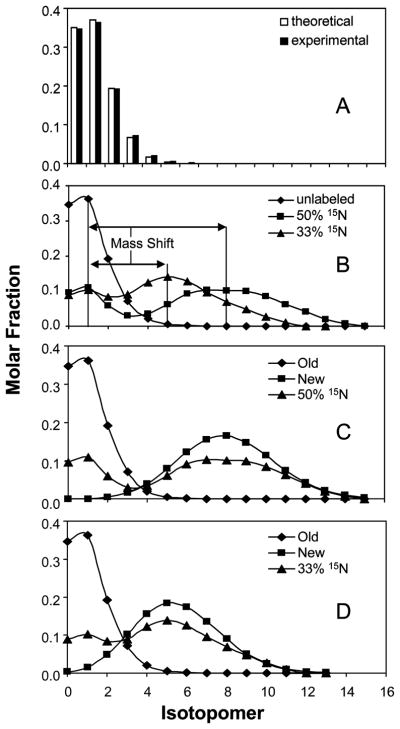
The data processing algorithm is illustrated in Figure 3. First, the intensity distribution of individual peaks in the peptide without labeling (control) was normalizedfootnote 2 (Figure 3A). After normalization, formula M1/M0=N×p/(1-p) (p = 0.0111) was used to calculate the carbon atoms by assuming 13C natural abundance to be 1.11%. Once the NC and p are known, a binomial distribution was set up following the formula: intensity of isotopomer (n) = (NC!/n!(NC-n)!)pn(1-p)Nc-n. In this formula, n stands for the number of 13C atoms and NC stands for the total number of carbon atoms in the peptide. The computation is shown in Table S-1. The near perfect fit between theoretical and experimental values are shown in Figure 3A. The theoretical carbon number is somewhat larger than the number calculated from peptide sequence due to other minor natural enrichments, like 2H, 18O, 15N, and 33S. In fact, the natural distribution is a concatenation of all these distributions. The approximation of the observed distribution by the theoretical distribution of 13C suggests that the mass isotopomer distribution is predominantly influenced by natural abundance of 13C (Figure 3A) and validate the use of binomial model for interpretation of peptide spectrum.
Figure 3.
Illustration of data analysis using example of 1699 m/z fragment from spot 6. Panel A: Theoretical and experimental distribution of the unlabeled fragment is compared. M0 is the monoisotopic peak. Panel B: Mass spectra of unlabeled and labeled peptides are aligned showing mass shifts, which was determined by curve fitting. Mass shift is larger with higher 15N enrichment. Panel C: Spectrum of peptide from MIA cells grown in 50% 15N enriched medium is shown with spectra of existing and newly synthesized peptide obtained by concatenation process. Panel D: Spectrum of peptide from MIA cells grown in 33% 15N enriched medium is shown with spectra of existing and newly synthesized peptide obtained by concatenation process. Fractional synthesis rate can be calculated using multiple linear regression analysis.
Both 50% and 33% of artificial enrichment of 15N in the medium caused obvious mass shift in spectrum distribution. The mass shift of the labeled spectrum can be determined by a simple curve fitting (Figure 3B). Based on the mass shift and the number of nitrogen atoms in the specific fragment (from sequence information), the average 15N enrichment can be deducedFootnote 3. After the NN and p′ are known, the theoretical 15N isotopomer distribution can be generated based on a binomial distribution function (Tables S-2 and S-3). The concatenation of 13C (Table S-1) and 15N distributions (Tables S-2 and S-3) represents isotopomer distribution of the newly synthesized peptide. The isotopomer distribution after 15N labeling which is represented by the concatenated distribution is shown in Figures 3C (50%) and 3D (33%). It is important to note that the sum of all isotopomers in any distribution, C-isotopomer, N-isotopomer or the concatenated isotopomer, is equal to 1.
The observed isotopomer distribution of a peptide is the linear combination of isotopomers of the natural (preexisting) and the labeled (new) peptide (Figure 3C&D). By multiple linear regression analysis using the observed distribution as the dependent variable and the preexisting and newly synthesized parts as the independent variables, the contribution of each of pre-existing and new peptides can be determined. Results based on 50% and 33% labeling are presented in Table 2 and 3, respectively. Variable I stands for the existing peptide before labeling while variable II represents the newly synthesized peptide. The regression coefficients of these variables are the fractions of existing peptide and newly synthesized peptide respectively. Theoretically, the coefficient of variable II is the turnover rate of the protein in 3 days, the duration of cell culture. The validity of the linear regression model is supported by the fact that most of the R-square values are >0.99 and all of them are >0.96 (Table 2 and 3). Ideally, the sum of the two coefficients should be equal to 1. Our results show that most of the sums are very close to 1 with a range between 0.96 and 1.04. To correct this minor deviation, all of the values for coefficient II were adjusted by restoring the sum of coefficient I and II to 1. The FSR values for each protein under two different 15N enrichments were calculated from three to four fragments.
Table 2.
Multiple regression analysis of turnover rate based on 50% 15N labeling in cell culture medium for 3 daysa.
| Spot | Fragment | Coefficient I b | SE c | Coefficient II d | SE | Sum | R2 | Adjusted coefficient II | Vogt I e | Vogt II f |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1410 | 0.59 | 0.015 | 0.41 | 0.024 | 1.00 | 0.99 | 0.41 | 0.41 | 0.40 |
| 1607 | 0.56 | 0.013 | 0.48 | 0.021 | 1.04 | 0.99 | 0.46 | 0.43 | 0.43 | |
| 1800 | 0.61 | 0.008 | 0.41 | 0.013 | 1.02 | 1.00 | 0.40 | 0.38 | 0.40 | |
| 2519 | 0.48 | 0.013 | 0.47 | 0.020 | 0.96 | 0.99 | 0.50 | 0.52 | 0.53 | |
| 2 | 1425 | 0.26 | 0.007 | 0.74 | 0.010 | 1.00 | 1.00 | 0.74 | 0.74 | 0.74 |
| 1556 | 0.28 | 0.014 | 0.71 | 0.022 | 1.00 | 0.99 | 0.72 | 0.71 | 0.73 | |
| 1804 | 0.28 | 0.012 | 0.70 | 0.019 | 0.98 | 0.99 | 0.71 | 0.71 | 0.74 | |
| 2509 | 0.24 | 0.008 | 0.75 | 0.012 | 0.99 | 1.00 | 0.76 | 0.76 | 0.77 | |
| 3 | 1234 | 0.33 | 0.015 | 0.65 | 0.027 | 0.98 | 0.99 | 0.66 | 0.67 | 0.69 |
| 1586 | 0.31 | 0.014 | 0.68 | 0.024 | 0.98 | 0.99 | 0.69 | 0.69 | 0.69 | |
| 1807 | 0.30 | 0.006 | 0.70 | 0.009 | 1.01 | 1.00 | 0.70 | 0.69 | 0.70 | |
| 2191 | 0.32 | 0.012 | 0.68 | 0.019 | 1.01 | 0.99 | 0.68 | 0.67 | 0.72 | |
| 4 | 1150 | 0.29 | 0.019 | 0.68 | 0.034 | 0.97 | 0.98 | 0.70 | 0.70 | 0.72 |
| 1868 | 0.28 | 0.006 | 0.73 | 0.010 | 1.01 | 1.00 | 0.72 | 0.72 | 0.73 | |
| 2130 | 0.26 | 0.019 | 0.74 | 0.030 | 1.00 | 0.98 | 0.74 | 0.73 | 0.80 | |
| 5 | 1149 | 0.50 | 0.019 | 0.47 | 0.031 | 0.97 | 0.99 | 0.48 | 0.51 | 0.52 |
| 1444 | 0.50 | 0.013 | 0.49 | 0.021 | 0.99 | 0.99 | 0.50 | 0.44 | 0.48 | |
| 1606 | 0.49 | 0.012 | 0.51 | 0.020 | 0.99 | 0.99 | 0.51 | 0.50 | 0.52 | |
| 1997 | 0.46 | 0.010 | 0.51 | 0.016 | 0.97 | 0.99 | 0.52 | 0.53 | 0.57 | |
| 6 | 1698 | 0.29 | 0.012 | 0.69 | 0.019 | 0.98 | 0.99 | 0.71 | 0.71 | 0.72 |
| 1740 | 0.31 | 0.014 | 0.70 | 0.023 | 1.01 | 0.99 | 0.69 | 0.68 | 0.73 | |
| 2632 | 0.29 | 0.017 | 0.71 | 0.026 | 1.00 | 0.98 | 0.71 | 0.68 | 0.74 | |
| 2996 | 0.29 | 0.005 | 0.71 | 0.007 | 1.00 | 1.00 | 0.71 | 0.70 | 0.72 | |
The observed isotopomer distribution was used as the dependent variable. The preexisting and calculated newly synthesized distributions were regarded as the independent variables in multiple linear regression analysis.
Coefficient I is the contribution of peptide from preexisting protein.
SE is standard error from regression analysis.
Coefficient II is the fraction of newly synthesized peptide or synthesis rate in 3 days.
Fractional synthesis rate calculated according to Vogt et al.16
Fractional synthesis rate calculated according to Vogt et al.17
Table 3.
Multiple regression analysis of turnover rate based on 33% 15N labeling in cell culture medium for 3 daysa
| Spot | Fragment | Coefficient I | SE | Coefficient II | SE | Sum | R2 | Adjusted coefficient II | Vogt I | Vogt II |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1410 | 0.49 | 0.013 | 0.51 | 0.020 | 1.01 | 1.00 | 0.51 | 0.51 | 0.51 |
| 1607 | 0.49 | 0.006 | 0.52 | 0.010 | 1.01 | 1.00 | 0.51 | 0.51 | 0.51 | |
| 1800 | 0.46 | 0.011 | 0.54 | 0.017 | 1.00 | 1.00 | 0.54 | 0.55 | 0.56 | |
| 2519 | 0.44 | 0.012 | 0.55 | 0.018 | 0.99 | 0.99 | 0.56 | 0.55 | 0.57 | |
| 2 | 1425 | 0.25 | 0.011 | 0.75 | 0.016 | 1.01 | 1.00 | 0.75 | 0.74 | 0.74 |
| 1556 | 0.25 | 0.008 | 0.76 | 0.013 | 1.00 | 1.00 | 0.76 | 0.75 | 0.76 | |
| 1804 | 0.28 | 0.010 | 0.73 | 0.015 | 1.00 | 1.00 | 0.73 | 0.72 | 0.72 | |
| 2509 | 0.18 | 0.010 | 0.79 | 0.014 | 0.98 | 1.00 | 0.81 | 0.82 | 0.84 | |
| 3 | 1234 | 0.30 | 0.007 | 0.71 | 0.011 | 1.00 | 1.00 | 0.70 | 0.71 | 0.71 |
| 1586 | 0.27 | 0.016 | 0.72 | 0.025 | 1.00 | 0.99 | 0.73 | 0.72 | 0.73 | |
| 1807 | 0.28 | 0.004 | 0.72 | 0.007 | 1.01 | 1.00 | 0.72 | 0.71 | 0.72 | |
| 2191 | 0.29 | 0.006 | 0.71 | 0.009 | 1.00 | 1.00 | 0.71 | 0.70 | 0.72 | |
| 4 | 1150 | 0.27 | 0.015 | 0.71 | 0.025 | 0.98 | 0.99 | 0.73 | 0.73 | 0.79 |
| 1868 | 0.21 | 0.020 | 0.80 | 0.030 | 1.01 | 0.99 | 0.79 | 0.78 | 0.80 | |
| 2130 | 0.24 | 0.012 | 0.73 | 0.018 | 0.98 | 0.99 | 0.75 | 0.77 | 0.78 | |
| 5 | 1149 | 0.45 | 0.016 | 0.54 | 0.025 | 0.99 | 0.99 | 0.54 | 0.55 | 0.55 |
| 1444 | 0.45 | 0.009 | 0.54 | 0.014 | 0.99 | 1.00 | 0.54 | 0.54 | 0.57 | |
| 1606 | 0.46 | 0.006 | 0.54 | 0.010 | 1.00 | 1.00 | 0.54 | 0.55 | 0.55 | |
| 1997 | 0.42 | 0.005 | 0.56 | 0.008 | 0.98 | 1.00 | 0.57 | 0.58 | 0.60 | |
| 6 | 1698 | 0.25 | 0.009 | 0.74 | 0.013 | 0.99 | 1.00 | 0.75 | 0.75 | 0.77 |
| 1740 | 0.26 | 0.008 | 0.74 | 0.012 | 1.00 | 1.00 | 0.74 | 0.73 | 0.75 | |
| 2632 | 0.27 | 0.031 | 0.72 | 0.045 | 0.99 | 0.96 | 0.73 | 0.75 | 0.77 | |
| 2996 | 0.25 | 0.006 | 0.74 | 0.009 | 0.99 | 1.00 | 0.75 | 0.75 | 0.76 | |
Analysis and interpretation are the same as in Table 2 except the observed distribution of 33% labeling was used as the dependent variable.
To further validate our method, the results were calculated based on fractional abundance (RIA) and change in average mass calculations of Vogt et al. 16;17 (Table 2 and 3). For the RIA method, we only used the average of molar fractions for the first two peaks based on their experience. The three sets of values are very close to each other (Table 2 and 3). However, our method differs from Vogt’s methods in that intensities of all isotopomers can be used in the determination of FSR and that reference protein with FSR close to 100% is not required. In their method, although the average mass approach utilizes the whole information of the spectra, the average enrichment k of each amino acid needs to be measured independently.
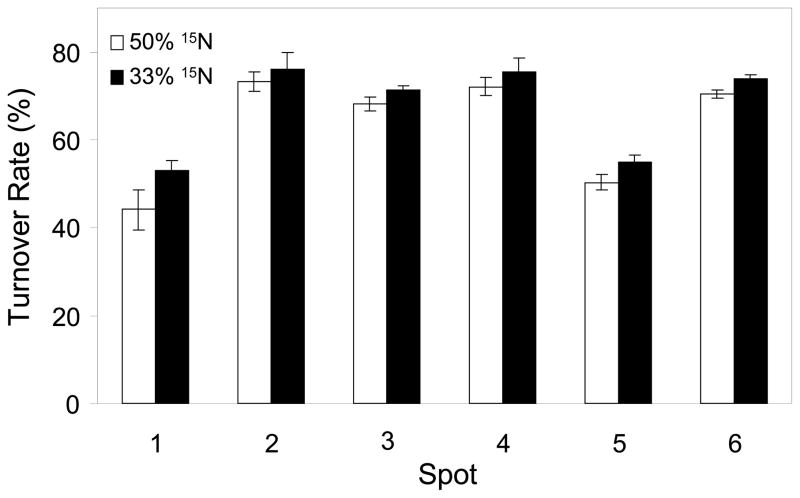
The average and standard deviations of the three to four FSR values for each protein calculated from these values in Tables 2 and 3 are shown in Figure 4. The FSRs of replicate measurements under two different 15N enrichment conditions were not very different from each other (Figure 4). Among the six proteins identified from these 2-D gel spots, we found that the turnover rates range between 44% and 76%. Based on 50% 15N labeling, these proteins could roughly be separated into two groups according to their turnover rates based on ANOVA analysis. The synthesis rates of calreticulin (spot 1) and prohibitin (spot 5) were similar, about 45% per 3 days (Figure 4). The synthesis rates of alpha-enolase (spot 2), triosephosphate isomerase (spot 3), phosphoglycerate mutase I (spot 4), and 40S ribosomal protein SA (spot 6) were about 70% per 3 days. The results based on 33% 15N labeling showed turnover of calreticulin and prohibitin of 50% while turnover of alpha-enolase, triosephosphate isomerase, phosphoglycerate mutase I, and 40S ribosomal protein SA of 70% per 3 days, respectively.
Figure 4.
Synthesis rate comparison among the six identified proteins after 72 hours. 1, Calreticulin precursor; 2, Alpha-enolase; 3, Triosephosphate isomerase; 4, Phosphoglycerate mutase I; 5, Prohibitin; 6, 40S ribosomal protein SA. Open columns represent 50% 15N labeling and filled columns represent 33% 15N labeling. The turnover rates of protein 1 and 5 are significantly lower than those of protein 2, 3, 4, and 6.
DISCUSSION
Study of protein synthesis or turnover has been a longstanding interest in biology. The basic measurement in such a study is the fraction of new protein (FSR) at a certain time after the introduction of labeled amino acids. FSR implicitly has the dimension of new fraction per unit time. When several FSRs are determined over time, turnover rate can be precisely calculated. The relationship between FSR and turnover rate can be appreciated using a single compartmental model in which FSR is given by the equation (1-e−kt). In this equation k is the fractional disappearance (degradation) rate of the protein. At metabolic steady state, the fractional disappearance rate is expected to be equal to the rate of appearance (synthesis). Absolute protein synthesis rate can be calculated when the concentration of the protein is known. The exponential equation (1-e−kt) justifies the approximation of turnover rate by FSR when FSR is low. However, FSR underestimates turnover rate when FSR is over 50% in that time period. For example, the same protein may have turned over several times in the time period for FSR of 100%. In the current experiments, the estimated FSR of 44–76% in the 72-hour period probably underestimates the true turnover rate and suggests the need for sampling at an earlier time point.
Previously, radioactive and stable isotope amino acid tracers were both used. Protein synthesis was determined from the ratio of specific activity in the product over the specific activity of the labeling agent.9 There are many shortcomings of such an approach including the need for purification of the protein and the determination precursor specific activity.9 We initiated the technique for quantitative proteomics15 using D2O as the labeling reagent. The labeling depends on amino acid metabolic pathways. It is thus named mSILAC, for modified stable isotope labeling with amino acids in cells. This method has been successfully applied to calculate protein turnover rate in vivo using deuterated water.15 Since deuterated water is not well tolerated at high enrichment, dueterated water with enrichment of less than 5% was used. Even at such low enrichment, we were able to determine fractional albumin synthesis in rat plasma. In order to calculate the contribution of the newly synthesized peptide, we had to independently measure the average enrichment of deuterium in the body, and use the inverse concatenation operation to determine the number of exchangeable hydrogen Nd. On the other hand, 15N amino acid mixture is well tolerated in animals at high enrichment.16 At 30–50% 15N enrichment, sufficient mass shift was observed for the calculation of 15N enrichment by curve fitting (Figure 3) using the concatenation model. Using the mass isotopomer distribution analysis approach, we determined FSR of 6 different proteins. The coefficient of variation of FSRs determined from 3–4 different peptides of the same protein was in the range of 1.2 to 10.4% with a median value of slightly less than 4%. There was general agreement between FSR of the same protein determined from lysate from cells cultured in 50% and 30% 15N enriched medium (Table 2 and 3) demonstrating the consistency within and between experiments.
Currently, there are several methods for the determination of protein synthesis. These methods vary in the choice of isotope, the duration and method of administration of the isotope depending on how precursor and product enrichment is determined. As a result, these methods vary in the sensitivity and precision in estimating turnover of proteins with widely different half-lives. In methods where precursor enrichment is separately determined, stable isotopes of essential amino acids having a large mass shift such as L-[ring-2,3,4,5,6 2H5] phenylalanine and [5,5,5-2H3]-leucine are often used. The list of commonly used stable isotopes is provided in a recent review by Beynon and Pratt. 21 The advantage of using specific labeled essential amino acid is that spectra of labeled and unlabeled peptides are well separated, and the range of FSR measurable depends on the amount of the labeled amino acid that can be safely infused per unit time. However, because of the dilution of the labeled amino acid due to recycling of unlabeled amino acid in proteolysis, the accuracy of precursor enrichment is often in doubt. Precursor enrichment can be determined from isotopomer ratio in the peptide. The methods of Papageorgopoulos et al.8 and that of Doherty et al.1 are examples of such a methodology. In order to determine isotopomer ratio accurately, a very high enrichment in labeled amino acid is required, e.g., 50% enriched [2H8] valine in the experiment of Doherty et al.1 Because of the high cost of fully labeled isotopes, it is not practical to measure proteins with a long half-life (low FSR) with such an approach.
There are other methods for the determination of protein synthesis without requiring the determination of precursor enrichment. The method of Cargile et al.11 introduces 13C carbon into protein by substituting natural glucose with [U13C6]-glucose (final enrichment >50%). In organisms, which can synthesize essential and non-essential amino acids from glucose and nitrogen, [U13C6]-glucose effectively replaces 12C by 13C in protein creating a heavy protein which can be separated by mass spectrometry. By quantitating the intensity of the labeled and the unlabeled peaks, a synthesis/degradation ratio can be calculated to represent relative dynamic protein turnover. Such a method is useful for the study of organisms such as bacteria and yeast, where 100% enriched 13C-glucose can be used. In the study by Cargile et al., protein turnover was expressed as the ratio of labeled over unlabeled protein. As protein synthesis increases, the denominator decreases rapidly and the synthesis/degradation ratio has the effect of magnifying the change, the scale for the ratio ranges from 0 to infinity. By contrast, in our current and previous studies, protein turnover is expressed as the fraction of new protein molecules which has an index ranging from 0 to 1. Recently Vogt et al.16 developed a method for estimation of FSR based on change in average mass of proteins with FSR of close to 100%. The observed change in average mass is the weighted sum of mass shift of the peptide with 100% FSR.16 In his recent paper, the change in average mass of a peptide was calculated as the sum of change in average mass of the individual amino acids.17 (See discussion in the Appendix of supplemental information). Our published method using deuterated water (D2O) and the current method using 15N amino acids illustrate the application of basic principle of spectral analysis in experiments using low (<5%) and high isotope enrichment (30–50%) for the determination of protein synthesis in vitro and in vivo. Such principle can be applied to design studies of protein dynamics using low enrichment (<10%) of low cost 13C or 15N isotopes in long infusion making such studies practical for clinical applications.
The dynamics of protein turnover reflects the regulation of protein expression in cell division and cell function. In tissue culture, as the cell divides its protein content is enriched with new protein. The lower limit of new protein fraction is bounded by cell division rate. On the other hand, proteins that are specific for a cell cycle have turnover rates above the cell division rate. In the six proteins chosen for this study, the turnover rate ranged from 44% to 76%. Calreticulin and prohibitin are among the proteins in the low turnover group. Calreticulin is a multifunctional calcium binding protein.22 Prohibitin is a negative regulator of cell proliferation and perhaps a cancer suppressor, which is ubiquitously expressed.23 Alpha-enolase, triosephosphate isomerase, and phosphoglycerate mutase I are all involved in the glycolytic pathway. Their turnover rates are similar and higher than those of the regulators of cell proliferation and cell death. Glycolytic pathway is the central pathway for glucose metabolism. Glycolysis provides substrates for the TCA cycle and intermediates for pentose cycle and other pathways. 40S ribosomal protein SA is known as human 34/67 kDa laminin receptor, which is involved in cellular adhesion.24 This protein also showed a fairly high turnover rate comparable to enzymes of the glycolytic pathway.
CONCLUSIONS
Current method for the determination of protein synthesis by tracer dilution principle requires the determination of isotope enrichment in specific protein and enrichment in the specific amino acid precursor. The development of newer mass spectrometers with the capability of de novo sequencing and accurate spectral data has enabled researchers to exploit labeling of proteins after extraction with stable isotope for the purpose of determining protein expression (quantitation) using LC/MS/MS or MALDI-TOFMS. However, the difficulty with introducing highly enriched isotopes of amino acids in vivo prevents the use of these methods for determining protein synthesis and turnover. Generally, it is feasible to label protein with low enrichment of 2H, 15N or 13C.16;17;25 Protein synthesis can be calculated from the shift in mass isotopomer distribution (mass shift). FSR can be determined using the concatenation model and curve fitting to arrive at the 100% labeling obviating the need for the use of a 100% newly synthesized protein as a reference as in Vogt’s methods.16;17 The concatenation function provides an ideal 100% labeled spectrum and multiple regression analysis uses all the information from the mass spectrum. Our mathematical algorithm represents a major improvement in the calculation of protein synthesis rate, permitting the use of isotope labeling of protein through the pathways of amino acid metabolism with low cost isotopes.
Supplementary Material
Acknowledgments
This work is jointly funded by the Bone Biology Program of the Cancer and Smoking Related Disease Research Program and the Nebraska Tobacco Settlement Biomedical Research Program (289104-845610 to GX), and partially supported by a grant awarded to WNPL from the UCLA Center of Excellence in Pancreatic disease (P01 AT003960-01) and Harbor-UCLA GCRC Mass Spectrometry Core (M01 RR00425-33).
Footnotes
Since a molecule of protein is made of many atoms of carbon, hydrogen, nitrogen, and oxygen, the probability of a protein molecule having one or more of the heavy isotope generates an isotopomer distribution reflected by the isotope envelope. Since the natural abundance of 13C is much greater than the natural abundance of either nitrogen or oxygen, the isotopomers of a natural (unlabeled) peptide is approaximated by 13C isotopomers (see RESULT) and are referred to as 13C isotopomers throughout this paper.
The normalization converts the individual intensity to relative intensity with the sum of intensities of all peaks equals to 1.
Since the amino acid composition of the algal mixture is different from the amino acid composition in the medium, the 15N enrichment of individual peptide is often different from the 50% or 33% according to the proportion of algal amino acids relative to those in the natural amino acids of the medium. The average 15N enrichment determined from NNp′ is an average depending on the peptide sequence.
SUPPORTING INFORMATION AVAILABLE
Mass shift, binomial distribution for 13C and 15N, concatenation process, fractional synthesis rate calculation. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Doherty MK, Whitehead C, McCormack H, Gaskell SJ, Beynon RJ. Proteomics. 2005;5:522–33. doi: 10.1002/pmic.200400959. [DOI] [PubMed] [Google Scholar]
- 2.Pratt JM, Petty J, Riba-Garcia I, Robertson DH, Gaskell SJ, Oliver SG, Beynon RJ. Mol Cell Proteomics. 2002;1:579–91. doi: 10.1074/mcp.m200046-mcp200. [DOI] [PubMed] [Google Scholar]
- 3.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Nat Biotechnol. 1999;17:994–99. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 4.Gan CS, Chong PK, Pham TK, Wright PC. J Proteome Res. 2007;6:821–27. doi: 10.1021/pr060474i. [DOI] [PubMed] [Google Scholar]
- 5.Busch R, Kim YK, Neese RA, Schade-Serin V, Collins M, Awada M, Gardner JL, Beysen C, Marino ME, Misell LM, Hellerstein MK. Biochim Biophys Acta. 2006;1760:730–44. doi: 10.1016/j.bbagen.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 6.Hellerstein MK, Neese RA. Am J Physiol. 1999;276:E1146–E1170. doi: 10.1152/ajpendo.1999.276.6.E1146. [DOI] [PubMed] [Google Scholar]
- 7.Papageorgopoulos C, Caldwell K, Shackleton C, Schweingrubber H, Hellerstein MK. Anal Biochem. 1999;267:1–16. doi: 10.1006/abio.1998.2958. [DOI] [PubMed] [Google Scholar]
- 8.Papageorgopoulos C, Caldwell K, Schweingrubber H, Neese RA, Shackleton CH, Hellerstein M. Anal Biochem. 2002;309:1–10. doi: 10.1016/s0003-2697(02)00253-1. [DOI] [PubMed] [Google Scholar]
- 9.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR., III Anal Chem. 2004;76:4951–59. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 10.Fern EB, Garlick PJ. Biochem J. 1974;142:413–19. doi: 10.1042/bj1420413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cargile BJ, Bundy JL, Grunden AM, Stephenson JL., Jr Anal Chem. 2004;76:86–97. doi: 10.1021/ac034841a. [DOI] [PubMed] [Google Scholar]
- 12.Previs SF, Fatica R, Chandramouli V, Alexander JC, Brunengraber H, Landau BR. Am J Physiol Endocrinol Metab. 2004;286:E665–E672. doi: 10.1152/ajpendo.00271.2003. [DOI] [PubMed] [Google Scholar]
- 13.Lee WN, Bergner EA, Guo ZK. Biol Mass Spectrom. 1992;21:114–22. doi: 10.1002/bms.1200210210. [DOI] [PubMed] [Google Scholar]
- 14.Lee WN, Bassilian S, Ajie HO, Schoeller DA, Edmond J, Bergner EA, Byerley LO. Am J Physiol. 1994;266:E699–E708. doi: 10.1152/ajpendo.1994.266.5.E699. [DOI] [PubMed] [Google Scholar]
- 15.Xiao GG, Garg M, Lim S, Wong D, Go VL, Lee WN. J Appl Physiol. 2008;104:828–36. doi: 10.1152/japplphysiol.00976.2007. [DOI] [PubMed] [Google Scholar]
- 16.Vogt JA, Schroer K, Holzer K, Hunzinger C, Klemm M, Biefang-Arndt K, Schillo S, Cahill MA, Schrattenholz A, Matthies H, Stegmann W. Rapid Commun Mass Spectrom. 2003;17:1273–82. doi: 10.1002/rcm.1045. [DOI] [PubMed] [Google Scholar]
- 17.Vogt JA, Hunzinger C, Schroer K, Holzer K, Bauer A, Schrattenholz A, Cahill MA, Schillo S, Schwall G, Stegmann W, Albuszies G. Anal Chem. 2005;77:2034–42. doi: 10.1021/ac048722m. [DOI] [PubMed] [Google Scholar]
- 18.Katz J, Lee WN, Wals PA, Bergner EA. J Biol Chem. 1989;264:12994–3004. [PubMed] [Google Scholar]
- 19.Lee WN, Byerley LO, Bergner EA, Edmond J. Biol Mass Spectrom. 1991;20:451–58. doi: 10.1002/bms.1200200804. [DOI] [PubMed] [Google Scholar]
- 20.Jennings ME, Matthews DE. Anal Chem. 2005;77:6435–44. doi: 10.1021/ac0509354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beynon RJ, Pratt JM. Mol Cell Proteomics. 2005;4:857–72. doi: 10.1074/mcp.R400010-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Williams DB. J Cell Sci. 2006;119:615–23. doi: 10.1242/jcs.02856. [DOI] [PubMed] [Google Scholar]
- 23.Merkwirth C, Dargazanli S, Tatsuta T, Geimer S, Lower B, Wunderlich FT, von Kleist-Retzow JC, Waisman A, Westermann B, Langer T. Genes Dev. 2008;22:476–88. doi: 10.1101/gad.460708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi YE, Torri J, Yieh L, Sobel ME, Yamada Y, Lippman ME, Dickson RB, Thompson EW. Clin Exp Metastasis. 1993;11:251–61. doi: 10.1007/BF00121168. [DOI] [PubMed] [Google Scholar]
- 25.Bouwman F, Renes J, Mariman E. Proteomics. 2004;4:3855–63. doi: 10.1002/pmic.200400861. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.