Abstract
Pulmonary cytochrome P450 2F3 (CYP2F3) catalyzes the dehydrogenation of the pneumotoxin 3-methylindole (3MI) to an electrophilic intermediate, 3-methyleneindolenine, which is responsible for the toxicity of the parent compound. Members of the CYP2F subfamily are the only enzymes known to exclusively dehydrogenate 3MI, without detectable formation of oxygenation products. Thus, CYP2F3 is an attractive model to study dehydrogenation mechanisms. The purpose of this study was to identify specific residues that could facilitate 3MI dehydrogenation. Both single and double mutations were constructed to study the molecular mechanisms that direct dehydrogenation. Double mutations in SRS 1 produced an inactive enzyme, while double mutants in SRS 4 did not alter 3MI metabolism. However, double mutations in SRS 5 and SRS 6 successfully introduced oxygenase activity to CYP2F3. Single mutations in SRS 5, SRS 6 and near SRS 2 also introduced 3MI oxygenase activity. Mutants S474H and D361T oxygenated 3MI but also increased dehydrogenation rates, while G214L, E215Q and S475I catalyzed 3MI oxygenation exclusively. A homology model of CYP2F3 was precisely consistent with specific dehydrogenation of 3MI via initial hydrogen atom abstraction from the methyl group. In addition, intramolecular kinetic deuterium isotope studies demonstrated an isotope effect (KH/KD) of 6.8. This relatively high intramolecular deuterium isotope effect confirmed the initial hydrogen abstraction step; a mutant (D361T) that retained the dehydrogenation reaction exhibited the same deuterium isotope effect. The results showed that a single alteration, such as a serine to isoleucine change at residue 475, dramatically switched catalytic preference from dehydrogenation to oxygenation.
Dehydrogenation or desaturation is considered an uncommon pathway among cytochrome P450-mediated reactions (1). In many instances, the products of the dehydrogenation reaction pathway are toxic and can form adducts with proteins and DNA (2-5). Bioactivation by a dehydrogenation pathway is also responsible for the toxicity of the pneumotoxicant 3-methylindole. Cytochrome P450-mediated oxygenation of 3MI produces the stable metabolites, 3-methyloxindole and 3-hydroxy-3-methyloxindole via a common precursor, the 2,3-epoxide (6). The epoxide precursor and 3-hydroxy-3-methylindolenine are reactive and can possibly form adducts with proteins and DNA. However, extensive studies have shown that dehydrogenation of 3MI, leading to the formation of the highly reactive imine methide, is the major bioactivation route that leads to the ultimate pneumotoxicity of the compound. (7-9). Covalent adducts of the 3-methyleneindolenine to both protein (10, 11) and DNA (12) have been identified.
In addition to the toxicity associated with dehydrogenation-dependent intermediates, this process may produce adverse drug/drug interactions. CYP1A2-catalyzed dehydrogenation of furafylline produces an imidazomethide intermediate that inactivates the enzyme by covalent attachment (13). This dehydrogenation-dependent, mechanism-based inactivation may also be operative in the inhibition of CYP2B6 by the anti-estrogenic compound tamoxifen (14). Indeed, dehydrogenation of 3MI to 3-methyleneindolenine by human CYP2F1 inhibited this enzyme at concentrations above 300 μM (15), and both CYP2F1 and CYP2F3 are inactivated by 3MI (16). The increasing importance of this unique cytochrome P450 pathway to the fields of toxicology and drug metabolism merits extensive research into the biochemical parameters that direct selective dehydrogenation. Because of its specificity for dehydrogenation, and subsequent bioactivation of the pneumotoxicant 3MI, CYP2F3 is an attractive choice to gain understanding of the dehydrogenation reaction pathway, but the mechanisms that govern this pathway have not been elucidated. Therefore, elucidation of the key structural elements responsible for catalytic specificity, leading to dehydrogenation by CYP2F3, is of considerable interest.
Site-directed mutagenesis along with homology modeling, based on the X—ray coordinates of P450 crystal structures, have been employed extensively to investigate the mechanisms of substrate-enzyme interactions, and to identify putative amino acids involved in substrate binding and catalysis (17-19). In 1992, Gotoh (20) proposed the existence of six substrate recognition sites (SRS) for the P450 family 2. These SRS were based on the alignment of 51 P450 family 2 sequences and 8 bacterial sequences. A vast majority of amino acid residues and chimeric fragments, identified as critical elements of substrate specificity for the CYP2 sub-family, fall within or near the putative SRS. Mutations in SRS regions 1, 4, 5 and 6 have been demonstrated to alter the selectivity and specificity of the CYP2 sub-family enzyme-catalyzed reactions (21). Because of the obvious importance of the SRS residues in substrate recognition and catalytic specificity, a sequence comparison focusing on the SRS regions was performed (Figure 1), and the results utilized to guide the rational design of mutants of CYP2F3.
Figure 1.
The aligned protein sequences of CYP2F3 and CYP2E1 and mutations. The numbers above the sequences correspond to the CYP2F3 sequence, and the numbers below the sequences correspond to the CYP2E1 sequence. The single and double mutations that were successfully mutated to produce active enzymes are shown in large bold type. The double mutations (both residues mutated simultaneously) are boxed. The six substrate recognition sites are indicated with bold type. All mutants were changed from the 2F3 sequence to the corresponding 2E1 residues.
CYP2E1 is an enzyme that is expressed primarily in hepatic tissues, but has been demonstrated to be present in lung as well (22). The enzyme plays an important role in the metabolism of drugs and is involved in the biotransformation of several xenobiotic compounds to toxic intermediates (23). Interestingly, 2E1 selectively catalyzes the oxygenation of 3MI (24) and therefore it offered a valuable comparative tool to interrogate the mechanisms of selectivity between P450-mediated oxygenation and dehydrogenation pathways. CYP2F1 selectively dehydrogenates 3MI, and is 84% identical to 2F3. Therefore, the sequence of the human homologue, CYP2F1, was also compared to 2E1 when the specific sites of substitution from the 2F3 sequence to 2E1 sequence were made. Both single and double mutants were chosen within the SRS regions to correspond to chemically significant differences between the 2F1 and 2E1 amino acid residues; that is, a strategy that changed the SRS residues in CYP2F3, to the corresponding homologous residues in 2E1, was employed to convert 2F3 into an enzyme that oxygenates 3MI. Specific residues and residue pairs were selected for replacement based on differences in chemical characteristics between 2F3 and 2E1 at the respective positions. Additional credibility for the choice of mutation sites was applied when literature precedence demonstrated the catalytic importance of the specific residue. The residue pairs in CYP2F3 that were targeted for substitution with the corresponding CYP2E1 residues were only chosen when they were conserved between CYP2F1 and CYP2F3.
Recent advances in the modification, expression, and crystallization of microsomal P450s have enabled the determination of structures of major human P450s by X-ray crystallography including 3A4 (25), 2C9 (26), 2C8 (27), 2A6 (28) and 1A2 (29). Sequence similarity between P450s of the same family are high, hence structures of the different P450 family 2 genes served as excellent templates for homology modeling studies with 2F3. Therefore, in addition to site-directed mutagenesis, a homology model for 2F3 was constructed based on previously published X-ray coordinates of 2C8, 2C9 and 2A6, and docking studies of 3MI within the 2F3 active site were conducted. The homology model helped us confirm the relative position of these mutations within the active site of CYP2F3, and if they were not within the active site, pointed to alterations that the specific mutations introduced to the mutant enzymes.
In addition to the elucidation of the key structural elements responsible for 3MI dehydrogenation, studies were also conducted to see if mutations altered the hydrogen abstraction step, a putative crucial component of CYP2F3-mediated 3MI dehydrogenation. Although lung selective cytochrome P450 isozymes, CYP2F1 (human form) and CYP2F3 (goat form), selectively catalyze the 3-methyl dehydrogenation of 3MI (24), the exact mechanism by which these enzymes dehydrogenate 3MI has yet to be firmly elucidated. Previous mechanistic studies using goat lung microsomes and 3MI-d2 demonstrated an isotope effect (kH/kD) of 5.5, suggesting that the mechanism of dehydrogenation does involve an initial hydrogen abstraction (30). However, goat lung microsomes are not pure preparations. In addition to CYP2F3, they certainly contain several other P450 enzymes, including CYP4B2 that was cloned from goat lung, and demonstrated to be an efficient dehydrogenase enzyme. Therefore, the isotope effect obtained through these studies could not be accurately attributed to any specific P450. Therefore, it was important to measure inter- and intra-molecular deuterium isotope effects with pure CYP2F3.
MATERIALS AND METHODS
Chemicals
The QuickChange mutagenesis system was purchased from Stratagene (La Jolla, CA). Restriction enzymes: Nde I, Xba I, EcoR I and Dpn I were obtained from Gibco, BRL (Gaithersburg, MD). Other enzymes: T4 DNA ligase was obtained from Gibco, BRL (Gaithersburg, MD) and Pfu-turbo DNA polymerase was obtained from Stratagene (La Jolla, CA). E. coli: DH5α were obtained from Gibco BRL (Gaithersburg, MD). NADPH reductase microsomes were purchased from Gentest. 3-[(N-acetyl-L-cystein-S-)methyl] indole (3MINAC) was synthesized as previously described (30). Bacterial Media: Lauria Bertani (LB) and Terrific Broth (TB) were obtained from Sigma (St. Louis, MO) as dry pre-mixtures. Tris-acetate, magnesium acetate, potassium phosphate, sucrose, EDTA, dithiothreitol, sodium chloride, potassium chloride, ethidium bromide, δ-aminolevulinic acid, ammonium acetate and 3-methylindole were obtained from Sigma (St. Louis, MO). Isopropyl thiogalactopyranoside (IPTG) and agarose for electrophoresis was obtained from Fisher (Pittsburgh, PA). Ampicillin was obtained from Boehringer Mannheim (Indianapolis, IN). DNA purification: large-scale plasmid purification kit (Midi), Ni-NTA beads and QIAEX II gel extraction kit were obtained from Qiagen (Santa Clarita, CA). Slide-A lyzer dialysis cassette was obtained from Pierce (Rockford, IL). All other chemicals were purchased from Sigma Aldrich.
Sequence Analysis and Rationale
All sequence alignments and comparisons were performed using the Accelrys GCG software package (Accelrys Inc., San Diego, CA) with the Seq Web interface, or the MODELLER program that was used to construct the molecular model of CYP2F3. Human CYP2E1 exclusively catalyzes oxygenation of 3MI to the stable metabolite 3-methyloxindole (31). These enzymes exhibit 50% sequence identity. However, within the SRS regions 4, 5 and 6 the sequence identity drops to 36%. It was therefore reasonable to hypothesize that differences existing between CYP2F3 and 2E1, contribute to, if not control the difference in, catalytic selectivity. A pair-wise global sequence alignment of CYP2F3 and CYP2E1 is presented in Figure 1. Focus was placed on differences in SRS regions 1, 4, 5 and 6. Mutations in these regions have been demonstrated to alter the selectivity and specificity of the CYP2 sub-family-catalyzed reactions (21). Specific single and pairs of adjacent CYP2F3 residues were selected from these regions that were different from the corresponding pair from CYP2E1. The residue pair from 2F3 was then replaced with the pair from 2E1. The specific residue pairs are indicated in Figure 1. The residue pairs were chosen based on their potential to produce a local structural and/or chemical change to the enzyme, and with consideration of similarities to the sequences of other 2-family enzymes for .which information from mutagenesis studies was available.
Consideration was also placed on the human homologue, CYP2F1, when choosing specific sites for substitution from 2E1 to 2F3. As described previously, 2F1 also selectively dehydrogenates 3-methylindole, and is 84% identical to 2F3. Therefore, differences between 2F1 and 2F3 may account for differences in catalytic efficiency, but not the catalytic selectivity for 3-methyldindole dehydrogenation. Therefore, we only replaced the residue pairs in CYP2F3 with the corresponding 2E1 residues when the pairs were conserved between 2F1 and 2F3.
Other Single Amino Acid Mutations
A sequence comparison of members of the CYP2 sub-family, of the region immediately downstream of SRS 2 that contained the SSPWGEM sequence, showed that residue 215 (glutamine) was highly conserved, except for P450 2F3, where residue 215 is glutamic acid. Therefore, residue 215 in 2F3 (glutamic acid) was targeted for site-directed mutagenesis. In addition, in 2F3 the adjacent residue (214) is a glycine, while in most other members of the CYP2 sub-family, including 2E1, this residue is leucine. Since changes in side-chain length have been shown to alter substrate binding and catalysis in 2B1 (32), residue 214 was also targeted for site-directed mutagenesis. The specific mutations in this region outside SRS 2 are shown in Figure 1.
Facilitation of Bacterial Expression of P450 2F3 and Mutants
Cytochrome P450s are membrane bound proteins. The hydrophobic membrane-spanning domain was removed (residues 3-21) and several positively charged residues were substituted at the N-terminus to increase expression and solubility. In addition, a C-terminal [His]4 tag was incorporated to aid in the purification of P450 2F3 by affinity chromatography. The truncated proteins are highly expressed in E coli and can be released from the membrane using high salt conditions (33). Modification of the N-terminal region of functional P450 significantly enhances the level of expression (34), but the enzyme’s catalytic properties are usually not altered (35). In addition to incorporating the N-terminal mutations, 5′ NdeI and 3′ XbaI restriction endonuclease sites were incorporated into P450 2F3 cDNA. This was done in order to facilitate cloning into the expression vector, pCWOri+ (a gift from Dr. Fred Guengerich, Vanderbilt University, Nashville, TN). Amplification of the modified P450 2F3 cDNA was performed by PCR and verified by gel electrophoresis. A double restriction digest using Nde I and Xba I was performed and P450 2F3 cDNA was subcloned into the same unique sites in the expression vector.
Introduction of Mutations to CYP2F3
Site-directed mutagenesis of CYP2F3 was performed by QuickChange™ mutagenesis kit, obtained from Stratagene (La Jolla, CA). Complementary mutagenic oligonucleotide primers were designed and ordered from Integrated DNA technologies, Inc. (IDT, Inc.). Mutagenesis reactions were carried out according to the manufacturer’s instruction. The reaction mixture contained 50 ng of pCW2F3, 125 ng of each mutagenic oligonucleotide, 1 μl of a dNTP pre-mixture, 5 μl of the manufacturer-supplied reaction buffer, 2.5 units of Pfu-turbo thermostable polymerase and water to a final volume of 50 μl. PCR reactions were performed under the following conditions: 1 cycle at 95 °C for 30 seconds, followed by 16 cycles of 95 °C for 30 seconds, 55 °C for 1 minute and 68 °C for 17 minutes (2 minutes/kb of plasmid length). The reaction mixtures were then cooled on ice for 2 minutes and the methylated parental strand was digested for an hour with 10 units of Dpn I. Approximately 3 μl of the Dpn I-treated DNA was used to transform library efficiency DH5α E. coli competent cells. Ampicillin resistant colonies were picked and grown in LB media and a plasmid miniprep was performed using established methods. A restriction digest analysis was performed with Nde I and Xba I in order to verify the correct insertion of the CYP2F3 cDNA insertion in the pCW vector. These enzymes excise the P450 cDNA from the expression vector. In addition, full-length sequencing of the P450 cDNA was performed in order to confirm the correct incorporation of the desired mutations. The mutant P450 enzymes were then utilized for expression studies.
Expression of P450 2F3 and Mutant Enzymes
Expression of the mutant enzymes was performed according to a previously established method for cytochrome P450 expression in bacterial systems (36). Bacterial clones containing the mutant enzymes were streaked on LB agar plates containing 100 μg/ml ampicillin and incubated at 37 °C for 12-16 hrs. Individual colonies were picked and used to inoculate 5 ml of LB media containing 100 μg/ml ampicillin. The starter cultures were then used to inoculate 500 ml of TB (Terrific Broth) media containing100 μg/ml ampicillin, bactopeptone (1g/0.5 L), 1 mM thiamine, and the trace elements, FeCl3, ZnCl2, Na2MoO4, CaCl2, CuCl2 and H3BO3. The culture was incubated at 37 °C for 2 hours at which time 0.5 mM δ-aminolevulinic acid was added. The culture was incubated until the absorbance at 600 nm (A600) was approximately 0.8-1.0. At this time, the culture was induced with 1 mM isopropyl-thiogalactopyranoside (IPTG) and both the temperature and rotation rate were reduced to 30 °C and 110 rpm, respectively. Incubation continued for 16-20 hours post-induction, at which time the bacterial cells were harvested by centrifugation at 4000 × g for 20 minutes, the supernatant was decanted, and the cells were stored at -70 °C until further use.
Preparation of Bacterial Membrane Fractions
Bacterial membranes were prepared by subtle modifications of previously established methods (33). The bacterial cell pellets were resuspended in 50 ml (10% total culture volume) 20 mM potassium phosphate buffer (pH 7.4) containing 20% glycerol. Lysozyme (0.2mg/ml) was added to the suspension and incubated for 30 minutes on ice. The suspension was centrifuged at 10,000×g (9500 rpm). The pellet containing the spheroblasts was then resuspended in 25 ml (5% total culture volume) 10 mM potassium phosphate buffer (pH 7.4) containing 10% glycerol and 0.5% CHAPS. A protease inhibitor cocktail was added in a 1000-fold dilution (25μl for 25ml resuspension). The suspension was then sonicated with four 25-second bursts at 50-60 % power from a 1.2-cm diameter probe. The suspension was transferred to a clean ultracentrifuge tube, and the membrane fraction was collected by ultracentrifugation at 105,000 × g (33,000 rpm) for 1 hr. The supernatant, which contained the protein, was stored at 4°C until further use. The P450 spectrum was checked at each step of the procedure as described below. The pellet left after ultracentrifugation containing the inclusion bodies and other undisrupted cells was resuspended in 5 ml 20 mM potassium phosphate buffer (pH 7.4) containing 20% glycerol and 0.5% CHAPS and the procedure repeated for extraction of any remaining P450. P450 content of prepared membrane fractions was determined by subtle modifications of a previously established method (37).
Purification of Mutant Enzymes
Purification of the histidine-tagged mutant enzymes was done using Ni-NTA beads purchased from QIAGEN. The purification protocol was based on slight modifications of the manufacturer’s instructions. To the supernatant from bacterial membrane preparation, 0.25 M NaCl, 10 mM β mercaptoethanol and 15 mM imidazole were added. The supernatant was then loaded onto a 5 ml polypropylene column (Qiagen) packed with 2 ml of Ni-NTA beads and allowed to flow-through. P450 content of the flow-through was checked to ensure optimal binding. If optimal binding was not achieved, the flow-through was reapplied to the column. The column was then washed three times with 10 mM potassium phosphate buffer containing 0.5% CHAPS, 0.5 M NaCl, 10% glycerol and increasing concentration of imidazole (10 mM, 20 mM and 40 mM). The flow-through from all three washes was analyzed for P450 content to ensure that the protein of interest was not lost during the washing steps. The protein in the column was then eluted with the buffer, but the imidazole content in the buffer was increased to 300 mM. The eluted protein was concentrated using centricon tubes (MW cut off 30,000) and dialyzed overnight in a Slide-A lyzer dialysis cassette (0.5-3 ml, MW 3500) in 2.5 L 10 mM potassium phosphate buffer containing 10% glycerol. The P450 spectrum and protein content were checked at each step and the specific activity of the protein was calculated. The purified protein was stored at -70°C for further use.
Incubations of CYP2F3 and Mutant Enzymes
CYP2F3 dehydrogenates 3MI to form the highly reactive 3-methyleneindolenine. This reactive methylene imine intermediate can be trapped with exogenously added nucleophiles, like N-acetyl-L-cysteine (NAC), and detected by mass spectrometry (Figure 2). The oxygenated metabolite, 3-methyloxindole, can be directly detected and quantified by mass spectrometry. Incubations were performed as follows; 100 pmol of P450 2F3 or mutant enzyme was warmed with 300 pmol cytochrome P450 NADPH reductase and 100 mM potassium phosphate buffer (pH 7.4) for 10 minutes at room temperature. After preincubation, 4 mM NAC and 3MI concentrations ranging from 20-500 μM were added to a total volume of 250 μl. The mixture was allowed to warm for an additional 3 minutes at 37°C before initiating the reaction with 2 mM NADPH. Incubations were carried out at 37°C for 30 minutes and the reaction was stopped by adding an equal volume of ice cold acetonitrile. Time-dependent analyses of products showed that the activities of CYP2F3 and its mutants were linear for more than 30 minutes. The proteins were precipitated by centrifugation at 7000 × g for 20 minutes. The samples were then concentrated by evaporation, to a volume of 200 μl using a Savant speed-vac, placed on the lowest temperature setting. Control incubations were performed in the absence of NADPH or enzyme. It has been previously demonstrated that thioglycolic acid can trap oxygenation-dependent intermediates of 3-methylindole (38). Thioglycolic acid alkylates the intermediate 3-hydroxy-3-methylindolenine at the C-2 position, and the conjugate then undergoes a cyclic esterification to form a thio-lactone adduct. The oxygen atom introduced by the P450 oxygenation remains trapped within the lactone ring (Figure 2). In order to determine if any NADPH-dependent oxygenated metabolites were formed in the metabolism of 3MI by the mutant enzymes, 4 mM thioglycolic acid was added to the incubations as an alternative trapping agent. The rest of the incubation conditions were unchanged.
Figure 2.
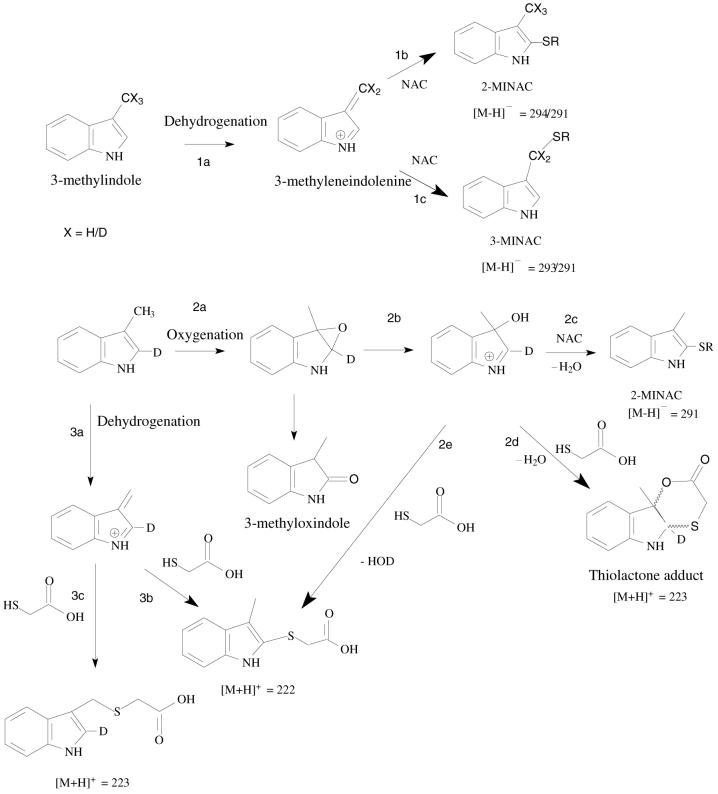
Thiol trapping of oxygenation- and dehydrogenation-dependent reactive intermediates of 3-methylindole. Pathways 1a, 1b, and 1c illustrate formation of the dehydrogenated reactive intermediate, 3-methyleneindolenine, and trapping of the electrophile with NAC. X = H corresponds to 3MI, X = D corresponds to 3MI-d3, and X = HD2 corresponds to 3MI-d2. [2-2H]-3-Methylindole is the deuterium-labeled substrate for pathways 2a and 3a. Pathways 2a, 2b, 2c, 2d, and 2e illustrate formation of the oxygenated reactive intermediate, 3-hydroxy-3-methylindolenine, and trapping of the electrophile with NAC or thioglycolic acid. 3-Methyloxindole is formed from ring opening and hydride shift of the epoxide (6). Pathways 3a, 3b, and 3c illustrate formation of the dehydrogenated intermediate, 3-methyleneindolenine, and trapping with thioglycolic acid.
Pathway 1a→1b - Formation of 3-methyleneindolenine from 3MI, followed by nucleophilic attack by N-acetyl-L-cysteine at the 2 position to form the C-2 adduct.
Pathway 1a→1c - Formation of 3-methyleneindolenine from 3MI, followed by nucleophilic attack by N-acetyl-L-cysteine at the 3 position to form 3MINAC.
Pathway 2a→2b→2c — Formation of the C-2 adduct by ring opening of the 2,3-epoxide to form 3-hydroxy-3-methylindolenine, followed by nucleophilic attack at the 2-position by N-acetyl-L-cysteine and subsequent dehydration with the loss of the C-2 deuterium.
Pathway 2a→2b→2d - Thioglycolic acid trapping of the oxygenation-dependent reactive intermediate 3-hydroxy-3-methylindolenine and cyclic esterification of the conjugate to form a thiolactone adduct. The cyclic thiolactone adduct retained the C-2 deuterium.
Pathway 2a→2b→2e — Trapping of the oxygenated metabolite, 3-hydroxy-3-methylindolenine with thioglycolic acid to form a 2-position adduct that dehydrates to aromatize with a loss of HOD.
Pathway 3a→3b - Thioglycolic acid trapping of the dehydrogenated intermediate and formation of the 2-position thioglycolic adduct. The adduct loses the C-2 deuterium during tautomerization to aromatize.
Pathway 3a→3c- Thioglycolic acid trapping of the dehydrogenated intermediate and formation of the 3-position thioglycolic adduct. The adduct retains the C-2 deuterium.
Liquid Chromatography/Mass Spectrometry (LC/MS)
Metabolites were analyzed on a Finnigan LCQ Advantage Max quadrupole ion trap mass spectrometer, interfaced with a Finnigan Surveyor LC pump and autosampler. Metabolites were separated on a Phenomenex Luna C18 column (5μm, 250 mm × 2.0 mm) using an organic phase of acetonitrile and an aqueous phase containing 0.1% (v/v) formic acid at a flow rate of 0.4 ml/min. The linear gradient was: 10% to 35% organic in 6 minutes, 50% organic at 10 minutes, 55% organic at 15 minutes, 95% organic at 19 minutes, isocratic for 2 minutes and back to 10% organic at 25 minutes. The column was equilibrated for 10 minutes between injections. Mass analysis was accomplished with ESI-SIM by monitoring ions at m/z 291 [M-H]- for the thiol adduct (3MINAC) of the dehydrogenated metabolite, 3-methyleneindolenine, and m/z 210 [M+H]+ for the internal standard phenyloxindole; and using ESI-SRM to monitor ions at m/z 147 [M+H]+ for the oxygenated metabolite, 3-methyloxindole under the following conditions: capillary temperature, 215°C; source voltage, 5.0 kV at 80 μA with the sheath gas pressure set at 50 psi N2. In addition, the amounts of 3MINAC and 3-methyloxindole were calculated by the ratio of their peak areas to the internal standard, compared to a standard curve. All quantifications were performed using Finnigan Xcaliber LC QUANT software.
LC/MS/MS of the Thioglycolic Acid Conjugate
Liquid chromatography was performed as previously described. Mass analysis was accomplished by monitoring ions using ESI-SIM/MS/MS for the parent ions of m/z 222 and 223. Positive daughter ions were monitored.
In addition, ionization was also accomplished by atmospheric pressure chemical ionization. Tandem mass spectrometry of the thioglycolic acid adduct was performed using the first quadrupole to select the parent ion, m/z 222, for collision-induced dissociation. The radio-frequency quadrupole was used as a collision cell, using Argon as the collision gas. Positive daughter ions were monitored by the third quadrupole.
Homology Modeling of P450 2F3 and 3MI Docking Studies
the P450 2F3 model was generated using MODELLER (8v2). Briefly, an alignment of the 2F3 sequence, based on the chain A of CYP2A6, CYP2C8 and CYP2C9 templates was generated using the command “salign” in MODELLER, and then the 3D model of 2F3 was produced automatically by MODELLER using its “automodel” class. A total of 15 models were generated. The final model was picked by using the value of the MODELLER objective function (an arbitrary structural energy number) with primary emphasis on the best stereochemical parameters. Both DOPE potential in MODELLER and PROCHECK (v.3.5.4) were used to evaluate the stereochemical quality of the model, and these results were compared with the three original templates.
AutoDock (version 3.05, Scripps Research Institute, La Jolla, CA) was used to predict the binding modes of 3MI in the active site of the P450 2F3 homology model. The 3MI structure was constructed by HyperChem 7.5 (Hypercube, Inc., Gainesville, FL), with the energy minimized using the molecular mechanics method (MM2), and then prepared by AutoDockTools (ADT, Scripps Research Institute, La Jolla, CA) with Gasteiger atomic charges assigned and flexible torsions defined. The P450 2F3 model was prepared by adding polar hydrogens, partial charges, and solvation parameters. AutoGrid (version 3.06) function in the AutoDock package was used to define the active site boundary of P450 2F3 by generating a grid box (30×30×30 Å) that was large enough to encompass the active site. Five hundred searches were executed by Lamarckian genetic algorithm (LGA) in AutoDock to optimize the 3MI conformation and orientation in the active site. The lowest-energy poses were chosen for analysis. Both AutoGrid and AutoDock were run within the Cygwin UNIX Shell (Red Hat, Inc., Raleigh, NC) on Microsoft Windows, and the results were visualized with ADT and PyMol software (DeLano Scientific LLC, Palo Alto, CA).
Synthesis of Deuterium Isotopologs of 3MI
the 3MI-d3 (CD3-methyl labeled 3MI) and 3MI-d2 (CHD2-methyl labeled 3MI) deuterated analogues of 3MI (see Figure 2 for structures) were synthesized by the general method of reduction of indole derivatives with LiAlD4 (39). 3-Methylindole with a deuterium substitution at the C-2 position, [2-2H]-3-methylindole, was previously synthesized in our laboratory (6). The isotopic purities of the synthesized deuterium compounds were determined using Brauman’s least-squares method for isotopic analysis, as described by Korzekwa et al. (40).
Noncompetitive Intermolecular Deuterium Isotope Effect
Separate experiments with 3MI and 3MI-d3 substrates were carried out. Briefly, 100 pmol of P450 2F3 or the D361T mutant with 300 pmol NADPH cytochrome P450 reductase microsomes (Gentest) and 100 mM potassium phosphate buffer (pH 7.4) for 10 minutes at room temperature. After preincubation, 4 mM NAC and 3MI or 3MI-d3 concentrations ranging from 10-200 μM were added to a total volume of 250 μl. NAC was added to the incubation in order to trap the highly reactive and unstable 3-methyleneindolenine intermediate as its thiol adduct, 3MINAC. The mixture was allowed to pre-incubate for an additional 3 minutes at 37°C before initiating the reaction with 2 mM NADPH. Incubations were carried out at 37°C for 20 minutes and the reaction was stopped by adding an equal volume of ice-cold acetonitrile. The proteins were precipitated by centrifugation at 7000 × g for 20 minutes. After protein precipitation, 4 nmole of the internal standard, 3-phenyloxindole (3POI), was added to each vial and the samples were then concentrated by evaporation to a volume of 200 μl using a Savant speed-vac, placed on the lowest temperature setting. The samples were then analyzed by both HPLC and LC/MS as described below. The experiment was repeated 2 more times on different days.
Intramolecular Deuterium Isotope Effects
the isotope effects were determined in a similar manner to those described above, with the following exception, 0.15 mM of 3MI-d2 was used as the substrate for the incubations. Concentrations higher than 0.15 mM inhibited the formation of 3MINAC, presumably because 3MI is a substrate inhibitor. In addition, the reactions were stopped after 12 minutes, within the linear portion of the rate of product formation. The samples were analyzed by LC-MS as described below.
High Pressure Liquid Chromatography Analysis
Initial analysis of the samples was conducted using high-pressure liquid chromatography, with UV detection. The samples were injected using a single sample injection port, into Beckman, 126 series binary pumps, at a flow rate of 1 ml per minute. The metabolites were separated by a Phenomenex (Torrance, CA) Luna column, 5μ, C18 IP (250 mm × 4 mm, 5 micron), and an instrument controlled gradient. The gradient conditions were as follows: 10% to 35% organic after 5 minutes, 50% organic at 10 minutes, 55% organic at 15 minutes, 95% organic at 19 minutes, isocratic for 2 minutes and back to 10% organic at 25 minutes. The phases were acetonitrile and ammonium acetate (10 mM), respectively. The organic and aqueous phase metabolites were detected by UV absorbance at 254 nm using a Beckman 166 detector.
Liquid Chromatography-Mass Spectrometry
Metabolites were analyzed on a Finnigan LCQ Advantage Max quadrupole mass spectrometer composed of a Finnigan Surveyor LC pump and autosampler and coupled to a diode-array UV-Vis detector. Separation was achieved on a Phenomenex Luna C18 column (5μm, 250 mm × 4.0 mm) using a linear gradient and 0.4 ml/min flow rate as described above. The column was re-equilibrated for 10 minutes between injections. Metabolites were detected and verified by UV absorbance at 280 nm, the maximum absorbance of 3MINAC. Mass analysis was accomplished by monitoring ions by ESI using the selective ion monitoring mode under the following conditions: capillary temperature, 215°C; source voltage, 5.0 kV at 80 μA with the sheath gas pressure set at 50 psi N2. The mass spectrometer was operated in the negative-ion mode to monitor the thiol adduct (3MINAC) of 3-methyleneindolenine at m/z 291 [M - H] - and m/z 293 [M - H] -, for the intermolecular deuterium isotope studies (incubations with 3MI and 3MI-d3). For the intramolecular deuterium isotope studies (incubations with 3MI-d2), ions with m/z 292 [M - H] - and m/z 293 [M-H] - were monitored. In addition, ions with m/z 210 [M + H]+ for the internal standard, phenyloxindole was monitored in all incubations. The isotopomers were quantified by peak area ratio comparison with a 3MINAC standard curve using 3POI as the internal standard. All quantifications were performed using Finnigan Xcaliber LC QUANT software.
Calculations
Kinetic Deuterium Isotope Effect
The velocity of 3MINAC formation (nmol 3MINAC/min/nmol P450) from separate incubations with 3MI and 3MI-d3 was calculated from a 3MINAC standard curve using 3POI as the internal standard. A Michaelis-Menten kinetic analysis with the velocity vs. substrate concentration was performed and the data points were plotted using KaleidaGraph. A Lineweaver-Burke graph of 1/nmol 3MINAC/min/nmol P450 vs. 1/[3MI] was also plotted and the Vmax and the Km determined from the y and x intercept respectively. In addition, the rate constant, k = Vmax/Km was calculated for each product and the intermolecular deuterium isotope effect (kH/kD) was then determined by dividing the rate constant kH, for the unlabeled 3MI by the rate constant kD, for the deuterated analog (3MI-d3).
The intramolecular deuterium isotope effect was calculated by dividing the ion intensity obtained from the metabolite peak of m/z 293 (di-deuterated metabolite that lost the hydrogen on the methyl group) by the ion intensity of the peak with m/z of 292 (mono-deuterated metabolite that lost one of the two deuteriums on the methyl group). The d2/d1 ratio thus obtained was corrected for the statistical probability for the removal of a hydrogen or deuterium by dividing by 0.5, the expected ratio in the absence of an isotope effect. The contribution of each isotopic peak, based on the mass scan of unlabeled standard was determined by monitoring the primary ion for each isotope species. The ion intensities were corrected for the ion overlap due to other isotopic species, by subtracting the percent contribution of each isotope from the metabolite of interest. This then gives a series of simultaneous equations, which was solved by the least-squares analysis method of Brauman (40).
RESULTS
Mutation Strategies
In order to maximize changes in product selectivity, we initially mutated two residues simultaneously for most of the SRS sites. The SRS 1 double mutation produced a holoenzyme, but the CO-reduced difference spectrum absorbed only at 420 nm, indicating that the enzyme was probably denatured during the expression process. Double mutations in SRS 4, 5, and 6 were viable, and the double mutations in SRS 5 and 6 regions introduced oxygenase activity. Therefore, we subsequently chose to mutate individual residues in SRS 5 and 6, along with a few additional sites that were potentially important residues according to our sequence analysis and molecular modeling studies. The single mutation of Y101L in SRS 1, and single mutations, G214L and E215Q, were also produced (see Discussion for mutation strategies). The results for the double and single mutants are presented together for clarity.
Construction and Expression of Mutant CYP2F3 Enzymes
Molecular models revealed that the tyrosine at position 101 was closely aligned with substrates in the active site of 2F3, and amino acid alignments divulged that the 2E1 enzyme substituted leucine, a residue with profoundly different chemistry. However, the SRS 1 single mutant, Y101L, did not produce a protein with a viable P450 spectrum, despite multiple attempts with varying temperatures, induction protocols, etc. Similarly, attempts to make Y101F mutant were not successful. Both double and single amino acid mutations to SRS regions 1, 4, 5 and 6 were successfully introduced, as confirmed by full-length sequence analysis within the P450 cDNA region of the expression vector pCW2F3. High P450 expression levels, similar to the wild type enzyme were achieved for single mutations, D361T (120 nmol/500ml culture) and E215Q (102 nmol/500ml culture) mutants. The P450 expression levels were approximately 30-40% lower for the SRS 6 mutants, S474H (80 nmol/500ml culture) and S475I (69 nmol/500ml culture) and for the G214L mutant (65 nmol/500ml culture). The levels of P450 expression achieved for the double mutants were typically lower than that achieved for the single mutations. Expression levels for the SRS 4 double mutant enzyme (H293A, N294D) was approximately 50 nmol/liter. Expression levels for the SRS 5 double mutant (A360I, D361T) were between 10 and 15 nmol/liter of culture, while the enzyme content for the SRS 6 double mutant (S474H, S475I) was between 6 and 10 nmol/liter culture.
Double mutations in SRS 1 did not produce a spectrally active enzyme. In addition, spectrally active P450 was not obtained for the SRS 5 single mutant (A360I). Since the mutants were fully sequenced and the sequence verified before expression, the lack of expression of spectrally active enzyme was not due to any unwanted mutations in the cDNA. Earlier P450 expression studies have shown that low P450 yields in cultures could be due to a decrease in the inherent levels of plasmid expression in cells. Such a decrease in plasmid expression levels could occur due to the toxicity and/or loss of the expression vector from cells, and therefore it was necessary to use freshly transformed cells for P450 expression. This phenomenon has been observed in P450 2B4 expression in JM109 (DES) pLys cells and P450 2E1 expression with the PJL2 plasmid (41). Extremely low expression levels were obtained when old DH5-α transformants were used. However, significant increases in expression levels were obtained by using freshly transformed cells (42). To determine if toxicity of the expression vector was a factor in the lack of P450 expression in the SRS 1 mutants and A360I mutant, we used fresh DH5-α transformants for the expression studies. However, in spite of several attempts with freshly transformed cells, we still did not see any P450 expression in these particular mutants. Therefore, lack of spectrally active P450 in these mutants is not an artifact of the toxicity and/or loss of the expression vector. It seems reasonable to assume that mutations to this region of CYP2F3 may introduce inherent instability to this specific isozyme. Additional study is required to explain the lack of expressed, functional enzyme for these mutants.
Characterization of Single Mutant Enzymes by LC/MS - Detection of Dehydrogenated Metabolites
The dehydrogenated metabolite, 3-methyleneindolenine, was trapped as its N-acetyl-L-cysteine adduct (3MINAC) and then analyzed. The detection and analysis of all metabolites were performed using LC/MS. As expected, the N-acetyl-L-cysteine adduct of the methylene imine (3MINAC, Figure 2, pathway 1a→1c) was detected in incubations containing CYP2F3. Interestingly, the formation of 3MINAC was increased in both the SRS 5 (D361T) and SRS 6 (S474H) mutants. No detectable formation of 3MINAC was observed with the S475I, G214L, and E215Q mutants. The detection and verification of this adduct was based on comparison with pure 3MINAC standard, previously synthesized in our laboratory.
In addition to the 3MINAC peak, another major peak with an m/z 291 was detected. Although the formation of this peak was seen with incubations of all of the enzymes, the intensity of the peak varied widely between wild type 2F3 and the different mutants. A 3.5-fold increase in the intensity of this peak, compared to the wild type P450 2F3 was observed for the S474H and the G214L mutants. In addition, 1.5-2 fold increases were observed for the D361T, S475I and E215Q mutants. In an attempt to identify this peak, incubations containing deuterium isotopologs of 3MI were carried out. When [2-2H]-3-methylindole was used as substrate, the major peak had a deprotonated m/z of 291, indicating the loss of the 2-position deuterium atom. Based on the loss of a deuterium from this adduct, this peak could either be the C-2 position adduct of NAC, formed through a dehydrogenation pathway (Figure 2, pathways 1a →1c), or an oxygenation metabolite formed via epoxidation at the 2, 3 position of 3MI, followed by a nucleophilic attack by NAC at C-2 and subsequent dehydration (Figure 2, 2a→2b→2c), (38). Both metabolites would lose the deuterium atom. The NAC could possibly add to either the epoxide or its ring-opened iminium electrophile; both pathways would produce the thioether, which would dehydrate to form an indole with its C-2 adduct. These pathways were described previously (38).
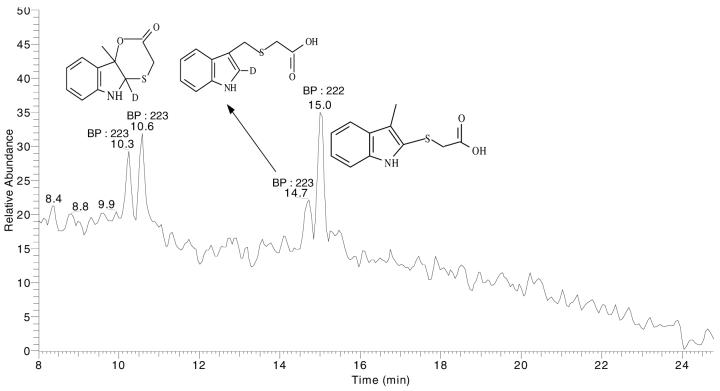
As described previously, it has been demonstrated that thioglycolic acid can trap oxygenation-dependent intermediates of 3MI (38). In further attempts to identify the C-2 position adduct of 3MI, and to track the pathway of its formation, incubations that contained thioglycolic acid and [2-2H]-3-methylindole were carried out. The adducts were analyzed with ESI. Figure 3 shows a representative ion chromatogram found in the incubation mixtures. Four NADPH-dependent peaks were formed, and two of these peaks eluted closely to each other, with retention times of 10.3 and 10.6 minutes. Based on their m/z ratios of 223, these peaks were presumed to be diastereomers of the cyclic thiol-lactone adduct of 3-hydroxy-3-methylindolenine. As shown in Figure 2, (pathway 2a→2b→2d), this adduct would retain the 2-position deuterium. These peaks had an identical MS/MS fragmentation pattern to the previously identified thiolactone adduct (38). Another major peak, with an m/z of 222, eluted at 15 minutes. This peak could be the 2-position adduct of the oxygenated metabolite, 3-hydroxy-3-methylindolenine (Figure 2, pathway 2a→2b→2e) or the 2-position adduct of the methylene imine, which would also have a m/z of 222 (Figure 2, pathway 3a→3b). In addition, a minor thioglycolic acid adduct with a retention time of 14.7 and m/z of 223 (deuterium retention) was detected. This peak was most likely the 3-position adduct of the methylene imine (Figure 2, pathway 3a→3c).
Figure 3.
Representative total ion chromatogram for incubations of CYP2F3 with [2-2H]-3-methylindole and thioglycolic acid. Closely eluting peaks with m/z ratios of 223 (RT: 10.3 and 10.6 minutes) were the diastereomers of the cyclic thiolactone adduct of 3-hydroxy-3-methylindolenine. The peak with an m/z of 222 (RT: 15 minutes) was the C-2 adduct, formed either by trapping of the oxygenated metabolite, 3-hydroxy-3-methylindolenine, or trapping of 3-methyleneindolenine. The thioglycolic acid adduct with an m/z of 223 (RT: 14.7) was most likely an adduct at the exocyclic carbon of the methylene imine. BP = molecular ion
Incubations with 3MI-d3 were also performed. The pair of peaks eluting at 10.3 and 10.6 minutes retained the 3 deuterium atoms (data not shown). This observation was consistent with the assignment of these peaks as the diastereomeric cyclic thiolactones, formed by addition of thioglycolic acid to 3-hydroxy-3-methylindolenine at the 2 position, followed by lactone ring closure, since this adduct should retain all three deuterium atoms. The adduct peak at 15 minutes also retained all three methyl deuterium atoms. These results suggest that this adduct peak was comprised of the thioglycolic acid adduct of the oxygenated metabolite, 3-hydroxy-3-methylindolenine, at the 2 position that failed to undergo ring closure, but was subsequently dehydrated. As explained previously, all the mutant enzymes showed increased formation of this adduct. Since the SRS mutations were based on a sequence alignment with P450 2E1, an enzyme that efficiently oxygenated 3MI (15), increased formation of this metabolite in the S474H and G214L mutants could be due to increased oxygenation efficiency, leading to increased formation of 3-hydroxy-3-methylindolenine. When 3MI-d3 was employed as the substrate, the thioglycolic acid adduct at the exocyclic methylene position (elution time of 14.7 minutes) disappeared. These results could have been caused by “metabolic switching” from dehydrogenation to oxygenation upon deuterium substitution at the methyl position.
Characterization of Single Mutant Enzymes by LC/MS - Detection of Oxygenated Metabolites
The oxygenated metabolite, 3-methyloxindole, is stable and was directly analyzed by LC/MS. As expected, 3-methyloxindole was not detected in incubations containing 2F3. Interestingly, 3-methyloxindole was detected in incubations containing the SRS 5 (D361T) and the SRS 6 (S474H and S475I) mutant enzymes. This peak was verified based on the retention time of pure 3-methyloxindole. No formation of 3-methyloxindole was seen in the G214L and E215Q mutants. This observation was surprising because it seems reasonable to assume that both 3-hydroxy-3-methylindolenine and 3-methyloxindole arise from rearrangements of the 2,3-epoxide (Figure 2). The formation of 3-methyloxindole was found to be both NADPH- and enzyme-dependent. These intriguing results showed that single amino acid mutations, D361T, S474H, and S475I, introduced a new catalytic ‘oxygenation’ function to the enzyme.
Characterization of Double Mutant Enzymes by LC/MS
Metabolite characterization was also performed in the double mutants. Incubations were performed as described above, using N-acetyl-L-cysteine as a trapping agent for the dehydrogenated metabolite 3-methyleneindolenine. Additional incubations were also performed using the alternative thiol trapping agent thioglycolic acid. The oxygenation-dependent reactive intermediate 3-hydroxy-3-methylindoline can be trapped with this agent as described previously. Similar to the results obtained from analysis of the single mutants, an NADPH-dependent adduct with a deprotonated m/z of 291 was detected in incubations of the wild type CYP2F3 and both SRS 5 (A360I, D361T) and SRS 6 (S474H, S475I) mutants. When incubations were performed using 3-(2H3-methyl)-indole as a substrate, an adduct with a deprotonated m/z of 293 was formed. This indicated that a shift of two mass units had occurred; therefore one of the deuterium atoms was lost to a dehydrogenation (de-deuteration in this case) process. These data strongly support the conclusion that the SRS 5 and SRS 6 mutant enzymes had retained the ability to dehydrogenate the substrate 3-methylindole at the methyl position. Thioglycolic acid-trapped adducts of oxygenation-dependent 3-methylindole reactive intermediates were analyzed using atmospheric pressure chemical ionization, monitoring positive ions. As seen in the case of single mutants, incubation mixtures containing SRS 5 (A360I, D361T) and SRS 6 (S474H, S475I) double mutant enzymes, showed the presence of an NADPH-dependent peak with a protonated molecular ion with m/z of 222. This was indicative of the cyclic thiol-lactone adduct of an oxygenation-dependent reactive intermediate of 3MI. The formation of this adduct was increased in the mutant enzymes, and was not formed in the absence of NADPH. These data show that double mutations to CYP2F3 in SRS 5 and SRS 6 introduced 3-methylindole-oxygenase catalytic function to the enzyme and that 3-hydroxy-3-methylindolenine was being formed.
Quantitative and Kinetic Analysis of Dehydrogenated and Oxygenated Metabolites
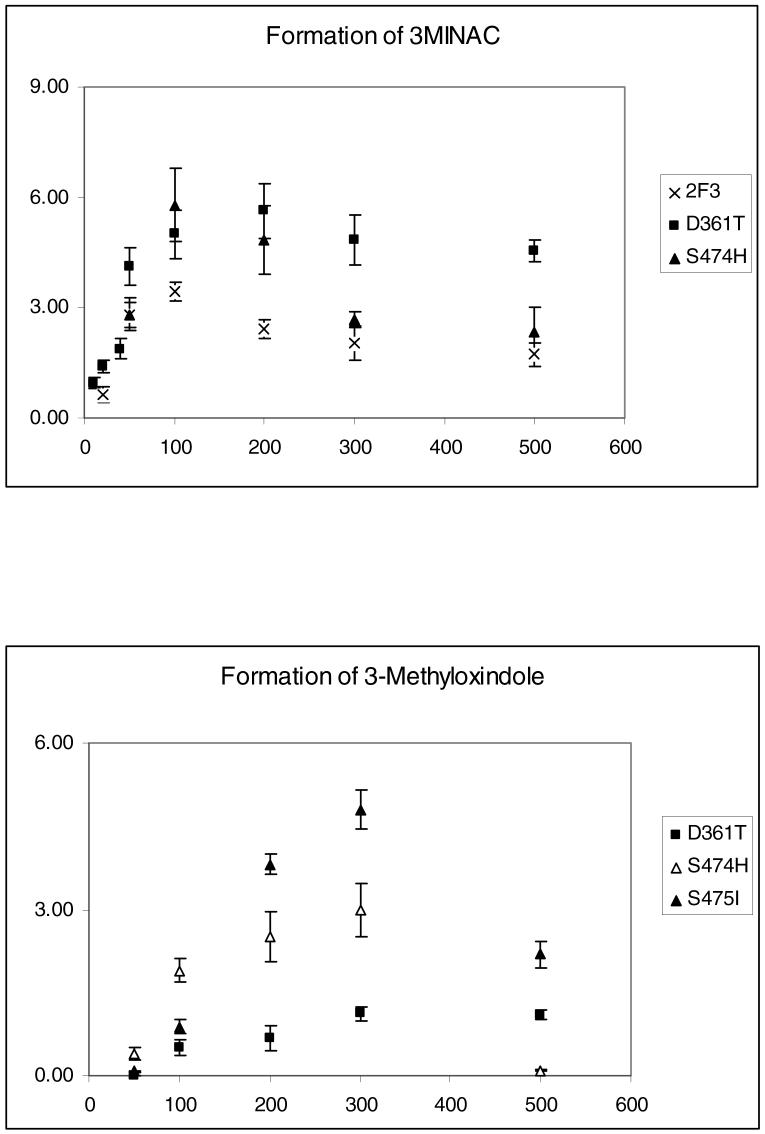
Quantitative analysis for the formation of 3MI metabolites was performed and the kinetic parameters Vmax and Km were determined. Figure 4 shows the relative formation of each metabolite in incubations with 2F3 and the mutant enzymes. The top panel showed the formation of the dehydrogenated metabolite, the 3-position adduct of NAC. Quantitation was performed based on a standard curve generated using the pure 3MINAC standard. The SRS 5 (D361T) and SRS 6 (S474H) mutants showed approximately a 2-fold increase in the formation of the dehydrogenated metabolite. Formation of 3MINAC was not detected in incubations of the S475I, G214L, and E215Q mutants. The bottom panel showed the formation of the oxygenated metabolite, 3-methyloxindole. This metabolite was quantified with a standard curve generated with a pure standard. As expected, no detectable formation of 3-methyloxindole was seen with 2F3. However, 3-methyloxindole formation was detected in the SRS 5 mutant (D361T) and both the SRS 6 (S474H and S475I) mutants, showing clearly that an oxygenase activity was introduced to the enzymes. Interestingly, 3-methyloxindole formation was not detected in the G214L and E215Q mutants, but ring-oxygenated metabolites of 3MI were produced by these mutant enzymes (data not shown). These data clearly indicate that mutations in the SRS 5 and 6 regions introduced an oxygenation function to the enzyme.
Figure 4.
Kinetics of formation of 3MI metabolites by 2F3 and the mutant enzymes. Data points represent the mean of 3 separate experiments. The X-axis is 3MI concentration in micromolar. The Y-axis is nmol product formed/min/nmol P450
Table 1 shows the kinetic constants, Vmax and Km for the formation of the metabolites. Since inhibition of enzyme activity was observed at higher substrate concentrations, to accurately estimate Vmax and Km, the data points were fitted to a substrate inhibition model and plotted using Graph Pad Prism Software. For the formation of 3MINAC peak, it was seen that the Vmax values didn’t change significantly for the D361T and S474H mutants, while the Km values were halved. The Vmax/Km values showed a 2-fold increase in the dehydrogenase activity for the D361T mutant. 3-Methyloxindole was not produced in incubations with 2F3, but it was detected in incubations with the SRS 5 (D361T) and SRS 6 (S475I) mutants and to a lesser extent with the S474H mutant. The dehydrogenase activity was completely abolished in incubations with the G214L and E215Q mutants.
Table 1.
Kinetic Constants of 3MI Metabolitesa
| 2F3 | D361T | S474H | S475I | G214L | E215Q | |
|---|---|---|---|---|---|---|
| 3MI Dehydrogenation to Form 3MINAC | ||||||
| Km (μM) | 70±13 | 36±6.0 | 35±8.4 | ND | ND | ND |
| Vmax | 6.2±1.9 | 5.8±0.5 | 6.9±0.8 | ND | ND | ND |
| V/K | 0.08±0.013 | 0.16±0.02 | 0.2±0.03 | ND | ND | ND |
| 3MI Oxygenation to Form 3-Methyloxindole | ||||||
| Km (μM) | ND | 138±16 | 122±13 | 118±20 | ND | ND |
| Vmax | ND | 1.44±0.09 | 4±0.72 | 4.9±0.74 | ND | ND |
| V/K | ND | 0.01±0.001 | 0.032±0.003 | 0.04±0.003 | ND | ND |
Purified CYP2F3 or mutant enzymes (100 pmol) and 300 pmol NADPH cytochrome P450 reductase were incubated with various concentrations of 3MI and NAC (4 mM) for 30 minutes, and the metabolites quantitated by LC/MS. The units of the Vmax values are nmol/min/nmol P450. The values represent the means ± S.D. of 3 separate experiments. ND = product not detected
Homology Modeling and 3MI Docking Studies
The purpose of the homology modeling and docking studies was predominantly to map the relative position of the investigated mutants within or around the enzyme active site, and to identify potentially important active site residues. Three-dimensional models of CYP2F3 were based on the X-ray crystal structures of P450 2A6 and 2C8 and 2C9. Initially, CYP2A6 was used as a single template to produce a model, but substrate docking was not commensurate with regioselective oxidation of 3MI because the B-C loop of CYP2F3 that contains several large aromatic residues (Y101, F104, F105 and F107) interfered with the packing of the amino acid side-chains of helices I and G. CYP2C9 was also used as a single template, but the ensuing models were incomplete, because the regions 38-46 and 215-222 of CYP2C9 were missing from the 1r9o PDB entry that was used as the template. Several active site residues, such as Y101 and E215 of CYP2F3, were not located in favored regions in the Ramachandran plot for the model. Therefore, a multiple template strategy using coordinates for 2A6, 2C9 and 2C8 was employed to improve the quality of the model by relaxing the restraints of the templates in regions where they varied. Models that exhibited the best values for the MODELLAR objective function were evaluated for good stereochemical parameters before a final model was chosen for molecular docking studies with substrate 3MI.
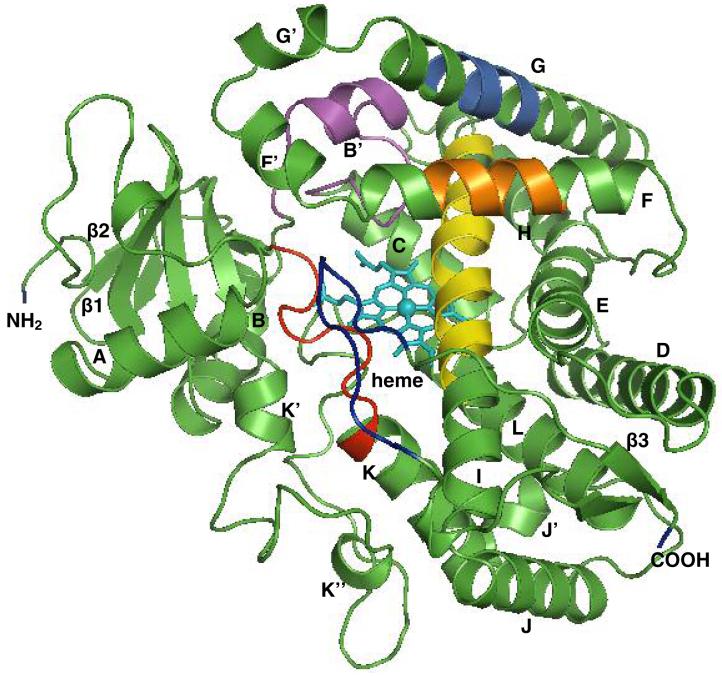
Figure 5A represents the overall fold of CYP2F3 molecule with all α-helices and β-sheets identified. The active site of CYP2F3 adapted its templates, which includes the heme forming the base, the B-C loop and helices F/G and I forming the roof, and SRS 5/6 forming the rest of the outer surfaces of the cavity. In the B-C loop of 2F3, a well structured B’ helix was observed, the similar structure as seen in P450 2A6, 2C8 and 1A2, rather than a less structured loop in P450 2C9 and 3A4. Measurement of the cavity demonstrated that the active site of CYP2F3 is smaller compared to CYP2C8/9, but larger than the active site of CYP2A6. This was expected because CPY2F3 showed a propensity to catalyze the oxidation of smaller molecules much more efficiently.
Figure 5.
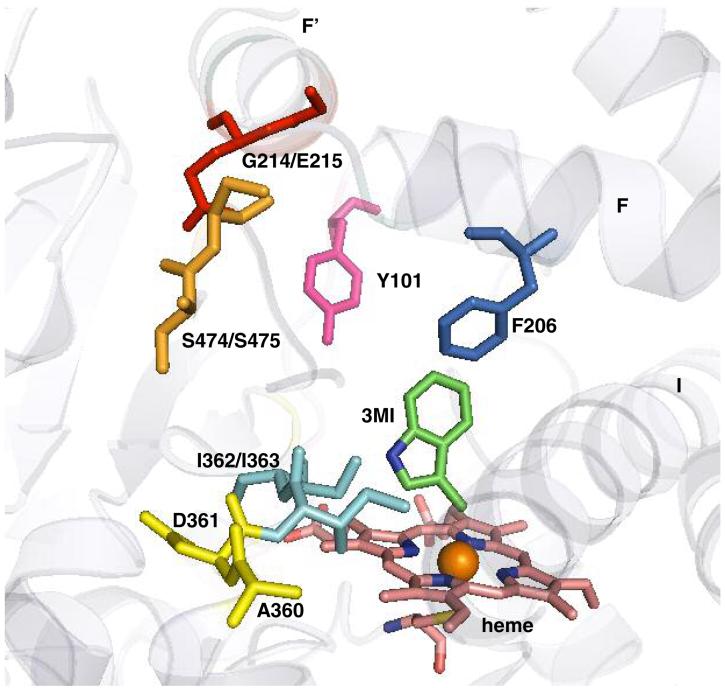
Homology model of P450 2F3 (A) and docking of substrate, 3MI, in the active site (B). A, all secondary structural α-helices and β-strands are shown in green and are labeled. Also shown are SRS 1 in magenta, SRS 2 in orange, SRS 3 in marine, SRS 4 in yellow, SRS 5 in red, and SRS 6 in blue. The heme is represented with sticks and is colored cyan. B, all secondary structures are portrayed with a transparent cartoon in the background, with the heme, 3MI, and important active site residues shown as colored sticks (the heme is pink, 3MI is green, SRS 5 mutated residues A360 and D361 are yellow, SRS 6 mutated residues S474 and S475 are orange, mutated residues G214 and E215 are red, residues I362 and I363 are cyan, Y101 is pink, and F206 is blue). The figures were produced with PyMOL.
Docking studies were performed, and the most energetically favorable 3MI orientation in the active site of the CYP2F3 model, obtained by automated molecular docking studies, is shown in Figure 5B. This pose positions the 3-methyl hydrogen atom of 3MI closest to the heme iron with a distance of 4.6 Å (Fig. 5B). This low energy orientation predicts preferential dehydrogenation at the methyl position for CYP2F3, a result identical to the experimental results with recombinant CYP2F3. Other 3MI orientations were also indicated, but they were less energetically favorable. These higher energy orientations would theoretically predict the oxidation of the benzene ring of 3MI to form phenols. Again, the docking studies precisely correlated with the experimental data, because phenols have not been identified from incubations of recombinant CYP2F3 with 3MI.
CYP2F3 residues Phe206 on helix F (SRS 2), residues Thr302, Gly298 and the adjacent peptide bond with Phe297 of Helix I (SRS 4), as well as residues Ile363 and Leu367 (SRS 5) provide van der Waals contacts that restrain the substrate in this position. The positions of the mutated residues in SRS 5 (D361), SRS 6 (S474 and S475) and residues G214 and E215 relative to the substrate binding site are depicted in Fig. 5. The D361 residue is approximately 12 angstroms away, while residues S474 and S475 are approximately 11 angstroms away from the bound 3MI.
In addition to the SRS 4 and 5 residues, mapping of an SRS 1 residue Y101 is also shown. This residue maps directly over the bound substrate and the catalytic heme. Tyrosine residues have been shown to participate in the P450-mediated radical transfer reactions, so it appeared that this phenolic group could play a vital role in stabilization of the putative benzylic indolylmethyl radical intermediate. Unfortunately, despite numerous attempts, mutation of the tyrosine residue produced a protein that did not form a reduced CO-binding spectrum. It is possible that Y101 serves a vital role to maintain the stability of the enzyme.
Synthesis of Deuterated Analogs of 3MI
The two methyl-deuterated analogs of 3MI were successfully synthesized by reduction of indole derivatives with LiAlD4. Isotopic purity of the deuterated analogs as determined by LC/MS and the least square analysis method was: 3MI-d2: 0.8% d1, 99.2% d2 and no detectable traces of d0 or d3; 3MI-d3: 0.59% d2, 99.4% d3 and no traces of d0 or d1.
Non-competitive Intermolecular Deuterium Isotope Studies
The non-competitive intermolecular deuterium isotope studies were conducted by separate incubations with 3MI and 3MI-d3 substrates. The dehydrogenated metabolite, 3-methyleneindolenine, was trapped by NAC, forming 3MINAC, as explained in the Materials and Methods Section and analyzed both by HPLC and LC/MS. The kinetic parameters, Vmax and Km for the formation of 3MINAC, from the d0 and 3MI-d3 substrates is shown in Table 2. Deuterium substitution decreased the Vmax from 3.6 ± 0.8 nmol MINAC/min/nmol P540 to 1.9 ± 0.31 3MINAC/min/nmol P450. Decreases in Vmax values upon deuterium substitution is an indication of an isotope effect. The Km values for the metabolism of 3MI-d3 were decreased from 42 ± 15 μM to 24 ± 8 μM. The catalytic efficiencies, Vmax/Km, for 3MINAC formation from both the unlabeled 3MI and the deuterated analog (3MI-d3) were calculated and the overall intermolecular kinetic deuterium isotope effects were obtained by dividing the rate constant for the unlabeled 3MI by the rate constant of the deuterated substrate (kH/kD). An intermolecular isotope effect of 1.1 was obtained for CYP2F3. As explained previously, the retention time of the 3MINAC peak was close to that of the C-2 adduct peak. The C-2 adduct peak is most likely formed through a combination of the oxygenation and dehydrogenation pathways and therefore the inter-molecular isotope effect was also calculated by analyzing the 3MINAC peaks using LC/MS. This was done in order to assure that the correct peak was being quantified. Similar results were obtained in these studies as well. The Vmax values decreased from 5.6 nmol 3MINAC/min/nmol P540 to 3.8 nmol/min/nmol P450 for CYP2F3 (data not shown). Although such a low isotope effect usually indicates that an initial hydrogen abstraction is not the rate limiting step in the dehydrogenation of 3MI, inter-molecular kinetic isotope effects are very prone to masking effects (43). Therefore, the results are not conclusive.
Table 2.
Deuterium Isotope Effects of CYP2F3
| Intermolecular Isotope Effect | |||||
|---|---|---|---|---|---|
| Substrate | Km1 | Vmax | VH/VD2 | k3 | kH/kD4 |
| 3MI-d0 | 42 ±15 | 3.6 ±0.8 | 1.9 | 0.09 | 1.1 |
| 3MI-d3 | 24 ±8 | 1.9 ±0.31 | 0.08 | ||
| Intramolecular Isotope Effect | |||||
|---|---|---|---|---|---|
| Enzyme | m/z 293 (Hydrogen abstraction) |
m/z 292 (Deuterium abstraction) |
Isotope Ratio5 |
Isotope Effect6 |
Isotope Effect7 |
| 2F3 | 21045 | 6963 | 3 | 6 | 6.8±0.75 |
| 28219 | 7480 | 3.8 | 7.5 | ||
| 17712 | 5209 | 3.4 | 6.8 | ||
Intermolecular Km and Vmax values were based on the rates of 3MINAC formation from separate incubations with unlabeled 3MI or 3MI-d3. The results show mean and standard deviations from 4 separate experiments. Intramolecular deuterium isotope effects were determined with 3MI-d2 as the substrate. The values are arbitrary peak areas for the respective ions. Each row corresponds to the results of one experiment.
Km in μM
Ratio of the Vmax (nmol 3MINAC/nmol P450/min) with 3MI as the substrate to Vmax with 3MI-d3 as the substrate
Catalytic efficiency k = Vmax/Km
Ratio of the intensity of the peak that lost the hydrogen to the peak that lost the deuterium
Isotope ratio/0.5 — the theoretical ratio in the absence of an isotope effect
Mean ± S.D. of the isotope effect from the isotope effects of each individual experiment
Intramolecular Deuterium Isotope Studies
Intramolecular deuterium isotope studies were carried out by incubations with 3MI-d2. This substrate has two positions that are equivalent in all respects except for deuterium substitution. The ion peak intensity of the di-deuterated metabolite (metabolite that lost the hydrogen on the methyl group), was divided by the ion peak intensity of the mono-deuterated metabolite (metabolite that lost the deuterium on the methyl group) and this value was also divided by 0.5 to obtain an intramolecular deuterium isotope effect of 6.8 for 3MINAC metabolism by 2F3 (Table 2). The intramolecular deuterium isotope effect values given above were obtained after correcting for the ion overlap, and percentage deuterium incorporation by the Brauman’s least square analysis method. This relatively high intramolecular isotope effect was observed for both the wild-type CYP2F3 and the D361T mutant. Metabolic switching caused by an isotope effect for the mutant was not found. Thus, it is possible that uncoupling of the catalytic cycle of the mutant enzyme occurred to produce excess water. These results indicate that C-H bond breakage through an initial hydrogen abstraction is most likely the rate limiting step for the wild-type and mutant enzymes. In addition, the D361T mutant catalyzed both dehydrogenation and oxygenation, but introduction of oxygenation did not appear to change the mechanism of dehydrogenation.
DISCUSSION
Three dimensional homology models of the major xenobiotic metabolizing P450 enzymes have been constructed and used in conjunction with site-directed mutagenesis studies to elucidate the molecular determinants of substrate specificity. In addition, substrate docking studies within the active site of the enzymes have helped identify key amino acid residues involved in substrate binding and catalysis and aided in the analysis of P450 inhibition and activation (44). The concept of substrate recognition sites by Gotoh (20) has provided an excellent roadmap to understand the basis of P450 specificity. Many of the site-directed mutagenesis studies today focus on the SRS regions. Molecular docking studies can predict changes in regio- and stereo-specificity of substrate binding upon site-directed mutagenesis and therefore, the results of site-directed mutagenesis studies are now being extensively validated using homology modeling and substrate docking studies (45, 46).
Mutations at positions 477 and 480 of SRS 6 region of 2B5 affected the metabolism of androstenedione and progesterone, while mutation at position 209 (SRS 4) altered the regiospecificity of progesterone hydroxylation (47). Studies on the metabolism of 7-alkoxycoumarins by CYP2B1 and its mutant enzymes showed that residues 363 (SRS 5) and 478 (SRS 6) were important in substrate orientation (32). In addition, residues from SRS 6 (477, 478, 480) were shown to be important for substrate selectivity (48, 49). Structure-activity relationships of CYP2C9 were also extensively characterized using mutagenesis and homology modeling. Mutations in SRS 1 (V113L/F114L) showed increased enzyme activity, while all mutations in SRS 5 (L362A/L362I) exhibited lower enzyme activity (50).
The site-directed mutagenesis presented in this work focused initially on double mutations in SRS 1, 4, 5, and 6 of P450 2F3. Double mutation to SRS 4 (H293A and N294D in the I helix) did not change the nature of the catalytic activity of 2F3, while double mutations in SRS 5 (A360I and D361T) and 6 (S474H and S475I) introduced 3-methylindole oxygenase activity to CYP2F3. This was demonstrated by the production of an oxygenation-dependent reactive metabolite by both mutant enzymes, and by the production of 3-methyloxindole.
Since double mutations in SRS 5 and 6 introduced 3MI oxygenation, we mutated each of these sites individually to determine which residues were critical for this dramatic alteration in substrate oxidation. The specific mutations were A360I, D361T, S474H and S475I. In addition, both double and single mutations in SRS 1 were also evaluated. As shown in Figure 5, the tyrosine residue (Y101) lies within the B-C loop and maps directly over the bound substrate, and it is conserved in CYP2F1 and CYP2F3.
The last two single mutations corresponded to two residues that mapped just to the C-terminal side of the SRS 2 region. The Q215 residue is highly conserved in all of the P450 family 2 genes, except for 2F3 and 2F1, where it is replaced by a glutamic acid, and we reasoned that introduction of an acidic functional group might have important consequences. Thus, we mutated the glutamate to the corresponding residue of CYP2E1, glutamine.
In addition, we chose to mutate the neighboring glycine (position 214). The corresponding residue in CYP2E1 and other CYP2 sub-family members is a highly conserved leucine. We opined that increases in the side chain length may alter/hinder substrate binding due to increased van der Waals overlap, because G214 is located in the F' helix, an important component of P450 enzymes. Changes in side-chain length have previously been shown to alter substrate binding and catalysis in 2B1 (32). The highly conserved glutamic acid and glycine residues of CYP2F1 and CYP2F3 could be vital to confer dehydrogenation activity to the enzymes.
In spite of several attempts, spectrally active holoenzymes were not obtained for the SRS 1 double (G110D, N111R), SRS 1 single (Y101L) and the SRS 5 single (A360I) mutants. It seems likely that perturbations to these regions affect enzyme stability. However, the site-directed single mutations of P450 2F3 in SRS regions 5 (D361T) and 6 (S474H and S475I), introduced significant oxygenase activities. Altered enzyme catalysis was indicated by the formation of the oxygenated reactive metabolite, 3-hydroxy-3-methylindolenine by these enzymes. In addition, the stable oxygenated metabolite, 3-methyloxindole, was formed in the three SRS 5 and 6 mutations but was not detected in incubations with 2F3. Thus, the introduction of oxygenase activity by the single mutants corresponded precisely to the results from the double mutant enzymes.
Docking studies with 3MI showed that although the SRS 5 and 6 residues were outside the normal sphere of active site residues, they produced profound changes in substrate catalysis, possibly through stereochemical interactions with their neighboring residues within the active site of CYP2F3, such as the downstream residues I362 and I363 in SRS 5. To better understand these results, molecular models of the mutant enzymes would be helpful, but unfortunately the process to rationalize and validate them is difficult using current homology modeling techniques. However, it can be postulated that these mutations change the substrate orientation in such a way so that additional metabolic sites are opened up, especially on the indole ring. Interestingly, in addition to an increase in the oxygenase function, the 3MI dehydrogenation capability was increased 2-fold in the D361T mutant. It is possible that this specific mutation increases the van der Waals contacts within the active site, causing the stabilization of the binding orientation and decreased substrate mobility, leading to the overall increase of enzyme activity. Another reasonable explanation for increased enzyme activity of the mutant is allosteric cooperativity. Binding of an allosteric effector — potentially a substrate molecule — at a site near the aspartate residue at 361 might be more facile with threonine at this residue, and might produce higher rates of dehydrogenation of 3MI at the active site of the mutant than the native enzyme. However, allosteric cooperativity would probably be operative for both the normal and mutant enzymes.
The S475I, G214L, and E215Q mutant enzymes completely abolished the dehydrogenase activity of P450 2F3. 3MI docking in the active site of CYP2F3 showed the 3-methyl group pointing directly at the heme, which is an orientation that would considerably favor the dehydrogenation pathway. Since the proximity of the methyl group to the catalytic heme may be required for efficient metabolism through a dehydrogenation pathway, it is likely that the orientation of the substrate changed in these mutant enzymes. The site-directed mutagenesis and homology modeling studies have identified specific amino acid residues within the SRS regions of 2F3 that direct the catalysis of 3MI. These studies clearly show mutations in SRS 5 (D361T) and 6 (S474H and S475I) introduced oxygenase activity. In addition, residues S475, G214 and E215 are probably important in maintaining optimum substrate orientation conducive for dehydrogenation, since site-directed mutagenesis of these residues completely abolished the dehydrogenase activity of the enzyme. Many recent studies have shown that P450 enzymes have highly dynamic tertiary structures, so it is probable that more distal residues such as the serine at 474, aspartate at 361, and glutamate at 215 would alter interactions of proximal residues such as tyrosine at 101 and phenylalanine at 206, to change dehydrogenation specificity.
The only mutation that simultaneously lost dehydrogenation and gained oxygenation was S475I. It is unlikely that the steric effects of an isoleucine side chain replacing a serine residue at a position outside the active site would lead to such a major catalytic change. Rather, the loss of a hydrogen bond donor at this site may have abolished a hydrogen bond network and decreased the stabilization of radical or cationic intermediates. We opine that stabilization of the 3-methylindolyl radical or cation, formed by hydrogen atom abstraction or by hydrogen atom abstraction followed by one-electron oxidation, could be a major mechanism for dehydrogenation specificity. Stabilization of the cationic benzylic intermediate by water molecules would be the most attractive and consistent mechanism. A similar mechanism of dehydrogenation, i.e. cationic intermediate stabilization, has been postulated (51).
The non-competitive intermolecular deuterium isotope studies revealed a decrease in the Vmax upon deuterium substitution for both CYP2F3 and the D361T mutant. Although a decrease in Vmax upon deuterium substitution is an indication of a kinetic isotope effect, the results cannot be considered conclusive. The value of Vmax depends not only on the rate constant for the isotopically sensitive step but also on the product release step. Any changes in product release due to deuterium substitution will affect the Vmax independent of the isotopically sensitive step and it has been shown previously that deuterium substitution can affect the rate of product release (52).
The Km values on deuterium substitution decreased from 42 μM to 24 μM for 2F3. However, the Km values also depend not only on the isotopically sensitive step, but also on the kinetics of substrate binding and rate of product release. Therefore, decreases in the Km value upon deuterium substitution in a multi-step enzyme reaction pathway cannot be attributed to one particular reaction step (43). The effect of deuterium substitution on the velocity of the reaction, VH/VD, was determined to be 1.9, while the effect of deuterium substitution on the catalytic efficiency, kH/kD [(V/K)H/(V/K)D] was 1.1. Although such low isotope effects suggest that cleavage of the carbon-hydrogen bond is probably not the rate limiting step (53), the results cannot be considered conclusive. The V/K value is dependent on not only the rate constant of the isotopically sensitive step, but on all rate constants up to and including the first irreversible step (43) and therefore, low isotope effects such as these could be due to an inherent masking of the isotope effect, and not because cleavage of the carbon-hydrogen bond is not rate limiting.
Due to the inherent isotope masking effects seen with inter-molecular deuterium isotope studies, we also performed intramolecular deuterium isotope studies with 3MI-d2, using purified CYP2F3. Intramolecular deuterium isotope studies are less susceptible to inherent isotope masking (43). The intramolecular isotope effect (kH/kD) of 6.8 was obtained for 2F3. Interestingly, the same intramolecular isotope effect for dehydrogenation was observed for the D361T mutant, despite the fact that this mutant produced both the dehydrogenated product and 3-methyloxindole. It seems feasible that alternative substrate orientations in the mutant enzyme could produce either the dehydrogenated product with essentially the same mechanism, which would be expected to exhibit a similar isotope effect as the native enzyme, or a different substrate orientation could direct formation of the epoxide and subsequently, 3-methyloxindole.
The relatively high intramolecular deuterium isotope effect suggests that the dehydrogenation of 3MI occurs through an initial hydrogen abstraction. The maximum inherent isotope effect for a carbon-centered oxidation, in the absence of tunneling is 9 (54), and therefore the intramolecular isotope effect observed in this case is lower. This lower isotope effect suggests that the formation of 3-methyleneindolenine shows a high commitment to forward catalysis, and although dehydrogenation of 3MI occurs through an initial hydrogen abstraction, the hydrogen abstraction is most likely only partially rate limiting.
In conclusion, the studies presented here have provided vital structural and mechanistic information that are highly relevant to the understanding of the biochemistry of CYP2F sub-family enzymes. Mutations to specific regions of CYP2F3 abolished its capacity to dehydrogenate 3MI and introduced the alternate oxygenation pathway. The kinetic isotope studies with purified 2F3 concur with goat lung microsomal results, (30) and strongly advocate that CYP2F3 dehydrogenation of 3MI is mediated through an initial hydrogen abstraction mechanism. Dehydrogenation specificity may be attributed to stabilization of the subsequent intermediate (the methylindolyl radical or cation) through the serine at residue 475.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the expert help of Ms. Diane Lanza with the metabolism experiments.
Abbreviations
- 3MI-d3
3-(2H3-methyl)-indole
- 3MI-d2
3-(2H2-methyl)-indole
- NAC
N-acetyl-L-cysteine
- 3MINAC
3-[(N-acetyl-L-cysteine-S-yl)methyl]indole
- 3POI
3-phenyloxindole
- SRS
substrate recognition sequence
Footnotes
This research was supported by NIH grants HL13645 from the National Heart, Lung, and Blood Institute and GM074249 from the National Institute of General Medical Sciences to G.S.Y. and GM031001 from the National Institute of General Medical Sciences to E.F.J.
REFERENCES
- 1.Guengerich FP. Uncommon P450-catalyzed reactions. Curr Drug Metab. 2001;2:93–115. doi: 10.2174/1389200013338694. [DOI] [PubMed] [Google Scholar]
- 2.Forkert PL, RP, Reid KI. Involvement of CYP2E1 and carboxylesterase enzymes in vinyl carbamate metabolism in human lung microsomes. Drug Metab Dispos. 2001;29:258–263. [PubMed] [Google Scholar]
- 3.Forkert PG, Lee RP. Metabolism of ethyl carbamate by pulmonary cytochrome P450 and carboxylesterase isozymes: involvement of CYP2E1 and hydrolase A. Toxicol Appl Pharmacol. 1997;146:245–254. doi: 10.1006/taap.1997.8233. [DOI] [PubMed] [Google Scholar]
- 4.Raucy JL, Lasker JM, Lieber CS, Black M. Acetaminophen activation by human liver cytochromes P450IIE1 and P450IA2. Arch Biochem Biophys. 1989;271:270–283. doi: 10.1016/0003-9861(89)90278-6. [DOI] [PubMed] [Google Scholar]
- 5.Rettie AE, Rettenmeier AW, Howald WN, Baillie TA. Cytochrome P-450--catalyzed formation of delta 4-VPA, a toxic metabolite of valproic acid. Science. 1987;235:890–893. doi: 10.1126/science.3101178. [DOI] [PubMed] [Google Scholar]
- 6.Skordos KW, Skiles GL, Laycock JD, Lanza DL, Yost GS. Evidence supporting the formation of 2,3-epoxy-3-methylindoline: a reactive intermediate of the pneumotoxin 3-methylindole. Chem Res Toxicol. 1998;11:741–749. doi: 10.1021/tx9702087. [DOI] [PubMed] [Google Scholar]
- 7.Huijzer JC, Adams JD, Jr., Yost GS. Decreased pneumotoxicity of deuterated 3-methylindole: bioactivation requires methyl C-H bond breakage. Toxicol Appl Pharmacol. 1987;90:60–68. doi: 10.1016/0041-008x(87)90306-1. [DOI] [PubMed] [Google Scholar]
- 8.Nocerini MR, Yost GS, Carlson JR, Liberato DJ, Breeze RG. Structure of the glutathione adduct of activated 3-methylindole indicates that an imine methide is the electrophilic intermediate. Drug Metab Dispos. 1985;13:690–694. [PubMed] [Google Scholar]
- 9.Yost GS, Nocerini MR, Carlson JR, Liberato DJ. Structure of the adduct of glutathione and activated 3-methylindole. Adv Exp Med Biol. 1986;197:373–380. doi: 10.1007/978-1-4684-5134-4_35. [DOI] [PubMed] [Google Scholar]
- 10.Thornton-Manning J, Appleton ML, Gonzalez FJ, Yost GS. Metabolism of 3-methylindole by vaccinia-expressed P450 enzymes: correlation of 3-methyleneindolenine formation and protein-binding. J Pharmacol Exp Ther. 1996;276:21–29. [PubMed] [Google Scholar]
- 11.Thornton-Manning JR, Ruangyuttikarn W, Gonzalez FJ, Yost GS. Metabolic activation of the pneumotoxin, 3-methylindole, by vaccinia-expressed cytochrome P450s. Biochem Biophys Res Commun. 1991;181:100–107. doi: 10.1016/s0006-291x(05)81387-7. [DOI] [PubMed] [Google Scholar]
- 12.Regal KA, Laws GM, Yuan C, Yost GS, Skiles GL. Detection and characterization of DNA adducts of 3-methylindole. Chem Res Toxicol. 2001;14:1014–1024. doi: 10.1021/tx0100237. [DOI] [PubMed] [Google Scholar]
- 13.Racha JK, Rettie AE, Kunze KL. Mechanism-based inactivation of human cytochrome P450 1A2 by furafylline: detection of a 1:1 adduct to protein and evidence for the formation of a novel imidazomethide intermediate. Biochemistry. 1998;37:7407–7419. doi: 10.1021/bi973011m. [DOI] [PubMed] [Google Scholar]
- 14.Sridar C, Kent UM, Notley LM, Gillam EM, Hollenberg PF. Effect of tamoxifen on the enzymatic activity of human cytochrome CYP2B6. J Pharmacol Exp Ther. 2002;301:945–952. doi: 10.1124/jpet.301.3.945. [DOI] [PubMed] [Google Scholar]
- 15.Lanza DL, Code E, Crespi CL, Gonzalez FJ, Yost GS. Specific dehydrogenation of 3-methylindole and epoxidation of naphthalene by recombinant human CYP2F1 expressed in lymphoblastoid cells. Drug Metab Dispos. 1999;27:798–803. [PubMed] [Google Scholar]
- 16.Kartha JS, Yost GS. Mechanism-based inactivation of lung-selective cytochrome P450 CYP2F enzymes. Drug Metab Dispos. 2008;36:155–162. doi: 10.1124/dmd.107.017897. [DOI] [PubMed] [Google Scholar]
- 17.Lewis DF, Lake BG, Dickins M, Goldfarb PS. Homology modelling of CYP2A6 based on the CYP2C5 crystallographic template: enzyme-substrate interactions and QSARs for binding affinity and inhibition. Toxicol In Vitro. 2003;17:179–190. doi: 10.1016/s0887-2333(02)00132-7. [DOI] [PubMed] [Google Scholar]
- 18.Lewis DF, Lake BG, Dickins M, Goldfarb PS. Homology modelling of CYP3A4 from the CYP2C5 crystallographic template: analysis of typical CYP3A4 substrate interactions. Xenobiotica. 2004;34:549–569. doi: 10.1080/00498250410001691325. [DOI] [PubMed] [Google Scholar]
- 19.Schoch GA, Attias R, Le Ret M, Werck-Reichhart D. Key substrate recognition residues in the active site of a plant cytochrome P450, CYP73A1. Homology guided site-directed mutagenesis. Eur J Biochem. 2003;270:3684–3695. doi: 10.1046/j.1432-1033.2003.03739.x. [DOI] [PubMed] [Google Scholar]
- 20.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 21.Lewis DF. The CYP2 family: models, mutants and interactions. Xenobiotica. 1998;28:617–661. doi: 10.1080/004982598239236. [DOI] [PubMed] [Google Scholar]
- 22.Runge DM, Stock TW, Lehmann T, Taege C, Bernauer U, Stolz DB, Hofmann S, Foth H. Expression of cytochrome P450 2E1 in normal human bronchial epithelial cells and activation by ethanol in culture. Arch Toxicol. 2001;75:335–345. doi: 10.1007/s002040100248. [DOI] [PubMed] [Google Scholar]
- 23.Gillam EM, Guo Z, Guengerich FP. Expression of modified human cytochrome P450 2E1 in Escherichia coli, purification, and spectral and catalytic properties. Arch Biochem Biophys. 1994;312:59–66. doi: 10.1006/abbi.1994.1280. [DOI] [PubMed] [Google Scholar]
- 24.Lanza DL, Yost GS. Selective dehydrogenation/oxygenation of 3-methylindole by cytochrome p450 enzymes. Drug Metab Dispos. 2001;29:950–953. [PubMed] [Google Scholar]
- 25.Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J Biol Chem. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 26.Williams PA, Cosme J, Ward A, Angove HC, Vinkovic D. Matak, Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–468. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 27.Schoch GA, Yano JK, Wester MR, Griffin KJ, Stout CD, Johnson EF. Structure of human microsomal cytochrome P450 2C8. Evidence for a peripheral fatty acid binding site. J Biol Chem. 2004;279:9497–9503. doi: 10.1074/jbc.M312516200. [DOI] [PubMed] [Google Scholar]
- 28.Yano JK, Hsu MH, Griffin KJ, Stout CD, Johnson EF. Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat Struct Mol Biol. 2005;12:822–823. doi: 10.1038/nsmb971. [DOI] [PubMed] [Google Scholar]
- 29.Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, Johnson EF. Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J Biol Chem. 2007;282:14348–14355. doi: 10.1074/jbc.M611692200. [DOI] [PubMed] [Google Scholar]
- 30.Skiles GL, Yost GS. Mechanistic studies on the cytochrome P450-catalyzed dehydrogenation of 3-methylindole. Chem Res Toxicol. 1996;9:291–297. doi: 10.1021/tx9501187. [DOI] [PubMed] [Google Scholar]
- 31.Lanza DL, Yost GS. Selective dehydrogenation/oxygenation of 3-methylindole by cytochrome p450 enzymes. Drug Metab Dispos. 2001;29:950–953. [PubMed] [Google Scholar]
- 32.Kobayashi Y, Fang X, Szklarz GD, Halpert JR. Probing the active site of cytochrome P450 2B1: metabolism of 7-alkoxycoumarins by the wild type and five site-directed mutants. Biochemistry. 1998;37:6679–6688. doi: 10.1021/bi9731450. [DOI] [PubMed] [Google Scholar]
- 33.Scott EE, Spatzenegger M, Halpert JR. A truncation of 2B subfamily cytochromes P450 yields increased expression levels, increased solubility, and decreased aggregation while retaining function. Arch Biochem Biophys. 2001;395:57–68. doi: 10.1006/abbi.2001.2574. [DOI] [PubMed] [Google Scholar]
- 34.Barnes HJ, Arlotto MP, Waterman MR. Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc Natl Acad Sci USA. 1991;88:5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larson JR, Coon MJ, Porter TD. Alcohol-inducible cytochrome P-450IIE1 lacking the hydrophobic NH2-terminal segment retains catalytic activity and is membrane-bound when expressed in Escherichia coli. J Biol Chem. 1991;266:7321–7324. [PubMed] [Google Scholar]
- 36.Gillam EM. Human cytochrome P450 enzymes expressed in bacteria: reagents to probe molecular interactions in toxicology. Clin Exp Pharmacol Physiol. 1998;25:877–886. doi: 10.1111/j.1440-1681.1998.tb02338.x. [DOI] [PubMed] [Google Scholar]
- 37.Omura T, Sato R. The Carbon Monoxide-binding Pigment of Liver Microsomes. J Biol Chem. 1964;239:2370–2385. [PubMed] [Google Scholar]
- 38.Skordos KW, Laycock JD, Yost GS. Thioether adducts of a new imine reactive intermediate of the pneumotoxin 3-methylindole. Chem Res Toxicol. 1998;11:1326–1331. doi: 10.1021/tx9801209. [DOI] [PubMed] [Google Scholar]
- 39.Leete E, Marion L. The hydrogenolysis of 3-hydroxymethylindole and other indole derevatives with lithium aluminium hydride. Canadian Journal of Chemistry. 1953;31:775–783. [Google Scholar]
- 40.Korzekwa K, Howald WN, Trager WF. The use of Brauman’s least squares approach for the quantification of deuterated chlorophenols. Biomed Environ Mass Spectrom. 1990;19:211–217. doi: 10.1002/bms.1200190402. [DOI] [PubMed] [Google Scholar]
- 41.Saribas AS, Gruenke L, Waskell L. Overexpression and purification of the membrane-bound cytochrome P450 2B4. Protein Expr Purif. 2001;21:303–309. doi: 10.1006/prep.2000.1377. [DOI] [PubMed] [Google Scholar]
- 42.Cheng D, Kelley RW, Cawley GF, Backes WL. High-level expression of recombinant rabbit cytochrome P450 2E1 in Escherichia coli C41 and its purification. Protein Expr Purif. 2004;33:66–71. doi: 10.1016/j.pep.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 43.Nelson SD, Trager WF. The use of deuterium isotope effects to probe the active site properties, mechanism of cytochrome P450-catalyzed reactions, and mechanisms of metabolically dependent toxicity. Drug Metab Dispos. 2003;31:1481–1498. doi: 10.1124/dmd.31.12.1481. [DOI] [PubMed] [Google Scholar]
- 44.Szklarz GD, Halpert JR. Molecular basis of P450 inhibition and activation: implications for drug development and drug therapy. Drug Metab Dispos. 1998;26:1179–1184. [PubMed] [Google Scholar]
- 45.Wang Q, Halpert JR. Combined three-dimensional quantitative structure-activity relationship analysis of cytochrome P450 2B6 substrates and protein homology modeling. Drug Metab Dispos. 2002;30:86–95. doi: 10.1124/dmd.30.1.86. [DOI] [PubMed] [Google Scholar]
- 46.Domanski TL, He YQ, Scott EE, Wang Q, Halpert JR. The role of cytochrome 2B1 substrate recognition site residues 115, 294, 297, 298, and 362 in the oxidation of steroids and 7-alkoxycoumarins. Arch Biochem Biophys. 2001;394:21–28. doi: 10.1006/abbi.2001.2504. [DOI] [PubMed] [Google Scholar]
- 47.Szklarz GD, He YQ, Kedzie KM, Halpert JR, Burnett VL. Elucidation of amino acid residues critical for unique activities of rabbit cytochrome P450 2B5 using hybrid enzymes and reciprocal site-directed mutagenesis with rabbit cytochrome P450 2B4. Arch Biochem Biophys. 1996;327:308–318. doi: 10.1006/abbi.1996.0127. [DOI] [PubMed] [Google Scholar]
- 48.He YA, Balfour CA, Kedzie KM, Halpert JR. Role of residue 478 as a determinant of the substrate specificity of cytochrome P450 2B1. Biochemistry. 1992;31:9220–9226. doi: 10.1021/bi00153a015. [DOI] [PubMed] [Google Scholar]
- 49.Liu J, He YA, Halpert JR. Role of residue 480 in substrate specificity of cytochrome P450 2B5 and 2B11. Arch Biochem Biophys. 1996;327:167–173. doi: 10.1006/abbi.1996.0105. [DOI] [PubMed] [Google Scholar]
- 50.Afzelius L, Zamora I, Ridderstrom M, Andersson TB, Karlen A, Masimirembwa CM. Competitive CYP2C9 inhibitors: enzyme inhibition studies, protein homology modeling, and three-dimensional quantitative structure-activity relationship analysis. Mol Pharmacol. 2001;59:909–919. doi: 10.1124/mol.59.4.909. [DOI] [PubMed] [Google Scholar]
- 51.Kumar D, De Visser SP, Shaik S. Oxygen economy of cytochrome P450: what is the origin of the mixed functionality as a dehydrogenase-oxidase enzyme compared with its normal function? J Am Chem Soc. 2004;126:5072–5073. doi: 10.1021/ja0318737. [DOI] [PubMed] [Google Scholar]
- 52.Ling KH, Hanzlik RP. Deuterium isotope effects on toluene metabolism. Product release as a rate-limiting step in cytochrome P-450 catalysis. Biochem Biophys Res Commun. 1989;160:844–849. doi: 10.1016/0006-291x(89)92511-4. [DOI] [PubMed] [Google Scholar]
- 53.Guengerich FP, Bocker RH. Cytochrome P-450-catalyzed dehydrogenation of 1,4-dihydropyridines. J Biol Chem. 1988;263:8168–8175. [PubMed] [Google Scholar]
- 54.Jones JP, Rettie AE, Trager WF. Intrinsic isotope effects suggest that the reaction coordinate symmetry for the cytochrome P-450 catalyzed hydroxylation of octane is isozyme independent. J Med Chem. 1990;33:1242–1246. doi: 10.1021/jm00166a024. [DOI] [PubMed] [Google Scholar]