Abstract
Studies in a broad spectrum of model organisms have reported that dietary restriction (DR) is associated with an increase in mitochondrial electron transport chain (ETC) function. However, the question of whether ETC function is required for DR-mediated longevity remains controversial. Here, we report that genetic and pharmacological interventions that target mitochondrial complex V affect Drosophila lifespan in a nutrient-dependent manner. These findings support a requirement for mitochondrial complex V in DR-mediated longevity in flies.
Keywords: Drosophila, mitochondria, dietary restriction, electron transport chain
Numerous correlative studies have suggested that alterations in mitochondrial electron transport chain (ETC) function may play a role in mediating the pro-longevity effects of dietary restriction (DR) (Lin et al. 2002; Nisoli et al. 2005; Bishop & Guarente 2007; Guarente 2008). At the same time, genetic studies in both invertebrate (Ishii et al. 1998; Feng et al. 2001; Dillin et al. 2002; Lee et al. 2003; Walker et al. 2006; Copeland et al. 2009) and vertebrate (Dell'agnello et al. 2007; Lapointe & Hekimi 2008) model systems have shown that inactivation of genes important for ETC function can affect animal aging. However, an understanding of the causal relationship between ETC function and DR remains controversial. In yeast (Lin et al. 2002) and nematodes (Bishop & Guarente 2007), it has been reported that ETC function is required for DR-mediated longevity. However, these findings have been challenged by the observation that respiratory–deficient yeast cells display a robust DR response (Kaeberlein et al. 2005). In this study, we investigate the interaction between ETC function and DR in the fruit fly Drosophila.
Dilution of brewer's yeast has been proposed as an effective regimen for DR studies in Drosophila (Bass et al. 2007). We confirmed that this yeast DR paradigm produced longevity effects in our control flies (Figure 1A; Table 1). Reduction of brewer's yeast concentration in the media from 6% (rich media) to 1% (DR) increased lifespan in control flies. Moreover, a further reduction in yeast concentration to 0.2% yeast resulted in a shortened lifespan. Recently, we reported that knock-down of a complex V (CG5389) subunit resulted in increased longevity under our standard laboratory food conditions (3% yeast) (Copeland et al. 2009). Here, we examined the dependence of this complex V-mediated lifespan extension upon nutritional conditions. We used a ubiquitous expression GAL4 line, daughterless (da)-GAL4 to activate the CG5389 RNAi transgene and compared survival of flies under rich, and DR media. Under rich media conditions, RNAi of CG5389 conferred an 18% increase in mean survival compared to isogenic control flies (Figure 1B; Table 1). However, this longevity effect was abolished under DR conditions.
Figure 1. Genetic and pharmacological manipulations that target mitochondrial complex V affect fly lifespan in a nutrient-dependent manner.
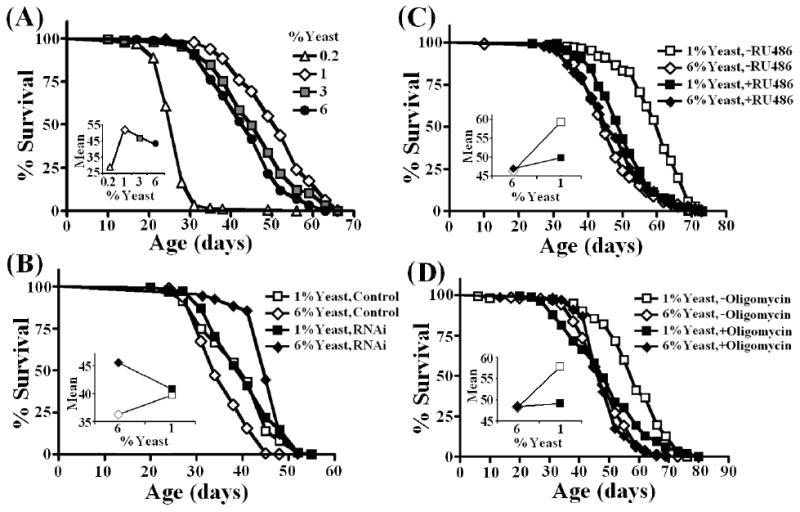
(A) Dietary restriction (1% yeast medium) extends Drosophila lifespan whereas malnutrition (0.2% yeast medium) shortens lifespan. (B, C, D) Perturbation of mitochondrial complex V affects DR-mediated extension of adult lifespan. Genotypes were as follows: (A) Tub-Gene-Switch/w1118; (B) daG32-GAL4/w1118 (Control) and daG32-GAL4/UAS-complexV-RNAi (RNAi); (C) Tub-Gene-Switch/UAS-complexV-RNAi fed with or without RU486 (50 μg/ml during adulthood); (D) Tub-Gene-Switch/w1118 fed with or without oligomycin (25 μg/ml during adulthood). Complete survival data including statistical analysis, replicate daG32-GAL4 experiments and developmental feeding of oligomycin and RU486 are given in Table 1.
Table 1.
The effects of yeast restriction on Drosophila longevity in the presence or absence of mitochondrial complex V inhibitors. UAS-RNAi targets the complex V subunit CG5389. Survival curves under yeast restriction (1% yeast) were compared to survival under rich media (6% yeast). The significance of the difference between survival curves was analyzed using log-rank test.
| Genotype | % Yeast | RU486 dosage (Dev/Adult) |
Oligomycin dosage (Dev/Adult) |
Mean LS (days) |
Mean LS extension (versus 6% yeast) |
Sample size (n =) |
p Value (log-rank) |
|---|---|---|---|---|---|---|---|
| TubGS/w1118 | 0.2 | 0 μg/ml | 0 μg/ml | 26.7 | -39.4 % | 196 | < 0.0001 |
| TubGS/w1118 | 1 | 0 μg/ml | 0 μg/ml | 51.28 | 16.4 % | 180 | < 0.0001 |
| TubGS/w1118 | 3 | 0 μg/ml | 0 μg/ml | 46.3 | 5.1 % | 190 | 0.0026 |
| TubGS/w1118 | 6 | 0 μg/ml | 0 μg/ml | 44.05 | 0 % | 185 | - |
| TubGS/w1118 | 1 | 0 μg/ml | 0 μg/ml | 58.18 | 20.5 % | 173 | < 0.0001 |
| TubGS/w1118 | 6 | 0 μg/ml | 0 μg/ml | 48.29 | 0 % | 173 | - |
| TubGS/w1118 | 1 | 0 μg/ml | 0/25 μg/ml | 49.18 | 1.4 % | 172 | 0.065 |
| TubGS/w1118 | 6 | 0 μg/ml | 0/25 μg/ml | 48.51 | 0 % | 175 | - |
| TubGS/w1118 | 1 | 0 μg/ml | 0 μg/ml | 49.4 | 19.8 % | 176 | < 0.0001 |
| TubGS/w1118 | 6 | 0 μg/ml | 0 μg/ml | 41.23 | 0 % | 181 | - |
| TubGS/w1118 | 1 | 0 μg/ml | 5/25 μg/ml | 40.98 | 0.1 % | 183 | 0.0636 |
| TubGS/w1118 | 6 | 0 μg/ml | 5/25 μg/ml | 40.94 | 0 % | 188 | - |
| TubGS/w1118 | 1 | 0 μg/ml | 0 μg/ml | 55.59 | 19.7 % | 182 | < 0.0001 |
| TubGS/w1118 | 6 | 0 μg/ml | 0 μg/ml | 46.44 | 0 % | 183 | - |
| TubGS/w1118 | 1 | 0/50 μg/ml | 0 μg/ml | 54.04 | 15.5 % | 186 | < 0.0001 |
| TubGS/w1118 | 6 | 0/50 μg/ml | 0 μg/ml | 46.8 | 0 % | 181 | - |
| TubGS/UAS-RNAi | 1 | 0 μg/ml | 0 μg/ml | 59.52 | 28 % | 174 | < 0.0001 |
| TubGS/UAS-RNAi | 6 | 0 μg/ml | 0 μg/ml | 46.5 | 0 % | 183 | - |
| TubGS/UAS-RNAi | 1 | 0/50 μg/ml | 0 μg/ml | 50 | 6 % | 180 | 0.0641 |
| TubGS/UAS-RNAi | 6 | 0/50 μg/ml | 0 μg/ml | 47.19 | 0 % | 180 | - |
| TubGS/UAS-RNAi | 1 | 0 μg/ml | 0 μg/ml | 60.1 | 13.3 % | 169 | < 0.0001 |
| TubGS/UAS-RNAi | 6 | 0 μg/ml | 0 μg/ml | 53.06 | 0 % | 180 | - |
| TubGS/UAS-RNAi | 1 | 10/50 μg/ml | 0 μg/ml | 56.57 | 3.9 % | 139 | < 0.0001 |
| TubGS/UAS-RNAi | 6 | 10/50 μg/ml | 0 μg/ml | 54.45 | 0 % | 190 | - |
| daG32-GAL4/w1118 | 1 | 0 μg/ml | 0 μg/ml | 39.61 | 9.5 % | 186 | < 0.0001 |
| daG32-GAL4/w1118 | 6 | 0 μg/ml | 0 μg/ml | 36.16 | 0 % | 187 | - |
| daG32-GAL4/UAS-RNAi | 1 | 0 μg/ml | 0 μg/ml | 40.7 | -10.4 % | 184 | < 0.0001 |
| daG32-GAL4/UAS-RNAi | 6 | 0 μg/ml | 0 μg/ml | 45.43 | 0 % | 187 | - |
| daG32-GAL4/w1118 | 1 | 0 μg/ml | 0 μg/ml | 43.42 | 13 % | 217 | < 0.0001 |
| daG32-GAL4/w1118 | 6 | 0 μg/ml | 0 μg/ml | 38.44 | 0 % | 210 | - |
| daG32-GAL4/UAS-RNAi | 1 | 0 μg/ml | 0 μg/ml | 39.21 | -13.2 % | 217 | < 0.0001 |
| daG32-GAL4/UAS-RNAi | 6 | 0 μg/ml | 0 μg/ml | 45.19 | 0 % | 211 | - |
| UAS-RNAi/w1118 | 1 | 0 μg/ml | 0 μg/ml | 45.8 | 9.6 % | 169 | < 0.0001 |
| UAS-RNAi/w1118 | 6 | 0 μg/ml | 0 μg/ml | 41.8 | 0 % | 188 | - |
While using the da-GAL4 driver activated the complex V RNAi transgene throughout development and adulthood, the DR regime was implemented exclusively during adulthood. To examine the impact of adult-specific RNAi of complex V on DR-mediated longevity, we used the mifepristone (RU486) inducible-GAL4 system (annotated P[Switch] or Gene-Switch (Osterwalder et al. 2001; Roman et al. 2001)). We used the ubiquitous tubulin (tub)-Gene-Switch (GS) driver line to examine the effect of activating RNAi of CG5389 only during adulthood when the flies were first exposed to DR and rich media conditions. Uninduced control flies again displayed a robust increase in lifespan under DR conditions (Figure 1C; Table 1). In contrast, RNAi of complex V in adult flies greatly impaired DR-mediated life extension. Inducer had no impact on longevity in control flies (w1118/tub-GS) under either nutritional regime (Table 1).
In agreement with our previous genetic results, flies exposed to oligomycin, a specific inhibitor of complex V, from the onset of adulthood did not show significant lifespan extension under DR conditions without shortening lifespan under rich media conditions (Figure 1D; Table 1). Taken together, our results indicate that ETC function plays an important role in mediating this DR paradigm (6% to 1% yeast). All three manipulations that target complex V (RNAi mediated by da-GAL4, RNAi mediated by tub-GS and oligomycin feeding) impaired the ability of DR to promote longevity. RNAi of complex V mediated by da-GAL4 resulted in increased longevity under rich media and a shortening of lifespan upon DR. However, RNAi of complex V mediated by tub-GS and/or oligomycin feeding had no major impact on longevity under rich media and prevented an increase in longevity under DR. It is possible that variation in the degree of complex V inactivation in different tissues may be responsible for these phenotypic differences.
For a number of technical reasons, we conducted this study using male flies: 1) RNAi of ETC genes can decrease fecundity in female flies (Copeland et al. 2009). As reproductive status can dramatically affect feeding behavior in female flies (Carvalho et al. 2006) this may be a confounding factor in a study that manipulates nutrient availability. 2) The presence of eggs and subsequently larvae in the media affects the texture of the food, which may affect mortality and/or result in complicated effects on feeding behavior and hence drug (RU486 or oligomycin) dosage. 3) Using this DR (yeast restriction) paradigm, we observed robust effects on longevity in male flies which are not subject to the above variables.
Our study strongly suggest that DR results in changes in ETC function important for longevity. It will be interesting to determine the nature of these changes and whether they are evolutionarily conserved. One attractive hypothesis is that DR delays the onset of the age-related decline in ETC activity reported across the animal kingdom (Wallace 2005).
While this manuscript was under review, Zid et al. reported that RNAi of subunits from complex I and complex IV diminished the lifespan extension obtained upon DR in flies (Zid et al. 2009).
Methods
Fly stocks
Tubulin-Gene-Switch was a gift from S. Pletcher (University of Michigan, Ann Arbor, MI). The CG5389 RNAi line was obtained from the Vienna Drosophila RNAi Center (Dietzl et al. 2007) and backcrossed into the white1118 background seven times. daG32-GAL4 was obtained from the Bloomington Stock Center. The age-related expression pattern of daG32-GAL4 was recently reported (Martin et al. 2009).
Fly food
One liter of stock medium consisted of 19 g of sucrose, 38 g of dextrose, 10 g of agar, 91 g of cornmeal, variable amount of Brewer's yeast (2, 10, 30, or 60 g), 11 mL of propionic acid, and 1.5 g of tegosept (1.5 g of tegosept dissolved in 15 mL of 95% ethanol).
Lifespan analysis
Lifespan studies were carried out at 25°C on a 12-hour light/dark cycle, with survivors transferred to fresh vials every 2-4 days. For Gene-Switch experiments, RU486 was administered by adding 200 μl of 0, 10, or 50 μg/ml RU486 dissolved in ethanol to the top of the fly food, as previously described. For adult-only induction, crosses were grown at 18°C during development. For oligomycin feeding, the drug was administered by adding 200 μl of 0, 5 or 25 μg/ml oligomycin dissolved in ethanol to the top of the fly food. Log-rank test was used to compare differences in survival curves.
Acknowledgments
The authors would like to thank Jeff Copeland and Margaret Wang for help with fly work and S. Pletcher, the Vienna Drosophila RNAi Center (Dietzl et al. 2007) and the Drosophila Stock Center (Bloomington) for fly stocks. D.W.W received support from the UCLA Older Americans Independence Center, NIH/NIA Grant P30-AG028748, and the content does not necessarily represent the official views of the National Institute on Aging or the National Institutes of Health. D.W.W also received support from the Ellison Medical Foundation, the American Federation for Aging Research, the UCLA Center for gene environment in Parkinson's Disease, and the Muscular Dystrophy Association. D.W.W is an Ellison Medical Foundation New Scholar in Aging.
Footnotes
Author Contributions: S.B, J.H, K.V and T.L designed and performed the experiments and analyzed the data. D.W.W designed experiments, supervised the work, analyzed the data and wrote the manuscript with helpful comments from S.B. and J.H.
References
- Bass TM, Grandison RC, Wong R, Martinez P, Partridge L, Piper MD. Optimization of dietary restriction protocols in Drosophila. J Gerontol A Biol Sci Med Sci. 2007;62:1071–1081. doi: 10.1093/gerona/62.10.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Carvalho GB, Kapahi P, Anderson DJ, Benzer S. Allocrine modulation of feeding behavior by the Sex Peptide of Drosophila. Curr Biol. 2006;16:692–696. doi: 10.1016/j.cub.2006.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland JM, Cho J, Lo T, Jr, Hur JH, Bahadorani S, Arabyan T, Rabie J, Soh J, Walker DW. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol. 2009;19:1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- Dell'agnello C, Leo S, Agostino A, Szabadkai G, Tiveron C, Zulian A, Prelle A, Roubertoux P, Rizzuto R, Zeviani M. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum Mol Genet. 2007;16:431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- Guarente L. Mitochondria--a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–176. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Fujii M, Hartman PS, Tsuda M, Yasuda K, Senoo-Matsuda N, Yanase S, Ayusawa D, Suzuki K. A mutation in succinate dehydrogenase cytochrome b causes oxidative stress and ageing in nematodes. Nature. 1998;394:694–697. doi: 10.1038/29331. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Hu D, Kerr EO, Tsuchiya M, Westman EA, Dang N, Fields S, Kennedy BK. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 2005;1:e69. doi: 10.1371/journal.pgen.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Hekimi S. Early mitochondrial dysfunction in long-lived Mclk1+/- mice. J Biol Chem. 2008;283:26217–26227. doi: 10.1074/jbc.M803287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Martin I, Jones MA, Rhodenizer D, Zheng J, Warrick JM, Seroude L, Grotewiel M. Sod2 knockdown in the musculature has whole-organism consequences in Drosophila. Free Radic Biol Med. 2009;47:803–813. doi: 10.1016/j.freeradbiomed.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman G, Endo K, Zong L, Davis RL. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2001;98:12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DW, Hajek P, Muffat J, Knoepfle D, Cornelison S, Attardi G, Benzer S. Hypersensitivity to oxygen and shortened lifespan in a Drosophila mitochondrial complex II mutant. Proc Natl Acad Sci U S A. 2006;103:16382–16387. doi: 10.1073/pnas.0607918103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]


