Abstract
The constitutive activation of Signal Transducer and Activator of Transcription 3 (STAT3) is frequently detected in most types of human cancer where it plays important roles in survival, drug-resistance, angiogenesis, and other functions. Targeting constitutive STAT3 signaling is thus an attractive therapeutic approach for these cancers. We have recently developed novel small molecule STAT3 inhibitors known as FLLL31 and FLLL32, which are derived from curcumin (the primary bioactive compound of turmeric). These compounds are designed to bind selectively to Janus Kinase 2 (JAK2) and the STAT3 SH2 domain, which serves crucial roles in STAT3 dimerization and signal transduction. Here we show that FLLL31 and FLLL32 are effective inhibitors of STAT3 phosphorylation, DNA binding activity, and transactivation in vitro, leading to the impediment of multiple oncogenic processes and the induction of apoptosis in pancreatic and breast cancer cell lines. FLLL31 and FLLL32 also inhibit colony formation in soft agar, cell invasion, and exhibit synergy with the anti-cancer drug doxorubicin against breast cancer cells. In addition, we show that FLLL32 can inhibit the induction of STAT3 phosphorylation by Interferon-α (IFNα) and Interleukin-6 (IL-6) in breast cancer cells. We also demonstrate that administration of FLLL32 can inhibit tumor growth and vascularity in chicken embryo xenografts as well as substantially reduce tumor volumes in mouse xenografts. Our findings highlight the potential of these new compounds and their efficacy in targeting pancreatic and breast cancers that exhibit constitutive STAT3 signaling.
Keywords: Pancreatic cancer, breast cancer, JAK2, curcumin analogues, STAT3
Introduction
The American Cancer Society estimated that over 200,000 new cases of invasive breast cancer and over 40,000 deaths are expected among women in the United States per year. In women, breast cancer remains the second most fatal form of cancer after lung cancer. Pancreatic cancer is one of the most serious of cancers. According to the American Cancer Society, there are an estimated 42,470 new cases and 35,240 deaths from pancreatic cancer in the United States in 2009. The prognosis is generally regarded as poor and the overall survival rate of all stages is less than 1% at 5 years with most patients dying within 1 year. There is a critical need to develop more effective treatments for pancreatic and breast cancer.
Signal Transducer and Activator of Transcription 3 (STAT3) is a member of a family of latent cytosolic transcription factors whose activation is contingent upon the phosphorylation of a conserved tyrosine residue (Y705) by upstream kinases such as Janus kinase 2 (JAK2) (1). This event promotes the dimerization of STAT3 monomers via their Src-homology 2 (SH2) domains, rendering them in a transcriptionally-active conformation (2). Persistent activation of the JAK2/STAT3 signaling pathway has been documented in a wide range of human solid and blood cancers, and is commonly associated with a worse prognosis (3, 4). Among the cancer-promoting activities ascribed to persistent STAT3 signaling are those involved with cell proliferation, metastasis, angiogenesis, host immune evasion, and resistance to apoptosis (5, 6). Interference with the STAT3 signaling pathway in cancer cells, such as by antisense oligonucleotides and dominant-negative STAT3 mutants, has been shown to result in growth inhibition and the induction of apoptosis (7–10). Although STAT3 serves critical and necessary roles in early embryogenesis, its presence in the majority of normal adult cell types is largely dispensable, making it an attractive target for cancer therapy (11–13). Numerous strategies have since been employed to inhibit constitutive STAT3 signaling in cancerous cells. The selective inhibition of JAK2 can prevent STAT3 phosphorylation and dimerization, and several JAK2 inhibitors including AG490, WP1066, and SD-1029 have been reported in recent literature (14–16). The necessity of an intact SH2 domain also makes it a rational target for the disruption of STAT3 signaling (17). Peptide-based SH2 inhibitors have previously been described, but they are hindered by limited cell permeability, poor in vivo stability and the potential for immunogenicity (18, 19). The development of non-peptide small molecular SH2 inhibitors, such as Stattic, STA-21, and S3I-201, has also been recently reported (17, 20, 21).
In conjunction with our collaborators, we have developed and evaluated a series of novel non-peptide JAK2/SH2 inhibitors based upon the structure of the plant phytochemical known as curcumin. Curcumin is the bioactive component of Curcuma longa and the subject of extensive research due to its broad spectrum of biologically-beneficial activities and its relative safety in large dosages (22, 23). The complex chemistry of curcumin allows it to inhibit multiple oncogenic processes, including those associated with the JAK2/STAT3 pathway (24). Poor bioavailability limits curcumin’s use as a cancer therapeutic agent, but it is potentially useful as a lead compound for the development of new JAK2/STAT3 inhibitors (25). In our initial testing, two of our compounds in particular, designated FLLL31 and FLLL32, were found to be especially potent at inhibiting the viability of breast and pancreatic cancer cells that feature constitutively-activated STAT3, thus warranting a closer look at their antitumor properties. Here we report our findings on the growth-suppressive activities of FLLL31 and FLLL32, their efficacy in inhibiting constitutive STAT3 signaling in vitro, and the effects of FLLL32 against the processes of tumor growth and angiogenesis in vivo.
Materials and Methods
Cell Culture
Pancreatic cancer cell lines (PANC-1, BXPC-3, HPAC, SW1990), breast cancer cell lines (MDA-MB-231, SK-BR-3, MDA-MB-468, MDA-MB-453), WI-38 normal human lung fibroblasts and Human Embryonic Kidney (HEK) 293 cells were purchased from the American Type Culture Collection. These cells were cultured in DMEM supplemented with 10% fetal bovine serum. SUM159 breast cancer cells were obtained from Dr. Max S. Wicha (University of Michigan). The normal human mammary epithelial cells (HMEC) and the normal human bladder smooth muscle cells (BdsMC) were purchased from Lonza Walkersville, Inc. SUM159 and HMEC were cultured in Ham’s F12 containing 5% FBS, 5 µg/ml insulin, 1 µg/ml hydrocortisone and 10 ng/ml epidermal growth factor. The BdsMC were cultured in medium recommended by the protocol from Lonza Walkersville, Inc. Human Pancreatic Duct Epithelial cells (HPDE) were provided by Dr. Ming-Sound Tsao and maintained in CnT-07CF epidermal keratinocyte medium supplemented with 0.07 mM CaCl2.
JAK2 and STAT3 Src homology 2 (SH2) inhibitors
FLLL31, FLLL32, and WP1066 (16), a JAK2 inhibitor, were synthesized in Dr. Pui-Kai Li’s laboratory. AG490 (15) and SD-1029 (14), JAK2 inhibitors and S3I-201 (21) and Stattic (20), STAT3 SH2 inhibitors, were purchased from Calbiochem. Curcumin was purchased from Sigma/Aldrich.
Western blot analysis
Cancer cells were treated with these agents or DMSO for 24 hours. For Interferon-α (IFNα) and interleukin-6 (IL-6) treatments, MDA-MB-453 breast cancer cells were serum starved for 24 hours. The cells were pre-treated with FLLL32 (10 µM) for 2 hours and 50 ng/ml of IFNα or IL-6 was then added for 30 minutes before the cells were collected. Membranes were probed with a 1:1000 dilution of antibodies (Cell Signaling Tech.) against phospho-specific STAT3 (hereafter referred to as P-STAT3) (Tyrosine 705), ERK1/2 (Threonine 202/Tyrosine 204), AKT (Serine473), EGFR (Tyrosine1068), P70S6K (Threonine389), PKC-δ (Threonine505), Src (Tyrosine 416), mTOR (Serine 2448), cleaved Poly (ADP-ribose) polymerase (PARP), cleaved caspase-3, cyclin D, Bcl-2, survivin, and GAPDH. Membranes were analyzed using enhanced chemiluminescence Plus reagents and scanned with the Storm Scanner (Amersham Pharmacia Biotech Inc). Integrated densities of the P-STAT3 (Y705), STAT3, cleaved caspase-3, and GAPDH bands in Western blots were quantified using ImageJ software. The integrated densities of the P-STAT3, STAT3, and cleaved caspase-3 bands were individually normalized to GAPDH band in each cancer cell line using ImageJ software. The relatively reduced or increased levels of P-STAT3, STAT3, and cleaved caspase-3 were compared with the DMSO control, which was set as 1.00.
JAK2 and other human kinase activity assay
JAK2 kinase activity was assessed with the HTScan® JAK2 Kinase Assay Kit per the manufacturer’s protocol (Cell Signaling Technologies). The possible effects of FLLL32 on other ten purified human protein kinases was determined by Reaction Biology Corp. (Malvern, PA) using a Kinase profiler assay.
STAT3 DNA-binding and STAT3-dependent transcriptional luciferase activity assays
Following treatment with FLLL31, FLLL32, curcumin, or DMSO for 24 hours, nuclear extracts of cells were analyzed for STAT3 and STAT1 DNA binding activities in triplicates using TransFactor Universal STAT3- and STAT1-specific kits following the manufacturer’s protocol (Clontech Inc.). To assess transcriptional activity, MDA-MB-231 cells were stably transfected with the pLucTKS3 luciferase reporter, which contains seven copies of the STAT3 binding site in a thymidine kinase minimal promoter (26). The cells were harvested in reporter lysis buffer after 24 hours of treatment. Luciferase activity is expressed as counts per second per µg protein. Luciferase activity is presented relative to a pLucTKS3-transfected sample treated with DMSO, arbitrarily set at 100%. The results of the luciferase assay represent the averages from three independent experiments.
Reverse transcription PCR (RT-PCR) analysis
RNA was collected from MDA-MB-231 and PANC-1 cells with RNeasy Kits (Qiagen) following 24 hours of treatment with FLLL31, FLLL32, and curcumin. cDNA was generated from 500ng samples of RNA using Omniscript RT (Qiagen). Primer sequences and source information can be found in supplemental Table 1.
MTT cell viability assay
Cells were seeded in 96-well plates (3,000 cells/well) in triplicate, treated with 0.5–5 µmol/L of FLLL31, FLLL32, or 0.5 to 30 µmol/L of curcumin (Sigma) for 72 hours. 25 µl of MTT (Sigma) was added to each sample and incubated for 3.5 hours. Then 100 µl of N,N-dimethylformamide solubilization solution was added to each well. The absorbance at 450 nm was read the following day. Half-Maximal inhibitory concentrations (IC50) were determined using Sigma Plot 9.0 software (Systat Software Inc.).
Soft agar colony formation assay
A 0.6% agar gel with 10% FBS in DMEM was prepared and added to 6-well culture dish as a base agar. 5,000 MDA-MB-231 cells per well were plated in 0.4% agar gel with 10% FBS in DMEM supplemented with the curcumin, FLLL31, or FLLL32 treatment on top of the base agar, and allowed to grow for two weeks. Colonies were stained with MTT dye. Numbers were normalized as a percentage of colonies formed in DMSO treatment. The results represent the averages from three independent experiments.
Cell invasion assay
Cell invasiveness was evaluated with the InnoCyte™ Laminin-based 96-well Cell Invasion Assay per the manufacturer’s instructions (EMD Biosciences). In brief, 5×104 MDA-MB-231 cells were seeded in a laminin-coated cell culture insert in serum-free DMEM and treated with 1 µM concentrations of DMSO, FLLL31, FLLL32, or curcumin. Cells were allowed to migrate/invade the laminin layer in the presence of DMEM supplemented with 10% FBS as a chemoattractant for 24 hours. Invading cells were stained with Calcein-AM and detected with a fluorimeter. The data were normalized as relative fluorescent units (RFU) per 104 cells. Results are from three independent experiments and compared as a percentage to the RFU of the DMSO-treated controls.
Tansfection with Constitutive active STAT3
HEK293 cells were transfected using Lipofectamine 2000 with a vector which encodes constitutive STAT3 (STAT3-C), which is tagged with a FLAG epitope (27). The second day, cells were treated with FLLL32 or DMSO. Twenty four hours later, the cells were harvested to for western blot analysis.
Mouse xenografts
All animal studies were conducted in accordance with the standard procedures approved by IACUC at the Research Institute at Nationwide Children’s Hospital. MDA-MB-231 breast cancer cells (1×107 in Matrigel) were implanted subcutaneously into the flank region of 4–5-week-old female athymic nude mice. After tumors developed (12 days), the mice were randomized into two groups and treated with 50 mg/kg FLLL32 or DMSO (5 mice per group) intraperitoneally daily for 19 days. Tumor growth was determined by measuring the major (L) and minor (W) diameter with a caliper. The tumor volume was calculated according to the formula: Tumor volume= 0.5236× L × W2.
CAM xenograft assay
The procedure used was a modification of a previously published procedure (28). Fertile Leghorn chicken eggs (CBT Farms) were incubated until 10 DI. The sample size was 8–14 embryos from two separate experiments. PANC-1 cells, 250,000 in inert extracellular matrix were implanted just under the chorioallantoic membrane (CAM). Four and 48 hr after tumor implantation, a maximal tolerated dose of 5 mg/kg gemcitabine (ChemieTek), 25 mg/kg FLLL32 or solvent (DMSO in Sterile water for injection) was placed onto the chorion. Four days after tumor implantation, CAMs were fixed as described (28). Images were captured at 6.25× magnification using a brightfield dissecting microcope (Wild M400 photomakroscop) connected to a cooled camera (QImaging, Burnaby). The number of vessels surrounding the tumor were manually counted and the size of the tumor determined using NIH Image J densitometry software.
Determination of combinatorial effects
The ability of doxorubicin, FLLL31, and FLLL32 to act in a synergistic manner with regards to growth inhibition was determined as described by Chou et al (29). Calcusyn software (Biosoft) was used to determine the combinational index (CI) for each concentration of drug mixture used. A value for the CI <1 represents a case where synergism of doxorubicin, FLLL31, and FLLL32 was present. CI values of 1 and >1 represent additive and antagonistic effects respectively.
Results
The JAK2/SH2 inhibitors FLLL31 and FLLL32
FLLL31 and FLLL32 are diketone analogues of curcumin (Figure 1A). The central β-dicarbonyl moiety of curcumin is subject to keto-enol tautomerization, which is hypothesized to influence its target selectivity and by virtue its biological activity (Fuchs et al., unpublished data). By replacing the two hydrogen atoms on the central carbon of curcumin with geminal dimethyl substituents (FLLL31) or a spiro-cyclohexyl ring (FLLL32), the ability of curcumin to enolize is eliminated. These modifications are also predicted to better interact with key binding sites of JAK2 and the SH2 dimerization domain of STAT3 than the keto-enol form of curcumin (Figure 1B). In addition, FLLL31 and FLLL32 feature 3,4-dimethoxy substituents to mimic those of dimethoxycurcumin, a curcumin analogue which has been shown to have increased stability, higher plasma concentration and greater efficacy against cancer cells than standard curcumin (30, 31).
Figure 1.
A. The structures of curcumin, FLLL31, and FLLL32. The chemical modifications made to the latter two compounds prevent enolization and are proposed to confer greater stability and better access to critical domains in JAK2 and STAT3. B. Computer-generated models comparing the interactions of curcumin and FLLL32 with JAK2 (left column) and the STAT3 SH2 domain (right column).
FLLL31 and FLLL32 inhibit JAK2 kinase activity
JAK2 serves as a docking site for the SH2 domain of STAT3 monomers. The subsequent JAK2- mediated phosphorylation of STAT3 activates the transcription factor, promoting its dimerization (32, 33). We evaluated the ability of FLLL31 and FLLL32 to inhibit JAK2 kinase activity. In our comparisons, 5 µM concentrations of FLLL31 and FLLL32 significantly inhibited JAK2 kinase activity (P<0.05) over a DMSO control, resulting in approximately 60% and 75% reductions in activity respectively (Supplemental Figure 1). In addition, our compounds were more effective than previously characterized JAK2 inhibitors, such as AG490 and WP1066, and curcumin itself.
FLLL31 and FLLL32 downregulate STAT3 phosphorylation and DNA-binding activity
To examine the efficacy of FLLL31 and FLLL32 at inhibiting STAT3 phosphorylation, a panel of breast and pancreatic cancer cell lines known to express high endogenous levels of constitutively activated STAT3 was used. FLLL31 and FLLL32 effectively reduced levels of PSTAT3 in MDA-MB-231 breast and PANC-1 pancreatic cancer cells (Figure 2). We observed similar results in the SK-Br-3, MDA-MB-468 and SUM159 breast and the HPAC, BXPC-3 and SW1990 pancreatic cancer cells (Supplemental Figure 2A–2C, 3A–3C respectively). The expression of STAT3 target genes, such as Cyclin D1, survivin, and Bcl-2 (3, 34, 35) were likewise downregulated, and neither compound had appreciable inhibitory effects on the expression or phosphorylation status of other kinases and transcription factors such as ERK1/2, mTOR, PKC-δ, p70S6K, AKT, and Src (Figure 2 and Supplemental Figure 2). Increased levels of P-ERK1/2 were observed in a few cancer cell lines upon FLLL31 and FLLL32 treatments. Decreases in P-STAT3 were concomitant with increased cleavage of pro-caspase-3 and poly-ADP ribose polymerase (PARP), one of the immediate substrates of this effector caspase.
Figure 2.
FLLL31 and FLLL32 are potent inhibitors of STAT3 phosphorylation. A. MDA-MB-231 breast cancer cells and B. PANC-1 pancreatic cancer cells were treated with 2.5 and 5 µM concentrations of FLLL31 and FLLL32 for 24 hours. The Integrated densities of the P-STAT3, STAT3 and cleaved caspase-3 expression were quantified and normalized to GAPDH using ImageJ software. The relatively levels of reduction or increasing of P-STAT3, STAT3, and cleaved caspase-3 were normalized to the DMSO control, which was set as 1.00. C. HMEC and HPDE normal human mammary and pancreatic cells tolerate relatively high dosages of FLLL31 and FLLL32 with no evidence of caspase-3 cleavage. D. FLLL32 inhibits STAT3 phosphorylation in interferon-α-stimulated MDA-MB-453 breast cancer cells. The phosphorylation of both STAT1 and STAT2 is unimpaired. Western blot analysis was analyzed as described in Materials and Methods.
We further examined whether FLLL32 inhibits other human kinase activity using a kinase profile assay. FLLL32 exhibits little inhibition (IC50 is greater than 100µM) on tyrosine kinases, containing SH2 or both SH2 and SH3 domains and other protein kinases such as AKT2, CDK2/Cyclin D1, EGFR, ErbB2/HER2, and Met (Supplemental table 2). These results support the specificity of FLLL32 to inhibit STAT3.
To support that induction of apoptosis is through the inhibition of endogenous STAT3, we transiently express a known constitutively active STAT3, STAT3-C (27) in cancer cells. We rationalize that if FLLL32 induces apoptosis through the inhibition of endogenous STAT3, the restoration of STAT3 activity by transfected STAT3-C should rescue FLLL32-mediated apoptosis. We chose to focus on FLLL32 on this experiment instead of FLLL31 because, based upon the computer modeling and free binding energy, FLLL32 is more selective for STAT3 (Figure 1) and is our leading compound to target STAT3. Our results show that the expression of STAT3-C can rescue the induction of apoptosis evidenced by cleaved caspase-3 in HEK293 cancer cells (Supplemental Figure 4). These results provide additional support that induction of apoptosis by FLLL32 is through the inhibition of endogenous STAT3 protein in cancer cells.
Furthermore, the induction of apoptosis was limited to the cancer cells, as concentrations of FLLL31 and FLLL32 as high as 10 µM were unable to induce caspase-3 cleavage in HMEC or HPDE cells, which represent normal breast and pancreatic cells respectively (Figure 2C). Additional normal cell lines, such as WI-38 lung and BdsMC bladder cells, likewise showed no evidence of apoptosis following FLLL31 and FLLL32 treatment (Supplemental Figure 5). We also found that the expressions of STAT3 phosphorylation levels are low and FLLL32 treatments slightly induce STAT3 phosphorylation levels in WI-38 and BdsMC cells (Supplemental Figure 5) but reduced STAT3 phosphorylation levels in HMEC cells (Data not shown). The selectivity of our compounds for STAT3 inhibition is evident when compared to closely related proteins, such as STAT1 and STAT2. The addition of IFNα results in the robust phosphorylation of STAT1, STAT2 and STAT3 (Figure 2D). Pre-treatment of these cells with FLLL32 effectively inhibited phosphorylation of STAT3 specifically, but had no impact on the extent of STAT1 and STAT2 phosphorylation. FLLL32 was similarly able to block induction of P-STAT3 in MDAMB-453 by interleukin-6 (Supplemental Figure 6).
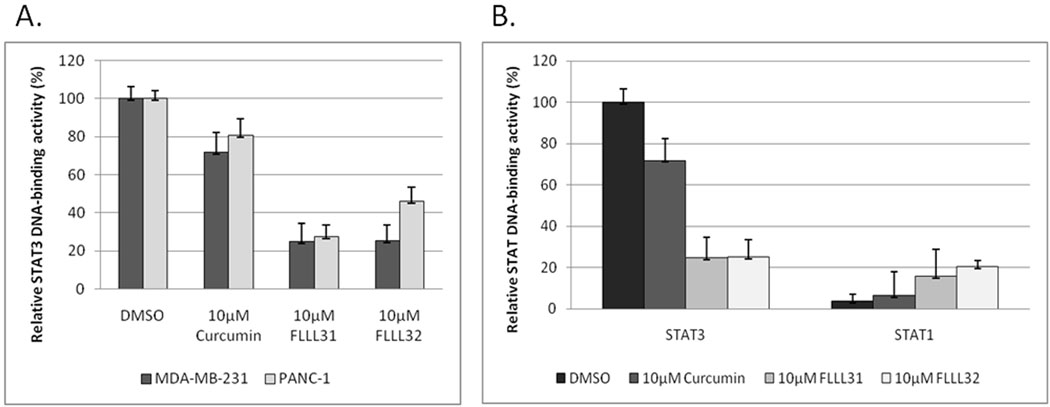
We then examined the ability of STAT3 to bind DNA in breast and pancreatic cancer cells following treatment with FLLL31 and FLLL32. Levels of STAT3 DNA-binding activity dropped sharply in MDA-MB-231 and PANC-1 cells following FLLL31 and FLLL32 treatment, decreases which were significantly lower than those elicited by equimolar concentrations of curcumin (Figure 3A). Similar results were observed in MDA-MB-468 and HPAC (Supplemental Figure 7A–B). It is again important to note that these effects were specific to STAT3 DNA-binding activity. STAT1 DNA-binding activity was not only unimpaired, but actually slightly enhanced by FLLL31 and FLLL32 (Figure 3B), which may have important consequences given the pro-apoptotic and anti-proliferative activities of STAT1 (36, 37).
Figure 3.
A. FLLL31 and FLLL32 reduce STAT3’s ability to bind DNA in nuclear extracts isolated from MDA-MB-231 breast cancer and PANC-1 pancreatic cancer cells. B. FLLL31-and FLLL32-mediated decreases in STAT3 DNA binding activity correlate with increases in STAT1 DNA binding activity in MDA-MB-231 cells.
FLLL31 and FLLL32 are potent inhibitors of STAT3-mediated gene transcription
We next assessed the impact of FLLL31 and FLLL32 on STAT3 transcriptional activity with luciferase assays. Due to its high endogenous levels of P-STAT3, the MDA-MB-231 breast cancer cell line was chosen for stable transfection with pLucTKS3 (Figure 4A). We found that both FLLL31 and FLLL32 reduced luciferase activity in a dose-dependent fashion following 24 hours of treatment (Figure 4B). Furthermore, the extents of these reductions were significantly greater than those induced by comparable dosages of curcumin (p<0.05).
Figure 4.
FLLL31 and FLLL32 inhibit STAT3-dependent transcriptional activation. A. The pLucTKS3 luciferase reporter construct. B. STAT3-mediated expression of luciferase activity is inhibited by FLLL31 and FLLL32 to a greater extent than curcumin. C. MDA-MB-231 breast cancer and D. PANC-1 pancreatic cancer cells show decreased expression of STAT3 downstream target genes following treatment with FLLL31 and FLLL32. RNA was collected and RT-PCR reactions were performed.
To further confirm these observations, we then looked at the expression of several STAT3 target genes, such as Cyclin D1, Bcl-2, Bcl-xl , Survivin and VEGF (38, 39). In MDAMB-231 (Figure 4C) and PANC-1 (Figure 4D) cancer cells, FLLL31 and FLLL32 visibly reduced the expression of the STAT3 target genes over a DMSO control and equimolar concentrations of curcumin.
FLLL31 and FLLL32 inhibit cell viability, cell invasion and generate synergy with doxorubicin
After establishing the efficacy of FLLL31 and FLLL32 as selective JAK2/STAT3 inhibitors, we compared their growth suppressive activities to those produced by previously reported inhibitors including the aforementioned JAK2 inhibitors WP1066, SD-1029, and AG490, and the STAT3 SH2 inhibitors known as Stattic and S3I-201. The IC50 values calculated for each compound and cell line are summarized in Supplemental Table 3. In our comparisons, FLLL31 and FLLL32 showed better growth suppressive activity relative to the other compounds, performing similarly with respect to one another in each of the cell lines tested.
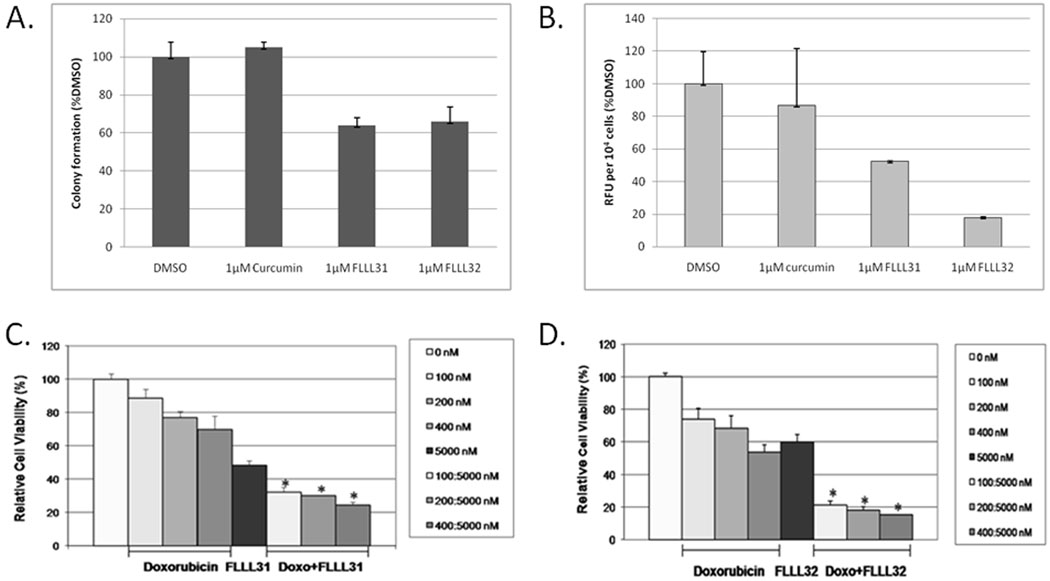
We next looked at the impact FLLL31 and FLLL32 had on the anchorage-independent growth and invasive capacity of the MDA-MB-231 cell line. We assessed anchorage-independent growth with soft-agar colony formation assays (40). As expected, FLLL31 and FLLL32 were both found to inhibit the formation of MDA-MB-231 colonies, each reducing overall colony numbers by approximately 40% over that of curcumin (Figure 5A).
Figure 5.
A. Anchorage-independent growth and colony formation in MDA-MB-231 breast cancer cells is impaired by FLLL31 and FLLL32. B. The invasive capacity of MDA-MB-231 breast cancer cells is reduced to a greater extent by FLLL31 and FLLL32 than curcumin. C. FLLL31 or D. FLLL32 exhibits synergy with doxorubicin (Doxo) in growth suppression of MDA-MB-231 breast cancer cells.
We also determined what effects FLLL31 and FLLL32 had on the invasive capacity of MDA-MB-231, which has been reported to have a highly metastatic phenotype (41). Consistent with our prior observations, FLLL31 and FLLL32 substantially reduced the invasive capacity of MDA-MB-231 over curcumin, which had comparatively minimal impact (Figure 5B).
We also examined whether FLLL31 or FLLL32 could act in an additive or synergistic manner with conventional chemotherapeutic drugs, such as doxorubicin. We found that 100–400 nM dosages of doxorubicin acted in a synergistic fashion with a 5000 nM dose of FLLL31 or FLLL32 (Figure 5C and 5D respectively) in MDA-MB-231 breast cancer cell line, resulting in a significant decrease in cell viability over either agent acting alone (p < 0.05).
FLLL32 suppresses tumor growth and vascularization in vivo
Mouse xenograft experiments were then performed to gauge the anti-tumor effects of our compounds in an in vivo system. We shifted our focus to FLLL32 for these experiments due to its higher predictive binding energy to JAK2 and STAT3 SH2 than FLLL31 (Data not shown). The administration of FLLL32 resulted in significantly reduced tumor burdens in the MDA-MB-231 xenografted mice relative to their DMSO-treated counterparts (Figure 6A). Western blots performed with tumor tissue samples harvested from these mice also showed decreases in total levels of STAT3 phosphorylation.
Figure 6.
A. Comparison of tumor volumes over time in mouse xenografts with MDA-MB-231 breast cancer cells. The mice were given daily intraperitoneal dosages of 50 mg/kg FLLL32 or DMSO. Tumor samples from mice were analyzed with P-STAT3 and one representative sample was shown here. B. Effect of FLLL32 on vascularity and tumor growth in CAMs. The pictures show blood vessel density around xenografted tumors (t). C. Relative tumor sizes of CAM xenografts. D. Relative CAM blood vessel density. Data are from 7–9 individual embryos and 2 separate experiments. * P>0.05, ** P<0.01, *** P<0.001 by one-way ANOVA with Neuman-Keuls post-test. Untreated (UnTx).
We next examined whether FLLL32 would affect tumor growth and vascularity in a chicken embryo chorioallantoic membrane (CAM) human tumor xenograft model (28) (Figure 6B–6D). It was found that gemcitabine, the standard chemotherapeutic agent for treatment of pancreatic cancer, reduced both tumor volume (Figure 6C) and tumor vascularity (Figure 6D) as compared to untreated embryos, but these decreases were statistically insignificant. FLLL32 administration resulted in a significant reduction in both tumor volume (Figure 6C) and tumor vascularity (Figure 6D) in the CAM xenograft assay.
Discussion
In the United States alone, breast and pancreatic cancer are estimated to have caused approximately 40,170 and 35,240 deaths respectively for the year ending in 2009 (42). The present work was prompted by the following observations: a large percentage of pancreatic and breast cancers feature aberrantly activated STAT3 (3), approaches that selectively target constitutive STAT3 signaling have been shown to be effective at inhibiting cancer-associated processes and cancer cell viability (38, 43), curcumin, a natural plant polyphenol, displays some capacity for inhibiting STAT3 without the dose-limiting toxicities inherent to other cancer therapeutic agents (22, 44, 45). Herein we investigated the effects of two novel curcumin-based compounds, designated FLLL31 and FLLL32, against breast and pancreatic cancer in both in vitro and in vivo systems.
FLLL31 and FLLL32 differ with respect to other curcumin analogues in that they were designed to preferentially interact with both the SH2 domain of STAT3 and its upstream activator, the JAK2 kinase. As our data can attest, this strategy produced potent and selective inhibitors of the STAT3 signaling pathway. The merits of targeting STAT3 as a potential cancer therapy have been thoroughly investigated, and a growing body of evidence has shown that the inhibition of constitutively active STAT3 leads to impaired survival and proliferation (3, 38). The ability of curcumin to inhibit the JAK2/STAT3 pathway was one of many factors that made it a desirable choice for a lead compound. In addition to its anti-proliferative and pro-apoptotic effects, curcumin also has antioxidant and anti-inflammatory properties (46). Although we have yet to evaluate FLLL31 and FLLL32 in these other regards, their close structural similarity to curcumin suggest that they may share its pleiotropic nature.
In this report, we have demonstrated that FLLL31 and FLLL32 are effective JAK2/STAT3 inhibitors, and in our testing each had higher efficacy against pancreatic and breast cancer cell viability than currently available commercial JAK2 and STAT3 inhibitors as well as curcumin. Moreover, FLLL31 and FLLL32 are selective for their intended targets, JAK2 and STAT3. FLLL31 and FLLL32 did not inhibit STAT1 DNA binding activity or the phosphorylation of mTOR, p70S6K, ERK1/2, AKT, EGFR, and Src. FLLL32 also had little effect on the kinase activity of five tyrosine kinases containing SH2 and /or SH3 domains and five other protein kinases. The ability of FLLL31 and FLLL32 to generate synergy with doxorubicin in breast cancer cells suggests that a combination of FLLL31 or FLLL32 with doxorubicin may be more effective to treat cancer patients than either drug alone. In addition, the capacity of FLLL32 to inhibit tumor growth and vascularity in a chicken embryo chorioallantoic membrane model using PANC-1 pancreatic cancer cells suggess that FLLL32 may have the potential to inhibit tumor angiogenesis in cancer patients. Furthermore, FLLL32 is capable of suppressing tumor growth in the MDA-MB-231 breast cancer (Figure 6A) and PANC-1 pancreatic cancer (Data not shown) xenografts in mouse tumor models. These results provide the supportive evidence that FLLL32 may be effective to suppress pancreatic and breast tumor cell growth in cancer patients with constitutive STAT3 signaling.
Although both FLLL31 and FLLL32 inhibit JAK2 and STAT3, FLLL31 is better by eliminating the enol form, thus enhancing effective inhibition concentration, while FLLL32 gains additional binding interactions and is more selective to STAT3 and JAK2 than FLLL31. FLLL32 has a docking free energy −8.5 Kcal/mol of binding to STAT3 SH2 compared to FLLL31 with a docking free energy −8.1 Kcal/mol. For binding to JAK2, FLLL32 has a docking free energy −10.3 Kcal/mol compared to FLLL31 has a docking free energy −9.6 Kcal/mol. To our knowledge, FLLL32 is the first small molecular inhibitor that was designed from curcumin to target both JAK2 and STAT3 SH2 domain. The basic requirements for potential chemopreventive and therapeutic agents require them to be: effective against tumor cells in vitro and in mouse tumor models in vivo, relatively non-toxic to the normal cells, and have better bioavailability. Our present investigation of FLLL32 suggests that this small molecular meets many of these criteria, and its further evaluation and optimization may yield even more effective antitumor agents in the future.
Supplementary Material
Acknowledgements
We thank Gregory Lesinski and Dale Hoyt for helpful advice and Yan Liu for technical assistance. This work was supported in part by the Pancreatic Cancer Action Network-AACR, National Foundation for Cancer Research grant, the Susan G. Komen Breast Cancer Foundation, and a NIHR21 grant (R21CA133652-01) to Jiayuh Lin.
References
- 1.Zhong Z, Wen Z, Darnell J. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 2.Sasse J, Hemmann U, Schwartz C, et al. Mutational analysis of acute-phase response factor/Stat3 activation and dimerization. Mol Cell Biol. 1997;17:4677–4686. doi: 10.1128/mcb.17.8.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 4.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8:945–954. [PubMed] [Google Scholar]
- 5.Real P, Sierra A, De Juan A, Segovia J, Lopez-Vega J, Fernandez-Luna J. Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene. 2002;21:7611–7618. doi: 10.1038/sj.onc.1206004. [DOI] [PubMed] [Google Scholar]
- 6.Wang T, Niu G, Kortylewski M, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004:48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 7.Aoki Y, Feldman G, Tosato G. Inhibition of STAT3 signaling induces apoptosis and decreases survivin expression in primary effusion lymphoma. Blood. 2003;101:1535–1542. doi: 10.1182/blood-2002-07-2130. [DOI] [PubMed] [Google Scholar]
- 8.Burke W, Jin X, Liu R, Huang M, Reynolds RK, Lin J. Inhibition of constitutively active Stat3 pathway in ovarian and breast cancer cells. Oncogene. 2001;20:7925–7934. doi: 10.1038/sj.onc.1204990. [DOI] [PubMed] [Google Scholar]
- 9.Calvin D, Nam S, Buettner R, Sekharam M, Torres-Roca J, Jove R. Inhibition of STAT3 activity with STAT3 antisense oligonucleotide (STAT3-ASO) enhances radiation-induced apoptosis in DU145 prostate cancer cells. Int J Radiat Oncol Biol Phys. 2003;57:S297. [Google Scholar]
- 10.Kaptein A, Paillard V, Saunders M. Dominant negative stat3 mutant inhibits interleukin-6-induced Jak-STAT signal transduction. J Biol Chem. 1996;271:5961–5964. doi: 10.1074/jbc.271.11.5961. [DOI] [PubMed] [Google Scholar]
- 11.Takeda K, Noguchi K, Shi W, et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akira S. Roles of STAT3 defined by tissue-specific gene targeting. Oncogene. 2000;19:2607–2611. doi: 10.1038/sj.onc.1203478. [DOI] [PubMed] [Google Scholar]
- 13.Aggarwal BB, Sethi G, Ahn KS, et al. Targeting signal-transducer-and-activator-oftranscription-3 for prevention and therapy of cancer: modern target but ancient solution. Ann N Y Acad Sci. 2006;1091:151–169. doi: 10.1196/annals.1378.063. [DOI] [PubMed] [Google Scholar]
- 14.Duan Z, Bradner JE, Greenberg E, et al. SD-1029 inhibits signal transducer and activator of transcription 3 nuclear translocation. Clin Cancer Res. 2006;12:6844–6852. doi: 10.1158/1078-0432.CCR-06-1330. [DOI] [PubMed] [Google Scholar]
- 15.Meydan N, Grunberger T, Dadi H, et al. Inhibition of acute lymphoblastic leukaemia by a Jak-2 inhibitor. Nature. 1996;379:645–648. doi: 10.1038/379645a0. [DOI] [PubMed] [Google Scholar]
- 16.Iwamaru A, Szymanski S, Iwado E, et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene. 2007;26:2435–2444. doi: 10.1038/sj.onc.1210031. [DOI] [PubMed] [Google Scholar]
- 17.Song H, Wang R, Wang S, Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci USA. 2005;102:4700–4705. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turkson J, Ryan D, Kim J, et al. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem. 2001;276:45443–45455. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- 19.Coleman DRt, Ren Z, Mandal PK, et al. Investigation of the binding determinants of phosphopeptides targeted to the SRC homology 2 domain of the signal transducer and activator of transcription 3. Development of a high-affinity peptide inhibitor. J Med Chem. 2005;48:6661–6670. doi: 10.1021/jm050513m. [DOI] [PubMed] [Google Scholar]
- 20.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol. 2006;13:1235–1242. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 21.Siddiquee K, Zhang S, Guida WC, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci U S A. 2007;104:7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatcher H, Planalp R, Cho J, Torti FM, Torti SV. Curcumin: From ancient medicine to current clinical trials. Cell Mol Life Sci. 2008;65:1631–1652. doi: 10.1007/s00018-008-7452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goel A, Kunnumakkara AB, Aggarwal BB. Curcumin as "Curecumin": from kitchen to clinic. Biochem Pharmacol. 2008;75:787–809. doi: 10.1016/j.bcp.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 24.Aggarwal BB, Sung B. Pharmacological basis for the role of curcumin in chronic diseases: an age-old spice with modern targets. Trends Pharmacol Sci. 2009;30:85–94. doi: 10.1016/j.tips.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Bioavailability of curcumin: problems and promises. Mol Pharm. 2007;4:807–818. doi: 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- 26.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Stat3 activation by src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bromberg J, Wrzeszczynska M, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 28.Lucarelli E, Sangiorgi L, Benassi S, et al. Angiogenesis in lipoma: An experimental study in the chick embryo chorioallantoic membrane. Int J Mol Med. 1999;4:593–596. doi: 10.3892/ijmm.4.6.593. [DOI] [PubMed] [Google Scholar]
- 29.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Advances in enzyme regulation. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 30.Lin L, Shi Q, Nyarko AK, et al. Antitumor agents. 250. Design and synthesis of new curcumin analogues as potential anti-prostate cancer agents. J Med Chem. 2006;49:3963–3972. doi: 10.1021/jm051043z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amolins MW, Peterson LB, Blagg BS. Synthesis and evaluation of electron-rich curcumin analogues. Bioorg Med Chem Lett. 2009;17:360–367. doi: 10.1016/j.bmc.2008.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 33.Reich NC, Liu L. Tracking STAT nuclear traffic. Nature reviews. 2006;6:602–612. doi: 10.1038/nri1885. [DOI] [PubMed] [Google Scholar]
- 34.Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer lett. 2007;251:199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 35.Kanda N, Seno H, Konda Y, et al. STAT3 is constitutively activated and supports cell survival in association with survivin expression in gastric cancer cells. Oncogene. 2004;23:4921–4929. doi: 10.1038/sj.onc.1207606. [DOI] [PubMed] [Google Scholar]
- 36.Bromberg J, Horvath C, Wen Z, Schreiber R, Darnell JJ. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci U S A. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephanou A, Latchman DS. STAT-1: a novel regulator of apoptosis. Int J Exp Pathol. 2003;84:239–244. doi: 10.1111/j.0959-9673.2003.00363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251:199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 39.Kanda N, Seno H, Konda Y, et al. STAT3 is constitutively activated and supports cell survival in association with survivin expression in gastric cancer cells. Oncogene. 2004;23:4921–4929. doi: 10.1038/sj.onc.1207606. [DOI] [PubMed] [Google Scholar]
- 40.Wylie PG, Bowen WP. Determination of cell colony formation in a high-content screening assay. Clinics in laboratory medicine. 2007;27:193–199. doi: 10.1016/j.cll.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 42.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 43.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 44.Chakravarti N, Myers J, Aggarwal B. Targeting constitutive and interleukin-6-inducible signal transducers and activators of transcription 3 pathway in head and neck squamous cell carcinoma cells by curcumin (diferuloylmethane) Int J Cancer. 2006;119:1268–1275. doi: 10.1002/ijc.21967. [DOI] [PubMed] [Google Scholar]
- 45.Bharti A, Donato N, Aggarwal B. Curcumin (diferuloylmethane) inhibits constitutive and IL-6-inducible STAT3 phosphorylation in human multiple myeloma cells. J Immunol. 2003;171:3863–3871. doi: 10.4049/jimmunol.171.7.3863. [DOI] [PubMed] [Google Scholar]
- 46.Aggarwal BB, Sundaram C, Malani N, Ichikawa H. Curcumin: the Indian solid gold. Adv Exp Med Biol. 2007;595:1–75. doi: 10.1007/978-0-387-46401-5_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.