Abstract
Systems biology based on integrative computational analysis and high technology is in a position to construct networks, to study the interactions between molecular components and to develop models of cardiac function and anatomy. Clinical cardiology gets an integrated picture of parameters that are addressed to ventricular and vessel mechanics, cardiac metabolism and electrical activation. The achievement of clinical objectives is based on the interaction between modern technology and clinical phenotype. In this review the need for more sophisticated realization of the structure and function of the cardiovascular system is emphasized while the incorporation of the systems biology concept in predicting clinical phenotypes is a promising strategy that optimize diagnosis and treatment in cardiovascular disease.
Keywords: systems biology, cardiac models, network biology, clinical phenotypes
In the latter part of the last century, biology was driven by reductionist concepts that concerned with information originating from the interactions of genes and molecules. A novel conceptual idea developed over the last few years with the purpose of understanding biological processes which were not explained fully by the properties of genes and molecules. Both evolutionary theories over many decades and systems biology recently have transformed the consensus of biology.
Systems biology (SB) is a new discipline which tries to explain the biological phenomenon not only with delineation of the function of genes and molecules but by reconstructing biochemical supramolecular networks that represent various cellular functions. Systems biology concern with biochemical networks is focused on their functional status and on the nature of the links connecting the molecules of the network1. This concept considers the biological networks as 'systems' of cooperating and interacting components with complex behaviors.
This novel approach is transforming the way of thinking in research, biology and disease. Network modeling could explain cardiovascular metabolism and thus design new diagnostic and therapeutic tools. Also the SB perspective could help to predict and treat complex diseases such as heart failure, metabolic syndrome and diabetes mellitus.
Biological components and systems analysis
In the second half of the previous century after the discovery of the DNA construction, attention focused on explaining the cellular function and the chemical composition of isolated biological components (gene, protein, pathways). This reductionist tendency was increased with the advent of genomics and proteomics. The DNA sequences in a large number of organisms are known but their function at the moment is incomplete. The proteomic technology is important in predicting the activation of some particular genes but remains still in its infancy.
The recent advent of integrative analysis of the different gene products, the new experimental technologies, and in particular the notion that the cells are organized in systems has forced biologists to give special attention to the systems properties. This shift of focus from individual cellular components to systems properties and tissue organization is an inexorable process that eventually would apply to the study of tissue and organ functions. The appearance of novel systems properties in cellular, tissue or organ level, are considered properties emerging from the complex metabolic and regulatory networks of tissues and organs and are not properties of distinct cellular components.
There is a hierarchical coordination that in steps or levels involves genes, genetic products, cellular functions, tissues and organs. There is not a simple relationship between genes but rich biological networks that transform genetic potential into phenotypic reality. The coordinated function at each level between the different components produces a kind of circuit. The term "genetic circuit" was used to designate a collection of different gene products executing a particular cellular function1. Genetic circuits are responsible for information processing, metabolism, cellular function and evolution. Therefore the integrated functions of many genetic circuits form the basic material for cellular function and tissue as well as organ coordinated operation.
In a similar way with the molecular biology and the complex metabolic and regulatory networks, the scientific pursuit can extend to tissue and organ functional coordination in the different levels of the hierarchical ladder. The advances in bioinformatics and clinical phenotype make plausible this kind of research and application2–4.
The proceeding steps of systems biology
The process leading to the emergence of SB is a new paradigm of holistic decoding of biological systems function and consists of four principal steps1: a) Enumeration of biological components participating in the process including the plurality of –omics (Genome, Transcriptome, Proteome, Metabolome). b) Interaction diagrams between these biological components are depicted and the constructed cellular networks are demonstrated. Such examples of biological networks are gene and proteomic networks, immunological networks, and metabolomic and disease networks. c) Reconstructed networks are analyzed and the biological functions are predicted. d) The above emerging models should predict experimental outcomes and explore the phenotypic space.
The universal theories of networks by Barabasi et al emphasized that the cell functional organization is based on biological similar networks that comply with universal laws ruling complex systems5–7.
It is likely that the SB from the level of genome-cell exploration would advance to the broader field of an organismic concept. This broader field of SB from the networks of cellular interactions would be directed to the diverse phenotypic space of tissues and organs. The previous mentioned hierarchical thinking starts from the DNA and continues to the systemic two-dimensional genome-scale stoichiometric matrix. The hierarchical structure extends from introns, exones, alleles and other components of DNA scale, through a network of chemical reactions, modules and pathways, to tissue functional organization and organ cooperation.
The cardiovascular (CV) disorders have the highest mortality rates in the world, change the quality of life and increase the economic burden of the society. Therefore there is great significance to understand the cellular and molecular alterations that influence the progression of cardiac pathologies. Systems biology is an important current area of biological research and the practical application of the accumulated molecular and cellular knowledge can give some answers to the field of cardiology. In this overview we summarize the most important areas of cardiac systems biology as a new approach to decode cardiovascular diseases. The tools of SB are ideal for analyzing the complex molecular programs involved in regulatory cardiac networks.
Systems biology components (-omics) in cardiology
The discipline of SB has evolved after progress was made in molecular biology, new technologies and understanding of complex metabolic and regulatory networks. Important studies have been directed at genome, proteome, transcriptome and metabolome connecting DNA with messenger RNA, intracellular proteins and metabolites. The accumulated and informative data should be integrated as the need has emerged for the construction of a functional cellular model8.
Genomic studies produced an extensive list of genes connected with the heart in various physiological and disease states and in a variety of temporal stages and spatial locations. In some studies genes have been identified and expressed in different diseases encoding special proteins9. These genes were responsible for sarcomeric and cytoskeletal proteins, stress proteins and calcium regulators10. The failure of the molecular and genetic research to explain fully a complex disease like heart failure or coronary heart disease soon became evident and thus the utilization of genomics directed at the functional genomics attempted to fill the existing gaps of our understanding11.
A review article was addressed to all Programs for Genomic Application' (PGAs) that was designed for the advancement of functional genomic research in the field of cardiovascular diseases12. The Cardio Genomics program was assigned to study the transcriptional network of the cardiovascular system under genetic, environmental and disease stresses. This program was addressed to the transcriptional features of murine models of cardiomyopathy and human myocardium, and to gene mutations that are responsible for familial hypertrophic cardiomyopathy.
Genes and their function are unable to explain the functional complexity of a whole organ or organism as multiple proteins are produced by decipherment of one gene while many proteins are modified. Therefore it seems that the proteomics can play a significant role in explaining many cardiac cellular phenotypes. The cellular genotype is regulated by the DNA system, while the cellular phenotype is arranged by the available protein content. The proteomics technology improved the cognition of the molecular mechanisms responsible for the protein emergence and investigated the pathologies in the cardiovascular system (CVS). The proteomics technology using many experimental techniques inquired into the diseased proteome and was most informative about cardiovascular biomarkers13–16.
The biomarkers provide more accurate and sensitive potential than clinical assessment in the identification and characterization of cardiovascular diseases17. Biomarkers are turned out as an important adjunct to clinical evaluation in the CVS and may be molecular in nature (hormone concentration, genome, protein expression) or reveal organ function (pulmonary or systemic artery pressure)18,19. While proteomics is still a new discipline, it is emerging as a technique for screening biomarkers in cells, tissues and fluids. Proteomics also identified candidate proteins altered in pathological states like atherosclerosis, dilated cardiomyopathy and ischemia/reperfusion injury.
Despite the above facts the wide clinical application of these biomarkers for preventive medicine is limited at the moment for various reasons: 1) absence of information on the prevalence and risk contribution of these markers across population; 2) inadequate data of biomarkers inheritance and how they affect individual's risk for various diseases; 3) limited knowledge on how genetic risk factors interact with environmental factors; and 4) further interventional studies are not considered to test the effect of genetic risk factors on common diseases4.
The use of metabolomics to genetic/chemical interventions and the way that it can be integrated with other –omics technologies demonstrated the importance of nuclear magnetic resonance (NMR) in defining metabolic profiles of intact cardiac tissues and linking them to phenotypes20. Grainger DJ studied the metabolic profiling generated by using NMR spectroscopy as a diagnostic test for cardiovascular diseases like identification of individuals who will suffer a myocardial infarction21.
Recently some novel cardiac genes were identified and characterized with the help of libraries of transcript data and use of integrative methods22–24. The newly discovered genes are involved in mitochondrial functions, calcium metabolism and gene transcription which are expected to be useful in future cardiovascular research. In cardiac research the SB concept could be realized only if data can be gathered in different levels-genome, proteome, metabolome. More information for higher functional and interactive levels depends on advanced molecular genetics that cannot be done in man due mainly to ethical reasons8. This kind of genetic studies can be performed in other mammals but up today the only cardiac mammalian cell line are atria-derived HL1 cells from the adult mouse25. Non-mammalian model organisms recently are involved in functional molecular studies with the intention of solving complex biological problems. This is a rational experimental strategy of systemic biology that could be extended to human cardiovascular research26.
Databases and integration
The genomic and proteomic data are voluminous but understanding their functions in a complex system like the heart is a difficult task often based on genetic manipulation and gene targeting in model organisms like the mouse or simpler organisms. This kind of simpler organisms are Escherichia coli, Caenorhabditis elegans, and Saccharomyces cerevesiae27,28. One of the differences of approach between the two kinds of organisms is the time of study. A single transgenic mouse takes months to develop and study while a knockout mouse takes over a year making studies very long and costly29. The simpler organisms are amenable to large scale mutagenesis and rapid phenotyping (genomic phenotyping) while the phenotypic characteristics can be analyzed for genome mutant libraries and a large number of alleles can be stored for further investigation29. The integration of pnenotypic information with genomic or proteomic data can detect novel pathways and identify new networks. In S.cerevesiae the identification of new alleles important for cellular recovery and the integration with phenotypic data on cell recovery managed to detect several protein networks significant for damage recovery28. Also it was discovered that some networks were associated with DNA metabolism and repair, but most of them were found to possess unexpected functions concerning the cytoskeletal remodeling and lipid metabolism.
Therefore the molecular diagnostics integrated with phenotypic characteristics give us the best chance of coming up with the right picture of SB. Probably this data integration will help in the study of cardiac biology with a holistic approach. There are several experimental data attempting to use Drosophila melanogaster for research in the field of cardiac dysfunction30,31. The fruit fly (Drosophila melanogaster) was the first organism with an active circulation with its genome sequenced32. Drosophila was proposed many times as a human disease model and several of these models were examined, particularly for neurological diseases, genetics of aging and adult cardiac dysfunction30–33. Human medicine and biology was enriched with many important findings emanated from studies in Drosophila or other invertebrates. Some important findings are the detection of genes regulating embryonal development in Drosophila, the identification of many components of the apoptotic machinery and the recognition of gene pathways involved in neurogenesis34–36. A Drosophila model suitable for studies in humans is the model with decline of the maximum heart rate with aging, a significant sign of cardiac dysfunction and a cause of restricted physical work in the elderly37.
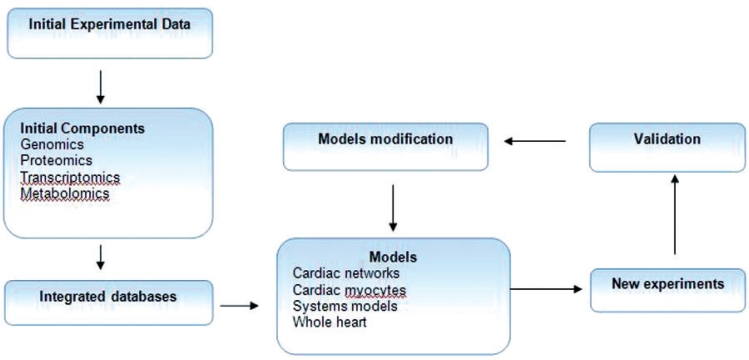
The heart is a complex system and the descriptive study of its molecular components and individual pathways is not adequate without integrating all the informations enclosed in different databases (Figure 1). The data integration is achieved with different proteomic and metabolomic techniques38,39. H-NMR spectroscopy is used to combine proteomics and metabolomic analyses while the joined use of proteomic and metabolomic techniques succeeded in the integrated assessment of enzymes and their corresponding metabolites. Worldwide there are various databases that accumulate impressive amount of biological material, but the use of these databases is nearly impossible as the included material must be gathered manually40–47. The only method for extraction of useful biological information is the integration of the data from different sources with cross-correlation, analysis and attainment of functional networks. The larger fraction of the biological information is included in general databases while the number of the databases addressed specifically to cardiology research is restricted48–50. At the present time another comprehensive database is required to integrate the gathered volume of –omics material with other information such as kinetic data and mathematical models8. The Cardiac Integrated Database Management System (CIDMS) is considered as a database for genes expressed in the heart51.
Figure 1. Systems biology schedule. Formulation of initial components from original experimental data and integration in higher-level databases. The construction of mathematical or biological models precipitates new experiments to acquire new biological parameters which are incorporated and simultaneously modify the models.
Cardiac networks
Previous theoretical studies have demonstrated the main aspects of evolution and deduced a simple relationship between genes and reproduction. This assumption neglected the important biological networks that operate during morphogenesis and development and the capacity to convert genetic potential into phenotypic actuality. Biological and mathematical methods and models have been developed to explore the interaction between networks, evolution and phenotypic variations. These methods were able to reconstruct the biochemical reaction networks of the cells and tissues. The reconstruction process of metabolic, regulatory and signaling networks are concerned with the chemical reactions or interactions that comprise these networks. All these networks are not independent but they interact between themselves and produce the complex biological phenotype1.
Different types and properties of biological networks have been recorded and published in the literature52,53. Extensive biological networks were constructed and studied in simple biological systems like bacteria and yeast but the cardiovascular networks in humans and other mammals are still under development as no large-scale analysis of these networks has been undertaken. That is due probably to inadequate knowledge about interactions between metabolic networks and signaling pathways. Metabolism includes the breakdown of molecules for energy and the synthetic ability for metabolites, while signaling conducts information. A cardiovascular metabolism network has been described that presented cardiovascular metabolism as an interactive and capable network of metabolites designed for regular and predictable energy delivery54. Drake et al produced a network modeling integrating the CVS genetic system of the mouse with gene expression data55. In a recent study it was demonstrated how a network analysis of human coronary artery In-Stent Restenosis (ISR) unraveled significant components of biological pathways that could be targeted for future therapeutic intervention56.
Cardiac myocyte models
The first endeavor for a cardiac myocyte model was the description of the cardiac action and pacemaker potentials based on the Hodgkin-Huxley equations57–58. This model evaluated the sodium and potassium currents during the action potential and described also the mechanistic basis of the plateau of the action potential. The more recent cardiac myocyte ionic models include twenty ionic fluxes and more than forty differential equations29. These models are actually functionally integrated systems models because they incorporate different separate cellular compartments such as the sarcoplasmic reticulum and the dyadic subsarcolemmal space.
Other systems models integrate functional components of cellular subsystems connected with calcium handling, energy production and myocardial filament interactions. Then more complex systems models are created with the functional integration of more than two of the above systems models. An example of this kind of complex interaction and integration is the excitation-contraction (E-C) coupling model correlating electrophysiological with energy metabolism models. Some of this kind of energy-mechanistic E-C coupling systems models have been described during the last thirty years59.
Recently a thermokinetic model was developed explaining the cardiac mitochondrial bioenergetic behavior incorporating information and differential equations for the oxidative phosphorylation, the tricarboxylic acid cycle and mitochondrial calcium handling60.
Cardiac systems models
The use of the classical experimental methodology to understand the biological systems often encounters difficulties particularly in complex pathophysiological circumstances. Then computational models and simulation can help to provide a quantitative interpretation of biological networks and signaling pathways. Simulation of normal and disease conditions performed on or via computer simulation ("in silico") can formulate new hypotheses and predict biological functional differences. These models include a wide range of biological systems in the level of genomics, proteomics, cells, tissues and organs. In cardiac biology and signaling networks this kind of models have been used with success in normal and diseased situations61,62.
The mechanistic system models are low abstraction models that typically codify biochemical reactions, with kinetic rate laws as differential equations, from experimental data. They incorporate quantitative information that flows from one biological component to another and can make quantitative predictions experimentally testable29.
These quantitative models proved efficient in explaining cardiac systems behavior which is confirmed with many such examples. Soulis et al investigated the wall shear stress oscillation in a normal human left coronary artery bifurcation computational model by applying non-Newtonian blood properties and phasic flow. They found that the lateral walls of the bifurcation, where low and oscillating wall shear stress is observed, are more susceptible to atherosclerosis63. Chatzizisis et al demonstrated that the geometrically correct three-dimensional reconstruction of human coronary arteries by integrating intravascular ultrasound and biplane angiography constitutes a promising imaging method for coronaries for either diagnostic or investigational purposes64. Hoffmann et al.predicted functional differences between IkB isoforms in computer simulation and this model tested experimentally proving the above prediction65. Bhalla and Iyengar demonstrated that feedback and crosstalk between neuronal signaling pathways could direct to emergent properties like bistability66. Luo et al used a model of an energetic metabolic network in mammalian myocardial cells and reconstructed it under normal and ischemic conditions. They indicated through simulation that the metabolic networks in myocardial cells under ischemic conditions do not maximize the production of ATP but attain a suboptimal level of energy production adapted to a moderate survival response67.
Models of whole heart phenotype
Computational multicellular models have been useful in formally explaining the propagation of AP in the intracellular and extracellular domains. Drug induced QT prolongation has been studied in model systems designed to clarify the mechanisms of malignant arrhythmias in the genetic long Q-T syndromes (LQTSS) associated with high mortality and mutations of genes encoding the fast and slow potassium channels (IKr and IKs) and the fast sodium channel(INa)68. Anatomical models of ventricular geometry and muscle fiber construction have been built up in animals and man improving the understanding of cardiac electrical impulse propagation and wall mechanics69–71. The cardiac models are structurally or functionally integrated models enquiring about structural organization, voltage-gated channels, membrane transportation, force generation and SB application to study the mitochondria and the functional significance of proteomics72–74.
A recent publication concerning the 4th Fairberg Workshop examined the various functional interactions in the cardiac system and explored the analytical approaches to the integration of the single and multiscale phenomena75. The goals of these Workshops are a) the interdisciplinary interaction: b) the relation of the basic phenomena to cardiac phenotype: c) the application of conceptual modeling: d) the interpretation of clinical data: and e) the enhancement of the international cooperation75.
Conclusions
Systems biology has transformed the understanding of biology and clinical cardiology. The simple relationship between genes and disease has been replaced by biological networks and/or tissue and organ potential in an endeavor to comprehend the clinical phenotypic reality. Therefore systems biology represents a very important contemporary area of biological research in the field of cardiology.
References
- 1.Palsson BO. Systems biology. New York: Cambridge University press; 2006. [Google Scholar]
- 2.Heidecker B, Hare JM. The use off transcriptomic bio-markers for personalized medicine. Heart Fail Rev. 2007;12:1–11. doi: 10.1007/s10741-007-9004-7. [DOI] [PubMed] [Google Scholar]
- 3.Arab S, Gramolini AO, Ping P, Kislinger T, Stanley B, van Eyk J. Cardiovascular proteomics: tools to develop novel biomarkers and potential applications. J Am Coll Cardiol. 2006;48:1733–1741. doi: 10.1016/j.jacc.2006.06.063. [DOI] [PubMed] [Google Scholar]
- 4.Feero WG, Guttmacher AE, Collins FC. The genome gets personal almost. J Am Med Assoc. 2008;299:1351–1352. doi: 10.1001/jama.299.11.1351. [DOI] [PubMed] [Google Scholar]
- 5.Barabasi AL, Bonabeau E. Scale-free networks. Sci Amer. 2003;288:60–69. doi: 10.1038/scientificamerican0503-60. [DOI] [PubMed] [Google Scholar]
- 6.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 7.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 8.Shreenivasaiah PK, Rho S-H, Kim T, Kim DH. An overview of cardiac systems biology. J of Molecular and Cellular Cardiol. 2008;44:460–469. doi: 10.1016/j.yjmcc.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Barrans JD, Liew CC. Chipping away at heart failure. Methods Mol Med. 2006;126:157–169. doi: 10.1385/1-59745-088-X:157. [DOI] [PubMed] [Google Scholar]
- 10.Liew CC, Dzau VJ. Molecular genetics and genomics of heart failure. Nat Rev Genet. 2004;5:811–825. doi: 10.1038/nrg1470. [DOI] [PubMed] [Google Scholar]
- 11.Donahue MP, Marchuk DA, Rockman HA. Redefining heart failure: the utility of genomics. J Amer Coll Cardiol. 2006;48:1289–1298. doi: 10.1016/j.jacc.2006.05.062. [DOI] [PubMed] [Google Scholar]
- 12.Winslow RL, Boguski MS. Genome informatics: current status and future prospects. Circ Res. 2003;92:953–961. doi: 10.1161/01.RES.0000072475.04373.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scobioala S, Klocke R, Michel G, Kuhlmann M, Nikol S. Proteomics: state of the art and its application in cardiovascular research. Curr Med Chem. 2004;11:3203–3218. doi: 10.2174/0929867043363767. [DOI] [PubMed] [Google Scholar]
- 3218.Arrell DK, Neverova I, Van Eyk JE. Cardiovascular proteomics: evolution and potential. Circ Res. 2001;88:763–773. doi: 10.1161/hh0801.090193. [DOI] [PubMed] [Google Scholar]
- 15.White MY, Van Eyk JE. Cardiovascular proteomics: past, present, and future. Mol Diagn Ther. 2007;11:83–95. doi: 10.1007/BF03256227. [DOI] [PubMed] [Google Scholar]
- 16.Matt P, Carrel T, White M, Lefkovits I, Van EykJ. Proteomics in cardiovascular surgery. J Thorc Cardiovasc Surg. 2007;133:210–214. doi: 10.1016/j.jtcvs.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Vasan RS. Biomarkers of cardiovascular disease: molecular basis and practical considerations. Circulation. 2006;113:2335–2362. doi: 10.1161/CIRCULATIONAHA.104.482570. [DOI] [PubMed] [Google Scholar]
- 18.Tang WH, Francis GS, Morrow DA, Newby LK, Cannon CP, Jesse RL. National Academy of Clinical Biochemistry Laboratory Medicine. National Academy of Clinical Biochemistry Laboratory Medicine practice guidelines: clinical utilization of cardiac biomarkers testing in heart failure. Circulation. 2007;116:e99–e109. doi: 10.1161/CIRCULATIONAHA.107.185267. [DOI] [PubMed] [Google Scholar]
- 19.Adams KF. Introduction: biomarkers in heart failure. Heart Fail Rev. 2009 doi: 10.1007/s10741-009-9145-y. Published online. [DOI] [PubMed] [Google Scholar]
- 20.Griffin J. Metabolic profiles to define the genome: can we hear the phenotypes? Philos Trans R Soc Lond B Biol Sci. 2004;359:857–871. doi: 10.1098/rstb.2003.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grainger DJ. Metabolic profiling in heart disease. Heart Metab. 2006;32:22–25. [Google Scholar]
- 22.Jenuth JP, The NCBI Publicly available tools and resources on the Web. Methods Mol Biol. 2000;132:301–312. doi: 10.1385/1-59259-192-2:301. [DOI] [PubMed] [Google Scholar]
- 23.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, et al. NCBI GEO: mining tens of millions of expression profiles-database and tools update. Nucleic Acids Res. 2007;35:D760–765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park I, Hong SE, Kim TW, Lee J, Oh J, Choi E. Comprehensive identification and characterization of novel cardiac genes in mouse. J Mol Cell Cardiol. 2007;43:93–106. doi: 10.1016/j.yjmcc.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 25.White SM, Constantin PE, Claycomb WC. Cardiac physiology at the cellular level: use of cultured HL-1 cardiomyocytes for studies of cardiac muscle cell structure and function. Am J Physiol Heart Circ Physiol. 2004;286:H823–829. doi: 10.1152/ajpheart.00986.2003. [DOI] [PubMed] [Google Scholar]
- 26.Hu N, Sedmera D, Yost HJ, Clark EB. Structure and function of the developing Zebrafish heart. The Anatomical Record. 2000;260:148–157. doi: 10.1002/1097-0185(20001001)260:2<148::AID-AR50>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 27.Singer JB, Hill AE, Burrage LC, et al. Genetic dissection of complex traits with chromosome substitution strains of mice. Science. 2004;304:445–448. doi: 10.1126/science.1093139. [DOI] [PubMed] [Google Scholar]
- 28.Begley TJ, Rosenbach T, Ideker T, Samson LD. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol Cancer Res. 2002;1:103–112. [PubMed] [Google Scholar]
- 29.McCulloch AD, Paternostro G. Cardiac systems biology. Ann NY Acad Sci. 2005;1047:283–295. doi: 10.1196/annals.1341.025. [DOI] [PubMed] [Google Scholar]
- 30.Lakatta EG. Heart aging: a fly in the ointment? Circ Res. 2001;88:984–986. doi: 10.1161/hh1001.091963. [DOI] [PubMed] [Google Scholar]
- 31.Paternostro G, Vignola C, Bartsch DU, et al. Age-associated cardiac dysfunction in Drosophila melanogaster. Circ Res. 2001;88:1053–1058. doi: 10.1161/hh1001.090857. [DOI] [PubMed] [Google Scholar]
- 32.Adams MD, Celniker SE, Holt RA, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 33.Fanny MB, Bender WW. A Drosophila model of Parkinson's disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 34.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 35.White K, Grether ME, Abrams JM, et al. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 36.Artavanis-Tsakonas S, Muskavitch MA, Yedvobnick B. Molecular cloning of Notch, a locus affecting neurogenesis in Drosophila melanogaster. Proc Natl Acad Sci USA. 1983;80:1977–1981. doi: 10.1073/pnas.80.7.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lakatta EG. Introduction: chronic heart failure in older persons. Heart Fail Rev. 2002;7:5–8. doi: 10.1023/a:1013708104410. [DOI] [PubMed] [Google Scholar]
- 38.Joyce AR, Palsson BO. The model organism as a system: integrating-' omics' data sets. Nat Rev Mol Cell Biol. 2006;7:198–210. doi: 10.1038/nrm1857. [DOI] [PubMed] [Google Scholar]
- 39.Mayr M, Madhu B, Xu Q. Proteomics and metabolomics combined in cardiovascular research. Trends in Cardiovasc Med. 2007;17:43–48. doi: 10.1016/j.tcm.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Couto FM, Silva MJ, Lee V, Dimmer E, Camon E, Apweiler R. GO annotator: linking protein GO annotations to evidence text. J Biomed Discov Collab. 2006;1:1–19. doi: 10.1186/1747-5333-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P. Human protein reference database – 2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong SE, Rho SH, Yeom YI, Kim DH. HCNet: a database of heart and calcium functional network. Bioinformatics. 2006;22:2053–2054. doi: 10.1093/bioinformatics/btl331. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Q, Lu M, Shi L, Rui W, Zhu X, Chen G. Cardio: a web-based knowledge resource of genes and proteins related to cardiovascular disease. Int J Cardiol. 2004;97:245–249. doi: 10.1016/j.ijcard.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 50.Cotter D, Guda P, Fahy E, Subramaniam S. MitoProteome: mitochondrial protein sequence database and annotation system. Nucleic Acids Res. 2004;32:D463–D467. doi: 10.1093/nar/gkh048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.CIDMS: Cardiac Integrated Database Management System, version 1.0. [online] Available from URL: http://cidms.org.
- 52.Zhu X, Gerstein M, Snyder M. Getting connected: analysis and principles of biological networks. Genes Dev. 2007;21:1010–1024. doi: 10.1101/gad.1528707. [DOI] [PubMed] [Google Scholar]
- 53.Bansal M, Belcastro V, Ambesi-Impiombato A, di Bernardo D. How to infer gene networks from expression profiles. Mol Syst Biol. 2007;3:78–88. doi: 10.1038/msb4100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weiss JN, Yang L, Qu Z. Systems biology approaches to metabolic and cardiovascular disorders: network perspectives of cardiovascular metabolism. J Lipid Res. 2006;47:2355–2366. doi: 10.1194/jlr.R600023-JLR200. [DOI] [PubMed] [Google Scholar]
- 55.Drake TA, Schadt EE, Lusis AJ. Integrating genetic and geneexpression data: application to cardiovascular and metabolic traits in mice. Mamm Genome. 2006;17:466–479. doi: 10.1007/s00335-005-0175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ashley EA, Ferrara R, King JY, Vailaya A, Kuchinsky A, He X. Network analysis of human in-stent restenosis. Circulation. 2006;114:2644–2654. doi: 10.1161/CIRCULATIONAHA.106.637025. [DOI] [PubMed] [Google Scholar]
- 57.Noble D. Cardiac action and pacemaker potentials based on the Hodgkin-Huxley equations. Nature. 1960;188:495–497. doi: 10.1038/188495b0. [DOI] [PubMed] [Google Scholar]
- 58.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rice JJ, de Tombe PP. Approaches to modeling crossbridges and calcium dependent activation in cardiac muscle. Prog Biophys Mol Biol. 2004;85:179–195. doi: 10.1016/j.pbiomolbio.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 60.Cortassa S, Aon MA, Marban E, et al. An integrated model of cardiac mitochondrial energy metabolism and calcium dynamics. Biophys J. 2003;84:2734–2755. doi: 10.1016/S0006-3495(03)75079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ideker T, Lauffenburger D. Building with a scaffold: emerging strategies for high- to low-level cellular modeling. Trends Biotechnol. 2003;21:255–262. doi: 10.1016/S0167-7799(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 62.Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signaling circuits in molecular interaction networks. Bioinformatics. 2002;18:S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 63.Soulis JV, Giannoglou GD, Chatzizisis YS, Farmakis TM, Giannakoulas GA, Parcharidis GE, et al. Spatial and phasic oscillation of non-Newtonian wall shear stress in human left coronary artery bifurcation: an insight to atherogenesis. Coronary Artery Dis. 2006;17:351–358. doi: 10.1097/00019501-200606000-00005. [DOI] [PubMed] [Google Scholar]
- 64.Chatzizisis YS, Giannoglou GD, Matakos A, Basdekidou C, Sianos G, Panagiotou A, et al. In-vivo accuracy of geometrically correct three-dimensional reconstruction of human coronary arteries: is it influenced by certain parameters? Coronary Artery Dis. 2006;17:545–551. doi: 10.1097/00019501-200609000-00008. [DOI] [PubMed] [Google Scholar]
- 65.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB -NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 66.Bhalla US, Iyengar R. Emergent properties of networks of biological signaling pathways. Science. 1999;283:381–387. doi: 10.1126/science.283.5400.381. [DOI] [PubMed] [Google Scholar]
- 67.Luo RY, Liao S, Tao GY, Li YY, Zeng S, Li YX. Dynamic analysis of optimality in myocardial energy metabolism under normal and ischemic conditions. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100071. 2006.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saucerman JJ, Healy SN, Belik ME, Puglisi JL, McCulloch AD. Proarrhythmic consequences of a KCNQ1 AKAP-binding domain mutation. Computational models of whole cells and heterogeneous tissue. Circ Res. 2004;95:1216–1224. doi: 10.1161/01.RES.0000150055.06226.4e. [DOI] [PubMed] [Google Scholar]
- 69.Stevens C, Hunter PJ. Sarcomere length changes in a 3D mathematical model of the pig ventricles. Prog Biophys Mol Biol. 2003;82:229–241. doi: 10.1016/s0079-6107(03)00023-3. [DOI] [PubMed] [Google Scholar]
- 70.Harrild D, Henriquez C. A computer model of normal conduction in the human atria. Circ Res. 2000;87:E25–E36. doi: 10.1161/01.res.87.7.e25. [DOI] [PubMed] [Google Scholar]
- 71.Xie F, Qu Z, Yang J, Baher A, Weiss JN, Garfinkel A. A simulation study of the effects of cardiac anatomy in ventricular fibrillation. J Clin Invest. 2004;113:686–693. doi: 10.1172/JCI17341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Noble D, Rudy Y. Models of cardiac ventricular action potentials: iterative interaction between experiment and simulation. Philos Trans: Mats, Phys and Engi Sci. 2001;359:1127–1142. [Google Scholar]
- 73.Winslow RL, Greenstein JL. The ongoing journey to understand heart function through integrative modeling. Circ Res. 2004;95:1135–1136. doi: 10.1161/01.RES.0000151330.81518.73. [DOI] [PubMed] [Google Scholar]
- 74.Vo TD, Palsson BO. Building the power house: recent advances in mitochondrial studies through proteomics and systems biology. Am J Physiol Cell Physiol. 2007;292:C164–177. doi: 10.1152/ajpcell.00193.2006. [DOI] [PubMed] [Google Scholar]
- 75.Sideman S. The challenge of cardiac modeling-interaction and integration. Ann NY Acad Sci. 2006;1080:xi–xxiii. doi: 10.1196/annals.1380.001. [DOI] [PubMed] [Google Scholar]