Every year in the United States, 160,000 cases of colorectal cancer are diagnosed, and 57,000 patients die of the disease, making it the second leading cause of death from cancer among adults.1 The disease begins as a benign adenomatous polyp, which develops into an advanced adenoma with high-grade dysplasia and then progresses to an invasive cancer.2 Invasive cancers that are confined within the wall of the colon (tumor–node–metastasis stages I and II) are curable, but if untreated, they spread to regional lymph nodes (stage III) and then metastasize to distant sites (stage IV).3-5 Stage I and II tumors are curable by surgical excision, and up to 73% of cases of stage III disease are curable by surgery combined with adjuvant chemotherapy.3,4,6 Recent advances in chemotherapy have improved survival, but stage IV disease is usually incurable.3,4
The clinical behavior of a colorectal cancer results from interactions at many levels (Fig. 1). The challenges are to understand the molecular basis of individual susceptibility to colorectal cancer and to determine factors that initiate the development of the tumor, drive its progression, and determine its responsiveness or resistance to antitumor agents. This review summarizes areas of current knowledge, recognizing that the topics presented are only fragments of the total picture.
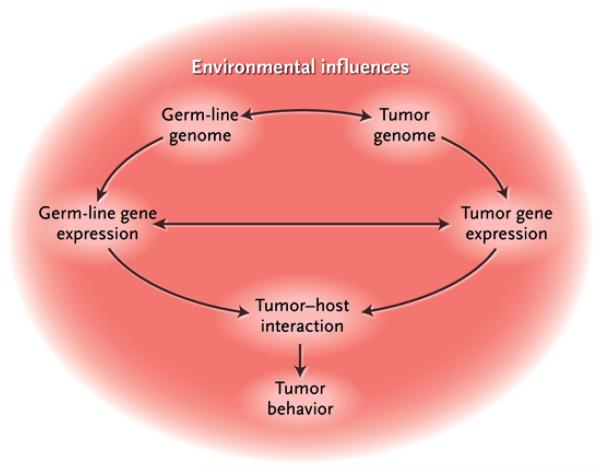
Figure 1. Multifactorial Colorectal Carcinogenesis.
The molecular events that drive the initiation, promotion, and progression of colorectal cancer occur on many interrelated levels. This dynamic process involves interactions among environmental influences, germ-line factors dictating individual cancer susceptibility, and accumulated somatic changes in the colorectal epithelium.
GENOMIC INSTABILITY
The loss of genomic stability can drive the development of colorectal cancer by facilitating the acquisition of multiple tumor-associated mutations. In this disease, genomic instability takes several forms, each with a different cause (Table 1).7-26
Table 1.
Patterns of Genomic Instability in Colorectal Cancer.*
| Type of Instability and Syndrome | Type of Defect |
Genes Involved | Phenotype |
|---|---|---|---|
| Chromosomal instability – loss of heterozygosity at multiple loci | Somatic | Loss of heterozygosity at APC, TP53, SMAD47,8 | Characteristic of 80 to 85% of sporadic colorectal cancers, depending on stage |
| DNA mismatch-repair defects | |||
| Hereditary nonpolyposis colon cancer | Germ-line | MLH1, MSH2, MSH6 germ-line gene mutations9-14 | Multiple primary colorectal cancers, accelerated tumor progression, and increased risk of endometrial, gastric, and urothelial tumors |
| Sporadic colorectal cancer with mismatch-repair deficiency | Somatic | MLH1 somatic methylation15-17 | Colorectal cancer with increased risk of poor differentiation, more commonly located in right colon, less aggressive clinical behavior than tumors without mismatch-repair deficiency |
| CpG island methylator phenotype – methylation target loci | Somatic | Target loci MLH1, MINT1, MINT2, MINT318-23 | Characteristic of 15% of colorectal cancers, with most showing mismatch-repair deficiency from loss of tumor MLH1 expression |
| Base excision repair defect – MYH-associated polyposis | Germ-line | MYH24-26 | Development of 15 or more colorectal adenomas with increased risk of colorectal cancer |
MYH denotes mutY homologue.
CHROMOSOMAL INSTABILITY
The most common type of genomic instability in colorectal cancer is chromosomal instability, which causes numerous changes in chromosomal copy number and structure.7 Chromosomal instability is an efficient mechanism for causing the physical loss of a wild-type copy of a tumor-suppressor gene, such as APC, P53, and SMAD family member 4 (SMAD4), whose normal activities oppose the malignant phenotype.2,27,28 In colorectal cancer, there are numerous rare inactivating mutations of genes whose normal function is to maintain chromosomal stability during replication, and in the aggregate, these mutations account for most of the chromosomal instability in such tumors.8 In contrast to some other cancers, colorectal cancer does not commonly involve amplification of gene copy number29 or gene rearrangement.
DNA-REPAIR DEFECTS
In a subgroup of patients with colorectal cancer, there is inactivation of genes required for repair of base–base mismatches in DNA, collectively referred to as mismatch-repair genes (Fig. 2 and 3). The inactivation can be inherited, as in hereditary nonpolyposis colon cancer (HNPCC), also known as the Lynch syndrome, or acquired, as in tumors with methylation-associated silencing of a gene that encodes a DNA mismatch-repair protein.
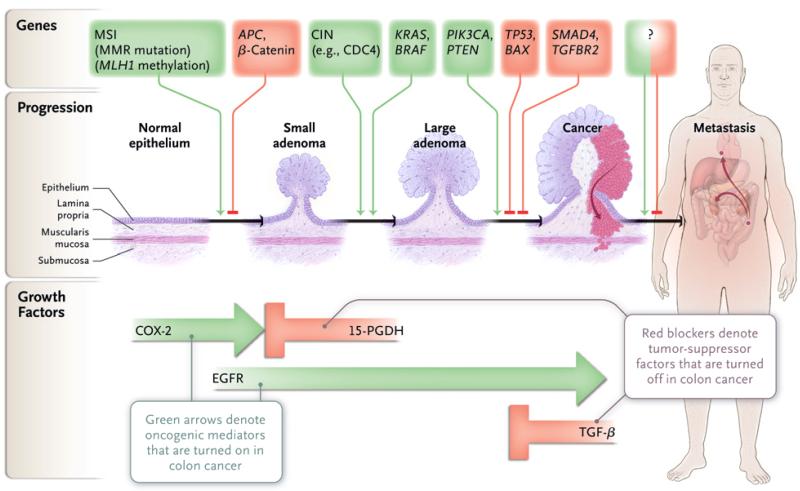
Figure 2. Genes and Growth Factor Pathways That Drive the Progression of Colorectal Cancer.
In the progression of colon cancer, genetic alterations target the genes that are identified at the top of the diagram. The microsatellite instability (MSI) pathway is initiated by mismatch-repair (MMR) gene mutation or by aberrant MLH1 methylation and is further associated with downstream mutations in TGFBR2 and BAX. Aberrant MLH1 methylation and BRAF mutation are each associated with the serratedadenoma pathway. The question mark indicates that genetic or epigenetic changes specific to metastatic progression have not been identified. Key growth factor pathways that are altered during colon neoplasia are shown at the bottom of the diagram. CIN denotes chromosomal instability, EGFR epidermal growth factor receptor, 15-PGDH 15-prostaglandin dehydrogenase, and TGF-β transforming growth factor β.
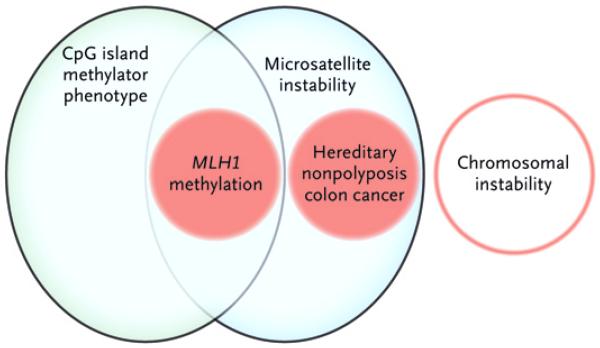
Figure 3. Genetic Instability Pathways That Drive Colon Neoplasias.
Shown are the overlapping relationships that define the major pathways of genomic instability in colon cancers: chromosomal instability, microsatellite instability caused by defects in DNA mismatch-repair genes that are either inherited as germ-line defects (e.g., in hereditary nonpolyposis colon cancer) or somatically acquired (e.g., by aberrant methylation and epigenetic silencing of MLH1), and the CpG island methylator phenotype.
In patients with HNPCC, germ-line defects in mismatch-repair genes (primarily MLH1 and MSH2) confer a lifetime risk of colorectal cancer of about 80%, with cancers evident by the age of 45 years, on average.10-13,30,31 The loss of mismatch-repair function in patients with HNPCC is due not only to the mutant germ-line mismatch-repair gene but also to somatic inactivation of the wild-type parental allele.31 Genomic instability arising from mismatch-repair deficiency dramatically accelerates the development of cancer in patients with HNPCC — some cancers arise within 36 months after normal results on colonos-copy.32 For this reason, yearly colonoscopy is recommended for carriers of an HNPCC mutation,30,32 and prophylactic colectomy should be considered for patients with high-grade lesions. Germ-line mutations of another mismatch-repair gene, MSH6, attenuates the predisposition to familial cancer.9,33,34 Somatic inactivation of mismatch-repair genes occurs in approximately 15% of patients with nonfamilial colorectal cancer. In these patients, biallelic silencing of the promoter region of the MLH1 gene by promoter methylation inactivates mismatch repair15-17 (Fig. 2 and 3).
The loss of mismatch-repair function is easy to recognize by the associated epiphenomenon of microsatellite instability, in which the inability to repair strand slippage within repetitive DNA sequence elements changes the size of the mononucleotide or dinucleotide repeats (micro-satellites) that are scattered throughout the genome. Mismatch-repair deficiency can also be detected by immunohistochemical analysis, which can identify the loss of one of the mismatch-repair proteins.14,35-37 Cancers characterized by mismatch-repair deficiency arise primarily in the proximal colon, and in sporadic cases, they are associated with older age and female sex.30 In mismatch-repair deficiency, tumor-suppressor genes, such as those encoding transforming growth factor β (TGF-β) receptor type II (TGFBR2) and BCL2-associated X protein (BAX), which have functional regions that contain mononucleo-tide or dinucleotide repeat sequences, can be inactivated.2,27,28
An alternative route to colorectal cancer involves germ-line inactivation of a base excision repair gene, mutY homologue (MUTYH, also called MYH).25,33 The MYH protein excises from DNA the 8-oxoguanine product of oxidative damage to guanine.24,25,33 In persons who carry two inactive germ-line MYH alleles, a polyposis phenotype develops, with a risk of colorectal cancer of nearly 100% by the age of 60 years.33 MYH-associated polyposis is increasingly recognized: one third of all persons in whom 15 or more colorectal adenomas develop have MYH-associated polyposis.33 The diagnosis requires genetic testing, which is facilitated by two mutations, Y165C and G382D, that together account for 85% of cases.33 Thus far, somatic inactivation of MYH has not been detected in colorectal cancer.
ABERRANT DNA METHYLATION
Epigenetic silencing of genes, mostly mediated by aberrant DNA methylation, is another mechanism of gene inactivation in patients with colorectal cancer.18,20 A methylated form of cytosine in which a methyl group is attached to carbon 5 (5-methylcytosine) defines a fifth DNA base, introduced by DNA methylases that modify cyto-sines within CpG dinucleotides.18 In the normal genome, cytosine methylation occurs in areas of repetitive DNA sequences outside of exons; it is largely excluded from the CpG-rich “CpG islands” in the promoter regions of approximately half of all genes.18 By comparison, in the colorectal-cancer genome, there is a modest global depletion of cytosine methylation but considerable acquisition of aberrant methylation within certain promoter-associated CpG islands.18 This aberrant promoter-associated methylation can induce epigenetic silencing of gene expression.18 In sporadic colorectal cancer with microsatellite instability, somatic epi-genetic silencing blocks the expression of MLH1.18
Among the loci that can undergo aberrant methylation in colorectal cancer, a subgroup seems to become aberrantly methylated as a group, a phenomenon called the CpG island methylator phenotype (CIMP, or CIMP-high).18,19 The molecular mechanism for CIMP remains unknown, but the phenomenon is reproducibly observed in about 15% of colorectal cancers and is present in nearly all such tumors with aberrant methylation of MLH118,19,21,38 (Fig. 2 and 3). The pathogenetic consequence of MLH1 silencing is well established, but the contribution of other epigenetic silencing events to colorectal carcino-genesis remains an area of ongoing study. An intermediate level of aberrant methylation in CIMP may define a subtype (i.e., CIMP2 and CIMP-low) that is thought to account for 30% of CIMP cases.22,23 A third pattern of aberrant methylation is exemplified by exon 1 of the gene encoding vimentin. Although this locus is not expressed by normal colon mucosa or colorectal cancer, it is aberrantly methylated in 53 to 83% of patients with colorectal cancer in a pattern that is independent of CIMP.39,40
MUTATIONAL INACTIVATION OF TUMOR-SUPPRESSOR GENES
APC
Colorectal cancers acquire many genetic changes, but certain signaling pathways are clearly singled out as key factors in tumor formation (Fig. 2 and Table 2).41-62 One of these changes, the activation of the Wnt signaling pathway, is regarded as the initiating event in colorectal cancer.2,28,43 Wnt signaling occurs when the oncoprotein β-catenin binds to nuclear partners (members of the T-cell factor–lymphocyte enhancer factor family) to create a transcription factor that regulates genes involved in cellular activation.2,28,43 The β-catenin degradation complex controls levels of the β-catenin protein by proteolysis. A component of this complex, APC, not only degrades β-catenin but also inhibits its nuclear localization.
Table 2.
Tumor-Suppressor Genes and Oncogenes Commonly Associated with Colorectal Cancer.*
| Affected Gene | Frequency % |
Nature of Defect | Comments |
|---|---|---|---|
| APC | 85 | Activation of Wnt signaling due to inability to degrade the β-catenin oncoprotein41,42 | Germ-line mutation in familial adenomatous polyposis; somatic inactivation found in 85% of sporadic colorectal cancers43 |
| MLH1, MSH2, MSH6 | 15–25 | DNA single-nucleotide mismatch-repair defect permitting the accumulation of oncogenic mutations and tumor-suppressor loss10-14,31,35 | Germ-line mutation in hereditary nonpolyposis colorectal cancer30; epigenetic silencing causes loss of tumor MLH1 protein expression |
| TP53 | 35–55 | Encoding a protein responsible for cell-cycle regulation44,45; inactivating missense mutations paired with loss of heterozygosity at 17p | Germ-line mutation in Li–Fraumeni syndrome46 |
| TGFBR2 | 25–30 | Receptor responsible for signaling pathways mediating growth arrest and apoptosis; inactivated by frameshift mutation in polyA repeat within TGFBR2 coding sequence in patients with mismatch-repair defects47 or by inactivating mutation of kinase domain48,49 | Mutation present in >90% of tumors with microsatellite instability and 15% of microsatellite stable colon cancers50 |
| SMAD4 | 10–35 | Critical components of transforming growth factor β pathway signaling, along with related proteins SMAD2 and SMAD3; SMAD4 and SMAD2 are located on chromosome 18q, a frequent site of loss of heterozygosity in colorectal cancers; inactivated by homozygous deletion or mutation51,52 | Germ-line mutations in familial juvenile polyposis, with a risk of colorectal cancer as high as 60% over three to four decades53 |
| KRAS | 35–45 | Encoding the KRAS G-protein, with constitutive activation resulting in activation of both the PI3K–PDK1–PKB and RAF–MEK–ERK1/2 signaling pathways, thereby promoting cell survival and apoptosis suppression54,55 | Germ-line mutation in the cardiofaciocutaneous syndrome56 |
| BRAF V600E | 8–12 | Activating mutation in the BRAF serine–threonine kinase, a downstream mediator of signaling through the RAF–MEK–ERK1/2 pathway, which mimics the biologic consequences of KRAS mutation38,57 | Associated with hyperplastic polyposis, with increased incidence in serrated adenomas58,59; like KRAS, germ-line mutation in the cardiofaciocutaneous syndrome56 |
| PTEN | 10–15 | Promotion of the activation of PI3K pathway signaling through loss of function by inactivating mutation, resulting in cell-survival signaling and apoptosis suppression | Germ-line mutation in Cowden's syndrome, which carries a high risk of breast cancer, with 10% increased risk of colorectal cancer; possible role in maintenance of chromosomal stability60-62 |
ERK denotes extracellular signal–regulated kinase, MAPK mitogen-activated protein kinase, MEK MAPK kinase, PDK1 pyruvate dehydrogenase kinase isozyme 1, PI3K phosphatidylinositol 3-kinase, and PKB protein kinase B.
The most common mutation in colorectal cancer inactivates the gene that encodes the APC protein. In the absence of functional APC — the brake on β-catenin — Wnt signaling is inappropriately and constitutively activated. Germ-line APC mutations give rise to familial adenomatous polyposis, an inherited cancer-predisposition syndrome in which more than 100 adenomatous polyps can develop; in carriers of the mutant gene, the risk of colorectal cancer by the age of 40 years is almost 100%.2,30,43 Somatic mutations and deletions that inactivate both copies of APC are present in most sporadic colorectal adenomas and cancers.2,43 In a small subgroup of tumors with wild-type APC, mutations of β-catenin that render the protein resistant to the β-catenin degradation complex activate Wnt signaling.2,41-43
TP53
The inactivation of the p53 pathway by mutation of TP53 is the second key genetic step in colorectal cancer. In most tumors, the two TP53 alleles are inactivated, usually by a combination of a missense mutation that inactivates the transcriptional activity of p53 and a 17p chromosomal deletion that eliminates the second TP53 allele.2,27,28,44,45 Wild-type p53 mediates cell-cycle arrest and a cell-death checkpoint, which can be activated by multiple cellular stresses.63 The inactivation of TP53 often coincides with the transition of large adenomas into invasive carcinomas.64 In many colorectal cancers with mismatch-repair defects, TP53 remains wild-type, though in these cancers the activity of the p53 pathway is probably attenuated by mutations in the BAX inducer of apoptosis.2,28
TGF-β TUMOR-SUPPRESSOR PATHWAY
The mutational inactivation of TGF-β signaling is a third step in the progression to colorectal cancer.50 In about one third of colorectal cancers, somatic mutations inactivate TGFBR2.47,49,50,65,66 In tumors with mismatch-repair defects, TGFBR2 is inactivated by distinctive frameshift mutations in a polyadenine repeat within the TGFBR2 coding sequence.47 In at least half of all colorectal cancers with wild-type mismatch repair, TGF-β signaling is abolished by inactivating missense mutations that affect the TGFBR2 kinase domain or, more commonly, mutations and deletions that inactivate the downstream TGF-β pathway component SMAD4 or its partner transcription factors, SMAD2 and SMAD3.29,47,49-51,65-68 Mutations that inactivate the TGF-β pathway coincide with the transition from adenoma to high-grade dysplasia or carcinoma.69
ACTIVATION OF ONCOGENE PATHWAYS
RAS AND BRAF
Several oncogenes play key roles in promoting colorectal cancer (Fig. 2 and Table 2). Oncogenic mutations of RAS and BRAF, which activate the mitogen-activated protein kinase (MAPK) signaling pathway, occur in 37% and 13% of colorectal cancers, respectively.21,55,57,70,71 RAS mutations, principally in KRAS, activate the GTPase activity that signals directly to RAF. BRAF mutations signal BRAF serine–threonine kinase activity, which further drives the MAPK signaling cascade.70,71 BRAF mutations are detectable even in small polyps,21 and as compared with RAS mutations, they are more common in hyperplastic polyps, serrated adenomas, and proximal colon cancers, particularly in those with the CIMP phenotype (Fig. 3). Patients with numerous and large hyper-plastic lesions, a condition termed the hyperplastic polyposis syndrome, have an increased risk of colorectal cancer, with disease progression occurring through an intermediate lesion with a serrated luminal border on histologic analysis.18,22,38,58,59
PHOSPHATIDYLINOSITOL 3-KINASE
One third of colorectal cancers bear activating somatic mutations in PI3KCA, which encodes the catalytic subunit of phosphatidylinositol 3-kinase (PI3K).72 Less common genetic alterations that may substitute for PI3KCA mutations include loss of PTEN, an inhibitor of PI3K signaling, as well as amplification of insulin receptor substrate 2 (IRS2), an upstream activator of PI3K signaling, and coamplification of AKT and PAK4, which are downstream mediators of PI3K signaling.73
SEQUENCING THE COLORECTAL-CANCER GENOME
Advances in DNA sequencing technology have made it possible to sequence the entire coding genome of a human cancer. Colorectal cancer provided the first example of the power of this technology, with high-throughput sequencing of 18,000 members of the Reference Sequence (RefSeq) database of the National Center for Biotechnology Information.65,66 Cancer-associated somatic mutations were identified in 848 genes. Of these, 140 were identified as candidate cancer genes that probably contributed to the cancer phenotype because they were mutated in at least two colorectal cancers and when corrected for gene size showed more mutations than expected by chance.
The average stage IV colorectal-cancer genome bears 15 mutated candidate cancer genes and 61 mutated passenger genes (very-low-frequency mutational events). The predominance of low-frequency mutations in candidate cancer genes implies enormous genetic heterogeneity among colorectal cancers, which mirrors the heterogeneity of the clinical behavior of colorectal cancers. it difficult to determine the clinical effect of individual mutational events. Moreover, these initial results are probably conservative, because some mutations, which were initially labeled as rare “passengers” in colorectal cancer, have subsequently emerged as common and are probably pathogenetic in other cancer types (e.g., an IDH1 mutation noted initially in one colorectal cancer but subsequently in many gliomas).65,66,74
High-throughput sequencing of the colorectal-cancer genome has identified new common mutational targets. These include the ephrin receptors EPHA3 and EPHB6 (receptor tyrosine kinases), which together are mutated in 20% of colorectal cancers, and FBXW7, which functions in a protein degradation pathway that regulates levels of cyclin E and is mutated in 14% of colorectal cancers.65,66,75 An important challenge is to reduce the complexity of the 140 candidate cancer genes by identifying the biologic pathways and processes that are common downstream targets of multiple mutational events.
GENOMIC CHANGES AND TUMOR PROGRESSION
The sequence of transformation from adenoma to carcinoma, as initially formulated,2,28,43 was a model of the development of colorectal cancer in which specific tumor-promoting mutations are progressively acquired. This model predicts the presence of mutations that dictate specific tumor characteristics, such as the presence of regional or distant metastases (Fig. 2). Unexpectedly, the examination of results of full-genome sequencing from primary colorectal cancers and distant metastases in the same patient showed no new mutations in the metastases,76 implying that new mutations are not required to enable a tumor cell to leave the primary tumor and seed a distant site. Because the ongoing generation of mutations serves as a molecular clock, the finding that all the mutations in a metastasis are also present in the primary tumor implies that metastatic seeding is rapid, requiring less than 24 months from the appearance of the final mutation in the primary tumor.76
GROWTH FACTOR PATHWAYS
ABERRANT REGULATION OF PROSTAGLANDIN SIGNALING
The activation of growth factor pathways is common in colorectal cancer (Fig. 2). An early and critical step in the development of an adenoma is the activation of prostaglandin signaling.77,78 This abnormal response can be induced by inflammation or mitogen-associated up-regulation of COX-2, an inducible enzyme that mediates the synthesis of prostaglandin E2, an agent strongly associated with colorectal cancer.78 Prostaglandin E2 activity can also be increased by the loss of 15-prostaglandin dehydrogenase (15-PGDH), the rate-limiting enzyme in catalyzing degradation of prostaglandin.79-81 Increased levels of COX-2 are found in approximately two thirds of colorectal cancers,78,82 and there is loss of 15-PGDH in 80% of colorectal adenomas and cancers.79 Clinical trials have shown that the inhibition of COX-2 by nonsteroidal antiinflammatory drugs prevents the development of new adenomas83-86 and mediates regression of established adenomas.87
EPIDERMAL GROWTH FACTOR RECEPTOR
Epidermal growth factor (EGF) is a soluble protein that has trophic effects on intestinal cells. Clinical studies have supported an important role of signaling through the EGF receptor (EGFR) in a subgroup of colorectal cancers.88-91 EGFR mediates signaling by activating the MAPK and PI3K signaling cascades. Recent clinical data have shown that advanced colorectal cancer with tumor-promoting mutations of these pathways — including activating mutations in KRAS,92-94 BRAF,95,96 and the p110 subunit of PI3K97 — do not respond to anti-EGFR therapy.
VASCULAR ENDOTHELIAL GROWTH FACTOR
Vascular endothelial growth factor (VEGF) that is produced in states of injury or during the growth of normal tissue drives the production of new stromal blood vessels (angiogenesis). Clinical studies have suggested a role for angiogenic pathways in the growth and lethal potential of colorectal cancer. Treatment with the anti-VEGF antibody bevacizumab added an average of 4.7 months to the overall survival of patients with advanced colorectal cancer (15.6 months with standard therapy).98 The identification of molecular distinctions between cancers that benefit from this treatment and those that do not remains a challenge.
STEM-CELL PATHWAYS
Stem cells in colorectal cancers are believed to be uniquely endowed with the capacity to renew themselves.99-102 Single colorectal-cancer stem cells, by definition, can lodge in a permissive site, such as the liver, and produce a metastasis. Currently, it is not possible to isolate individual colorectal-cancer stem cells, although certain cell-surface proteins (e.g., CD133, CD44, CD166, and aldehyde dehydrogenase 1) are promising markers. Normal stem cells that reside in the colonic crypt rely on adhesive and soluble stromal–epithelial interactions to maintain division and differentiation. The extent of alterations in these regulatory mechanisms in colorectal-cancer stem cells is a promising area of investigation, since agents that control the growth of colorectal-cancer stem cells could theoretically be used for cancer prevention and treatment.
PREDICTIVE AND PROGNOSTIC MARKERS
One ongoing challenge is to translate the wealth of knowledge regarding colorectal-cancer genomics into clinically applicable predictive or prognostic tests (Table 3). The relation between mutations in EGFR signaling components RAS and BRAF and anti-EGFR therapy is currently the only application of colorectal-cancer genomics to treatment.92-96 A few genomic markers are useful for prognosis. For example, germ-line mutations in tumor-suppressor genes, such as APC, MLH1, and MSH2, indicate a very high risk of colorectal cancer and guide the frequency of colorectal-cancer surveillance and recommendations for prophylactic surgery. Other somatic markers have modest or unconfirmed prognostic value and are not currently used to direct care. Sporadic colorectal cancers with a mismatch-repair deficiency generally have a favorable prognosis35,103,105,108; poor survival in stage II and III colon cancers is associated with the loss of p27 (a proapoptotic regulator of the cell cycle109) or the loss of heterozygosity at chromosomal location 18q.105
Table 3.
Prognostic and Predictive DNA Markers in Colorectal Cancer.*
| DNA Marker | Comments |
|---|---|
| Prognostic | |
| APC | A germ-line mutation defines the colorectal-cancer predisposition syndrome, familial adenomatous polyposis, with an 80 to 100% lifetime risk of colorectal cancer. Patients with germ-line APC mutations undergo prophylactic colectomy or proctocolectomy. |
| MLH1,MSH2, MSH6 | A germ-line mutation in these and, less commonly, in other mismatch-repair genes defines hereditary nonpolyposis colon cancer, with a 40 to 80% lifetime risk of colorectal cancer, as well as an increased risk of endometrial cancer. Patients with germ-line mismatch-repair gene mutations undergo frequent colonoscopic surveillance and may be considered for prophylactic colectomy and hysterectomy. |
| MLH1 methylation– associated silencing | The somatic inactivation of MLH1 in primary colorectal cancers is evidenced by either detection of DNA microsatellite instability or loss of tumor MLH1 protein expression on immunohistochemical analysis, and is more frequent in early-stage colorectal cancers than in advanced disease. Such inactivation may be a marker of more indolent disease or a better prognosis in the absence of adjuvant chemotherapy.103,104 |
| 18q Loss of heterozygosity | The somatic loss of heterozygosity at chromosomal location 18q, a site containing genes associated with colorectal cancer (e.g., SMAD4 and SMAD2), is associated with a poorer outcome in patients with stage II or stage III colon cancer than that in patients with tumors retaining both parental alleles at 18q.105 |
| Predictive | |
| KRAS | The somatic mutation produces unrestricted activity of signaling through the MAPK and PI3K cascades. Patients with stage IV colorectal cancer and activating mutations in KRAS do not have a response to EGFR-inhibitor therapy.92-94 |
| BRAF V600E | The somatic mutation activating this kinase causes unrestricted MAPK pathway signaling. Patients with stage IV colorectal cancer and the activating BRAF V600E mutation do not have a response to EGFR-inhibitor therapy.95 |
| MLH1 methylation-associated silencing | The loss of the mismatch-repair function contributes to the loss of other tumor suppressors (e.g., TGFBR2 and BAX). Patients with mismatch-repair–deficient tumors may not have a response to fluorouracil and may have an improved response to irinotecan-containing regimens.106,107 |
BAX denotes BCL2-associated X protein, EGFR epidermal growth factor receptor, MAPK mitogen-activated protein kinase, PI3K phosphatidylinositol 3-kinase, and TGFBR2 transforming growth factor receptor β type II.
NONINVASIVE MOLECULAR DETECTION
The development of molecular diagnostics for the early detection of colorectal cancer is an important translation of colon-cancer genetics into clinical practice. One example is the development of assays to detect mutations that are specific to colorectal cancer and cancer-associated aberrant DNA methylation in fecal DNA from patients with colorectal cancer or advanced adenomas. These assays have a sensitivity of 46 to 77% for detecting early-stage colorectal cancer, which is superior to the sensitivity of testing for fecal occult blood although their superiority in preventing death from cancer has not been shown.39,110-113 Stool DNA testing for colorectal cancer has been added to the cancer-screening guidelines of the American Cancer Society114 and appears to be equally sensitive for detecting advanced adenomas.115 Although still in the developmental stage, assays for detecting plasma cell-free DNA may also be clinically useful,115 and assays for tumor-specific plasma protein or RNA profiles also remain targets of research. Questions that remain to be resolved are the optimal interval between serial tests and the performance and cost-effectiveness of stool DNA testing as compared with those of newer immuno-chemical fecal occult-blood tests.116
GENETIC INFLUENCES IN POPULATION SUSCEPTIBILITY
Genetic epidemiology and twin studies indicate that 35 to 100% of colorectal cancers and adenomas develop in persons with an inherited susceptibility to the disease.117-119 In addition, an HNPCC-like syndrome occurs in some families without any evidence of defects in mismatch repair.120 Several genomic loci that could harbor highly penetrant susceptibility genes have been identified with the use of linkage approaches,121-123 but the underlying mutations are unknown. Genomewide association studies have also identified germ-line DNA variants that are strongly associated with susceptibility, but the clinical use of these results is probably limited, since the relative risk associated with these variants is low.124-129
CONCLUSIONS
Studies that aid in the understanding of colorectal cancer on a molecular level have provided important tools for genetic testing for high-risk familial forms of the disease, predictive markers for selecting patients for certain classes of drug therapies, and molecular diagnostics for the noninvasive detection of early cancers. In addition, biologic pathways that could form the basis of new therapeutic agents have been identified. Although some high-frequency mutations are attractive targets for drug development, common signaling pathways downstream from these mutations may also be tractable as therapeutic targets. Recent progress in molecular assays for the early detection of colorectal cancer indicates that understanding the genes and pathways that control the earliest steps of the disease and individual susceptibility can contribute to clinical management in the near term.
An understanding of the signals that dictate the metastatic phenotype will provide the information necessary to develop drugs to control or prevent advanced disease. The considerable recent advances encourage us to believe that improvements in our knowledge of the molecular basis of colorectal cancer will continue to reduce the burden of this disease.
Footnotes
No other potential conflict of interest relevant to this article was reported.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Kinzler KW, Vogelstein B. Colorectal tumors. In: Vogelstein B, Kinzler KW, editors. The genetic basis of human cancer. 2nd ed McGraw-Hill; New York: 2002. pp. 583–612. [Google Scholar]
- 3.Libutti SK, Saltz LB, Tepper JE. Colon cancer. In: DeVita VT Jr, Lawrence TS, Rosenberg SA, editors. DeVita, Hellman, and Rosenberg's cancer: principles and practice of oncology. Vol. 1. Lippincott Williams & Wilkins; Philadelphia: 2008. pp. 1232–84. [Google Scholar]
- 4.Compton C, Hawk ET, Grochow L, Lee F, Ritter M, Niederhuber JE. Colon cancer. In: Abeloff MD, Armitage J, Niederhuber JE, Kastan MB, McKenna GW, editors. Abeloff's clinical oncology. Churchill Livingstone; Philadelphia: 2008. pp. 1477–534. [Google Scholar]
- 5.Markowitz SD, Dawson DM, Willis J, Willson JK. Focus on colon cancer. Cancer Cell. 2002;1:233–6. doi: 10.1016/s1535-6108(02)00053-3. [DOI] [PubMed] [Google Scholar]
- 6.André T, Boni C, Mounedji-Boudiaf L, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004;350:2343–51. doi: 10.1056/NEJMoa032709. [DOI] [PubMed] [Google Scholar]
- 7.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–7. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 8.Barber TD, McManus K, Yuen KW, et al. Chromatoid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:3443–8. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyaki M, Konishi M, Tanaka K, et al. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet. 1997;17:271–2. doi: 10.1038/ng1197-271. [DOI] [PubMed] [Google Scholar]
- 10.Leach FS, Nicolaides NC, Papadopoulos N, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–25. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 11.Papadopoulos N, Nicolaides NC, Wei YF, et al. Mutations of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–9. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 12.Fishel R, Lescoe MK, Rao MR, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–38. doi: 10.1016/0092-8674(93)90546-3. [Erratum, Cell 1994;77:167.] [DOI] [PubMed] [Google Scholar]
- 13.Bronner CE, Baker SM, Morrison PT, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–61. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 14.Ionov Y, Peinado M, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–61. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 15.Kane MF, Loda M, Gaida GM, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–11. [PubMed] [Google Scholar]
- 16.Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–5. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veigl ML, Kasturi L, Olechnowicz J, et al. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci U S A. 1998;95:8698–702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–93. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 19.Toyota M, Ahuja N, Ohe-Toyota M, Herman J, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–6. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004;23:29–39. doi: 10.1023/a:1025806911782. [DOI] [PubMed] [Google Scholar]
- 21.Nosho K, Irahara N, Shima K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3(11):e3698. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen L, Toyota M, Kondo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A. 2007;104:18654–9. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barault L, Charon-Barra C, Jooste V, et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68:8541–6. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- 24.Jones S, Emmerson P, Maynard J, et al. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C→T:A mutations. Hum Mol Genet. 2002;11:2961–7. doi: 10.1093/hmg/11.23.2961. [DOI] [PubMed] [Google Scholar]
- 25.Al-Tassan N, Chmiel NH, Maynard J, et al. Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–32. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 26.Cleary SP, Cotterchio M, Jenkins MA, et al. Germline MutY human homologue mutations and colorectal cancer: a multisite case-control study. Gastroenterology. 2009;136:1251–60. doi: 10.1053/j.gastro.2008.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grady WM, Markowitz S. Colorectal cancer: genetic alterations. In: Kelsen D, Daly J, Kern S, Levin B, Tepper J, editors. Gastrointestinal oncology: principles and practice. Lippincott Williams & Wilkins; Philadelphia: 2002. pp. 685–702. [Google Scholar]
- 28.Fearon ER, Bommer GT. Molecular biology of colorectal cancer. In: DeVita VT Jr, Lawrence TS, Rosenberg SA, editors. De-Vita, Hellman, and Rosenberg's cancer: principles & practice of oncology. Vol. 1. Lippincott Williams & Wilkins; Philadelphia: 2008. pp. 1218–31. [Google Scholar]
- 29.Leary RJ, Lin JC, Cummins J, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:16224–9. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynch HT, Lynch JF, Lynch PM, Attard T. Hereditary colorectal cancer syndromes: molecular genetics, genetic counseling, diagnosis and management. Fam Cancer. 2008;7:27–39. doi: 10.1007/s10689-007-9165-5. [DOI] [PubMed] [Google Scholar]
- 31.Boland CR, Koi M, Chang DK, Carethers JM. The biochemical basis of microsatellite instability and abnormal immunohistochemistry and clinical behavior in Lynch syndrome: from bench to bedside. Fam Cancer. 2008;7:41–52. doi: 10.1007/s10689-007-9145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Järvinen HJ, Aarnio M, Mustonen H, et al. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology. 2000;118:829–34. doi: 10.1016/s0016-5085(00)70168-5. [DOI] [PubMed] [Google Scholar]
- 33.Kastrinos F, Syngal S. Recently identified colon cancer predispositions: MYH and MSH6 mutations. Semin Oncol. 2007;34:418–24. doi: 10.1053/j.seminoncol.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolodner RD, Tytell JD, Schmeits JL, et al. Germ-line msh6 mutations in colorectal cancer families. Cancer Res. 1999;59:5068–74. [PubMed] [Google Scholar]
- 35.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–9. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 36.Aaltonen LA, Peltomäki P, Leach FS, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–6. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 37.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–60. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 38.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 39.Chen WD, Han ZJ, Skoletsky J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–32. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 40.Zou H, Harrington JJ, Shire AM, et al. Highly methylated genes in colorectal neoplasia: implications for screening. Cancer Epidemiol Biomarkers Prev. 2007;16:2686–96. doi: 10.1158/1055-9965.EPI-07-0518. [DOI] [PubMed] [Google Scholar]
- 41.Korinek V, Barker N, Morin PJ, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–7. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 42.Morin PJ, Sparks AB, Korinek V, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 43.Goss KH, Groden J. Biology of the adenomatous polyposis coli tumor suppressor. J Clin Oncol. 2000;18:1967–79. doi: 10.1200/JCO.2000.18.9.1967. [DOI] [PubMed] [Google Scholar]
- 44.Baker SJ, Fearon ER, Nigro JM, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–21. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 45.Baker SJ, Markowitz S, Fearon ER, Willson JK, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–5. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 46.Malkin D, Li FP, Strong LC, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–8. doi: 10.1126/science.1978757. [Erratum, 1993;259:878.] [DOI] [PubMed] [Google Scholar]
- 47.Markowitz S, Wang J, Myeroff L, et al. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–8. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 48.Parsons R, Myeroff LL, Liu B, et al. Microsatellite instability and mutations of the transforming growth factor β type II receptor gene in colorectal cancer. Cancer Res. 1995;55:5548–50. [PubMed] [Google Scholar]
- 49.Grady WM, Myeroff LL, Swinler SE, et al. Mutational inactivation of transforming growth factor β receptor type II in microsatellite stable colon cancers. Cancer Res. 1999;59:320–4. [PubMed] [Google Scholar]
- 50.Grady WM, Markowitz SD. TGF-β signaling pathway and tumor suppression. In: Derynck R, Miyazano K, editors. The TGF-β family. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2008. pp. 889–938. [Google Scholar]
- 51.Thiagalingam S, Lengauer C, Leach FS, et al. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet. 1996;13:343–6. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 52.Takagi Y, Kohmura H, Futamura M, et al. Somatic alterations of the DPC4 gene in human colorectal cancers in vivo. Gastroenterology. 1996;111:1369–72. doi: 10.1053/gast.1996.v111.pm8898652. [DOI] [PubMed] [Google Scholar]
- 53.Howe JR, Roth S, Ringold JC, et al. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science. 1998;280:1086–8. doi: 10.1126/science.280.5366.1086. [DOI] [PubMed] [Google Scholar]
- 54.Downward J. Ras signalling and apoptosis. Curr Opin Genet Dev. 1998;8:49–54. doi: 10.1016/s0959-437x(98)80061-0. [DOI] [PubMed] [Google Scholar]
- 55.Bos JL, Fearon ER, Hamilton SR, et al. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327:293–7. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- 56.Aoki Y, Niihori T, Narumi Y, Kure S, Matsubara Y. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum Mutat. 2008;29:992–1006. doi: 10.1002/humu.20748. [DOI] [PubMed] [Google Scholar]
- 57.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 58.O'Brien MJ. Hyperplastic and serrated polyps of the colorectum. Gastroenterol Clin North Am. 2007;36:947–68. doi: 10.1016/j.gtc.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 59.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–30. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 60.Liaw D, Marsh DJ, Li J, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–7. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 61.Marsh DJ, Dahia PL, Zheng Z, et al. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat Genet. 1997;16:333–4. doi: 10.1038/ng0897-333. [DOI] [PubMed] [Google Scholar]
- 62.Yin Y, Shen WH. PTEN: a new guardian of the genome. Oncogene. 2008;27:5443–53. doi: 10.1038/onc.2008.241. [DOI] [PubMed] [Google Scholar]
- 63.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7:979–87. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 64.Baker SJ, Preisinger AC, Jessup JM, et al. p53 Gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 1990;50:7717–22. [PubMed] [Google Scholar]
- 65.Sjöblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 66.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 67.Eppert K, Scherer SW, Ozcelik H, et al. MADR2 maps to 18q21 and encodes a TGFβ-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86:543–52. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 68.Riggins GJ, Thiagalingam S, Rozenblum E, et al. MAD-related genes in the human. Nat Genet. 1996;13:347–9. doi: 10.1038/ng0796-347. [DOI] [PubMed] [Google Scholar]
- 69.Grady WM, Rajput A, Myeroff L, et al. Mutation of the type II transforming growth factor-β receptor is coincident with the transformation of human colon adenomas to malignant carcinomas. Cancer Res. 1998;58:3101–4. [PubMed] [Google Scholar]
- 70.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 71.Siena S, Sartore-Bianchi A, Di Nicolantonio F, Balfour J, Bardelli A. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst. 2009;101:1308–24. doi: 10.1093/jnci/djp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 73.Parsons DW, Wang TL, Samuels Y, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 74.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rajagopalan H, Jallepalli PV, Rago C, et al. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 76.Jones S, Chen WD, Parmigiani G, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–8. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Markowitz SD. Aspirin and colon cancer — targeting prevention? N Engl J Med. 2007;356:2195–8. doi: 10.1056/NEJMe078044. [DOI] [PubMed] [Google Scholar]
- 78.Cha YI, DuBois RN. NSAIDs and cancer prevention: targets downstream of COX-2. Annu Rev Med. 2007;58:239–52. doi: 10.1146/annurev.med.57.121304.131253. [DOI] [PubMed] [Google Scholar]
- 79.Yan M, Rerko RM, Platzer P, et al. 15-Hydroxyprostaglandin dehydrogenase, a COX-2 oncogene antagonist, is a TGF-β-induced suppressor of human gastrointestinal cancers. Proc Natl Acad Sci U S A. 2004;101:17468–73. doi: 10.1073/pnas.0406142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Myung SJ, Rerko RM, Yan M, et al. 15-Hydroxyprostaglandin dehydrogenase is an in vivo suppressor of colon tumorigenesis. Proc Natl Acad Sci U S A. 2006;103:12098–102. doi: 10.1073/pnas.0603235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Backlund MG, Mann JR, Holla VR, et al. 15-Hydroxyprostaglandin dehydrogenase is down-regulated in colorectal cancer. J Biol Chem. 2005;280:3217–23. doi: 10.1074/jbc.M411221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chan AT, Ogino S, Fuchs CS. Aspirin use and risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–42. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 83.Baron JA, Cole BF, Sandler RS, et al. A randomized trial of aspirin to prevent colorectal adenomas. N Engl J Med. 2003;348:891–9. doi: 10.1056/NEJMoa021735. [DOI] [PubMed] [Google Scholar]
- 84.Sandler RS, Halabi S, Baron JA, et al. A randomized trial of aspirin to prevent colorectal adenomas in patients with previous colorectal cancer. N Engl J Med. 2003;348:883–90. doi: 10.1056/NEJMoa021633. [Erratum, N Engl J Med 2003;348:1939.] [DOI] [PubMed] [Google Scholar]
- 85.Bertagnolli MM, Eagle CJ, Zauber AG, et al. Celecoxib for the prevention of sporadic colorectal adenomas. N Engl J Med. 2006;355:873–84. doi: 10.1056/NEJMoa061355. [DOI] [PubMed] [Google Scholar]
- 86.Arber N, Eagle CJ, Spicak J, et al. Celecoxib for the prevention of colorectal adenomatous polyps. N Engl J Med. 2006;355:885–95. doi: 10.1056/NEJMoa061652. [DOI] [PubMed] [Google Scholar]
- 87.Steinbach G, Lynch PM, Phillips RKS, et al. The effect of celecoxib, a cyclooxygenase-2 inhibitor, in familial adenomatous polyposis. N Engl J Med. 2000;342:1946–52. doi: 10.1056/NEJM200006293422603. [DOI] [PubMed] [Google Scholar]
- 88.Saltz LB, Meropol NJ, Loehrer PJ, Needle MN, Kopit J, Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004;22:1201–8. doi: 10.1200/JCO.2004.10.182. [DOI] [PubMed] [Google Scholar]
- 89.Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–45. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 90.Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–87. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 91.Van Cutsem E, Peeters M, Siena S, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol. 2007;25:1658–64. doi: 10.1200/JCO.2006.08.1620. [DOI] [PubMed] [Google Scholar]
- 92.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–34. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 93.Lièvre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–9. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 94.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 95.Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–12. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 96.Wong R, Cunningham D. Using predictive biomarkers to select patients with advanced colorectal cancer for treatment with epidermal growth factor receptor antibodies. J Clin Oncol. 2008;26:5668–70. doi: 10.1200/JCO.2008.19.5024. [Erratum, J Clin Oncol 2009;27:3070.] [DOI] [PubMed] [Google Scholar]
- 97.Jhawer M, Goel S, Wilson AJ, et al. PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab. Cancer Res. 2008;68:1953–61. doi: 10.1158/0008-5472.CAN-07-5659. [Erratum, Cancer Res 2008;68:6859.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hurwitz H, Fehrenbacher L, Novotny W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–42. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 99.Boman BM, Huang E. Human colon cancer stem cells: a new paradigm in gastrointestinal oncology. J Clin Oncol. 2008;26:2828–38. doi: 10.1200/JCO.2008.17.6941. [DOI] [PubMed] [Google Scholar]
- 100.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 101.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 102.Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–63. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Samowitz WS, Curtin K, Ma KN, et al. Microsatellite instability in sporadic colon cancer is associated with an improved prognosis at the population level. Cancer Epidemiol Biomarkers Prev. 2001;10:917–23. [PubMed] [Google Scholar]
- 104.Kim GP, Colangelo LH, Wieand HS, et al. Prognostic and predictive roles of high-degree microsatellite instability in colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J Clin Oncol. 2007;25:767–72. doi: 10.1200/JCO.2006.05.8172. [DOI] [PubMed] [Google Scholar]
- 105.Watanabe T, Wu T-T, Catalano PJ, et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med. 2001;344:1196–206. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–57. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bertagnolli MM, Niedzwiecki D, Compton CC, et al. Microsatellite instability predicts improved response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: Cancer and Leukemia Group B Protocol 89803. J Clin Oncol. 2009;27:1814–21. doi: 10.1200/JCO.2008.18.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–6. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bertagnolli MM, Warren RS, Niedzwiecki D, et al. p27Kip1 in stage III colon cancer: implications for outcome following adjuvant chemotherapy in Cancer and Leukemia Group B protocol 89803. Clin Cancer Res. 2009;15:2116–22. doi: 10.1158/1078-0432.CCR-08-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704–14. doi: 10.1056/NEJMoa033403. [DOI] [PubMed] [Google Scholar]
- 111.Itzkowitz SH, Jandorf L, Brand R, et al. Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol. 2007;5:111–7. doi: 10.1016/j.cgh.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 112.Itzkowitz S, Brand R, Jandorf L, et al. A simplified, noninvasive stool DNA test for colorectal cancer detection. Am J Gastroenterol. 2008;103:2862–70. doi: 10.1111/j.1572-0241.2008.02088.x. [DOI] [PubMed] [Google Scholar]
- 113.Ahlquist DA, Sargent DJ, Loprinzi CL, et al. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann Intern Med. 2008;149:441–50. doi: 10.7326/0003-4819-149-7-200810070-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Levin B, Lieberman DA, McFarland B, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J Clin. 2008;58:130–60. doi: 10.3322/CA.2007.0018. [DOI] [PubMed] [Google Scholar]
- 115.Li M, Chen WD, Papadopoulos N, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol. 2009;27:858–63. doi: 10.1038/nbt.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hundt S, Haug U, Brenner H. Comparative evaluation of immunochemical fecal occult blood tests for colorectal adenoma detection. Ann Intern Med. 2009;150:162–9. doi: 10.7326/0003-4819-150-3-200902030-00005. [DOI] [PubMed] [Google Scholar]
- 117.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer: analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 118.Hemminki K, Mutanen P. Genetic epidemiology of multistage carcinogenesis. Mutat Res. 2001;473:11–21. doi: 10.1016/s0027-5107(00)00162-7. [DOI] [PubMed] [Google Scholar]
- 119.Cannon-Albright L, Skolnick M, Bishop T, Lee R, Burt R. Common inheritance of susceptibility to colonic adenomatous polyps and associated colorectal cancers. N Engl J Med. 1988;319:533–7. doi: 10.1056/NEJM198809013190902. [DOI] [PubMed] [Google Scholar]
- 120.Lindor NM, Rabe K, Petersen GM, et al. Lower cancer incidence in Amsterdam-I criteria families without mismatch repair deficiency: familial colorectal cancer type X. JAMA. 2005;293:1979–85. doi: 10.1001/jama.293.16.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wiesner GL, Daley D, Lewis S, et al. A subset of familial colorectal neoplasia kindreds linked to chromosome 9q22.2-31.2. Proc Natl Acad Sci U S A. 2003;100:12961–5. doi: 10.1073/pnas.2132286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Papaemmanuil E, Carvajal-Carmona L, Sellick GS, et al. Deciphering the genetics of hereditary non-syndromic colorectal cancer. Eur J Hum Genet. 2008;16:1477–86. doi: 10.1038/ejhg.2008.129. [DOI] [PubMed] [Google Scholar]
- 123.Neklason DW, Kerber RA, Nilson DB, et al. Common familial colorectal cancer linked to chromosome 7q31: a genome-wide analysis. Cancer Res. 2008;68:8993–7. doi: 10.1158/0008-5472.CAN-08-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 125.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 126.Broderick P, Carvajal-Carmona L, Pittman AM, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–7. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 127.Tomlinson IP, Webb E, Carvajal-Carmona L, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–30. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 128.Tenesa A, Farrington SM, Prendergast JG, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–7. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Houlston RS, Webb E, Broderick P, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–35. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]