Abstract
Cellular homeostasis is linked tightly to mitochondrial functions. Some damage to mitochondrial proteins and nucleic acids can lead to the depolarization of the inner mitochondrial membrane, thereby sensitizing impaired mitochondria for selective elimination by autophagy. Mitochondrial dysfunction is one of the key aspects of the pathobiology of neurodegenerative disease. Parkin, an E3 ligase located in the cytosol and originally discovered as mutated in monogenic forms of Parkinson’s disease (PD), was found recently to translocate specifically to uncoupled mitochondria and to induce their autophagy.
Keywords: Parkin, Parkinson’s disease, mitophagy, mitochondrial quality control, PINK1
Mitochondria and mitochondrial autophagy, mitophagy
One of the cellular organelles, mitochondria, can be paraphrased as a master regulator for the life of cell, due to mitochondrial functions in energy consumption and promotion of cell death, apoptosis [1]. Recent work on the biology of the autophagy has demonstrated that autophagy is one of the regulating mechanisms for mitochondrial quality control, especially as an active mechanism for elimination of damaged or excess mitochondria from the cell [2] [3] [4].
Recent studies on mammalian systems have suggested that mitochondrial elimination by autophagy is essential for variety cellular events, including the maturation of erythroid cells [5] [6] [7] and the maintenance of neuronal tissues, which may be targeted in neurodegenerative diseases [8]. Mitochondria also have a risk of causing normal cell dysfunction at any time due to problems, such as oxidative stress [9] or mutations in the mitochondrial genome [10], occurring along the metabolic pathway mediated by mitochondrial respiratory chain complexes. Thus, mitochondria need to remain functional by several mechanisms since the function is tightly linked to the homeostasis of cells (Figure 1). Cells may compensate for mitochondrial defects by the function of antioxidant enzymes, DNA repair, or complementing the damage through the fusion of a healthy mitochondrion with a damaged mitochondrion. Alternatively, proteins on the damaged mitochondria may be selectively degraded [11]. Furthermore, in order to prevent the problem of unsalvageable damaged mitochondria spreading within the cell, there is a mechanism to eliminate the dysfunctional mitochondria by autophagy, called mitophagy [2] [3] [4].
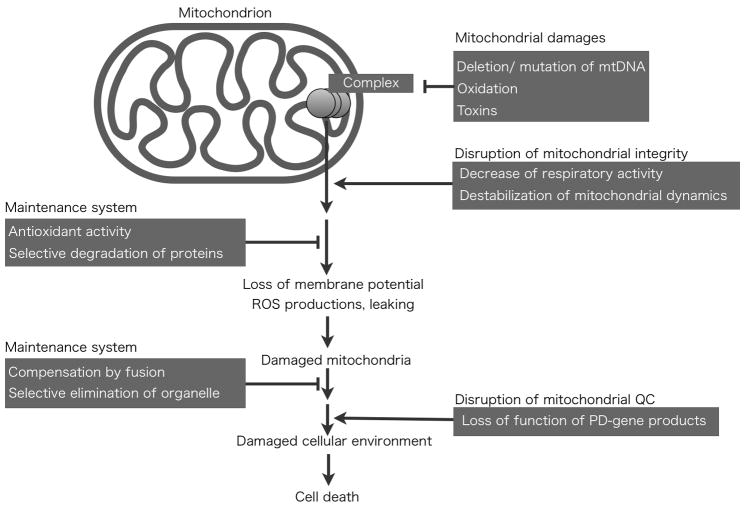
Fig 1. Mitochondria suffer cellular stress and require the maintenance systems.
Many stress generated from inner (mtDNA mutations, deletions) or outer (oxidative stress, toxins) of mitochondria damage mitochondrial functions. If mitochondria failed to maintain their functions by their maintenance system, mitochondria proceed to dysfunctional state. Accumulation of dysfunctions, mitochondria should be eliminated by autophagy system, named mitophagy.
Mitophagy was originally found under starvation conditions, which are a trigger for the bulk autophagy process. Bulk autophagy captures cytoplasmic components simultaneously, and then degrades them to recycle amino acids resources. Organelle specific mitophagy can be seen in the maturation process of erythroid cells, which requires mitochondrial elimination. Although some observations suggest the presence of the elimination system for this dynamic organelle, the molecular mechanisms are poorly understood [12].
Recent works [4,13–16] explore how the maintenance of a functional mitochondrial network, i.e. mitochondria quality control [17], is mediated by Parkin, which is an E3 ligase originally discovered as mutated in monogenic forms of Parkinson’s disease (PD) [18]. Parkin is working to selectively recognize and eliminate damaged mitochondria from the cell by autophagy [4]. These findings illuminate the possibility of clinical application by providing a working model for the pathological state and pathogenic mechanism of PD.
1. Mitochondrial dysfunctions and Parkinson’s disease
PD is one of the neurodegenerative diseases characterized by degeneration of dopaminergic neurons of the substantia nigra in the midbrain which leads to the principal symptoms: progressive movement dysfunctions [19]. A relationship between mitochondrial dysfunction and neurodegenerative disorders has been suggested in numerous studies, especially for PD [20]. The dysfunction of mitochondria in PD patients or PD animal models is due to at least one of the following: deletion of mitochondrial DNA, accumulation of mitochondrial DNA mutations [10] or the increase of oxidative stress from reactive oxygen species (ROS) which is generated through the mitochondria-mediated metabolic pathways [9]. Further demonstrating the relationship between mitochondrial function and PD, some of the chemical toxins that induce parkinsonism [21] are inhibitors of the mitochondrial respiratory chain complexes. Additional evidence for a link between PD and mitochondria is supported by molecular studies using PD-associated genes (PARK genes) products. Some of the PD-associated gene products are located on or linked to mitochondria [22,23], especially PINK1 (PARK6) and Parkin (PARK2). A Drosophila model using pink1 and parkin strains has been thoroughly characterized [24–30]. Mutant fly strains of pink1 and parkin phenocopy one another with phenotypes including the disruption of dopaminergic neurons and swollen mitochondrial morphology. Recent work suggests that PINK1 and Parkin function in the same pathway for the maintenance of mitochondrial integrity [24–26]. From genetic approaches, it was revealed that PINK1 functions upstream of Parkin in the regulation of mitochondrial dynamics [27–30].
Taken together, these lines of evidence demonstrate that the relationship between mitochondrial dysfunction and PD-pathogenic mechanism strongly suggests that the accumulation of mitochondrial damage might cause PD pathogenesis; however, the detailed mechanisms are not completely understood.
2. Cellular localization and function of Parkin
PARK2, an autosomal recessive-juvenile PD (AR-JP)-causing gene, was identified from Japanese patients with PD [18], and it is now known to be the causative gene in 10 to 15% of juvenile Parkinson’s disease. PARK2 gene encodes Parkin, which is an E3 ubiquitin ligase [31]. Since many substrate proteins in various subcellular locations are ubiquitinated by Parkin [32], there was no unified theory on the cellular localization of Parkin. Based on the idea that Parkin functions in the maintenance of mitochondrial integrity, we compared mitochondria in a healthy state to those in a damaged state induced by a chemical reagent, CCCP (carbonyl cyanide m-chlorophenyl hydrazone), which causes depolarization of mitochondria. Surprisingly, Parkin had drastic accumulation on the depolarized, fragmented mitochondria [4]. Parkin can be selectively recruited to individual electrochemically compromised mitochondria, which display greater Parkin accumulation than electrochemically active mitochondria (figure 2). This finding led to the hypothesis that impaired mitochondria are selectively targeted by Parkin [4].
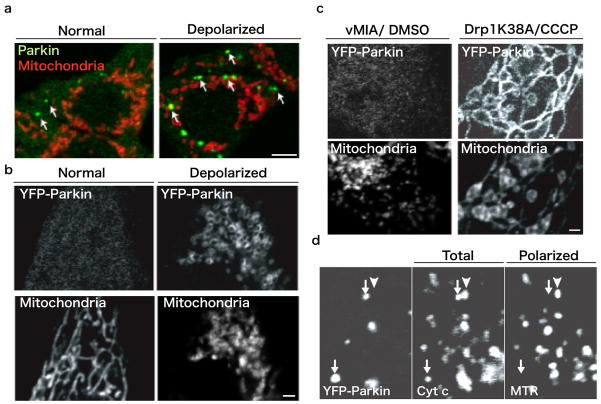
Fig 2. Parkin selectively translocates to the depolarized mitochondria.
(a) Under normal conditions (left), most of the Parkin (green) is in cytosol, whereas some (arrows) are found on the fragmented mitochondria (red). When mitochondria were depolarized by CCCP (right), the cytoplasmic Parkin accumulated on the fragmented mitochondria. HEK293T cells, Scale: 5 μm. (b) In HeLa cells expressing YFP-Parkin, Parkin also accumulated on depolarized and fragmented mitochondria after CCCP treatment (right). Scale: 1 μm. (c) Only mitochondrial fragmentation by vMIA expression, whereas mitochondria maintain their membrane potential, does not signal for the Parkin translocation (left). Mitochondria blocked their division by Drp1K38A recruit YFP-Parkin upon depolarization (right). HeLa cells, Scale: 1 μm. (d) Mitochondria in MEF cells derived from Mfn1−/−, Mfn2−/−[53] mice, which are showing heterogenic mitochondrial membrane potential. Cytochrome c immunostaining indicates total mitochondrial images in a cell, Mitotracker Red indicates polarized (healthy, arrowhead) mitochondria. YFP-parkin accumulates only on depolarized (Mitotracker red-negative, damaged) mitochondria (arrows).
It was unclear whether mitochondrial depolarization or the fragmentation that results from the depolarization was the signal for the translocation of Parkin to the mitochondria. Since these occur very close in time, we addressed the role of fragmentation alone. To cause fragmentation without mitochondrial depolarization in HeLa cells stably expressing YFP-Parkin, we overexpressed vMIA (viral mitochondrial inhibitor of apoptosis), a human cytomegalovirus structural protein that fragments mitochondria without inducing mitochondrial depolarization [33]. YFP-Parkin in these cells did not show mitochondrial localization, suggesting that mitochondrial fragmentation alone does not induce Parkin translocation. Next, we addressed if mitochondrial depolarization without fragmentation could target Parkin to the mitochondria. We used overexpression of Drp1Lys38Ala [34] (a mutation in dynamin-related protein 1, Drp1, which causes a inhibition of mitochondrial division) to block mitochondrial division and then induced mitochondrial depolarization with CCCP (Figure 2). Even in the absence of mitochondrial fragmentation, YFP-Parkin accumulated on the elongated mitochondria treated with CCCP. Taken together these results indicate that mitochondrial depolarization, but not fragmentation, is a signal for Parkin translocation to the mitochondria. Since CCCP also induces the disruption of a chemical potential (ΔpH) of mitochondria, we further examined the condition of Parkin recruitment. The treatment with valinomycin, which dissipates membrane potential loss, but not ΔpH, recruits Parkin onto mitochondria, whereas nigericin, which lowers pH but not membrane potential, does not recruit Parkin onto mitochondria [13]. These evidences also indicate that the translocation of Parkin is strictly regulated in a response to the electrochemical conditions of the mitochondria.
What is the function of Parkin after translocation to damaged mitochondria? If the idea that Parkin mediates the maintenance of mitochondrial integrity is true, Parkin may translocate to the damaged mitochondria to eliminate problems. Healthy mitochondria create ATP by the respiratory chain complexes during which they generate the membrane potential. Membrane potential is indispensable to the membrane structure and the functional maintenance of mitochondria, disruption of the membrane potential results in mitochondrial fragmentation [35]. Recent work by Twig et al. demonstrated that a subpopulation of depolarized and fragmented mitochondria that are removed from the mitochondrial network, are captured by autophagosomal structures which then undergo the autophagosomal/lysosomal cellular digestion system [3]. Indeed, after Parkin translocation to the damaged mitochondria, Parkin-labelled mitochondria could be eliminated from cells through an autophagy/lysosome dependent manner [4]. Co-localization of mitochondria and autophagosomes was found under mitochondrial depolarization conditions in cells expressing Parkin. In cells not expressing Parkin, most cells had little co-localization between mitochondria and autophagosomes (Figure 3). Parkin may promote autophagosome recruitment to Parkin-labelled mitochondria, which are depolarized and likely have accumulated damages. These autophagosomes, which include Parkin-labelled mitochondria, proceed to the lysosomes to be degraded. This sequential process was significantly blocked by the addition of autophagosome or lysosome inhibitors. Furthermore, Parkin failed to eliminate depolarized mitochondria [4] in mouse embryonic fibroblasts derived from ATG5 gene knockout mice [36], which due to the elimination of the key component, Atg5, in the autophagy process cannot form autophagosomes. From all of these results, it was clear that the mechanism for the selective, Parkin-mediated elimination of damaged mitochondria is autophagy-dependent.
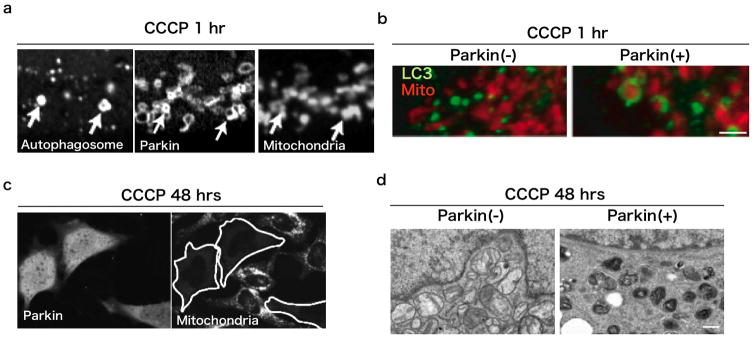
Fig 3. Parkin eliminates damaged mitochondria.
(a) 1 hour after mitochondrial depolarization with CCCP, mitochondria (right) are surrounded by Parkin (center) and autophagosomes (LC3, left). (b) The recruitment of autophagosomes to mitochondria is induced by Parkin. In the absence of Parkin expression (left), depolarized mitochondria (red) are not associated with autophagosomes (LC3, green), whereas autophagosomes are more associated with mitochondria in the presence of Parkin (right). Scale: 1 μm. (c) 48 hours after mitochondrial depolarization with CCCP, mitochondria are not detectable with immunostaining. Only cells expressing YFP-Parkin (left), mitochondria (right) are completely eliminated. (d) 48 hours after mitochondrial depolarization with CCCP, mitochondria were taken up by lysosomes only in the HeLa cells expressing YFP-Parkin (right). Scale: 500 nm.
3. Remaining questions and perspective
Others and we have confirmed experiments identifying the mechanisms of Parkin-mediated selective mitophagy in mammalian system [4,13–16]; 1) Parkin has the ability to specifically recognize and localize to the damaged mitochondria, and 2) Parkin localized on damaged mitochondria induces the elimination of mitochondria using autophagy (mitophagy). The molecular mechanism of mitochondrial quality maintenance mediated by Parkin is intriguing, especially when considering the perspective of the relationship between neurodegenerative disorders and mitochondria.
The point to emphasize here is not the mitochondrial elimination by means of bulk autophagy previously observed in the starvation-state in which cytoplasmic components were simultaneously taken up (bulk mitophagy), but rather the key point is that Parkin induces the specific elimination of damaged mitochondria (selective mitophagy). This likely explains the signification of mitochondrial dysfunction and the increase in intracellular oxidative stress in PD patients and animal model. What is the significance of Parkin-mediated elimination of damaged mitochondria from the cell? We propose that Parkin naturally contributes to protection of the cell from the adverse effects of the intracellular spread of damaged mitochondria by eliminating severely damaged mitochondria from within the cell (Figure 4). Further supporting a protective function for Parkin, there are also reports that increasing the intracellular overexpression of Parkin suppresses cell death [37–39]. Parkin could protect preemptively against cell death, the worst scenario for a cell, by keeping the cell healthy through mitochondrial quality control. However, further studies described as followed for the insight of mitophagy are still on going and required.
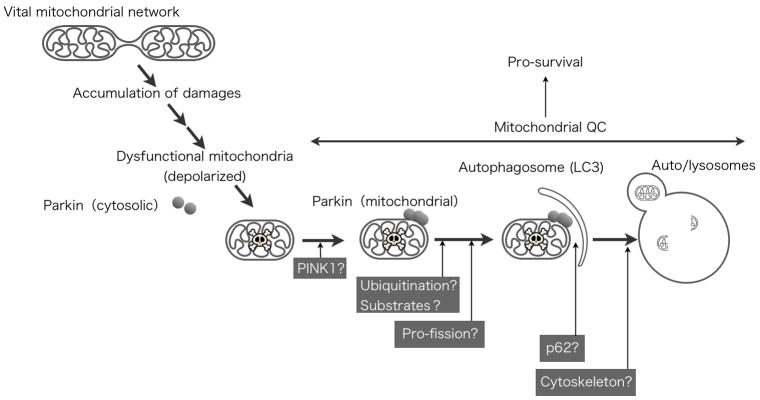
Fig 4. Working model for the Parkin-mediated mitochondrial quality control.
Depolarized mitochondria are sensed by Parkin. After Parkin recruitment to the damaged mitochondria (PINK1 dependent), Parkin may ubiquitinate some substrates to degrade or tagging to proceed following process. After translocation, Parkin also recruits autophagosomes to promote mitophagy.
1) Translocation mechanism of Parkin
For the mechanism of Parkin translocation to mitochondria, it has been shown that PINK1 overexpression induces Parkin translocation [30]. As mentioned above, PINK1 and Parkin function together in a same pathway to maintain mitochondrial integrity. It is logical that if PINK1, which localizes to mitochondria and functions upstream of Parkin, might be required for the Parkin function, specifically translocation to damaged mitochondria. Very recent interesting works are predicting this point [14–16,30]. These groups found independently that PINK1 is required for the Parkin recruitment to mitochondria. Moreover, PINK1 overexpression suffices to recruit Parkin to mitochondria with normal membrane potential. These observations are suggesting that the physical interaction of PINK1 and Parkin promotes the redistribution of Parkin from the cytosol to the mitochondria.
2) Parkin as an E3 ubiquitin ligase
Many patient mutations within the PARK2 (Parkin) gene cause a decrease or complete loss of E3 ubiquitin ligase activity of Parkin protein [40]. In the animal model for PD deletion of PARK2 causes mitochondrial dysfunction and the increase of oxidative stress.
To link this evidence to ubiquitin ligase activity of Parkin and Parkin-mediated mitophagy, we also can expect that some substrates for Parkin exist on the mitochondria. One possible candidate for mitochondrial substrate comes from Drosophila studies [27–29]. The mutant strains of parkin or pink1 can be partially complemented by the suppression of mitochondrial fusion proteins, such as Opa1 and Mitofusin/Marf. This suggests that the pro-fission state of mitochondria is required for mitophagy and that Parkin can ubiquitinate and degrades these mitochondrial fusion proteins. Thus, a PINK1/Parkin pathway may regulate mitophagy process by changing mitochondrial dynamics [17], especially forcing mitochondria to an excessive fission state, which allows mitochondria to be captured by autophagosomes. Moreover, recent work supports this idea by demonstrating that the fission of mitochondria is required for the autophagic degradation of mitochondria [3]. Conflicting with above, it was shown that the suppression of PINK1 results in the fragmentation of mitochondria [41] and induces mitochondrial autophagy, however we suggest based on an our working model that loss of PINK1 or Parkin function exacerbates accumulation of mitochondrial damage, due to defective removal of damaged mitochondria, and the ensuing excessive damage results in mitochondrial fragmentation.
3) Autophagosomes recruitment to the damaged mitochondria
Although we clearly showed that autophagic structures are recruited to Parkin-labelled, damaged mitochondria, the molecular mechanism of this is still unknown. Targeting of ubiquitin onto organelle or inclusion bodies surface is sufficient to recruit autophagic structures [42–44], and it has been suggested that p62 acts as a bridging protein between the ubiquitinated proteins/structures and autophagosomes. A recent work suggests that p62 may involve with Parkin-mediated clearance of the depolarized mitochondria; cells silenced p62 expression decrease the mitochondrial clearance upon depolarization [15]. Further prediction proposed by Vives-Bauza et al. suggests that PINK1/Parkin mediated damaged mitochondria clearance is regulated by transportation of damaged organelles to the lysosomes in a microtubule dependent manner [14]. Since many evidences are suggesting the sequential steps of mitochondrial clearance by PINK1/Parkin pathway followed by autophagic pathway, we still poorly understand the molecular mechanisms. Parkin ubiquitinates various substrates not only for the degradation by ubiquitin-proteasome pathway, but also for signal transduction [32]. Most ubiquitinated proteins that are degraded by the proteasome system are tagged by Lys48 type ubiquitin chains, whereas ubiquitinated proteins that have a Lys63 linked ubiquitin function in signal transduction [45]. Based on our findings, Parkin may function simultaneously to ubiquitinate mitochondrial dynamics proteins to induce pro-fission state by Lys48 ubiquitination and degradation, and tagging for the autophagosomes recruitment by Lys63 ubiquitination of unidentified mitochondrial proteins. More experiments are required to distinguish what role the Parkin-mediated ubiquitination plays in mitophagy.
4) Lessons from mitochondrial elimination by other pathways
Studies in yeast identified autophagy-related genes [ATG genes) and uncovered the mechanisms of several types of autophagy process; macroautophagy for non-selective autophagy, cytoplasm to vacuole targeting (Cvt) pathway, pexophagy, and mitophagy for selective autophagy of several proteins or organelles [46]. Very recent studies with excellent genetic screens by Okamoto et al. and Kanki et al. identified independently a mitophagy-related gene in yeast, named ATG32 [47,48]. Atg32 protein is identified as an outer mitochondrial membrane protein, and a receptor molecule for Atg11 proteins, which is a key component for the recruitment of pre autophagic structures (PAS) to mitochondria. Atg32 also contains a conserved WXXI/L/V motif for the interaction with Atg8, which is a yeast homologue of a mammalian autophagic initiator, LC3 protein. Mammalian p62 protein also contains this motif to interact with LC3. Thus, these lines of evidence suggest that yeast and mammal may share common components for the selective mitophagy, although any mammalian homologue of yeast Atg-proteins for the selective mitophagy have not yet been identified. Recent studies reported that an outer mitochondrial membrane protein Nix/BNIP3 is required for the selective mitochondrial elimination in an autophagy dependent manner during mammalian reticulocytes maturation[5] [6] [7]. Thus Nix/BNIP3 may represent a functional homologue or a counterpart for Atg32, whereas many details in the process of mitophagy in mammalian system are still open to future studies.
5) Endogenous mitophagy: “Does Parkin allow cells to survive by maintaining mitochondrial health? ”
Mitochondria can promote cell death by initiating apoptosis. If cell had a serious problem that could not be resolved by autophagy, they could execute cell suicide by apoptosis to prevent the inflammation of entire tissues. What would be the case for a tissue that cannot choose cell death? One key example would be differentiated nerve cells. In patients with PD the lack of dopaminergic neurons in the substantia nigra is probably due to the relative stress placed on the mitochondria by the generation of reactive oxygen species as a byproduct in the dopaminergic route of these neurons [49] and/or the resulting toxicity of the mutated Parkin protein which may form aggregates in the cytoplasm itself [50]. This increased stress without functional mitophagy, would lead to death of the dopaminergic neurons. Moreover, since dopaminergic neurons appear to require increased Parkin function for mitochondrial quality and functionality more so than other neurons, when the Parkin system for eliminating damaged mitochondria breaks down and no longer functions the neurons probably die quickly. The above explanation does not completely explain the dopaminergic neurons dropout by PD. The ubiquitin ligase activity of Parkin is also required for the clearance of some aggregately proteins, for example alpha synuclein [51], Pael-R [52] and Parkin itself by ubiquitin proteasome system [50]. Disruption of turnover or functions of these proteins by mutated Parkin or PINK1 may also cause the demolition of cell homeostasis. This possibility also clouds studies on PD pathobiology, due to the possibility that dysfunction of protein turnover may also lead to mitochondrial dysfunctions. However these complex possible mechanisms need to be resolved by future investigations, we may have good explanations to tackle them based on our current model of a Parkin-related mitochondrial quality control. As mentioned above, mitochondria need to be maintained as functional in a cell. When cells accumulate damage (i.e. oxidized proteins, aggregates of misfolded proteins, ROS), mitochondria also gain a risk of malfunction caused by this damages. Mitochondria compensate this damage, whereas if damage overwhelms the compensation, cells may have serious problems to keep their functionality. Dysfunction of mitochondrial quality control system, such as impaired Parkin/PINK1 mediated system, also accumulates damaged mitochondria. Then if damaged mitochondria overcome the healthy mitochondrial network, cells also fall into fatal scenario. These proposing models may suggest the pathobiology of various neurodegenerative diseases including PD (Figure 5).
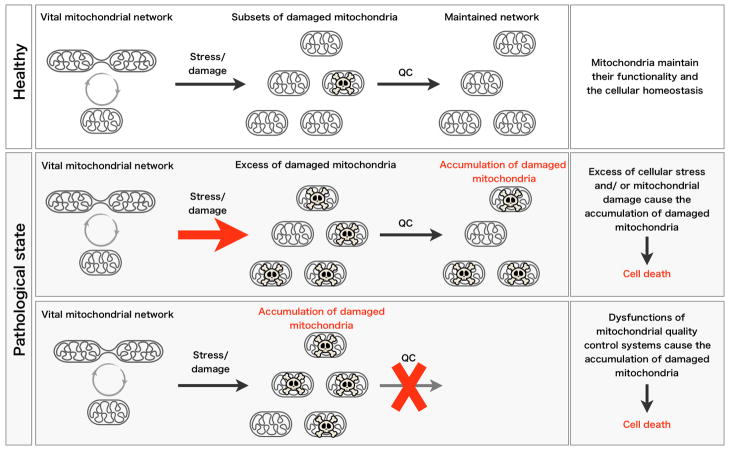
Fig 5. Working model for the Pathogenesis of PD.
The vital mitochondrial network is maintained by the quality control (QC) system (top, also see in Figure 1). In pathological states of PD, cellular stress or damage cause the excess of damaged mitochondria, then overwhelm the QC system (middle). Disruption of mitochondrial QC system also accumulates damaged mitochondria (bottom). Both pathological states may cause the collapse of the cellular environment following by the cell death.
4. Concluding remarks
Recent progress on neurodegenerative disease and mitochondrial dysfunctions are being extended to a variety of scientific fields. Other neurodegenerative disease, such as Alzheimer disease and Huntington disease, also suggest there is a relationship between mitochondrial function and the pathobiology mechanisms. Many unsolved issues remain in this field. Nevertheless, our findings could provide future targets for the therapeutic treatments of PD as well as other neurodegenerative disorders.
Acknowledgments
I would like to thank Professor Noboru Mizushima (Tokyo Med. Dent. Univ.) for giving me the opportunity of this article. I also thank R. Youle for support and M. Cleland for the critical reading for the manuscript. Our studies are supported by NINDS/NIH intramural program (to RJY) and JSPS (to AT)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–90. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–53. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo J. 2008;27:433–46. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schweers RL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–5. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kundu M, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–5. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yue Z, Friedman L, Komatsu M, Tanaka K. The cellular pathways of neuronal autophagy and their implication in neurodegenerative diseases. Biochim Biophys Acta. 2009;1793:1496–507. doi: 10.1016/j.bbamcr.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hald A, Lotharius J. Oxidative stress and inflammation in Parkinson’s disease: is there a causal link? Exp Neurol. 2005;193:279–90. doi: 10.1016/j.expneurol.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Nakada K, Inoue K, Ono T, Isobe K, Ogura A, Goto YI, Nonaka I, Hayashi JI. Inter-mitochondrial complementation: Mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat Med. 2001;7:934–40. doi: 10.1038/90976. [DOI] [PubMed] [Google Scholar]
- 11.Neutzner A, Youle RJ, Karbowski M. Outer mitochondrial membrane protein degradation by the proteasome. Novartis Found Symp. 2007;287:4–14. discussion 14–20. [PubMed] [Google Scholar]
- 12.Dengjel J, Kristensen AR, Andersen JS. Ordered bulk degradation via autophagy. Autophagy. 2008;4:1057–9. doi: 10.4161/auto.6824. [DOI] [PubMed] [Google Scholar]
- 13.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin-induced mitophagy in the pathogenesis of Parkinson disease. Autophagy. 2009;5:706–8. doi: 10.4161/auto.5.5.8505. [DOI] [PubMed] [Google Scholar]
- 14.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 107:378–83. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 12:119–31. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 16.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway: a mitochondrial quality control system? J Bioenerg Biomembr. 2009;41:499–503. doi: 10.1007/s10863-009-9253-3. [DOI] [PubMed] [Google Scholar]
- 18.Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 19.Maetzler W, Liepelt I, Berg D. Progression of Parkinson’s disease in the clinical phase: potential markers. Lancet Neurol. 2009;8:1158–71. doi: 10.1016/S1474-4422(09)70291-1. [DOI] [PubMed] [Google Scholar]
- 20.Corti O, Hampe C, Darios F, Ibanez P, Ruberg M, Brice A. Parkinson’s disease: from causes to mechanisms. C R Biol. 2005;328:131–42. doi: 10.1016/j.crvi.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci. 2000;3:1301–6. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Guo M. Protein degradation in Parkinson disease revisited: it’s complex. J Clin Invest. 2009;119:442–5. doi: 10.1172/JCI38619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bueler H. Impaired mitochondrial dynamics and function in the pathogenesis of Parkinson’s disease. Exp Neurol. 2009;218:235–46. doi: 10.1016/j.expneurol.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Park J, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–61. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci U S A. 2006;103:10793–8. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark IE, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–6. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 27.Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105:1638–43. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, Lu B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci U S A. 2008;105:7070–5. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng H, Dodson MW, Huang H, Guo M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci US A. 2008;105:14503–8. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park J, Lee G, Chung J. The PINK1-Parkin pathway is involved in the regulation of mitochondrial remodeling process. Biochem Biophys Res Commun. 2009;378:518–23. doi: 10.1016/j.bbrc.2008.11.086. [DOI] [PubMed] [Google Scholar]
- 31.Shimura H, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–5. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 32.Moore DJ. Parkin: a multifaceted ubiquitin ligase. Biochem Soc Trans. 2006;34:749–53. doi: 10.1042/BST0340749. [DOI] [PubMed] [Google Scholar]
- 33.McCormick AL, Skaletskaya A, Barry PA, Mocarski ES, Goldmacher VS. Differential function and expression of the viral inhibitor of caspase 8-induced apoptosis (vICA) and the viral mitochondria-localized inhibitor of apoptosis (vMIA) cell death suppressors conserved in primate and rodent cytomegaloviruses. Virology. 2003;316:221–33. doi: 10.1016/j.virol.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–56. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Legros F, Lombes A, Frachon P, Rojo M. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol Biol Cell. 2002;13:4343–54. doi: 10.1091/mbc.E02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 37.Darios F, et al. Parkin prevents mitochondrial swelling and cytochrome c release in mitochondria-dependent cell death. Hum Mol Genet. 2003;12:517–26. doi: 10.1093/hmg/ddg044. [DOI] [PubMed] [Google Scholar]
- 38.Hasegawa T, Treis A, Patenge N, Fiesel FC, Springer W, Kahle PJ. Parkin protects against tyrosinase-mediated dopamine neurotoxicity by suppressing stress-activated protein kinase pathways. J Neurochem. 2008;105:1700–15. doi: 10.1111/j.1471-4159.2008.05277.x. [DOI] [PubMed] [Google Scholar]
- 39.Rawal N, Corti O, Sacchetti P, Ardilla-Osorio H, Sehat B, Brice A, Arenas E. Parkin protects dopaminergic neurons from excessive Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2009;388:473–8. doi: 10.1016/j.bbrc.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 40.Matsuda N, Kitami T, Suzuki T, Mizuno Y, Hattori N, Tanaka K. Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J Biol Chem. 2006;281:3204–9. doi: 10.1074/jbc.M510393200. [DOI] [PubMed] [Google Scholar]
- 41.Dagda RK, Zhu J, Chu CT. Mitochondrial kinases in Parkinson’s disease: converging insights from neurotoxin and genetic models. Mitochondrion. 2009;9:289–98. doi: 10.1016/j.mito.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 43.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105:20567–74. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan JM, Wong ES, Dawson VL, Dawson TM, Lim KL. Lysine 63-linked polyubiquitin potentially partners with p62 to promote the clearance of protein inclusions by autophagy. Autophagy. 2007:4. [Google Scholar]
- 45.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–78. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 46.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–67. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 47.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 48.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hastings TG. The role of dopamine oxidation in mitochondrial dysfunction: implications for Parkinson’s disease. J Bioenerg Biomembr. 2009 doi: 10.1007/s10863-009-9257-z. [DOI] [PubMed] [Google Scholar]
- 50.Schlehe JS, Lutz AK, Pilsl A, Lammermann K, Grgur K, Henn IH, Tatzelt J, Winklhofer KF. Aberrant folding of pathogeni Parkin mutants: aggregation versus degradation. J Biol Chem. 2008;283:13771–9. doi: 10.1074/jbc.M707494200. [DOI] [PubMed] [Google Scholar]
- 51.Burke RE. alpha-Synuclein and parkin: coming together of pieces in puzzle of Parkinson’s disease. Lancet. 2001;358:1567–8. doi: 10.1016/S0140-6736(01)06668-5. [DOI] [PubMed] [Google Scholar]
- 52.Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell. 2001;105:891–902. doi: 10.1016/s0092-8674(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 53.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–92. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]