Abstract
Autophagy, or cellular self-digestion, is activated in cancer cells in response to multiple stresses and has been demonstrated to promote tumor cell survival and drug resistance. Nonetheless, genetic evidence supports that autophagy functions as a tumor suppressor mechanism. Hence, the precise role of autophagy during cancer progression and treatment is both tissue and context dependent. Here, we discuss our current understanding of the biological functions of autophagy during cancer development, overview how autophagy is regulated by cancer-associated signaling pathways, and review how autophagy inhibition is being exploited to improve clinical outcomes.
Keywords: Autophagy, cancer, genome instability, cell death, chemotherapy
1. Introduction
Macroautophagy (hereafter called autophagy) is an evolutionarily conserved catabolic process where a cell self-digests its cytoplasmic contents (literally “eats itself”). In eukaryotic cells, autophagy is a key mechanism for long-lived protein degradation and organelle turnover, and serves as a critical adaptive response that recycles energy and nutrients during starvation or stress [1]. Autophagy is tightly regulated by a limited number of highly conserved genes called ATGs (for AuTophaGy gene). Originally identified in yeast, these landmark results have led to numerous recent breakthroughs in mammals demonstrating how autophagy critically regulates key physiological and pathological processes, including cancer initiation and progression. Autophagy is activated in response to multiple stresses during cancer progression, such as nutrient starvation, the unfolded protein response (ER stress), and hypoxia; in addition, it is observed upon treatment of cancers with a wide spectrum of cytotoxic and targeted chemotherapeutic agents [2]. The role of autophagy as a survival mechanism in response to these diverse stressors has been well established. Moreover, it has become increasingly clear that a basal level of autophagy serves housekeeping functions vital for maintaining cellular homeostasis; specifically, the failure to clear protein aggregates or damaged organelles via autophagy has been implicated in multiple pathological conditions, including cancer [3].
Based on these studies, autophagy has been proposed to promote the cellular fitness of cancer cells; thus self-eating should function unitarily as a tumor-promoting mechanism. However, genetic evidence supports otherwise. Rather, autophagy can exert important tumor suppressive functions. Clearly, several fundamental issues must be addressed to most effectively exploit autophagy for therapeutic purposes against cancer. In this review, we summarize the current understanding of autophagy in the context of tumorigenesis and oncogenic transformation. We discuss the potential mechanisms by which autophagy suppresses tumor formation, how autophagy is regulated by oncogenes and tumor suppressors, and overview how autophagy is being exploited to improve clinical outcomes.
2. Autophagy as a Tumor Suppressor Mechanism
The role of autophagy as a tumor suppressor was first broached through genetic studies of Beclin1, the mammalian orthologue of yeast Atg6 [4]. Beclin1 was mapped to a tumor susceptibility locus that is monoallelically deleted in a high percentage of human breast, ovarian, and prostate cancers; furthermore, analysis of human tissue samples revealed decreased Beclin1 expression in human breast carcinomas compared to normal breast tissue. Ectopic Beclin1 expression reduced cancer cell proliferation in vitro and decreased tumorigenic potential in vivo, further suggesting a role for autophagy in tumor suppression [4]. In support of this notion, two groups demonstrated that heterozygous disruption of beclin1 promoted tumorigenesis in mice, albeit over an extended latency [5,6]. Mice lacking one copy of beclin1 suffered from a high incidence of spontaneous tumors, including B cell lymphoma, hepatocellular carcinoma, and lung adenocarcinoma. These reports provided the first direct genetic evidence that beclin1/atg6 functions as a haploinsufficient tumor suppressor.
In addition to Beclin1, the genetic alteration of other autophagy-related genes has been found in various types of cancer. These include the deletion of atg5, an essential autophagy gene, in natural killer (NK) cell malignancies [7]; nonsense mutations in UV irradiation Resistance-Associated Gene (UVRAG), a Beclin1 binding protein that positively regulates autophagy, in colon cancer cells [8], and gastric carcinomas [9], and the downregulation of Bax-binding protein-1 (Bif-1), another positive regulator of autophagy, in colon adenocarcinomas [10]. In animal models, Atg4c knockout mice exhibit increased susceptibility to fibrosarcomas [11]. These findings support that tumor suppression is a shared property of different components of the autophagic pathway, rather than a phenotype unique to disrupting any individual autophagy regulator.
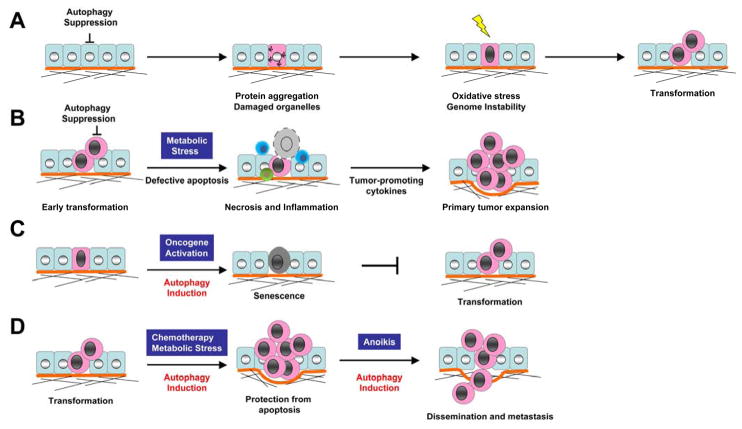
How the suppression of autophagy leads to cancer remains unclear, although several potential mechanisms have emerged. First, a mounting body of work from Eileen White and colleagues demonstrates an important role for autophagy in protecting cells from genotoxic stress and maintaining genome integrity. Mouse epithelial cells that are defective in both autophagy and apoptosis, due to the loss of one copy of beclin1 combined with the overexpression of Bcl-2, are more prone to necrosis during metabolic stress, yet paradoxically, these cells exhibit enhanced tumorigenic potential [12]. Notably, these studies demonstrate that autophagy defective cells show a remarkable increase in DNA double strand breaks and gene amplification in response to metabolic stress compared to their autophagy competent counterparts [13]. In a 3D morphogenesis assay using mammary epithelial cells derived from Beclin1 heterozygous mice, accelerated cell death and increased DNA damage was also observed in central acinar cells, where metabolic stress occurred [14]. Based on these studies, White and colleagues raised the hypothesis that defective autophagy may enhance DNA damage, thus causing an increased mutation rate in cells that survive metabolic stress, which ultimately leads to tumorigenesis. One major caveat is that these experiments have been conducted in cells harboring multiple genetic abnormalities, including the inactivation of the tumor suppressor p53, which is well-known for its ability to maintain genomic integrity [15]. As a result, one can argue that defective autophagy most likely functions as a secondary modifier, rather than a fundamental driver, of genomic damage during tumor progression. Nonetheless, additional studies point to the molecular mechanism by which defective autophagy induces genome damage. In response to metabolic stress, autophagy incompetent cells have defects in protein turnover, resulting in the accumulation of the scaffold protein p62/SQSTM, ER chaperones, damaged mitochondria, and elevated reactive oxygen species (ROS) [16]. Notably, the accumulation of p62/SQSTM, an ubiquitin-binding scaffold protein specifically degraded by autophagy [17], is a significant contributor to tumorigenesis. Taken together, these findings delineate novel connections linking decreased autophagy to tumorigenesis; autophagy suppression causes the accumulation of damaged mitochondria and p62 protein aggregation, resulting in a cascade of events involving increased oxidative stress, DNA damage, and chromosomal instability, and ultimately leading to cancer development (Figure 1A).
Figure 1. Diverse roles for autophagy in cancer progression and therapy.
(A) The suppression of autophagy induces protein aggregates and the accumulation of damaged organelles, resulting in a cascade of increased oxidative stress, genome instability, and ultimately, malignant transformation. (B) Tumor cells with combined defects in apoptosis and autophagy are prone to necrosis in response to metabolic stress. Necrosis promotes inflammatory responses, notably the recruitment of macrophages, that favors primary tumor growth. (C) Autophagy promotes oncogene-induced senescence, a barrier to malignant transformation. (D) Autophagy induction promotes tumor cell survival in response to diverse stresses, including chemotherapy, metabolic stress, and anoikis, which may facilitate drug-resistance and metastasis.
The second potential mechanism of autophagy-mediated tumor suppression is also based on the observation that when apoptosis is blocked, autophagy incompetent cells are more prone to undergo necrosis in response to metabolic stress whereas autophagy-competent cells are more resistant to cell death [12]. Remarkably, inflammatory cells infiltrate tumor sites in response to necrosis resulting from hypoxia and metabolic stress, both of which commonly affect solid tumors. Although certain inflammatory cells, such as cytotoxic T cells and NK cells are anti-metastatic, chronic tumor inflammation associated with severe hypoxia and metabolic stress generally favors pro-tumor immunity [18–20]. Importantly, infiltration of pro-tumor inflammatory mediators, like macrophages, correlates with poor clinical prognosis, underscoring the importance of understanding the biological mechanisms by which tumor cells tip the balance in favor of pro-tumor immunity over tumor suppressive immunity [20]. Thus, by limiting tumor cell necrosis, autophagy may serve as a non-autonomous mechanism for tumor suppression by preventing leukocyte infiltration of the primary tumor site (Figure 1B).
In addition, a recent study reports that autophagy is required for the establishment of oncogene-induced senescence (OIS), providing an alternative mechanism of autophagy mediated tumor suppression [21]. Senescence is a state of irreversible cell cycle arrest that limits the proliferation of damaged cells. OIS is induced by aberrant hyper-proliferation signals from oncogenes, thus serving as an early barrier to cancer development. Recent work demonstrates that autophagy is activated during oncogene- and DNA damage-induced senescence; RNAi-mediated depletion of atg5 or atg7 facilitates the ability of cells to bypass OIS and delays senescence-associated cytokine production, suggesting that autophagy contributes to the establishment of senescence arrest (Figure 1C). These results also implicate that basal autophagy plays an important role in restricting cell growth and proliferation during oncogenic stress as well as precluding further genomic insults.
Despite these exciting experimental findings in cell-based and mouse models, it is important to recognize that no direct evidence exists showing that reduced or defective autophagy is a common requirement for human cancer initiation. This is primarily because the direct and quantitative detection of autophagy levels in human tumor samples remains technically intractable.
3. Autophagy and Tumor Cell Survival
Although reduced autophagy promotes certain aspects of tumor formation, a minimal level of autophagy does appear to contribute to the cellular fitness and survival of established tumor cells in response to various stresses (Figure 1D). Because oncogenic cells have a high demand for nutrients and oxygen to facilitate their high proliferation rate, cells often encounter metabolic stress and hypoxia, especially in poorly vascularized solid tumors. Accordingly, cells in the interior of a tumor have higher levels of autophagy compared to those at the margin, which protects them from both apoptosis and necrosis [12]. In addition, a pro-survival effect of autophagy may promote the later stages of cancer progression, such as dissemination and metastasis, the principal cause of cancer mortality. For instance, detachment from extracellular matrix (ECM) induces autophagy in epithelial cells, which protects them from cell death, commonly termed anoikis. Tumor cells must overcome anoikis in order to metastasize [22]. Furthermore, autophagy is upregulated by the tumor suppressor ARHI (aplasia Ras homolog member I) in ovarian cancer cells, which has been shown to promote in vivo survival of dormant cells in the context of a tumor microenvironment [23]. Finally, a large body of work also demonstrates that autophagy serves as an important survival mechanism for cancer cells during anti-cancer therapies, which we discuss in detail below.
4. Molecular Regulation of Autophagy In Cancer
mTORC1 dependent regulation of autophagy
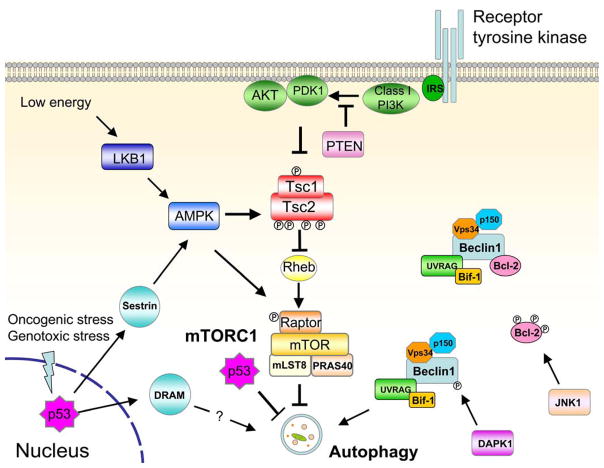
The best known regulator of autophagy is mammalian target of rapamycin (mTOR), a serine/threonine kinase conserved throughout eukaryotes [24]. mTOR forms two complexes in mammalian cells; of these, mTOR complex 1 (mTORC1) plays a central role in sensing nutrient availability, mitogenic signals, and energy status. Thus, it controls cell growth, protein translation, and cell proliferation [25]. The activity of mTORC1 is inversely correlated with autophagy induction; in fact, rapamycin, a mTORC1 inhibitor, potently induces autophagy, even in the presence of abundant nutrients [24]. Furthermore, a variety of pathways that direct mTORC1 activation converge on the Tuberous Sclerosis Complex (TSC) and the Ras Homolog Enriched in Brain (Rheb), a small GTPase that activates mTORC1 in its GTP-bound form [25]. TSC, which is comprised of two primary components, TSC1 (harmartin) and TSC2 (tuberin), has a GTPase activating protein (GAP) function that hydrolyzes GTP, thereby negatively regulating Rheb and inhibiting mTORC1 activity [26]. Importantly, TSC is primarily regulated via phosphorylation downstream of diverse upstream pathways; these phosphorylation events can be activating or deactivating, depending on specific residues that are phosphorylated on TSC1 or 2 (Figure 2).
Figure 2. Cancer-associated signaling pathways that regulate autophagy in mammalian cells.
See text for details.
As the principal regulator of cell growth, mTORC1 is deregulated in most human cancers [25]. Many of the signaling molecules that regulate mTORC1 are found to be oncogenes and tumor suppressors. Rheb is overexpressed in prostate cancer and co-operates with PTEN haplo-insufficiency to promote tumorigenesis [27]. High Rheb activity, due to loss of TSC1, inhibits autophagy and sensitizes cells to cell death induced by misfolded-proteins [28]. Most importantly, activation of the class I phosphotidylinositol 3-kinase (PI3K) and downstream components, such as the kinase AKT, have been demonstrated to activate mTORC1. Enhanced PI3K activity is often found through activating kinase mutations or gene amplification in cancers; such events should inhibit autophagy. In support, the lipid products of class I PI3K, PI (3,4) P2 or PI (3,4,5) P3, suppress autophagy induction in colon cancer cells [29]. Furthermore, mutational activation of AKT suppresses autophagy in response to metabolic stress [12] whereas AKT kinase inhibition potently induces autophagy through inactivating mTORC1 [30]. On the other hand, overexpression of the tumor suppressor PTEN, the negative regulator of PI3K/AKT pathway, induces autophagy [31]. Similarly, ARHI, another tumor suppressor commonly lost in ovarian cancers, induces autophagy via negatively regulating the PI3K pathway [23].
LKB1-AMPK
The activation of autophagy has been proposed to play a critical role in cell survival in response to reduced energy (ATP). The LKB1-AMPK pathway links metabolism to cell growth control, and increasing evidence supports a role for this signaling axis in controlling autophagy [32]. LKB1-AMPK monitors intracellular energy levels and sends signals to mTORC1 through TSC dependent and independent mechanisms. During metabolic stress, the reduction in ATP levels results in an elevated AMP/ATP ratio and activates the energy-sensing kinase, LKB1, which subsequently phosphorylates and activates AMPK. An established connection to autophagy downstream of these signals is through AMPK-mediated phosphorylation of TSC, which results in the inactivation of mTORC1 and induction of autophagy [33,34]. Still, various other TSC-independent pathways may promote autophagy. First, AMPK has been found to directly inhibit mTORC1 in a TSC independent manner, via direct phosphorylation of Raptor, an activating component of mTORC1; this effect is LKB1-dependent and promotes cell survival in response to energy stress, presumably through autophagy induction [35]. In addition, the LKB1-AMPK axis can phosphorylate and stabilize p27kip, a cyclin-dependent kinase inhibitor that induces cell cycle arrest [36]. These studies also show that p27 is required for glucose starvation-induced autophagy and cell survival downstream of LKB-AMPK. Because the relationship between p27 phosphorylation and mTORC1 in response to energy stress was not evaluated in this study, it remains unknown whether p27 regulates autophagy through mTORC1 dependent or independent pathways. Furthermore, cells expressing the oncogene simian virus 40 small T antigen are more resistant to glucose deprivation induced cell death through activation of AMPK and autophagy [37].
Beclin1 complexes
Mammalian Beclin 1 (ATG6) was originally identified as a Bcl-2 interacting protein [38]. Subsequent studies indicated that Beclin1 forms multi-molecular complexes with numerous proteins that impact the induction or inhibition of autophagy (Figure 2). Nonetheless, the core functions of Beclin1 in autophagy induction result from its interactions with Vps34 and p150. Vps34 is a class III PI3K that is regulated by p150 [39]. In addition, Beclin1/Vps34/p150 interacts with UVRAG and Atg14 [40]. Although the exact mechanism by which these complexes promote autophagy is unclear, it has been shown that the interaction between Beclin1 and Vps34 is required for engaging Vps34 kinase activity in the autophagic pathway [41,42].
Notably, UVRAG is a candidate tumor suppressor, which like Beclin1, is monoallelically deleted in human colon cancers [43]. Although expression of UVRAG alone has little effect on cell death, over-expression of wild type, but not Beclin1-binding deficient UVRAG, suppresses colon cancer cells growth in vitro and tumor formation in vivo, suggesting the tumor suppressor function of UVRAG requires its interaction with Beclin1. Nonetheless, another binding partner of UVRAG, the C-Vps (class C Vps complex), a key component of the endosomal fusion machinery, has also been implicated in the regulation of autophagy, independent of Beclin1. The interaction of the C-Vps complex with UVRAG promotes autophagosome maturation and enhances the degradation of autophagic cargo [44]. Thus, UVRAG may also positively regulate autophagy by enhancing autophagosome maturation.
In addition to stimulating Beclin1/Vps34 activity, UVRAG also facilitates Beclin1 interaction with other proteins implicated in autophagy. One such protein is Bif-1, a potential tumor suppressor that belongs to the endophilin family of proteins [45]. Loss of Bif-1 expression reduces Vps34 kinase activity and suppresses autophagy induction in response to starvation. At a structural level, an N-terminal Bin/Amphiphysin/Rvs (BAR) domain of Bif-1 is critical for its role in autophagy regulation. Because BAR-domain containing proteins have been implicated in membrane binding and membrane curvature, it has been speculated that Bif-1 may facilitate autophagosome isolation membrane building [46]. In line with this notion, Bif-1 co-localizes with Atg5 and LC3, suggesting its involvement in early autophagosome formation.
Moreover, another binding partner of Beclin1 has been identified as mammalian Atg14/Atg14L/Barkor, the mammalian orthologue of yeast Atg14 [40,47–49]. Studies from four groups show that mammalian Atg14 interacts with the Beclin1 complex and positively regulates autophagy. Accordingly, the RNAi-mediated depletion of Atg14 impairs both autophagosome formation and autophagic flux in response to starvation. Studies reveal that Beclin1/Vps34/p150 forms two distinct complexes with Atg14 and UVRAG in a mutually exclusive manner. These complexes regulate autophagy at different steps, namely autophagosome formation and maturation, respectively [40,47,48]. In addition, two groups identified another molecule, called Rubicon, as a negative regulator of autophagy, in association with Beclin1 complexes. Over-expression of Rubicon causes the aberrant expansion of late endosomes and lysosomes, resulting in defects in autophagosome maturation. Given that other Beclin1 complex components function as tumor suppressors, dissecting the potential roles of Atg14 and Rubicon in cancer development remains an important subject for future study.
Members of Bcl-2 family
Bcl-2, the first protein found to physically interact with Beclin1, has been shown to possess an anti-autophagy function in addition to its well-established role in apoptosis inhibition [38]. In addition to Bcl-2, other anti-apoptotic Bcl-2 family proteins, such as Bcl-xL and Mcl-1, are also capable of interacting with Beclin1 and suppressing autophagy [50,51]. Because these anti-apoptotic proteins are commonly over-expressed in cancer cells, autophagy inhibition may have an additive effect on the oncogenic properties of these Bcl-2 proteins. In support of this notion, RNAi-mediated depletion of Bcl-2 is sufficient to trigger autophagy and apoptosis in tumor cells [52,53]. At a structural level, Beclin1 interacts with multi-domain Bcl-2 proteins via its BH3 domain and a BH3-receptor domain on Bcl-2 proteins. Disruption of such interaction by a BH3-mimetic agent, ABT737, or expression of “BH3-only” protein, such as Bad, induces autophagy [50]. Importantly, the phosphorylation status of either protein can interfere with this autophagy-regulating interaction (Figure 2). During nutrient starvation, JNK1 mediated Bcl-2 phosphorylation dissociates Beclin1/Bcl-2 complex and induces autophagy [54]. On the other hand, the tumor suppressor DAPK1, directly phosphorylates the Beclin1 BH3 domain, promoting the dissociation of the Beclin1/Bcl-xL complex and thus inducing autophagy [55]. Moreover, the tumor suppressor ARF, a well-established cell cycle regulator, can induce autophagy by binding to Bcl-xL and thereby releasing Beclin1 [56].
In addition, certain pro-apoptotic proteins of the Bcl-2 family have been implicated in autophagy regulation in different conditions. Expression of BNIP3, a “BH3-only” Bcl-2 family protein, has been found to disrupt the Beclin1/Bcl-2 interaction and induce the autophagic degradation of mitochondria (mitophagy) in response to hypoxia [57]. Furthermore, Puma, another “BH3-only” protein, induces mitochondrial autophagy in response to mitochondrial perturbations in a Bax/Bak dependent manner [58].
p53-related regulators of autophagy
The transcription factor p53 is the most commonly mutated tumor suppressor. p53 levels increase in multiple cancer-related stress conditions, including DNA damage, oncogenic stress and hypoxia. In mammalian cells, p53 shuttles between the nucleus and the cytoplasm; activated p53 translocates to the nucleus and induces target gene expression, resulting in cell cycle arrest, DNA repair or apoptosis. Although p53 is best known as a nuclear transcription factor, recent work indicates that cytoplasmic p53 mediates mitochondrial outer membrane permeabilization and apoptosis [59].
Similarly, p53 plays a dual role in autophagy regulation depending on its subcellular location. The role of nuclear p53 in inducing autophagy appears to primarily arise from the transcriptional control of mTOR pathway regulators (Figure 2). Indeed, multiple p53 target genes, such as AMPKβ, TSC2, and PTEN, are all known negative regulators of mTORC1 [60]. Remarkably, two p53 target genes, sestrin1 and sestrin2, have been identified as a critical link between p53 activation and mTORC1 activity [61]. Sestrin is induced by p53 in response to DNA damage and oxidative stress, which inhibits mTORC1 activity by stimulating AMPK-mediated TSC activation. Interestingly, Sestrin expression is able to override oncogene-mediated mTOR activation and cell growth, providing another mechanism of p53 mediated tumor suppression. Subsequent work corroborates that Sestrin2 is required for autophagy induction in response to various stress inducers, including nutrient starvation and rapamycin [62].
Another target of p53, DRAM (damage-regulated autophagy modulator), has been shown to positively regulate autophagy [63]. DRAM is essential for p53-mediated autophagy and apoptosis in response to DNA damage agents, and the ectopic expression of DRAM is sufficient to activate autophagy but has little effect on apoptosis. The role of DRAM as a tumor suppressor is suggested by the finding that DRAM expression is downregulated in cancer cells due to promoter hyper-methylation [63]. The exact mechanism of DRAM-mediated autophagy activation remains unclear.
Paradoxically, p53 has also been found to suppress autophagy independently of its transcriptional function in the nucleus [64]. Rather, cytoplasmic p53 inhibits autophagy in response to multiple autophagy-inducing agents. Many stresses, such as nutrient starvation, rapamycin, or ER stress, promote p53 degradation; hence, p53 stabilization suppresses autophagy mediated by these stressors. Importantly, the genetic deletion, RNAi-mediated depletion, or pharmacological inhibition of p53 can all elicit autophagy induction; moreover, p53 inhibition promotes cell survival in response to glucose starvation through autophagy. Based on these results one can hypothesize that, upon p53 deletion in tumors, elevated autophagy will provide a survival advantage to malignant cells in response to unfavorable conditions found in the tumor microenvironment.
The above studies demonstrate how multiple cancer-associated pathways regulate autophagy. Importantly, one must recognize that these signaling pathways do not function independently. Instead, a signaling network is formed by constant crosstalk among the different pathways; furthermore, it is likely that certain signals will be dominant over others during specific aspects of oncogenic transformation and tumor progression. For example, although oncogenes such as activated PI3K have inhibitory effects on autophagy induction, when tumor cells encounter stresses, such as lack of nutrients or energy, the inhibitory effects of this oncogenic PI3K on autophagy may be overridden by signals targeting downstream effectors (such as mTORC1). Similarly, p53 can also induce autophagy despite the presence of oncogenes, for example, through Sestrin expression. In this case, AMPK-mediated TSC activation overrides oncogene-induced TSC inhibition, resulting in decreased mTORC1 activity. Thus, the ultimate induction or inhibition of autophagy in cancer cells reflects the integration of signals resulting from intracellular perturbations mediated by individual oncogenes and tumor suppressors as well as extracellular cues, both micro-environmental and iatrogenic, impinging on the tumor cell.
5. Autophagy and cancer therapy
Increased autophagy is often observed in tumor cells in response to chemotherapy and radiation. The majority of studies indicate that autophagy inhibition sensitizes tumor cells to a wide spectrum of cancer therapies, while others show that treatment-induced tumor cell death requires intact autophagic machinery. In this section, we will overview how the manipulation of autophagy potentially influences treatment outcomes in cancer.
To inhibit autophagy, studies commonly employ pharmacological inhibitors such as 3-methyladenine (3-MA), Bafilomycin A1 (BafA), and Chloroquine (CQ). 3-MA is purported as an inhibitor of class III PI3K, Vps34 [29]. Together with its regulator p150, Vps34 is involved in a variety of intracellular membrane modeling events, including autophagosome isolation membrane formation [39]. Hence, 3-MA inhibits autophagy at an early stage, the sequestration step. In contrast, BafA, a vacuolar H+-ATPase inhibitor, prevents lysosomal function, thus blocking autophagosome degradation, a late stage of the process. [65]. Similarly, CQ, a weak base that can be trapped in acidic vesicles and elevate intralysosomal pH [66], also blocks cargo degradation [67,68]. Although these compounds can effectively suppress autophagy and are widely touted as autophagy inhibitors, none are specific for autophagy. Instead, these drugs have multiple off-target effects and modulate other cellular activities, such as endocytosis, intracellular trafficking, and lysosomal biogenesis and function. A more direct evaluation of autophagy-dependent effects in response to therapies can be achieved by loss-of-function approaches directed at essential autophagy genes (e.g. atg7, atg5, beclin-1/atg6).
Autophagy inhibition promotes chemotherapeutic efficacy
High levels of autophagy observed in tumor cells following anti-cancer treatment commonly represent an adaptive response that enables tumor cells to survive the therapeutic insult [2]. Thus, it is logical to hypothesize that autophagy inhibition will synergize with other chemotherapies and more effectively eliminate cancer cells. Indeed, abundant recent work supports this notion in multiple tumor types and in response to diverse chemotherapeutic agents, highlighting the possibility of targeting autophagy as adjuvant therapy for cancer. The effect of inhibiting autophagy in combination with current anti-cancer therapies has been tested in multiple tumor models, including glioma [30,69–71], multiple myeloma [72], breast [73,74], colon[75,76], and prostate cancer [77]. Table 1 provides a partial list of such studies. Readers are also referred to recent reviews for additional information [78,79].
Table 1.
Autophagy Inhibition During Cancer Chemotherapy.
| Cancer type | Primary Treatment | Target | Method(s) of Autophagy Inhibition | Reference |
|---|---|---|---|---|
| Breast | Trastuzumab | Her2 | 3-MA, BafA, RNAi (LC3) | [73] |
| CPT | DNA topoisomerase | 3-MA, BafA, RNAi (BECN1, ATG7) | [74] | |
| CML | Imatinib | BCR/ABL | CQ, BafA, RNAi (ATG5, ATG7) | [80] |
| INNO-406 | BCR/ABL | CQ | [84] | |
| SAHA | HDAC | CQ, 3-MA | [81] | |
| Colon | Vorinostat | HDAC | CQ, RNAi (ATG7) | [75] |
| Radiation | DNA damage | RNAi (BECN1, ATG3, ATG4B, ATG5) | [76] | |
| Glioma | AKTi-1/2 | AKT | CQ | [30] |
| PI-103 | PI3K, mTOR | CQ | [30] | |
| Imatinib | Tyrosine kinases | BafA | [71] | |
| Radiation | DNA damage | 3-MA, BafA RNAi (BECN1 ATG5) | [70] | |
| Radiation | DNA damage | 3-MA, BafA | [69] | |
| Malignant Rhabdoid Tumor | FK228 | HDAC | CQ | [94] |
| Multiple myeloma | 8-aminoadenosine | DNA synthesis | CQ | [72] |
| Prostate | ADI-PEG20 | Arginine in blood | CQ, 3-MA, RNAi (BECN1) | [77] |
Abbreviations: BafA, Bafilomycin A1; CPT, Camptothecin; CQ, Chloroquine. HDAC, Histone deacetylases; MNNG N-methyl-N′-nitro-N-nitrosoguanidine; mTOR, mammalian target of rapamycin; PI3K, Phosphoinositide 3-kinase; RNAi, RNA interference (ATG target is in parentheses).
Among these studies, one striking example demonstrating the efficacy of autophagy inhibition is in chronic myelogenous leukemia (CML), a major subtype of leukemia in adults. The disease is driven by a chromosomal translocation that produces the fusion protein BCR/ABL, resulting in a constitutively active tyrosine kinase that promotes both cell proliferation and survival. CML patients have been successfully treated with imatinib (Gleevac), a tyrosine kinase inhibitor (TKI) that inhibits BCR/ABL. However, imatinib resistance is a common problem in CML patients, which is partly due to development of mutations in the Abl kinase domain, and the relative insensitivity of CML stem cells to TKIs [80,81]. Even in patients with robust response to imatinib treatment, residual disease is still detectable, indicating CML stem cells are not eliminated [82].
Imatinib has been shown to induce autophagy in a dose dependent manner in various cell lines [83]. Recently, the co-treatment of imatinib with CQ was demonstrated to significantly augmented cell death in CML cells [80,84]. Similar results were also obtained using RNAi against essential autophagy genes, atg5 and atg7, proving a direct link between autophagy inhibition and sensitization to cell death. More importantly, imatinib plus CQ enhanced cell death in primary cells isolated from patients at three distinct disease stages, including an individual that was newly diagnosed, one undergoing imatinib treatment, and a relapse patient carrying an imatinib-resistant mutation. Whereas a previous study showed imatinib alone does not efficiently kill CML progenitor cells [85], imatinib/CQ co-treatment almost completely eradicated CML stem cells in vitro, suggesting autophagy promoted CML stem cell survival during imatinib treatment. In support, an earlier study also showed that targeting autophagy augmented the effect of SAHA, an HDAC inhibitor, to overcome BCR/ABL mediated drug resistance [81].
Two additional studies assessing the effect of CQ on lymphomagenesis in vivo demonstrate that CQ-mediated lysosomal inhibition enhances p53-mediated apoptosis [67,68]. In a Myc-driven B cell lymphoma model using cells derived from p53ERTAM/p53ERTAM mice, p53 expression can be switched on in lymphoma cells when tamoxifen (TAM) is administered. CQ treatment alone modestly impaired Myc-driven tumor growth, but did not elicit regression. Similarly, inducing p53 expression with TAM induced rapid tumor regression initially, but all tumors resumed growth several days later despite continued TAM administration. On the other hand, TAM+CQ co-treatment significantly delayed tumor recurrence. A more detailed analysis implicated that CQ enhanced p53-mediated apoptosis via inhibiting autophagy, as atg5 knockdown recapitulated the effect of CQ on cell death upon p53 activation. Furthermore, the combined treatment of CQ and cyclophosphamide, the first-line therapy for human lymphomas, also significantly delayed tumor recurrence.
A second study tested the idea of using CQ as a cancer prevention agent in Eμ-Myc mice, a transgenic mouse model of human Burkitt’s lymphoma. Interestingly, the epidemiological association of CQ with reduced Burkitt’s lymphoma had been reported two decades ago [86]. In the Eμ-Myc model, continuous treatment of CQ alone significantly delayed lymphoma development, an effect that was dependent on p53 expression. The cells death observed in CQ-treated lymphoma cells exhibited hallmarks of both apoptosis and autophagy. Interestingly, blocking either apoptosis or autophagy alone had little effect on CQ mediated death; however, when both processes were simultaneously blocked, CQ-mediated cell death was suppressed. Collectively, these studies suggest a pro-survival role for autophagy that reduces p53-mediated apoptosis. Because p53 expression is often induced by diverse anticancer treatments, autophagy may serve as an adaptive response to apoptosis stimulus and promote tumor cell survival. Thus, inhibiting autophagy in this context may sensitize tumor cells to p53-mediated cell death.
Among the autophagy inhibitors tested, the lysosomal inhibitor CQ and its derivatives have gained special attention due to their favorable pharmacological properties and because of their long history of use as anti-malarial agents and in diseases such as rheumatoid arthritis. Currently, multiple clinical trials using CQ as a sensitizing reagent in combination with standard cancer therapies are under evaluation in different tumor types, including lung cancer, glioblastoma multiforme, multiple myeloma, breast cancer, melanoma, colon cancer, prostate cancer, and advanced solid tumors unresponsive to chemotherapy (http://clinicaltrials.gov). However, as discussed above, the cytotoxic effects of chloroquine and similar agents may involve processes other than autophagy; hence, further studies are needed to dissect the precise contributions of autophagy inhibition when employing these anti-malarials in diverse clinical settings.
Autophagy-dependent cell death in cancer therapy
Although the concept of “autophagic cell death” in mammalian cells remains highly controversial [87], studies do show that autophagy is required for the efficient killing of tumor cells in certain circumstances. In pursuit of new drugs to treat renal cell carcinoma (RCC), Giaccia and colleagues identified compound STF-62247, which selectively kills von Hippel-Lindau (VHL) deficient (a tumor suppressor gene lost in 75% of RCCs) cells in a synthetic lethal manner [88]. STF-62247 caused autophagy induction and massive vacuolization in VHL-deficient RCC cells with no apparent DNA damage or apoptosis. Blocking autophagy with 3-MA or silencing atg5, atg7, or atg9 increased cell survival, supporting that STF-62247 induces an autophagy-dependent cell death in these cells.
In addition, Δ9-tetrahydrocannabinol (THC) can also induce cell death in glioma cells through the stimulation of autophagy [89]. In this study, THC treatment induced ER stress and inhibited the AKT/mTOR pathway. Genetic deletion of atg5 or RNAi-mediated depletion of atg1 partially protected cells from death during THC treatment. Interestingly, blocking apoptosis also prevented cell death to a degree similar to atg5 deletion, which led the authors to speculate that autophagy is required for robust apoptosis induction in response to THC. In support, knockdown of atg genes prevented caspase-3 activation, and xenograft tumors formed by transformed atg5−/− fibroblasts were resistant to THC-induced apoptosis. Thus, THC-induced autophagy may promote the apoptosis of cancer cells rather than directly cause their death.
The coordination of autophagy and apoptosis was also observed in a recent study with melanoma cells [90]. The classic double-stranded RNA mimic polyinosine-polycytidylic acid complexed with polyethyleneimine ([PIC]PEI) potently killed melanoma cells both in vitro and in vivo, and both autophagy and apoptosis were required for inducing efficient cell death. Notably, [PIC]PEI induced rapid and persistent endocytic flux in melanoma cells. When autophagy was inhibited via BafA, CQ or atg5 deletion, cell death was suppressed. Interestingly, cell death was also significantly reduced in melanoma cells treated with caspase-inhibitors or in cells doubly deficient for Bax and Bak, two multi-domain Bcl-2 family members required for apoptosis, without affecting autophagy induction. Based on these results and the observed time lag between autophagy induction (2–5h post-treatment) and final cell collapse (at 24–48h), the authors speculated that early and persistent autophagy induction is required for apoptosis activation at a subsequent stage. These observations illustrate the complex interplay between autophagy and apoptosis.
How then do we reconcile the seemingly conflicting roles of autophagy on cell death in response to cancer therapy? Multiple factors should be considered before drawing conclusions. For example, although 3-MA and BafA are both considered autophagy inhibitors, when used with temozolomide (TMZ) to treat glioma cells, two opposite outcomes occurred. 3MA suppressed whereas BafA enhanced TMZ-mediated cell death [91]. This indicates a stage-specific consequence for autophagy inhibition, since 3-MA and BafA inhibit the early and late stages of autophagy, respectively [71]. Also, autophagy can exert both pro- or anti-apoptotic effects in the same cells depending on the death stimulus administered [92]. Furthermore, when genetically modified cells (e.g. ATG deficient cells) are used, other forms of autophagy, such as chaperone-mediated autophagy or microautophagy, may compensate for the loss of macroautophagy and thus, influence tumor cell killing in unexpected ways [93]. Moreover, in limited circumstances, it does appear that both autophagy and apoptosis are needed to kill cells; in such situations, stimulating autophagy may enhance drug cytotoxicity [90]. Finally, due to the lack of autophagy specific inhibitors, the majority of studies have been conducted with pharmacological agents with known off-target effects beyond autophagy. Development of autophagy specific inhibitors, such as small molecules targeting kinases (e.g. ULK1) or proteases (e.g. Atg4B) required for autophagy induction, may help determine the precise role of autophagy in cancer treatment.
In summary, the papers discussed above support that targeting autophagy in combination with current cancer treatments is a promising therapeutic strategy. However, due to the contrasting roles autophagy plays during cancer initiation and progression, the use of autophagy inhibitors should be tailored to specific tumor contexts and in combination with specific therapeutics.
6. Concluding remarks
Numerous links between deregulated autophagy and human diseases have emerged. In cancer, current evidence supports that autophagy suppresses tumorigenesis, particularly during the early stages of tumor initiation. However, in established tumors, autophagy can function as a survival pathway in response to stresses imposed during cancer progression and due to chemotherapy. Accordingly, autophagy inhibition as an adjuvant to standard cancer therapy may increase treatment efficacy in some tumor types. Although such experimental findings have therapeutic implications, the clinical utility of modulating autophagy in human cancers still remains unclear. Notably, the efficacy of lysosomal agents that inhibit autophagy, such as CQ, is probably tumor-type specific, and it remains unclear if long-term autophagy inhibition may elicit untoward side effects in real patients. Ongoing clinical trials should provide valuable information on whether and how to manipulate autophagy during cancer treatment.
Acknowledgments
JD is supported by grants from the NIH (RO1CA126792; CA126792-S1 ARRA), the California Tobacco Related Disease Research Program (18XT-0106), and an HHMI Physician Scientist Early Career Award.
Abbreviations
- ARHI
aplasia Ras homolog member I
- ATG
autophagy regulator
- BafA
bafilomycin A1
- Bif-1
Bax-binding protein 1
- C-Vps
class C Vps complex
- CML
chronic myelogenous leukemia
- CQ
chloroquine
- DRAM
damage-regulated autophagy modulator
- ER
Endoplasmic reticulum
- ECM
extracellular matrix
- GAP
GTPase activating protein
- mTOR
mammalian target of rapamycin
- NK
natural killer
- OIS
oncogene-induced senescence
- PI3K
Phosphoinositide 3-kinase
- [PIC]PEI
polyinosine-polycytidylic acid complexed with polyethyleneimine
- RCC
renal cell carcinoma
- Rheb
Ras Homolog Enriched in Brain
- ROS
reactive oxygen species
- TAM
tamoxifen
- THC
Δ9-tetrahydrocannabinol
- TKI
tyrosine kinase inhibitor
- 3-MA
3-methyladenine
- TMZ
temozolomide
- TSC
Tuberous Sclerosis Complex
- UVRAG
UV irradiation Resistance-Associated Gene
- VHL
von Hippel-Lindau
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 2.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–34. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 5.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iqbal J, et al. Genomic analyses reveal global functional alterations that promote tumor growth and novel tumor suppressor genes in natural killer-cell malignancies. Leukemia. 2009;23:1139–51. doi: 10.1038/leu.2009.3. [DOI] [PubMed] [Google Scholar]
- 8.Ionov Y, Nowak N, Perucho M, Markowitz S, Cowell JK. Manipulation of nonsense mediated decay identifies gene mutations in colon cancer Cells with microsatellite instability. Oncogene. 2004;23:639–45. doi: 10.1038/sj.onc.1207178. [DOI] [PubMed] [Google Scholar]
- 9.Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ. Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Hum Pathol. 2008;39:1059–63. doi: 10.1016/j.humpath.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Coppola D, Khalil F, Eschrich SA, Boulware D, Yeatman T, Wang HG. Down-regulation of Bax-interacting factor-1 in colorectal adenocarcinoma. Cancer. 2008;113:2665–70. doi: 10.1002/cncr.23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–83. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 12.Degenhardt K, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathew R, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–35. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer. 2009;9:714–23. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 16.Mathew R, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeNardo D, Johansson M, Coussens L. Immune cells as mediators of solid tumor metastasis. Cancer and Metastasis Reviews. 2008;27:11–18. doi: 10.1007/s10555-007-9100-0. [DOI] [PubMed] [Google Scholar]
- 19.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM. CD4+ T Cells Regulate Pulmonary Metastasis of Mammary Carcinomas by Enhancing Protumor Properties of Macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. The Journal of Pathology. 2002;196:254–265. doi: 10.1002/path.1027. [DOI] [PubMed] [Google Scholar]
- 21.Young AR, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Z, et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest. 2008;118:3917–29. doi: 10.1172/JCI35512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He C, Klionsky DJ. Regulation Mechanisms and Signaling Pathways of Autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–68. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 27.Nardella C, et al. Aberrant Rheb-mediated mTORC1 activation and Pten haploinsufficiency are cooperative oncogenic events. Genes Dev. 2008;22:2172–7. doi: 10.1101/gad.1699608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou X, Ikenoue T, Chen X, Li L, Inoki K, Guan KL. Rheb controls misfolded protein metabolism by inhibiting aggresome formation and autophagy. Proc Natl Acad Sci U S A. 2009;106:8923–8. doi: 10.1073/pnas.0903621106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–8. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 30.Degtyarev M, et al. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008;183:101–16. doi: 10.1083/jcb.200801099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, Codogno P, Ogier-Denis E. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem. 2001;276:35243–6. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- 32.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–75. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corradetti MN, Inoki K, Bardeesy N, DePinho RA, Guan KL. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 2004;18:1533–8. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoki K, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–68. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 35.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–26. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang J, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–24. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 37.Kumar SH, Rangarajan A. Simian virus 40 small T antigen activates AMPK and triggers autophagy to protect cancer cells from nutrient deprivation. J Virol. 2009;83:8565–74. doi: 10.1128/JVI.00603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–39. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–82. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–72. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–70. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 42.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 43.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–99. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 44.Liang C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–87. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–51. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta. 2006;1761:897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 47.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–6. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–96. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 49.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–76. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maiuri MC, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. Embo J. 2007;26:2527–39. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Erlich S, et al. Differential interactions between Beclin 1 and Bcl-2 family members. Autophagy. 2007;3:561–8. doi: 10.4161/auto.4713. [DOI] [PubMed] [Google Scholar]
- 52.Saeki K, Yuo A, Okuma E, Yazaki Y, Susin SA, Kroemer G, Takaku F. Bcl-2 down-regulation causes autophagy in a caspase-independent manner in human leukemic HL60 cells. Cell Death Differ. 2000;7:1263–9. doi: 10.1038/sj.cdd.4400759. [DOI] [PubMed] [Google Scholar]
- 53.Akar U, et al. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy. 2008;4:669–79. doi: 10.4161/auto.6083. [DOI] [PubMed] [Google Scholar]
- 54.Wei Y, Weng D, Li F, Zou X, Young DO, Ji J, Shen P. Involvement of JNK regulation in oxidative stress-mediated murine liver injury by microcystin-LR. Apoptosis. 2008;13:1031–42. doi: 10.1007/s10495-008-0237-2. [DOI] [PubMed] [Google Scholar]
- 55.Zalckvar E, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–92. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pimkina J, Humbey O, Zilfou JT, Jarnik M, Murphy ME. ARF induces autophagy by virtue of interaction with Bcl-xl. J Biol Chem. 2009;284:2803–10. doi: 10.1074/jbc.M804705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–46. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yee KS, Wilkinson S, James J, Ryan KM, Vousden KH. PUMA- and Bax-induced autophagy contributes to apoptosis. Cell Death Differ. 2009;16:1135–45. doi: 10.1038/cdd.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–4. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 60.Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–53. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 61.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–60. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maiuri MC, Malik SA, Morselli E, Kepp O, Criollo A, Mouchel PL, Carnuccio R, Kroemer G. Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle. 2009;8:1571–6. doi: 10.4161/cc.8.10.8498. [DOI] [PubMed] [Google Scholar]
- 63.Crighton D, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–34. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 64.Tasdemir E, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 66.Luiken JJ, Aerts JM, Meijer AJ. The role of the intralysosomal pH in the control of autophagic proteolytic flux in rat hepatocytes. Eur J Biochem. 1996;235:564–73. doi: 10.1111/j.1432-1033.1996.00564.x-i2. [DOI] [PubMed] [Google Scholar]
- 67.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maclean KH, Dorsey FC, Cleveland JL, Kastan MB. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008;118:79–88. doi: 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ito H, Daido S, Kanzawa T, Kondo S, Kondo Y. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int J Oncol. 2005;26:1401–10. [PubMed] [Google Scholar]
- 70.Lomonaco SL, Finniss S, Xiang C, Decarvalho A, Umansky F, Kalkanis SN, Mikkelsen T, Brodie C. The induction of autophagy by gamma-radiation contributes to the radioresistance of glioma stem cells. Int J Cancer. 2009;125:717–22. doi: 10.1002/ijc.24402. [DOI] [PubMed] [Google Scholar]
- 71.Shingu T, et al. Stage-specific effect of inhibition of autophagy on chemotherapy-induced cytotoxicity. Autophagy. 2009;5:537–9. doi: 10.4161/auto.5.4.8164. [DOI] [PubMed] [Google Scholar]
- 72.Shanmugam M, et al. Targeting glucose consumption and autophagy in myeloma with the novel nucleoside analogue 8-aminoadenosine. J Biol Chem. 2009;284:26816–30. doi: 10.1074/jbc.M109.000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA. Autophagy facilitates the development of breast cancer resistance to the anti-HER2 monoclonal antibody trastuzumab. PLoS One. 2009;4:e6251. doi: 10.1371/journal.pone.0006251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–10. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- 75.Carew JS, et al. Autophagy inhibition enhances vorinostat-induced apoptosis via ubiquitinated protein accumulation. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–94. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 77.Kim RH, et al. Arginine deiminase as a novel therapy for prostate cancer induces autophagy and caspase-independent apoptosis. Cancer Res. 2009;69:700–8. doi: 10.1158/0008-5472.CAN-08-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoyer-Hansen M, Jaattela M. Autophagy: an emerging target for cancer therapy. Autophagy. 2008;4:574–80. doi: 10.4161/auto.5921. [DOI] [PubMed] [Google Scholar]
- 79.Eisenberg-Lerner A, Kimchi A. The paradox of autophagy and its implication in cancer etiology and therapy. Apoptosis. 2009;14:376–91. doi: 10.1007/s10495-008-0307-5. [DOI] [PubMed] [Google Scholar]
- 80.Bellodi C, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest. 2009;119:1109–23. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carew JS, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–22. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Copland M, et al. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML but does not eliminate the quiescent fraction. Blood. 2006;107:4532–9. doi: 10.1182/blood-2005-07-2947. [DOI] [PubMed] [Google Scholar]
- 83.Ertmer A, Huber V, Gilch S, Yoshimori T, Erfle V, Duyster J, Elsasser HP, Schatzl HM. The anticancer drug imatinib induces cellular autophagy. Leukemia. 2007;21:936–42. doi: 10.1038/sj.leu.2404606. [DOI] [PubMed] [Google Scholar]
- 84.Kamitsuji Y, et al. The Bcr-Abl kinase inhibitor INNO-406 induces autophagy and different modes of cell death execution in Bcr-Abl-positive leukemias. Cell Death Differ. 2008;15:1712–22. doi: 10.1038/cdd.2008.107. [DOI] [PubMed] [Google Scholar]
- 85.Jorgensen HG, Allan EK, Jordanides NE, Mountford JC, Holyoake TL. Nilotinib exerts equipotent antiproliferative effects to imatinib and does not induce apoptosis in CD34+ CML cells. Blood. 2007;109:4016–9. doi: 10.1182/blood-2006-11-057521. [DOI] [PubMed] [Google Scholar]
- 86.Geser A, Brubaker G, Draper CC. Effect of a malaria suppression program on the incidence of African Burkitt’s lymphoma. Am J Epidemiol. 1989;129:740–52. doi: 10.1093/oxfordjournals.aje.a115189. [DOI] [PubMed] [Google Scholar]
- 87.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–10. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Turcotte S, Chan DA, Sutphin PD, Hay MP, Denny WA, Giaccia AJ. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell. 2008;14:90–102. doi: 10.1016/j.ccr.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Salazar M, et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119:1359–72. doi: 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tormo D, et al. Targeted activation of innate immunity for therapeutic induction of autophagy and apoptosis in melanoma cells. Cancer Cell. 2009;16:103–14. doi: 10.1016/j.ccr.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11:448–57. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- 92.Wang Y, et al. Loss of macroautophagy promotes or prevents fibroblast apoptosis depending on the death stimulus. J Biol Chem. 2008;283:4766–77. doi: 10.1074/jbc.M706666200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kaushik S, Massey AC, Mizushima N, Cuervo AM. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell. 2008;19:2179–92. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Watanabe M, et al. Induction of autophagy in malignant rhabdoid tumor cells by the histone deacetylase inhibitor FK228 through AIF translocation. Int J Cancer. 2009;124:55–67. doi: 10.1002/ijc.23897. [DOI] [PubMed] [Google Scholar]