Abstract
We compared hepatic expression of genes that regulate lipid biosynthesis and metabolic signaling in liver biopsy specimens from women who were undergoing gastric bypass surgery (GBP) for morbid obesity (MO) to that in women undergoing ventral hernia repair who had experienced massive weight loss (MWL) following prior GBP. Comprehensive metabolic profiles of MO (22 subjects) and MWL (9 subjects) were also compared. Analyses of gene expression in liver biopsies from MO and MWL were accomplished by Affymetrix microarray, real-time PCR and Western blotting techniques. Following GBP, MWL subjects had lost on average 102 pounds as compared to MO subjects. This was accompanied by effective reversal of the dyslipidemia and insulin resistance that was present in MO. As compared with MWL, livers of MO subjects exhibited increased expression of Sterol Regulatory Element Binding Protein (SREBP-1c) and its downstream lipogenic targets, fatty acid synthase (FAS) and acetyl-CoA-carboxylase-1 (ACC-1). Livers of MO subjects also exhibited enhanced expression of suppressor of cytokine signaling -3 (SOCS-3) protein and attenuated Janus kinase signal transducer and activator of transcription (JAK/STAT) signaling. Consistent with these findings, we found that the human SREBP-1c promoter was positively regulated by insulin and negatively regulated by STAT3. These data support the hypothesis that SOCS-3 mediated attenuation of the STAT signaling pathway and resulting enhanced expression of SREBP-1c, a key regulator of de novo lipid biosynthesis, are mechanistically related to the development of hepatic insulin resistance and dyslipidemia in morbidly obese women.
1. Introduction
Both insulin insensitivity and the accompanying hyperinsulinemia and dsylipidemia are key features of obesity (1). In obesity, dyslipidemia associated with insulin resistance results, in large part, from increased hepatic synthesis and secretion of very-low-density-lipoprotein (VLDL) (2). Although increased availability of fatty acids from adipose tissue as a result of impaired ability of insulin to inhibit lipolysis is, in part, responsible for enhanced hepatic triglyceride synthesis in obesity, enhanced hepatic de novo lipogenesis also appears to play an important role. In addition, in obesity, enhanced production of triglycerides (TG) by the liver often exceeds its ability to secrete TG thereby leading to the development of non-alcoholic fatty liver disease, steatohepatitis, fibrosis and cirrhosis (3, 4).
Although de novo hepatic lipogenesis is a relatively minor pathway in the production of VLDL triglyceride in lean humans (5), metabolic turnover studies performed in obese subjects (6) and in those consuming high carbohydrate diets indicate that de-novo lipogenesis becomes a major source of VLDL-TG (7–9). In obese hyperinsulinemic rodents, increased hepatic lipid synthesis results from up-regulation of the lipogenic regulator Sterol Regulatory Element Binding Protein-1c (SREBP-1c) and its downstream lipogenic enzyme gene targets (10, 11). SREBP-1c expression in the liver is directly enhanced by insulin both at the transcriptional and posttranslational level (12–15). The rodent SREBP-1c promoter contains a multi-component insulin response element that mediates rapid induction of SREBP-1c transcription (12). Conversely, insulin also enhances the transport of nascent SREBP-1c and its chaperone, sterol cleavage activating protein (SCAP), to the Golgi where it undergoes proteolytic cleavage to release the transcriptionally active n-terminal SREBP-1c fragment (15, 16). Enhanced hepatic expression of SREBP-1c in response to hyperinsulinemia in obesity seems somewhat paradoxical, however, given the attenuation of other effects of insulin including suppression of gluconeogenesis and lipolysis as a result of obesity related insulin resistance (17).
The mechanisms of insulin resistance in obesity involve many factors, including elevated levels of pro-inflammatory adipokines such as tumor necrosis factor-alpha (TNF-α) and interleukin-6 (IL-6) (18). Recent studies have begun to shed light on the mechanisms by which adipokines oppose insulin action; specifically, the suppressor of cytokine signaling (SOCS) proteins have been implicated in the induction of insulin resistance in obese rodents (19). Ueki and coworkers demonstrated that increased expression of SOCS-1 and SOCS-3 in liver and muscle of obese mice was associated with decreased tyrosine phosphorylation of insulin receptor substrate (IRS) proteins (20). In these animal studies, attenuation of JAK/STAT-3 signaling by SOCS-1 and SOCS-3 was associated with enhanced expression of SREBP-1c (21).
It is unclear whether enhanced hepatic de novo lipogenesis in obese humans also arises from aberrant regulation of SREBP-1c and the JAK/STAT signaling pathway. Examination of the effect of obesity on hepatic lipid gene expression in humans has been limited by the inherent difficulty in obtaining non-diseased liver tissue from obese and non-obese controls. In this regard, morbidly obese patients who are undergoing gastric bypass surgery (GBP) for weight loss provide a unique opportunity to study hepatic gene expression in the human. Furthermore, a subset of these individuals subsequently undergo ventral hernia repair following massive weight loss as a result of prior GBP. This provides an opportunity to sample liver tissue in MO subjects who have experienced reversion to the pre-obese state following massive weight loss. These individuals can serve as a post-obese control for MO insofar as massive weight loss following GBP is accompanied by effective reversal of both obesity-associated insulin resistance and dyslipidemia (22). To gain insight into the altered metabolic signaling and lipid homeostasis that accompanies morbid obesity, we compared gene expression profiles in liver biopsy samples from MO subjects undergoing GBP as compared to women undergoing ventral hernia repair as a result of MWL following GBP. Using gene ontology and hierarchical clustering analysis of microarray based gene expression, we recently reported altered expression of many genes involved in wound healing, bile acid transport, and xenobiotic metabolism in morbidly obese women (23). Since only a limited number of genes related to lipid and energy homeostasis met the stringent criteria for differential gene expression set in this previous analysis (23), in the present analysis we specifically mined the microarray data to address the question of altered expression of many key genes related to lipid synthesis and metabolic signaling. The microarray analyses were further supplemented by real-time PCR measurements of selected mRNAs and western blotting measurements of the putative protein products encoded by these mRNAs. We report here that expression of SREBP-1c and its downstream regulatory targets, fatty acid synthase (FAS), Diacylglycerol Acyltransferase (DGAT), stearoyl-CoA desaturase-1 (SCD-1), acetyl-CoA carboxylase-1 (ACC-1), and malic enzyme (ME), is increased in livers of morbidly obese women. Furthermore, morbid obesity is also associated with enhanced expression of the signaling inhibitors SOCS-1 and SOCS-3 and concomitant attenuation of JAK/STAT signaling.
2. Experimental procedures
2.1 Treatment of study subjects, tissue sample collection and clinical laboratory assessments
Institutional Review Boards of the University of Tennessee Health Science Center (UTHSC) and Memphis Baptist Memorial Hospital approved all study procedures. All participants gave written informed consent. Subjects were excluded from participating in the study if they had a history of triglyceride-related pancreatitis, or elevated creatinine, liver enzymes, bilirubin or hypoalbuminemia (23). Subjects were also excluded if there was a history of malignancy, diabetes mellitus, recent major illness or current treatment with corticosteroids, androgens, or lipid-lowering agents. The study participants were admitted to the UTHSC General Clinical Research Center (GCRC) one day prior to surgery for pre-operative care and clinical profiling. Participants were provided a diet diary and were instructed by the GCRC dietician to record their food intake for the 3 days prior to admission. The GCRC nutritionist reviewed the three-day diet diary and dietary composition was analyzed by computer program (Nutritionist Plus).
Blood samples were obtained in the fasting state for baseline determinations of plasma lipoproteins, glucose, insulin, and free fatty acid (FFA). Serum glucose, insulin, and FFA were assessed in MO and WL patients over a 3-hour period following initiation of oral glucose tolerance test (OGTT). Insulin sensitivity was estimated using an index of whole body insulin sensitivity based on OGTT derived glucose and insulin levels as previously described (24). The formula used was: 10,000/square root of (fasting glucose X fasting insulin) X (mean OGTT glucose X mean OGTT insulin). Plasma lipoproteins were isolated from fasting plasma samples; apolipoprotein and lipid content were analyzed using HPLC and enzymatic assays following separation of lipoprotein fractions by ultracentrifugation as described previously (25). Following completion of the OGTT, subjects were allowed intake of clear liquids ad libitum as part of their preoperative care. The patients were then transferred to the surgical units of the participating hospitals and remained fasting overnight until completion of surgery.
Gastric bypass surgery in the morbidly obese women was accomplished by an extended Roux-en-Y gastric bypass coupled with a horizontal gastric pouch as described previously (23). A separate group of patients who were, on average, one-year post-GBP and who had lost greater than 80 pounds of body weight who were undergoing abdominoplasty to repair ventral hernias and diastasis recti served as post-obese controls (MWL). A wedge biopsy specimen of liver was removed under direct operative visualization from patients undergoing GBP and from MWL subjects undergoing ventral hernia repair. A portion of the liver biopsy sample was immediately placed in RNAzol (Tel-Test, Friendswood, Texas) solution and the remainder of the tissue was snap-frozen in liquid nitrogen and stored at −70°C for subsequent protein analysis.
Samples were analyzed by the GCRC core laboratory and by commercial laboratory. Serum chemistry was determined by standard auto-analyzer techniques in a commercial laboratory. Insulin and C-peptide were measured using commercially available radioimmunoassay.
2.2 Determination of hepatic gene expression by microarray analysis
Total RNA was isolated by chloroform-phenol extraction and isopropanol precipitation following the manufacturers recommended protocol. RNA samples were re-dissolved in diethyl pyrocarbonate-treated water and were processed for microarray analysis using Affymetrix oligonucleotide arrays (HG-133A) as previously described (23). The expression data were analyzed according to published methods (26). Genes were arranged in decreasing order of their mean detection p-value and all of the genes possessing a value of 95% or greater (p≤ 0.05) were copied into a new file. The resulting 3576 genes were matched with their identification numbers from Unigene, Locus Link and GenBank databases that were used to confirm the identity of the gene. This probe set database was then queried to identify specific genes related to fatty acid metabolism and lipid related signaling and mean signal ratios for expression of these genes in livers of MO versus MWL were generated.
2.3 Determination of hepatic gene expression by real-time PCR
The expression of a subset of differentially expressed genes, as revealed by microarray analysis, along with a number genes involved in lipid metabolism that were not detected by microarray-based analyses, was determined by real-time PCR. Real-time PCR was performed with a LightCycler 480 System (Roche Diagnostics, Indianapolis, Indiana) using SYBR Green 1 dye (LightCycler 480 SYBR Green 1 Master Mix; Roche Diagnostics) intercalation to detect DNA amplification. Expression of both the target gene and control gene (cyclophilin D) within each sample were quantified based on their respective threshold cycle (Ct) values. Target gene Ct values were normalized to cyclophilin D control gene Ct values. The ratios of target gene expression of MO vs. MWL were derived. The Universal Probe Library from Roche Applied Science (Indianapolis, Indiana) was used to design the primers used for real-time PCR (Table 1).
Table 1.
Sequences of primers used for Real-Time PCR.
| Gene | Oligonucleotide Sequence | |
|---|---|---|
| FASN | Forward | 5’-ACAGGGACAACCTGGAGTTCT-3’ |
| Reverse | 5’-CTG TGG TCC CAC TTG ATG AGT-3’ | |
| SREBP-1 | Forward | 5' - GCTCCTCCATCAATGACAAAA - 3' |
| Reverse | 5' - TGCGCAAGACAGCAGATTTA - 3' | |
| SREBP-1c | Forward | 5' - GGAGGGGTAGGGCCAACGGCCT - 3' |
| Reverse | 5' - CATGTCTTCGAAAGTGCAATCC - 3' | |
| SREBP-2 | Forward | 5' - ATCTGGATCTCGCCAGAGG - 3' |
| Reverse | 5' - CCAGGCAGGTTTGTAGGTTG - 3' | |
| HMG-R | Forward | 5’-CTGAAGCTGGCAAATCAAAAG-3’ |
| Reverse | 5’-CTTTGCATGCTCCTTGAACA-3’ | |
| DGAT-1 | Forward | 5’-ACTACCGTGGCATCCTGAAC -3’ |
| Reverse | 5’-ATAACCGGGCATTGCTCA-3’ | |
| PGC-1a | Forward | 5’-GCAGGAGCAGAGCAAAGAGG-3’ |
| Reverse | 5’-AAAGTTGTTGGTTTGGCTTGTAAG T-3’ | |
| SCD-1 | Forward | 5’-GTACCGCTGGCACATCAACTT-3’ |
| Reverse | 5’-TTGGAGACTTTCTTCCGGTCAT-3’ | |
| ACC-1 | Forward | 5’-GGACAACACCTGTGTGGTAGAA-3’ |
| Reverse | 5’-CGTGGGGATGTTCCCTCT-3’ | |
| DGAT-1 | Forward | 5’-ACTACCGTGGCATCCTGAAC-3’ |
| Reverse | 5’-ATAACCGGGCATTGCTCA-3’ | |
Primers were designed using the Universal Probe Library from Roche Applied Science. Primers were obtained from Integrated DNA Technologies. Primers for SOCS3 and MAP2K6 were purchased from Qiagen Corporation. QuantiTect Primer Assay (proprietary)
2.4 Western blot analysis of liver proteins
Liver biopsy samples (2 gm) were homogenized in a protein lysis buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl pH8.0, 2 mM DTT) containing protease and phosphatase inhibitors and then centrifuged at 10,000xg for 10 min at 4°C. The supernatants were collected and their protein concentrations were determined. Equal aliquots of hepatic proteins were size-fractionated by SDS-PAGE, subject to western blot analysis and quantified according to published protocols (15). The anti-SREBP-1 (SC-13551), anti-SREBP-2 (SC-13552), anti-FAS (SC-20140), anti-ACC-1 (SC-26817), anti-SOCS-3 (SC-9023), anti-SOCS-1 (SC-9021), anti-STAT-3 (SC-482) anti-ApoA (SC-13549), and anti-ApoE (SC-13521) were purchased from Santa Cruz Biotechnology Inc (Santa Cruz, Ca.). Antiphospho-STAT-3 (9131S) was from Cell Signaling Technology (Beverly, MA). Anti-β-actin antibody (SC-1616) was used to determine protein loading.
2.5 Determination of response of the human SREBP-1c promoter-luciferase construct response to insulin and STAT3
To evaluate the potential significance of altered hepatic SREBP-1c and SOCS/STAT3 expression in MO, the response of the human SREBP-1c promoter to both insulin and STAT3 was evaluated in vitro. The BAC-CTD2502C3 clone (Open Biosystems, Huntsville, AL) containing the human SREBP-1c genomic DNA was used to amplify promoter DNA encompassing -1498 to +1 (SREBP-1c transcription start point denoted as +1) by PCR primers terminated with KpnI and BglII sites. The amplified DNA was ligated into the polylinker upstream of the luciferase coding sequence of pGL3 vector (Promega; Madison, WI) to generate the hSREBP-1c-luc plasmid. Rat primary hepatocytes were transfected with 1 µg of hSREBP1c-luc plus 1 µg of pRL-TK (Promega) and luciferase expression were quantified as reported previously (12, 27) following treatment with either insulin (100 nM) or the LXR agonist T0901317 (10 µM). To determine the effect of altered STAT3 expression on hSREBP-1c promoter activity, primary rat hepatocytes were transfected with the hSREBP1c-luc construct along with plasmids expressing wild type or dominant negative STAT3 proteins, or empty vector. Twenty-four hours after transfection cells were harvested and the luciferase activity measured. Cells were co-transfected with a control Renilla expression plasmid, pRL-TK.
2.6 Statistical Analyses
Significance of differences between plasma analytes, mRNA and protein expression and anthropometric variables between MWL and MO was determined by Student’s T-Test (continuous variables) or C2 (Fisher’s Exact Test for non-continuous variables) using JMP Statistical Software (Version 5, SAS, Cary NC).
3. Results
3.1 Clinical and metabolic characteristics of morbidly obese women compared to those with massive weight loss following gastric bypass surgery
Metabolic profiling was performed prior to GBP and ventral hernia repair in 22 MO and 9 MWL subjects, respectively. The MWL group was on average 102 pounds lighter (199 pounds versus 301 for MO), reflecting weight loss as a result of prior GBP (Table 2). Similarly, BMI was significantly lower in MWL (32.0 versus 54.5 kg/m2 for MO) (Table 2). Total plasma cholesterol and triglyceride levels were higher and HDL-cholesterol was lower than expected in MO subjects, given their age and sex (Table 2). Correspondingly, in women with MWL following prior GBP, plasma levels of total and LDL-cholesterol and triglyceride were significantly lower and HDL-Cholesterol was significantly higher compared to their pre-GBP (MO) counterparts (Table 2). Similarly, plasma apoB was markedly lower in MWL subjects, as was the VLDL apoprotein ApoCIII (Table 2). On the other hand, plasma levels of the HDL apoproteins, ApoAI and ApoAII, were not significantly different between MWL and MO (Table 2). Thus, reduced HDL-C appeared to result from reduced cholesterol content of HDL particles rather than reduced particle number. In summary, the comparison between MWL and MO indicated a significant elevation in plasma levels of ApoB containing lipoproteins, in particular TG-rich particles (VLDL, IDL), and LDL and decreased cholesterol content of HDL in MO.
TABLE 2.
Clinical and metabolic characteristics of morbidly obese (MO) women undergoing gastric bypass surgery (GBP) and of those experiencing massive weight loss (MWL) following GBP.
| Variable | MO (N= 22) | MWL (N=9) | ¶ P = |
|---|---|---|---|
| Age (Years) | 36.2 ± 1.4 | 39.2 + 2.5 | 0.27 |
| African American (%) | 22.7 % | 44.0 % | 0.38 |
| Weight (pounds) | 301 ± 17 | 199 ± 13 | 0.002 |
| Body Mass Index (BMI) | 54.5 ± 3.9 | 32.0 ± 1.8 | 0.001 |
| Cholesterol (Total) | 171 ± 7 | 114 ± 9 | 0.0002 |
| LDL-Cholesterol | 101 ± 6 | 65 ± 12 | 0.008 |
| HDL-Cholesterol | 40 ± 2 | 50 ± 2 | 0.002 |
| Triglyceride (Total) | 157 ± 26 | 63 ± 9 | 0.04 |
| ApoAI (mg/dl) | 146 ± 9 | 151 ± 7 | 0.74 |
| ApoAII (mg/dl) | 44 ± 3 | 41 ± 6 | 0.66 |
| ApoB (mg/dl) | 73 ± 3 | 44 ± 6 | 0.0002 |
| ApoCIII (mg/dl) | 12 ± 1 | 7 ± 1 | 0.016 |
| Glucose (mg/dl) | 105 ± 5 | 84 ± 4 | 0.03 |
| Insulin (µU/ml) | 20 ± 2 | 5 ± .9 | 0.0002 |
| FFA (µEQ/ml) | 889 ± 54 | 674 ± 98 | 0.047 |
| FFA (Post-OGTT) | 611 ± 44 | 285 ± 75 | 0.002 |
| Insulin Sensitivity Index* | 0.027 ± 0.008 | 0.220 ± 0.094 | <0.001 |
| AST (U/L) | 23.0 ± 3.5 | 26.2 ± 3.0 | 0.57 |
| ALT (U/L) | 35.2 ± 5.0 | 32.4 ± 4.6 | 0.73 |
Anthropometric measures, plasma lipoproteins, hepatic transaminase and an index of insulin sensitivity were assessed in MO prior to GBP and in another group of women undergoing ventral hernia repair after massive weight loss (MWL) following GBP. Data are mean ± SEM for demographic, anthropometric, and metabolic parameters.
Significance of differences between obese and lean participants determined by Student’s T-Test for continuous variables and Chi-Square (Fishers Exact Test) for non-continuous variables (SAS-JMP, Cary NC).
Whole-body insulin sensitivity (glucose utilization) was estimated based on OGTT insulin and glucose determinations as described by Matsumoto et al (24).
Dietary analysis confirmed higher daily intake of total calories in MO women, compared to those who had previously undergone GBP (2,268 vs. 1,604 Cal/day respectively; P = 0.012). Although total intake of protein, fat and carbohydrate was lower in MWL subjects, the most striking change in their diet was a marked reduction in carbohydrate intake (157 gm/day in MWL vs. 272 gm/day in MO; P=0.003). In particular, daily intake of simple sugars was dramatically reduced in MWL from 19.7% of total calories in MO to 10.9% (P= 0.02).
3.2 Gastric bypass surgery effectively reverses insulin resistance and hyperinsulinemia in MO women
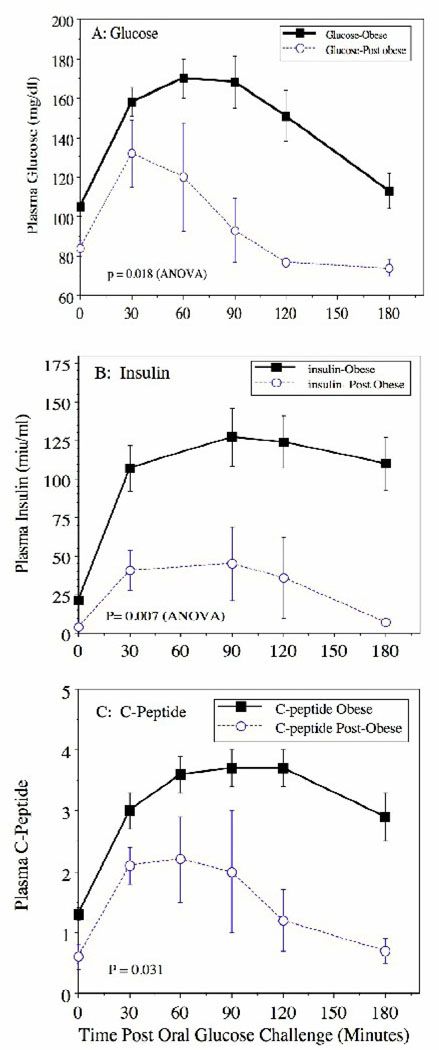
We assessed the glycemic status and insulin levels in MO and MWL patients both in the fasting state and via an OGTT that was administered after overnight fasting. As shown in Table 2, both fasting plasma glucose and insulin levels were higher in MO subjects and insulin sensitivity was significantly reduced (Table 2). Following oral glucose challenge, MO subjects exhibited significant postprandial hyperglycemia and hyperinsulinemia with elevated 2-hour levels of glucose vs. MWL (165 vs. 75 mg/dl respectively) and insulin (125 vs. 30 miµ/ml respectively) (Figure 1). Both fasting and postprandial levels of C-peptide were also higher in MO indicating increased insulin secretion (Figure 1). There was an almost 10-fold difference in insulin sensitivity (an index of whole body glucose disposal) between MO and MWL groups as estimated by the method of Matsuda et al (24) (0.028 ± 0.008 versus 0.220 ± 0.094 respectively) (Table 2). Fasting plasma levels of FFA were higher and the reduction in plasma FFA following OGTT was attenuated in MO subjects indicating impaired suppression of lipolysis by insulin (Table 2).
Figure 1. Weight loss following GBP effectively reverses abnormal glucose and insulin tolerance following oral glucose challenge in morbidly obese women.
Data are mean ± S.E.M. of (A) plasma glucose, (B) insulin and (C) C-peptide following oral glucose challenge (OGTT) in morbidly obese (Obese) women undergoing gastric bypass surgery (GBP) and in those undergoing ventral hernia repair after experiencing significant weight loss (Post-obese) following prior GBP. Significance of differences between responses to oral glucose challenge was assessed by 2-way Analysis of Variance (Group versus Time).
3.3 Hepatic expression of genes related to fatty acid metabolism and metabolic signaling: results of microarray analysis
We have recently defined the hepatic transcriptome of MO and MWL women using Affymetrix gene arrays and identified a number of genes that potentially linked the susceptibility of morbidly obese women to a wide range of diseases (23). Due to the stringent selection criteria needed for this initial transcriptome-wide analysis, this prior analysis did not provide information on key genes related to lipid synthesis and metabolic signaling. For the present study, we re-interrogated the hepatic transcriptome database to focus on a number of genes with well-known involvement in the regulation of metabolic signaling and fatty acid and lipid biosynthesis. This analysis indicated that a panel of genes regulating fatty acid oxidation was expressed at a higher level in livers of MO subjects (Table 3). These included enzymes mediating mitochondrial b-oxidation including hydroxyacyl-CoA dehydrogenase (HADH), and enoyl CoA hydratase (ECHS1). Similarly, Peroxisomal 3-oxoacyl-Coenzyme A thiolase (ACAA1), and the ketogenic enzyme 3-hydroxybutyrate dehydrogenase, type 2 (BDH2) were also up-regulated in MO (Table 3). Expression of the lipogenic enzyme malic enzyme 1 (ME1) was increased in MO (Table 3).Conversely, expression of the fatty acid modifying enzyme ELOVL2 was reduced in MO as compared to MWL (Table 3). Although expression of IRS1 was modestly increased in MO livers, expression of IRS2 and expression of mRNA encoding the insulin receptor was unaltered (data not shown). On the other hand, expression of key genes underlying signal transduction pathways evoked by IGF (Insulin-like-growth-factor-1 or somatomedin C) leptin (Leptin receptor), and pro-inflammatory cytokines (SOCS2 and STAT6) were differentially regulated in livers of MO and MWL subjects (Table 3).
Table 3.
Hepatic expression of genes related to fatty acid metabolism and metabolic signaling in liver biopsy samples of MO and MWL women as assessed by Affymetrix Microarray analysis.
| Affymetrix ID | Symbol | Gene Name | Fold Change (MO/MWL) |
P = |
|---|---|---|---|---|
| 218285_s_at | BDH2 | 3-hydroxybutyrate dehydrogenase type 2 | 1.35 | 0.014 |
| 211569_s_at | HADH | Hydroxyacyl-Coenzyme A dehydrogenase | 1.67 | 0.009 |
| 203658_at | SLC25A20 | carnitine/acylcarnitine translocase | 1.26 | 0.037 |
| 201135_at | ECHS1 | enoyl Coenzyme A hydratase, short chain, 1 | 1.51 | 0.001 |
| 202025_x_at | ACAA1 | peroxisomal 3-oxoacyl-Coenzyme A thiolase | 1.38 | 0.031 |
| 204059_s_at | ME1 | malic enzyme 1 | 1.47 | 0.038 |
| 220029_at | ELOVL2 | elongation of very long chain fatty acids | 0.34 | 0.021 |
| 204686_at | IRS1 | insulin receptor substrate 1 | 1.31 | 0.023 |
| 209541_at | IGF1 | insulin-like growth factor 1 (somatomedin C) | 0.64 | 0.028 |
| 209542_x_at | IGF1 | insulin-like growth factor 1 (somatomedin C) | 0.41 | 0.001 |
| 211354_s_at | LEPR | leptin receptor | 0.49 | 0.002 |
| 211355_x_at | LEPR | leptin receptor | 0.50 | 0.005 |
| 203372_s_at | SOCS2 | Suppressor of Cytokine Signaling-2 | 0.32 | 0.003 |
| 203373_at | SOCS2 | Suppressor of Cytokine Signaling -2 | 0.26 | 0.002 |
| 201331_s_at | STAT6 | signal transducer and activator of transcription 6 | 0.71 | 0.008 |
| 205698_s_at | MAP2K6 | Mitogen Activated Protein Kinase-Kinase-6 | 1.9 | 0.011 |
| 201627_s_at | INSIG1 | Insulin induced gene-1 | 1.1 | 0.722 |
| 209566_at | INSIG2 | Insulin induced gene-2 | 0.7 | 0.165 |
| 212329_at | SCAP | SREBP cleavage activating protein | 0.9 | 0.277 |
Gene expression was assessed in liver biopsy samples from morbidly obese (MO) women undergoing gastric bypass surgery (GBP) (N= 13) and in women undergoing ventral hernia repair following massive weight loss (MWL) after previous GBP (N=5) using Affymetrix oligonucleotide gene arrays (HG133A). Genes related to fatty acid metabolism and metabolic signaling were identified from the gene expression data set and assessed for significance of differences in signal intensity between MO and MWL by unpaired Students T-Test (SAS-JMP, Cary NC). Genes detected by multiple probe sets are presented individually.
3.4. Hepatic expression of genes related to fatty acid metabolism and metabolic signaling: results of real-time PCR analysis
We noted that a number of genes known to be critically involved in lipid synthesis and metabolic signaling were absent from our Affymetrix microarray-derived transcriptome. Therefore, to extend the microarray experiments, we further analyzed expression of a subset of genes by real-time PCR. This analysis demonstrated that in addition to ME1 an entire panel of insulin and SREBP-1c responsive genes that mediate both de novo lipogenesis and triglyceride synthesis were expressed at higher levels in livers of MO subjects (Table 4). These include acetyl-CoA-carboxylase-1 (ACC-1), fatty acid synthase (FASN) Diacylglycerol Acyltransferase (DGAT) and stearoyl-CoA-desaturase (SCD-1). Based on the observed up regulation of these SREBP-1c dependent genes and previous findings of increased SREBP-1c mRNA expression in livers of obese rodents (10, 11), we expected to see increased SREBP-1c mRNA expression in livers of MO subjects. Unexpectedly, neither expression of total SREBP-1 (1a plus 1c) mRNA nor that encoding the SREBP-1c isoform was increased in livers of MO subjects (Table 4). Expression of mRNA encoding the cholesterol regulating SREBP isoform SREBP-2 was higher in MO, however, expression of its target gene HMG-CoA-Reductase (HMGR) was not elevated (Table 4). Although expression of a panel of Peroxisomal proliferator activator-alpha (PPARα) dependent genes related to fatty acid oxidation was somewhat increased (Table 3), that of PPARα itself was not (Table 4). Conversely, expression of PPARγ and that of the co-activator PGC-1α was reduced in livers of MO subjects (Table 4).
Table 4.
Hepatic expression of genes related to fatty acid metabolism and metabolic signaling in liver biopsy samples of MO and MWL women as assessed by quantitative Real-Time PCR.
| Gene Name | Symbol | Fold Change (MO/MWL) |
|---|---|---|
| Diacylglycerol Acyltransferase | DGAT | 2.51 |
| Stearoyl-CoA-desaturase-1 | SCD-1 | 2.66 |
| Acetyl-CoA carboxylase-1 | ACC-1 | 1.34 |
| Fatty acid synthase | FASN | 2.67 |
| Hydroxymethylglutaryl CoA Reductase | HMGR | 0.87 |
| Sterol regulatory element binding protein-1 | SREBP-1 | 0.91 |
| Sterol regulatory element binding protein-1c | SREBP-1c | 0.82 |
| Sterol regulatory element binding protein-2 | SREBP-2 | 1.89 |
| Peroxisomal proliferator activated receptor-α | PPARα | 0.89 |
| Peroxisomal proliferator activated receptor-γ | PPARγ | 0.66 |
| PPARγ Coactivator-1α | PGC-1α | 0.67 |
| Mitogen activated protein kinase-kinase-6 | MAP2K6 | 6.11 |
| Suppressor of cytokine signaling-3 | SOCS-3 | 0.96 |
Data are relative expression of selected genes related to fatty acid metabolism and metabolic signaling determined by real-time PCR in liver biopsy samples derived from women undergoing gastric bypass surgery (GBP) for morbid obesity (MO) (N = 7 – 12) and in those undergoing ventral hernia repair after massive weight loss (MWL) following GBP (N = 3 – 6). Real-time PCR was carried out using primer sets outlined in table 1. Primer sets used to analyze MAP2K6 and SOCS-3 was obtained from Qiagen Corporation (Valencia, Ca). For SREBP-1c, primers were used that identified both 1a and 1c isoforms (SREBP-1) and that were specific for the 1c isoform (SREBP-1c).
In the microarray analysis (Table 3) expression of mRNA encoding the signaling proteins SOCS2 and STAT6 was significantly lower and expression of MAP2K6 was higher in MO livers, however SOCS-3 was not detected on the microarray. Based on the observations of Ueki et al (28) and Bode et al (29), both SOCS-3 and MAP2K6 appear to play a central role in development of hepatic insulin resistance in obesity. Therefore we examined mRNA expression of SOCS-3 and MAP2K6 using real-time PCR (Table 4). Concordant with the microarray findings, expression of MAP2K6 was 6 fold higher in livers of MO subjects (Table 4). Conversely, mRNA expression of the SOCS-3 isoform, a known negative regulator of insulin signaling (20), was unaltered in MO as compared to MWL subjects (Table 4).
3.5. Effect of morbid obesity on expression of genes related to fatty acid metabolism and metabolic signaling: results of Western blot protein expression analyses
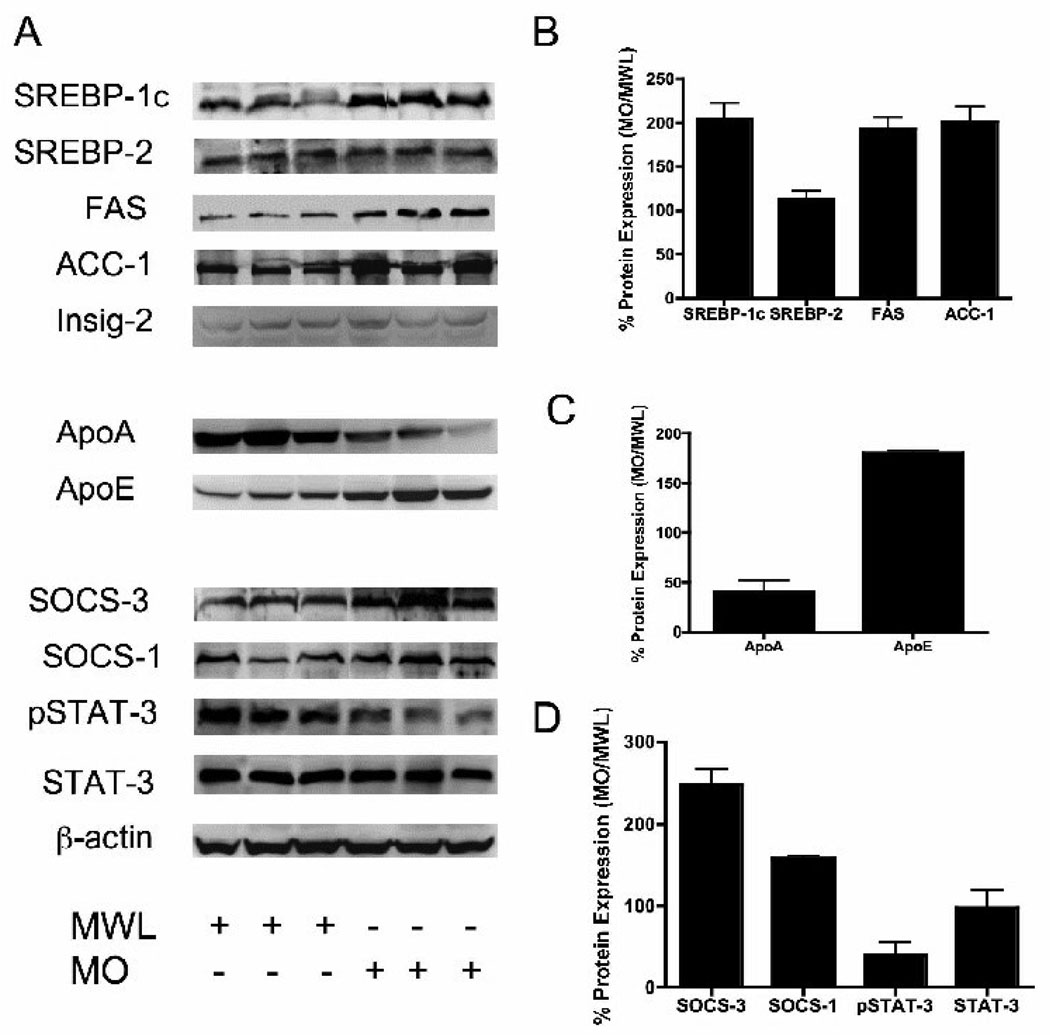
Since many genes are regulated by post-transcriptional mechanisms, we extended the microarray and real-time PCR analysis by assessing expression of proteins transcribed from these key genes by Western blotting of liver proteins from MO and MWL subjects. Insulin is known to accelerate proteolytic processing of nascent SREBP-1c thereby increasing nuclear content of its transcriptionally active n-terminal fragment (14, 15). Although we were not able to detect differential expression of SREBP-1c mRNA in livers of MO and MWL, expression of the n-terminal protein fragment of SREBP-1c (nSREBP-1c) was 2-fold higher in livers of MO. Similarly, protein expression of the downstream lipogenic enzyme targets of SREBP-1c, FAS and ACC-1, was also increased in MO (Figure 2 panels A and B). In contrast, protein expression of the cholesterol regulating isoform, SREBP-2, was unaltered in MO subjects (Figure 2, panels A and B).
Figure 2. Expression of hepatic proteins related to lipid metabolism and metabolic signaling in livers of Morbidly Obese (MO) and Massive Weight Loss (MWL) patients.
Panel A. Data are Western blots of total cell lysate from liver of MO (N=11) and MWL (N=4) subjects. Representative Western blots from six subjects are shown. Blots show expression of Sterol-Regulatory-Protein-1c and 2 (SREBP-1c / 2), Fatty acid synthase (FAS), Acetyl-CoA Carboxylase-1 (ACC-1), Insulin-induced gene-2 (INSIG-2), Apolipoprotein A1 (ApoA), Apolipoprotein E (ApoE), Suppressor of Cytokine Signaling -1 and 3 (SOCS-1, 3), STAT-3 and phospho-STAT-3 (pSTAT3). Panels B–D show results of densitometry scanning for Lipogenic Genes (panel B), Apolipoprotein genes (Panel C), and STAT signaling genes (Panel D). *P< 0.05 MO vs. MWL.
Prior to its transport to the Golgi and release of its n-terminal fragment by proteolytic processing, ER bound nascent full-length SREBP-1c is associated with the chaperone protein SCAP (SREBP cleavage activating protein) and the ER retention protein Insig-2a (16). Insulin is thought promote the ER to Golgi transport and subsequent proteolytic processing of SREBP-1c by reducing levels of Insig-2a (30, 31). The observation of increased nSREBP-1c in livers of MO prompted examination of the microarray database for expression of these SREBP-1c associated proteins. Although expression of the insulin-responsive gene isoform INSIG-2 trended lower in MO a condition that would be expected to favor enhanced proteolytic processing, this did not achieve statistical significance (Table 3). Further, expression of Insig-2a protein was unaltered in livers of MO subjects (Figure 2, Panel A). Further, expression of mRNA encoding SCAP and Insig-1 was comparable in livers of MO and MWL subjects (Table 3).
Importantly, we found that hepatic expression of SOCS-1 and SOCS-3 proteins was increased in MO (1.8-fold and 2.5-fold higher respectively) (Figure 2 Panel A and D). Although STAT-3 protein levels were unaltered in MO, STAT-3 signaling was, in fact, attenuated as evidenced by reduced levels of active (phosphorylated) form pSTAT-3 (Figure 2, Panels A and D).
Of additional interest, although microarray analysis showed that expression of mRNA encoding major apoproteins including ApoAI, ApoAII, ApoB, apoCII, apoE, and ApoCIII was unchanged (data not shown), protein expression of two apoproteins, ApoAI and ApoE, was reduced and increased respectively in livers of MO (Figure 2, Panels A and B). Despite repeated attempts, we were not able to measure expression of apoB or ApoCs by Western analysis.
3.6. the human SREBP-1c promoter is responsive to insulin and LXR agonists
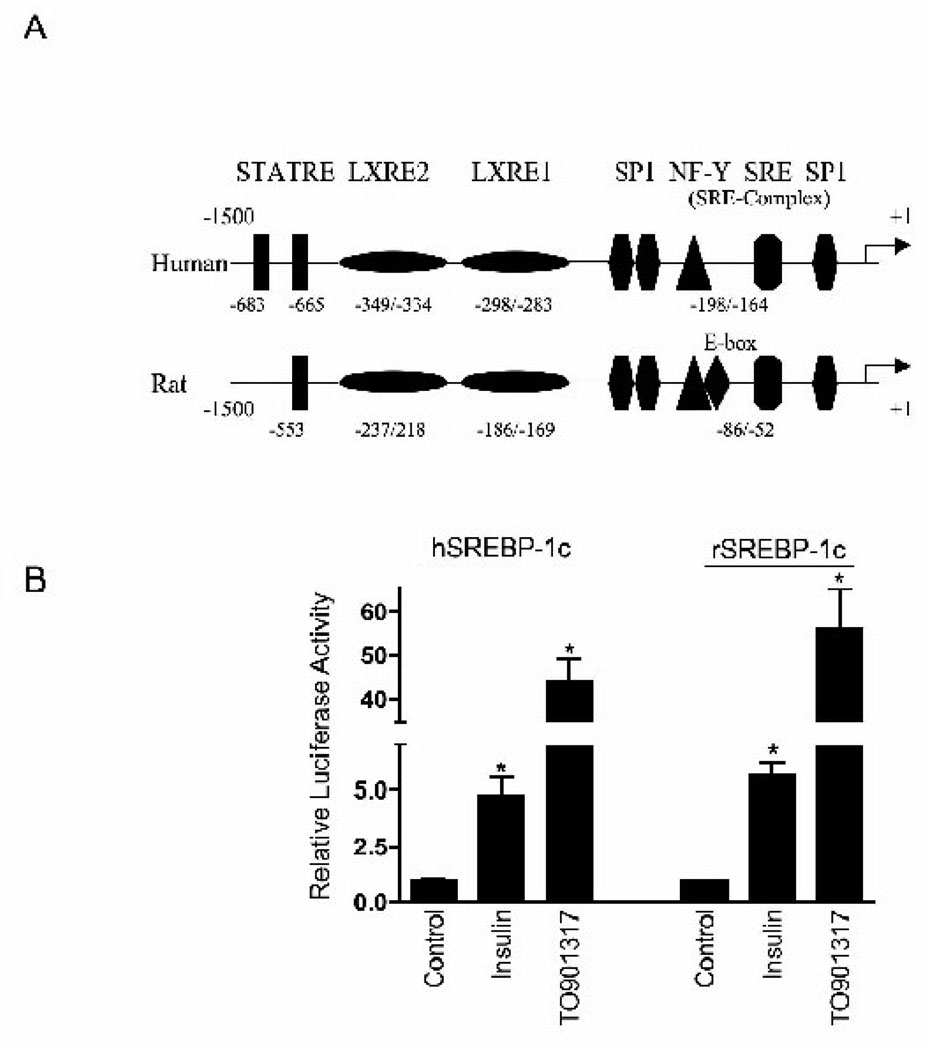
As stated earlier, the finding of comparable levels of mRNA encoding SREBP-1c in livers of MO versus MWL subjects was both unexpected and discordant with both prior animal studies and with the Western blot findings of enhanced expression of nSREBP-1c protein and both mRNA and protein of its downstream lipogenic targets in MO. Although this may reflect rapid decay of SREBP-1c mRNA during the obligatory overnight fast required as preparation for surgery in our subjects, we also considered the possibility that these findings may reflect attenuated response of the SREBP-1c promoter to insulin in the human as compared to the rodent. The human SREBP-1c promoter has been cloned and found to be responsive to insulin treatment when transfected into HEK-293 cells (13), however, its response to insulin has not been examined relative to the rodent ortholog in primary hepatocyte cultures. We therefore compared the response of human and rat SREBP-1c-Luciferase reporter constructs to insulin and the LXR ligand T0901317 in transient transfection of primary (rat) hepatocytes. Although there are minor differences in spatial organization of the rat and human SREBP-1c promoter, the major response elements are preserved (Figure 3, panel A). Accordingly, the human SREBP-1c promoter exhibited comparable response to both insulin (5–8 fold induction) and T0901317 (45–55 fold induction) as the rat promoter (Figure 3, panel B). Thus, the human SREBP-1c promoter is highly responsive to insulin.
Figure 3. The human SREBP-1c promoter is responsive to insulin treatment in vitro.
A) A schematic representation of the major cis-acting elements of the human and rat SREBP-1c promoter. The transcription initiation site of the human promoter, determined by TRANSFAC analysis, was designated as +1. Insulin-mediated activation of the human SREBP-1c promoter may involve a number of binding sites known to regulate the rat and mouse promoter. These include: STATRE, signal transducer and activator of transcription response element; LXRE, liver X receptor response element; SRE, sterol regulatory element; SP1, specificity protein-1; NF-Y, nuclear factor-Y. The locations of these cis-acting elements are not depicted to scale on the promoter models.
B) Primary rat hepatocytes were either transfected with human (hSREBP-1c) or rat (rSREBP-1c) SREBP-1c promoter-luciferase reporter constructs and incubated with and without insulin (100 nM) or LXR agonist TO901317 (10 µM) for 24 h. Luciferase activity was measured as described in Methods. Both human and rat SREBP-1c promoter constructs responded comparably to both insulin and TO901317. Data are mean ± SEM of luciferase activity relative to untreated (control) hepatocytes (N=12 determinations from 4 separate hepatocyte preparations). P<0.05 vs. control
3.7. The human SREBP-1c promoter is negatively regulated by STAT3
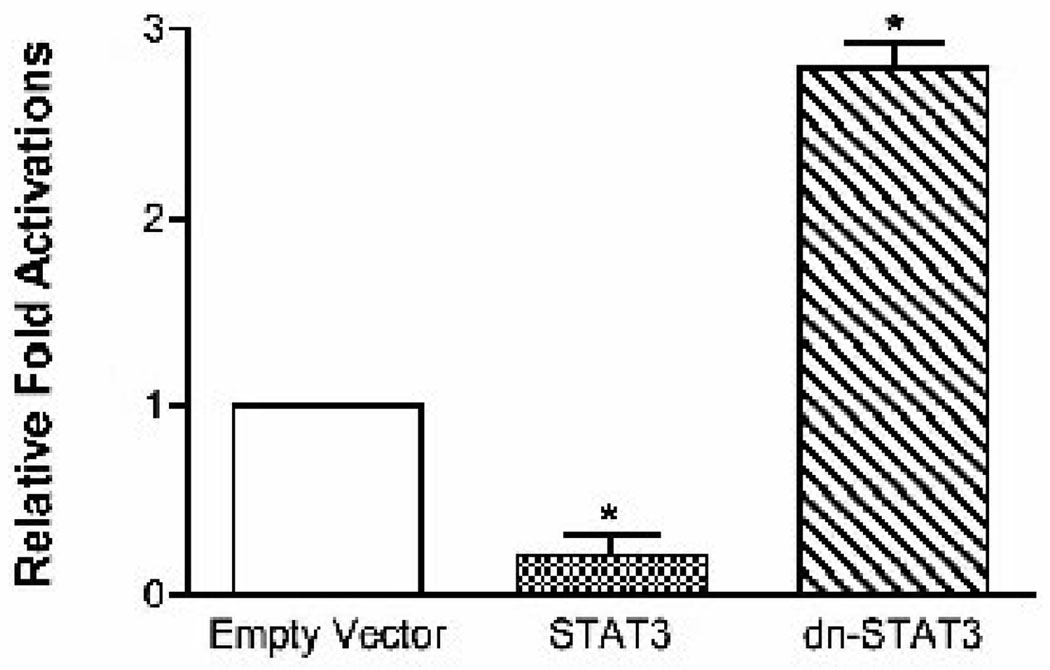
Based on our in vivo observations of differential hepatic regulation of SOCS1 and 3 as well as pSTAT3 in MO humans, together with the findings of the animal studies of Ueki et al (28) as well as those of Endo et al (32), we reasoned that the corresponding changes in the expression of SREBP-1c protein and attenuated JAK/STAT signaling in MO and WL patients may be mechanistically related. To directly test this hypothesis, we transfected primary hepatocytes with full-length hSREBP-1c-luc either alone or co-transfected with vectors designed to express wild type STAT3 or dominant negative STAT3. As shown in Figure 4, co-expression of wild type STAT3 led to a 5-fold decrease in luciferase expression driven by the SREBP1c promoter indicating repression of SREBP-1c transcription by STAT3. Conversely, mimicking the effect of SOCS-3 to suppress pSTAT3 via exogenous expression of a dominant negative form of STAT3 significantly increased hSREBP-1c promoter activity (Figure 4). These data support the hypothesis that SOCS mediated down-regulation of JAK/STAT signaling may be mechanistically related to up-regulation of SREBP-1c expression in obesity.
Figure 4. STAT3 represses the human SREBP-1c promoter.
Primary rat hepatocytes were transfected with hSREBP1c-luc DNA along with plasmids expressing wild type or dominant negative STAT3 proteins, or empty vector. Twenty-four hours after transfection cells were harvested and the luciferase activity was measured. Cells were co-transfected with a control Renilla expression plasmid, pRL-TK, and transfection results were normalized to Renilla luciferase activity. The results represent the mean ±S.E.M. for five independent experiments, with fold induction over the level observed with the reporter construct alone.
DISCUSSION
Morbid obesity is associated with insulin resistance and hypertriglyceridemia that results, at least in part, from increased synthesis and secretion of VLDL by the liver (33, 34). We report here that expression of the lipogenic regulator SREBP-1c and its downstream targets that mediate both de novo lipogenesis and triglyceride synthesis is significantly increased in livers of morbidly obese women. These findings support the hypothesis that morbid obesity leads to a widespread up-regulation of enzymes mediating synthesis of fatty acids and triglyceride, as previously observed in obese animal models (11, 35).
Until recently, the prevailing opinion was that under most circumstances plasma FFA were the predominant source of FA’s for VLDL-TG, and that the rate of delivery of FA’s to the liver from exogenous sources (dietary and lipolysis) was the major determinant of VLDL-TG secretion in the human (36). However, recent in vivo metabolic turnover studies have shown that enhanced de novo lipogenesis assumes a greater role in overproduction of VLDL-TG in the obese human (6–9, 33, 37). Increased de novo lipogenesis and enhanced VLDL-TG secretion was also seen in hyperinsulinemia caused by hyper caloric carbohydrate feeding in humans (38). We show here that both mRNA and protein expression of enzymes mediating de novo lipogenesis is, in fact, increased in livers of obese women. Thus, enhanced VLDL-TG production in obesity and carbohydrate feeding in humans also results from enhanced de novo lipogenesis. Reminiscent of findings in the obese rat (35), we observed modest increases in hepatic expression of mRNAs encoding enzymes mediating fatty acid oxidation in MO as compared to MWL. This likely represents an adaptive response to increased dietary intake of fat and carbohydrate. Since SREBP-1c strongly up regulates the enzymes that incorporate fatty acid into triglyceride (39), one would predict, however, that the predominant effect would be preferential utilization of fatty acids for esterification into triglyceride. Increased expression of these PPARα dependent genes was not related to increased PPARα expression but rather appears to result from increased ligand-dependent activation of PPARα. Conversely, expression of PPARγ and PGC-1α was reduced in livers of MO, similar to the previous observations of reduced levels of PGC-1α in skeletal muscle of insulin-resistant diabetic humans (40). The significance of reduced expression of PPARγ in livers of MO is not entirely clear.
Interestingly, we did not detect increased expression of mRNA encoding SREBP-1c in MO. This is in striking contrast to prior findings of enhanced SREBP-1c expression in liver of obese rodents (10, 11, 35). This finding may be related to a significant difference in nutritional state of the subjects in the present study as compared to that in prior animal studies. In the present study, liver biopsy samples were obtained following an overnight fast required as part of preparation for surgery in humans, whereas in prior animal studies liver samples were collected in the fed state. Under fasting conditions mRNA transcripts with short half-life such as SREBP-1c (6–8 hours) will have rapidly declined. Insofar as the human and rodent SREBP-1c promoters are structurally similar and exhibit comparable response to insulin in vitro, it is likely that our findings reflect the nutritional state of our subjects. On the other hand, it is possible that up-regulation of SREBP-1c in the human may be more tightly linked to enhanced posttranslational proteolytic processing, which is also insulin-dependent (15). In addition, lipogenic enzymes (i.e. FAS) may be induced in the presence of hyperglycemia independent of the effects of insulin and SREBP-1c via the carbohydrate response element (CHO-RE) (41).
A second major finding of the present study was the presence of increased expression of the inhibitory signaling protein SOCS-3, and reduced levels of its target, phosphorylated STAT3 in MO. These findings are highly significant in light of studies of Ueki et al (21) who reported that hepatic expression of SOCS-1 and SOCS-3 was elevated in livers of obese, insulin resistant mice, and that exogenous expression of either SOCS-1 or SOCS-3 proteins induced both insulin resistance and SREBP-1c expression. Our findings of concomitant up-regulation of SOCS-3 with decreased STAT-3 phosphorylation in livers of MO humans combined with the demonstration that the human SREBP-1c promoter is negatively regulated by STAT3 strengthen the putative mechanistic link between SOCS-3 induced impaired JAK/Stat signaling and enhanced SREBP-1c expression in obesity. In this regard, our finding of increased hepatic expression of the dual specificity protein kinase MAP2K6 in MO subjects is particularly intriguing since MAP2K6 phosphorylates p38 MAP kinase in response to inflammatory cytokines or environmental stress and may induce insulin resistance by a similar mechanism in obesity (29).
Although some alterations in expression of genes related to lipid metabolism in MO may be attributed to hyperinsulinemia, it is likely that many observed differences may also arise from alterations in the nutritional and hormonal milieu that accompanies morbid obesity. For example, the dietary intake of the two groups was very different. In addition to reduced overall caloric intake, MWL subjects exhibited a markedly reduced intake of sucrose and fructose as has been reported previously (42). Higher intake of simple sugars in general and fructose in particular in MO is significant in view of the known effects of fructose on hepatic lipogenesis and insulin resistance (43) and SREBP-1c gene expression (44).
Altered expression of enzymes related to modification of fatty acids, stearoyl-CoA-desaturase-1 (SCD-1) and the fatty acid elongase ELOVL2 in livers of MO is also of considerable interest. Our finding of higher expression of SCD-1 is reminiscent of earlier observations of increased SCD expression in livers of obese hyperinsulinemic ob/ob mice (10) and may be important in view of the postulated role of this enzyme in insulin insensitivity and hepatic lipid synthesis in obesity (45). Hepatic expression of the gene ELOVL2 that encodes an enzyme that mediates elongation of 20- and 22-carbon long chain polyunsaturated fatty acids (46) was reduced in MO subjects. This, together with the reported linkage of the related gene, ELOV16, with insulin resistance in rodents (47) suggests a potential role for altered ELOVL2 in the metabolic complications of obesity in humans.
The general implications of the data presented here must be tempered by a number of caveats arising from the inherent limitations of our study. First, it must be noted that GBP surgery is reserved for individuals with an extreme form of obesity. Second, although the MWL subjects had experienced significant weight loss and resolution of metabolic abnormalities following GBP, their BMI was not normal. Third, although many changes in gene expression can be attributed to amelioration of insulin resistance and hyperinsulinemia following GBP, many changes may be due to additional effects of weight loss and altered dietary intake.
This cautionary note notwithstanding, we conclude that enhanced de novo synthesis of lipids is critically involved in the dyslipidemia of obesity in humans. We further postulate that enhanced expression of SREBP-1c and its downstream gene targets is mechanistically linked to SOCS-mediated attenuation of STAT signaling in morbid obesity.
ACKNOWLEDGEMENTS
These studies were supported by Award Number R01DK075504 from the National Institute of Diabetes and Digestive And Kidney Diseases, and by Merit Review grants from the Department of Veterans Affairs (MBE, RR), the University of Tennessee General Clinical Research Center, Grant #MO1-RR0021, NIDDK RO1-DK75504-01 (MBE, RR, CY) and by and by postdoctoral fellowship awards to GEH (NIDDK F#DK083210 and USPHS GR HL 07641-14). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases or the National Institutes of Health. We thank Elizabeth Schreiner for technical assistance and Dr Grant Somes for statistical consultation. We wish to thank the staff of the UT Obesity Wellness Center and the operating room staff of UT-Bowld Hospital and Baptist Memorial Hospital Memphis and Dr William L Taylor of the UTCHS Molecular Resource Center for their invaluable assistance. The STAT3 expression vectors were a kind gift of Dr Larry Pfeffer UTHSC. RR is a Senior Research Career Scientist of the Department of Veterans Affairs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE STATEMENT: The authors have no conflicts of interest to disclose.
INSTITUTIONAL APPROVAL: This study was approved by the Institutional Review Boards of the University of Tennessee Health Sciences Center and Baptist Memorial Hospital-Memphis.
BIBLIOGRAPHY
- 1.Grundy SM. Metabolic complications of obesity. Endocrine. 2000 Oct;13(2):155–165. doi: 10.1385/ENDO:13:2:155. [DOI] [PubMed] [Google Scholar]
- 2.Castro Cabezas M, Halkes CJ, Erkelens DW. Obesity and free fatty acids: double trouble. Nutr Metab Cardiovasc Dis. 2001 Apr;11(2):134–142. [PubMed] [Google Scholar]
- 3.Festi D, Colecchia A, Sacco Sacco T, Bondi M, Roda E, Marchesini G. Hepatic steatosis in obese patients: clinical aspects and prognostic significance. Obes Rev. 2004 Feb;5(1):27–42. doi: 10.1111/j.1467-789x.2004.00126.x. [DOI] [PubMed] [Google Scholar]
- 4.Marchesini G, Marzocchi R, Agostini F, Bugianesi E. Nonalcoholic fatty liver disease and the metabolic syndrome. Curr Opin Lipidol. 2005 Aug;16(4):421–427. doi: 10.1097/01.mol.0000174153.53683.f2. [DOI] [PubMed] [Google Scholar]
- 5.Hellerstein MK, Christiansen M, Kaempfer S, Kletke C, Wu K, Reid JS, et al. Measurement of de novo hepatic lipogenesis in humans using stable isotopes. J Clin Invest. 1991 May;87(5):1841–1852. doi: 10.1172/JCI115206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faix D, Neese R, Kletke C, Wolden S, Cesar D, Coutlangus M, et al. Quantification of menstrual and diurnal periodicities in rates of cholesterol and fat synthesis in humans. J Lipid Res. 1993 Dec;34(12):2063–2075. [PubMed] [Google Scholar]
- 7.Schwarz JM, Neese RA, Turner S, Dare D, Hellerstein MK. Short-term alterations in carbohydrate energy intake in humans. Striking effects on hepatic glucose production, de novo lipogenesis, lipolysis, and whole-body fuel selection. J Clin Invest. 1995 Dec;96(6):2735–2743. doi: 10.1172/JCI118342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hudgins LC, Hellerstein M, Seidman C, Neese R, Diakun J, Hirsch J. Human fatty acid synthesis is stimulated by a eucaloric low fat, high carbohydrate diet. J Clin Invest. 1996;97(9):2081–2091. doi: 10.1172/JCI118645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lammert O, Grunnet N, Faber P, Bjornsbo KS, Dich J, Larsen LO, et al. Effects of isoenergetic overfeeding of either carbohydrate or fat in young men. Br J Nutr. 2000;84(2):233–245. [PubMed] [Google Scholar]
- 10.Shimomura I, Bashmakov Y, Horton JD. Increased Levels of Nuclear SREBP-1c Associated with Fatty Livers in Two Mouse Models of Diabetes Mellitus. J Biol Chem. 1999 October 15;274(42):30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- 11.Elam MB, Wilcox HG, Cagen LM, Deng X, Raghow R, Kumar P, et al. Increased hepatic VLDL secretion, lipogenesis, and SREBP-1 expression in the corpulent JCR:LA-cp rat. J Lipid Res. 2001 Dec;42(12):2039–2048. [PubMed] [Google Scholar]
- 12.Cagen LM, Deng X, Wilcox HG, Park EA, Raghow R, Elam MB. Insulin activates the rat sterol-regulatory-element-binding protein 1c (SREBP-1c) promoter through the combinatorial actions of SREBP, LXR, Sp-1 and NF-Y cis-acting elements. Biochem J. 2005 Jan 1;385(Pt 1):207–216. doi: 10.1042/BJ20040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dif N, Euthine V, Gonnet E, Laville M, Vidal H, Lefai E. Insulin activates human sterolregulatory-element-binding protein-1c (SREBP-1c) promoter through SRE motifs. Biochem J. 2006 Nov 15;400(1):179–188. doi: 10.1042/BJ20060499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegarty BD, Bobard A, Hainault I, Ferre P, Bossard P, Foufelle F. Distinct roles of insulin and liver X receptor in the induction and cleavage of sterol regulatory element-binding protein-1c. Proc Natl Acad Sci U S A. 2005 Jan 18;102(3):791–796. doi: 10.1073/pnas.0405067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yellaturu CR, Deng X, Cagen LM, Wilcox HG, Mansbach CM, 2nd, Siddiqi SA, et al. Insulin enhances posttranslational processing of nascent SREBP-1c by promoting its phosphorylation and association with COPII vesicles. J Biol Chem. 2009 Jan 21; doi: 10.1074/jbc.M805746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raghow R, Yellaturu C, Deng X, Park EA, Elam MB. SREBPs: the crossroads of physiological and pathological lipid homeostasis. Trends Endocrinol Metab. 2008 Mar;19(2):65–73. doi: 10.1016/j.tem.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Shimomura I, Matsuda M, Hammer RE, Bashmakov Y, Brown MS, Goldstein JL. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol Cell. 2000;6(1):77–86. [PubMed] [Google Scholar]
- 18.Biddinger SB, Kahn CR. From mice to men: insights into the insulin resistance syndromes. Annu Rev Physiol. 2006;68:123–158. doi: 10.1146/annurev.physiol.68.040104.124723. [DOI] [PubMed] [Google Scholar]
- 19.Emanuelli B, Peraldi P, Filloux C, Chavey C, Freidinger K, Hilton DJ, et al. SOCS-3 inhibits insulin signaling and is up-regulated in response to tumor necrosis factor-alpha in the adipose tissue of obese mice. J Biol Chem. 2001 Dec 21;276(51):47944–47949. doi: 10.1074/jbc.M104602200. [DOI] [PubMed] [Google Scholar]
- 20.Ueki K, Kondo T, Kahn CR. Suppressor of cytokine signaling 1 (SOCS-1) and SOCS-3 cause insulin resistance through inhibition of tyrosine phosphorylation of insulin receptor substrate proteins by discrete mechanisms. Mol Cell Biol. 2004 Jun;24(12):5434–5446. doi: 10.1128/MCB.24.12.5434-5446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ueki K, Kadowaki T, Kahn CR. Role of suppressors of cytokine signaling SOCS-1 and SOCS-3 in hepatic steatosis and the metabolic syndrome. Hepatol Res. 2005 Oct;33(2):185–192. doi: 10.1016/j.hepres.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 22.Buchwald H, Avidor Y, Braunwald E, Jensen MD, Pories W, Fahrbach K, et al. Bariatric surgery: a systematic review and meta-analysis. Jama. 2004 Oct 13;292(14):1724–1737. doi: 10.1001/jama.292.14.1724. [DOI] [PubMed] [Google Scholar]
- 23.Elam MB, Cowan GS, Jr, Rooney RJ, Hiler ML, Yellaturu CR, Deng X, et al. Hepatic Gene Expression in Morbidly Obese Women: Implications for Disease Susceptibility. Obesity (Silver Spring) 2009 Mar 5; doi: 10.1038/oby.2009.49. [DOI] [PubMed] [Google Scholar]
- 24.Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999 Sep;22(9):1462–1470. doi: 10.2337/diacare.22.9.1462. [DOI] [PubMed] [Google Scholar]
- 25.Hughes TA, Elam MB, Applegate WB, Bond MG, Hughes SM, Wang X, et al. Postprandial lipoprotein responses in hypertriglyceridemic subjects with and without cardiovascular disease. Metabolism. 1995 Aug;44(8):1082–1098. doi: 10.1016/0026-0495(95)90108-6. [DOI] [PubMed] [Google Scholar]
- 26.Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996 Dec;14(13):1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 27.Deng X, Yellaturu C, Cagen L, Wilcox HG, Park EA, Raghow R, et al. Expression of the rat sterol regulatory element-binding protein-1c gene in response to insulin is mediated by increased transactivating capacity of specificity protein 1 (Sp1) J Biol Chem. 2007 Jun 15;282(24):17517–17529. doi: 10.1074/jbc.M702228200. [DOI] [PubMed] [Google Scholar]
- 28.Ueki K, Kondo T, Tseng YH, Kahn CR. Central role of suppressors of cytokine signaling proteins in hepatic steatosis, insulin resistance, and the metabolic syndrome in the mouse. Proc Natl Acad Sci U S A. 2004 Jul 13;101(28):10422–10427. doi: 10.1073/pnas.0402511101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bode JG, Ludwig S, Freitas CA, Schaper F, Ruhl M, Melmed S, et al. The MKK6/p38 mitogen-activated protein kinase pathway is capable of inducing SOCS3 gene expression and inhibits IL-6-induced transcription. Biol Chem. 2001 Oct;382(10):1447–1453. doi: 10.1515/BC.2001.178. [DOI] [PubMed] [Google Scholar]
- 30.Yabe D, Komuro R, Liang G, Goldstein JL, Brown MS. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc Natl Acad Sci U S A. 2003 Mar 18;100(6):3155–3160. doi: 10.1073/pnas.0130116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yellaturu CR, Deng X, Cagen LM, Wilcox HG, Park EA, Raghow R, et al. Posttranslational processing of SREBP-1 in rat hepatocytes is regulated by insulin and cAMP. Biochem Biophys Res Commun. 2005 Jun 24;332(1):174–180. doi: 10.1016/j.bbrc.2005.04.112. [DOI] [PubMed] [Google Scholar]
- 32.Endo M, Masaki T, Seike M, Yoshimatsu H. TNF-alpha induces hepatic steatosis in mice by enhancing gene expression of sterol regulatory element binding protein-1c (SREBP-1c) Exp Biol Med (Maywood) 2007 May;232(5):614–621. [PubMed] [Google Scholar]
- 33.Chan DC, Watts GF, Redgrave TG, Mori TA, Barrett PH. Apolipoprotein B-100 kinetics in visceral obesity: associations with plasma apolipoprotein C-III concentration. Metabolism. 2002 Aug;51(8):1041–1046. doi: 10.1053/meta.2002.33339. [DOI] [PubMed] [Google Scholar]
- 34.Mittendorfer B, Patterson BW, Klein S. Effect of weight loss on VLDL-triglyceride and apoB-100 kinetics in women with abdominal obesity. Am J Physiol Endocrinol Metab. 2003 Mar;284(3):E549–E556. doi: 10.1152/ajpendo.00379.2002. [DOI] [PubMed] [Google Scholar]
- 35.Deng X, Elam MB, Wilcox HG, Cagen LM, Park EA, Raghow R, et al. Dietary olive oil and menhaden oil mitigate induction of lipogenesis in hyperinsulinemic corpulent JCR:LA-cp rats: microarray analysis of lipid-related gene expression. Endocrinology. 2004 Dec;145(12):5847–5861. doi: 10.1210/en.2004-0371. [DOI] [PubMed] [Google Scholar]
- 36.Havel RJKJ, Segel EO, Basso LV. Splanchnic metabolism of free fatty acids and production of triglyceride of very low density lipoproteins in normotriglyceridemic and hypertriglyceridemic humans. J Clin Invest. 1970;49:2017–2035. doi: 10.1172/JCI106422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egusa G, Beltz WF, Grundy SM, Howard BV. Influence of obesity on the metabolism of apolipoprotein B in humans. J Clin Invest. 1985 Aug;76(2):596–603. doi: 10.1172/JCI112011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aarsland A, Chinkes D, Wolfe RR. Contributions of De Novo Synthesis of Fatty Acids to Total VLDL-Triglyceride Secretion during Prolonged Hyperglycemia/Hyperinsulinemia in Normal Man. J Clin Invest. 1996 November 1;98(9):2008–2017. doi: 10.1172/JCI119005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002 May;109(9):1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003 Jul 8;100(14):8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rufo C, Teran-Garcia M, Nakamura MT, Koo SH, Towle HC, Clarke SD. Involvement of a unique carbohydrate-responsive factor in the glucose regulation of rat liver fatty-acid synthase gene transcription. J Biol Chem. 2001 Jun 15;276(24):21969–21975. doi: 10.1074/jbc.M100461200. [DOI] [PubMed] [Google Scholar]
- 42.Kenler HA, Brolin RE, Cody RP. Changes in eating behavior after horizontal gastroplasty and Roux-en-Y gastric bypass. Am J Clin Nutr. 1990 Jul;52(1):87–92. doi: 10.1093/ajcn/52.1.87. [DOI] [PubMed] [Google Scholar]
- 43.Havel PJ. Dietary fructose: implications for dysregulation of energy homeostasis and lipid/carbohydrate metabolism. Nutr Rev. 2005 May;63(5):133–157. doi: 10.1301/nr.2005.may.133-157. [DOI] [PubMed] [Google Scholar]
- 44.Takemoto T, Nishio Y, Sekine O, Ikeuchi C, Nagai Y, Maeno Y, et al. RBMX is a novel hepatic transcriptional regulator of SREBP-1c gene response to high-fructose diet. FEBS Lett. 2007 Jan 23;581(2):218–222. doi: 10.1016/j.febslet.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 45.Dobrzyn A, Ntambi JM. Stearoyl-CoA desaturase as a new drug target for obesity treatment. Obes Rev. 2005 May;6(2):169–174. doi: 10.1111/j.1467-789X.2005.00177.x. [DOI] [PubMed] [Google Scholar]
- 46.Leonard AE, Kelder B, Bobik EG, Chuang LT, Lewis CJ, Kopchick JJ, et al. Identification and expression of mammalian long-chain PUFA elongation enzymes. Lipids. 2002 Aug;37(8):733–740. doi: 10.1007/s11745-002-0955-6. [DOI] [PubMed] [Google Scholar]
- 47.Matsuzaka T, Shimano H, Yahagi N, Kato T, Atsumi A, Yamamoto T, et al. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat Med. 2007 Nov;13(10):1193–1202. doi: 10.1038/nm1662. [DOI] [PubMed] [Google Scholar]