Abstract
Autophagy is a highly conserved, ubiquitous process that is responsible for the degradation of cytosolic components in response to starvation. Autophagy is generally considered to be nonselective; however, there are selective types of autophagy that use receptor and adaptor proteins to specifically isolate a cargo. One type of selective autophagy in yeast is the cytoplasm to vacuole targeting (Cvt) pathway. The Cvt pathway is responsible for the delivery of the hydrolase aminopeptidase I to the vacuole; as such, it is the only known biosynthetic pathway that utilizes the core machinery of autophagy. Nonetheless, it serves as a model for the study of selective autophagy in other organisms.
Keywords: aminopeptidase I, cytoplasm to vacuole targeting, selective autophagy
Introduction
Autophagy is a ubiquitous process that is highly conserved in all eukaryotes. It is responsible for the degradation of cytosolic components and organelles in response to nutrient deprivation [1,2]. In addition to playing a role in the cellular response to stress, autophagy also plays a role in development [3], tumor suppression [4], pathogen resistance [5], and aging [6]. We now know that this process is involved in several human pathologies including cancer [7], diabetes [8,9], cardiomyopathy [10], and neurodegenerative disorders such as Alzheimer and Parkinson diseases [11].
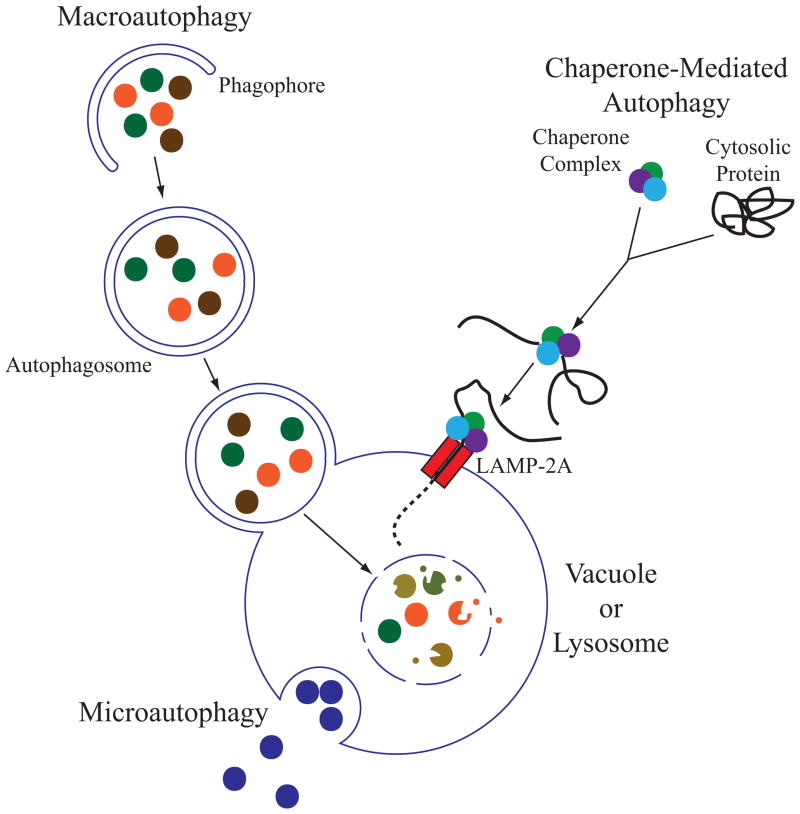
There are three main types of autophagy; chaperone-mediated autophagy, microautophagy, and macroautophagy (figure 1). Chaperone-mediated autophagy has only been characterized in higher eukaryotes, and currently there is no knowledge of a similar process in yeast. In this process, a chaperone protein binds to a specific target protein, causing it to unfold, allowing for direct transport across the lysosomal membrane [12]. Microautophagy sequesters cytoplasmic components, and delivers them for degradation by the direct invagination or protrusion/septation of the lysosomal or vacuolar membrane [13]. Macroautophagy, hereafter referred to as autophagy, is the most well characterized process of the three. Autophagy was first studied in mammalian cells [14], but the majority of the molecular components were initially elucidated in yeast [15], and this review focuses on the yeast model system.
Figure 1. Three main types of autophagy.
There are three main types of autophagy: chaperone-mediated autophagy, microautophagy, and macroautophagy. The schematic depicts a mixture of these processes in lower and higher eukaryotes. For example, the lysosome is much smaller than the fungal vacuole. Also, chaperone-mediated autophagy has only been characterized in higher eukaryotes, whereas microautophagy and macroautophagy are evolutionarily conserved. Macroautophagy is the best-characterized pathway out of the three and the hallmark of this process is the formation of a double-membrane vesicle that non-selectively sequesters cytoplasmic components and delivers them to the lysosome or vacuole for degradation and recycling of the cargo.
The morphological hallmark of autophagy is the de novo formation of the autophagosome, a double-membrane vesicle that sequesters cytosol and organelles. The outer membrane of the autophagosome fuses with the vacuole, releasing the inner membrane, termed the autophagic body, into the lumen where it and its contents are degraded [16]. Although autophagy’s primary role in yeast is to respond to cellular stress, homeostatic and biosynthetic functions have also been elucidated. For example, excess peroxisomes are degraded by pexophagy, a selective type of autophagy, when conditions make them superfluous [17]. In addition to pexophagy, there are other organelle-specific autophagy pathways, such as mitophagy for mitochondria [18], and reticulophagy for the endoplasmic reticulum [19]. Yeast cells also have the cytoplasm to vacuole targeting (Cvt) pathway. The Cvt pathway is responsible for the delivery of at least two hydrolases, α-mannosidase (Ams1) and aminopeptidase I (Ape1), to the vacuole. Ape1 is synthesized in the cytosol as a proenzyme that is relatively inactive. The Cvt pathway sequesters the precursor Ape1 into a Cvt vesicle at the phagophore assembly site or PAS (“phagophore” is the term that describes the initial sequestering vesicle used in autophagy-related pathways), using much of the same machinery as autophagy, and delivers it into the vacuole where it is activated by the removal of the propeptide [20,21]. Because of the similarity between autophagy and the Cvt pathway, the Cvt pathway is considered to be a selective type of autophagy. This paper will review the Cvt pathway and its principal cargo, precursor Ape1, in depth.
Ape1 and the Cvt pathway
Ape1 is one of four aminopeptidases that hydrolyze leucine substrates, identified in the yeast Saccharomyces cerevisiae [22]. Further characterization of the protein by subcellular fractionation revealed it to be localized to the vacuole [23,24], and it appeared to be a glycoprotein containing 12% carbohydrate [25]; Metz and Röhm (1976) originally concluded from western blot migration patterns that Ape1 was glycosylated and thus transported to the vacuole through a portion of the secretory pathway. However, the presence of carbohydrates on Ape1 was never confirmed and Klionsky et al. (1992) show that it is not glycosylated; treatment with the glycosylation inhibitor tunicamycin does not change the migration pattern of Ape1 during SDS-PAGE, and the protein does not bind the lectin concanavalin A [26]. Similar to many other vacuolar hydrolases, Ape1 is synthesized as a zymogen (prApe1) and is processed in a Pep4-dependent manner [22]. Precursor Ape1 maturation is normal in certain sec mutants, which block the secretory pathway, further indicating that prApe1 does not enter the endoplasmic reticulum [26]. Yoshihisa and Anraku (1990) had indicated that another vacuolar hydrolase, α-mannosidase (Ams1), enters the vacuole directly from the cytosol [27], and subsequent studies show that Ams1 uses the same delivery mechanism as prApe1 [28]. This alternative route is named the cytoplasm to vacuole targeting pathway to distinguish it from the canonical pathway used by most vacuolar hydrolases that transit through a portion of the secretory pathway.
Precursor Ape1 has a half-life of maturation of approximately 45 min, which, coupled with its activation by cleavage of the propeptide in the vacuole, makes it an ideal marker for the Cvt pathway [26]. A mutagenesis screen used Ape1 to identify components of this alternative vacuole transport pathway by isolating mutants that accumulate prApe1. The authors found two complementation groups that are allelic to the previously identified vps class B mutants that lack vacuolar acidification. One complementation group is allelic to the gene encoding proteinase B, which acts along with Pep4 in removal of the prApe1 propeptide, and the remaining five groups were determined to be phenotypically distinct from other known mutants. These mutants show a defect in prApe1 maturation, but no defect in Prc1 maturation. The mutants are named atg7, atg8, atg9 and atg11 (the original names were cvt2, cvt5, cvt6/cvt7 and cvt3/cvt9, respectively) [29]. Cellular fractionation experiments show that in those mutants prApe1 is blocked in delivery to the vacuole, so that it accumulates in the cytosolic fraction [29]. The cvt mutants were further analyzed using an in vitro import assay for prApe1 uptake. This assay radiolabels spheroplasts in vivo prior to being lysed, so that the maturation of newly synthesized prApe1 can be followed in vitro. From this assay it is determined that prApe1 maturation is time- and temperature-dependent. It is also shown that the Cvt pathway requires ATP, a functional vacuolar ATPase, and a GTP binding protein [30]. Site-directed mutagenesis of the APE1 gene indicates an important role for the propeptide region of prApe1 in the proper targeting of the protein to the vacuole [31,32]. The predicted secondary structure of the propeptide region includes two α-helices separated by a β-turn [33]. The first α-helix is amphipathic with both acidic and basic amino acids [31]. Random and site directed mutations in the first α-helix result in a defect in prApe1 maturation and localization. Mutations in the propeptide region do not prevent proper folding of prApe1, but mutations in the first α-helix do prevent association with a membrane. The mutagenesis screen identified one key residue, lysine 12, which is especially important for proper vacuolar localization. Mutations in the second α-helix do not affect the kinetics of prApe1 maturation [31]. These data indicate that the first helix is responsible for targeting prApe1 to the vacuole.
Two hypotheses were suggested for the vacuolar import of prApe1. The first suggested that prApe1 is transported across the vacuole membrane in an unfolded or partially unfolded state through a vacuolar pore or channel, similar to the mechanism used in chaperone-mediated autophagy. The problem with this hypothesis is that there is no morphological evidence for a pore complex in the vacuole limiting membrane. The second hypothesis proposed a vesicle-mediated transport mechanism [34]. This hypothesis was supported by a study that examined the oligomeric state of prApe1. Pulse-chase analysis shows that precursor Ape1 rapidly oligomerizes into a homododecamer, which then assembles into a higher order complex composed of multiple dodecamers (the Ape1 complex) [35]. The oligomerization of prApe1 occurs in the cytosol prior to associating with a membrane, and occurs independent of the Atg proteins. Truncations in the C terminus inhibit oligomerization indicating that this region is responsible for generating this higher order structure. Once assembled, the dodecamer is not disassembled, eliminating the possibility of prApe1 entering the vacuole through a small pore in the vacuole membrane, and ruling out translocation of the unfolded protein across the membrane as occurs in chaperone-mediated autophagy [35]. Interestingly, later studies show that Ams1, the other identified cargo of the Cvt pathway, also exists as an oligomer [28], suggesting that one function of this pathway is the transport of hydrolases that assemble into oligomeric complexes.
Vesicle-mediated transport of prApe1 is confirmed using fractionation and immunoelectron microscopy experiments. Subcellular fractionation reveals a pool of prApe1 that is associated with a non-vacuolar membrane compartment [34,36]. In the vps18 mutant, that inhibits fusion at the vacuole, prApe1 accumulates in a non-vacuolar membrane compartment, suggesting that prApe1 is transported to the vacuole inside a vesicle [34]. The prApe1P22L mutant is mutated in the β-turn of the propeptide region. This mutant has increased membrane-binding affinity, but inhibits subsequent steps of the transport process. In cells expressing this mutant, prApe1 co-fractionates with membrane that lacks vacuole markers [34]. Immunoelectron microscopy further confirms that prApe1 first binds to, and is then encapsulated by, a double-membrane vesicle before delivery to the vacuole. In atg15Δ (originally cvt17Δ) cells that are defective in the breakdown of Cvt bodies, cytosolic and subvacuolar prApe1-containing vesicles are visualized [34,36].
Once prApe1 is oligomerized and bound to a membrane is must be sequestered inside a Cvt vesicle of between 140 and 160 nm in diameter [37]. Electron micrographs illustrate that the Cvt vesicle forms near the vacuole at the PAS and apparently excludes all cytosolic components to selectively isolate prApe1 [36]. The formation of the Cvt vesicle requires the t-SNARE Tlg2 and the Sec1 homologue Vps45 [38]. Baba et al. (1997) were able to visualize the steps leading to fusion of the Cvt vesicle with the vacuole membrane using immunoelectron microscopy of pep4Δ cells [36]. The Cvt vesicle’s outer membrane fuses with the vacuole, leaving the inner membrane exposed to the lumen. Fusion requires the same machinery needed for other vesicle fusion events at the vacuole including the Q-SNAREs Vam3 and Vti1, the docking protein Vps18, and the rab GTPase Ypt7 [37]. This process is morphologically similar to nonselective autophagy. Both processes require a double-membrane vesicle to sequester cargo and the vesicle fusion machinery to fuse the outer membrane of the sequestering vesicle with the vacuole, which results in the release of the inner membrane and its contents in the vacuole lumen; in the case of autophagy, the contents are degraded and the breakdown products are released back into the cytosol, whereas the cargo of the Cvt pathway are resident hydrolases that function in the vacuole lumen [20]. The genetic screens for autophagy and Cvt pathway mutants reveal substantial overlap between the molecular components [20,21]. This knowledge changed the direction of study of the Cvt pathway to focus more on the differences and similarities it has with autophagy.
Core Machinery Required for Selective and Nonselective Autophagy
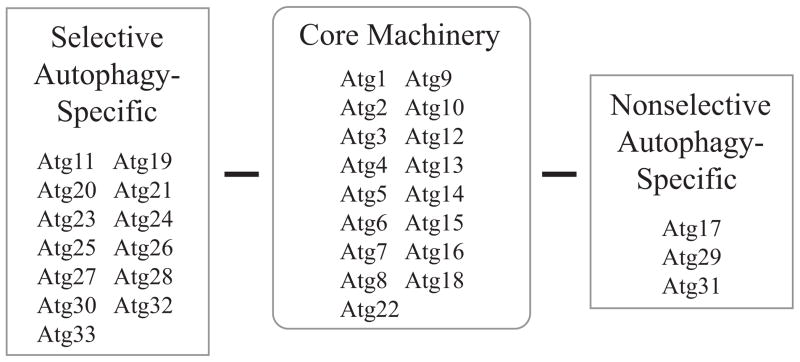
Several screens were carried out almost simultaneously for mutants defective in selective and nonselective types of autophagy, which resulted in the use of multiple names for the corresponding genes. In 2003, these genes were unified into the autophagy-related (ATG) nomenclature [39]. Currently, there are 33 ATG genes, with 17 making up the core machinery required for both the Cvt pathway and autophagy. The 17 core ATG genes can be classified into four groups based upon their function. The first group includes Atg9 and factors involved in its cycling, which particularly include the Atg1 protein kinase complex [40], the second includes the phosphatidylinositol 3-kinase (PtdIns3K) complex [41], the third group includes the ubiquitin-like (Ubl) protein system [42,43], and the fourth group is comprised of proteins that act at the last stages of autophagy, vesicle breakdown and efflux of the cargo degradation products back into the cytosol. The core Atg proteins function at various stages during the autophagy-related pathways, which can be broken down into several steps: 1) the phagophore nucleates at the PAS, a perivacuolar structure that is the site of sequestering vesicle formation in yeast. 2) The phagophore expands to sequester the cargo. 3) The phagophore closes creating the double-membrane autophagosome or Cvt vesicle. 4) The autophagosome or Cvt vesicle fuses with the vacuole, releasing the inner vesicle that is now termed an autophagic body or Cvt body. 5) The autophagic body and its contents are degraded by vacuolar hydrolases, and the products are released into the cytosol by various permeases; the Cvt body is also broken down, and its cargo are matured (in the case of prApe1) and carry out their functions in the vacuole lumen. The differences and similarities in the Atg proteins needed for autophagy and selective autophagy are shown in figure 2.
Figure 2. Classification of Atg proteins according to function.
Autophagy-related (Atg) proteins can be classified according to their role in selective and nonselective autophagy. There are 17 Atg proteins that are required for both types, and these are named the core machinery. Selective autophagy is represented by the cytoplasm to vacuole targeting (Cvt) pathway, mitophagy and pexophagy.
The Atg1 protein kinase complex acts at an initial step of autophagosome formation (and probably at later steps as well). In addition to Atg1, the kinase complex consists of Atg11, Atg13, Atg17, Atg20, Atg24, Atg29, and Atg31, not all of which are considered “core” components. Atg1 is a Ser/Thr kinase whose activity increases upon starvation, and this protein is essential for autophagy [40,44]. In addition, the Tor signaling pathway negatively regulates autophagy through the Atg1 kinase complex [45]. The Tor complex 1 (TORC1) is a nutrient sensor that is active during periods of readily available nutrients [46]. Parallel to TORC1 is the cyclic AMP (cAMP)-dependent protein kinase A (PKA) [47,48]. Both are responsible for the hyperphosphorylation of Atg13 during nutrient rich conditions. In response to starvation conditions, TORC1 is inhibited, resulting in the partial dephosphorylation of Atg13, which may allow the protein to associate with Atg1 with greater affinity, subsequently resulting in an increase in Atg1 kinase activity [40,49,50]. A second regulatory complex that modulates Atg1 kinase activity is the Atg17-Atg29-Atg31 complex. Atg17, Atg29 and Atg31 form a ternary complex in response to starvation conditions. The Atg17-Atg29-Atg31 complex has dual roles; it associates with Atg1, inducing Atg1 kinase activity, and it is also responsible for recruiting other core Atg proteins to the PAS by acting as an organizing scaffold [51,52]. The functions of Atg20 and Atg24 are not understood, whereas Atg11 serves as a scaffold protein that is required for most types of selective autophagy [53,54].
Saccharomyces cerevisiae has only one phosphatidylinositol 3-kinase, Vps34 [55]. Vps34 associates with two different complexes. The first complex consists of Vps34, Atg6 and Atg14 and is specific for autophagy, whereas the second complex contains Vps38 instead of Atg14, and is required for vacuolar protein sorting by the endosome [41,56]. Atg14 targets the Vps34 kinase complex to the PAS [56]. Accordingly, Atg14 is responsible for localizing PtdIns(3)P-binding proteins, including Atg18 (which also binds Atg2), to the PAS [57]. Atg18 binds PtdIns(3)P through an FRRY motif. When this motif is mutated to FTTY, Atg18 no longer binds to PtdIns(3)P resulting in a block in both the Cvt pathway and autophagy. This mutant does not affect binding to Atg2, but both Atg2 and the PtdIns(3)P binding capability of Atg18 are required to localize the complex to the PAS [58]. Relatively little is known about the functions of the Atg18-Atg2 complex, but it is involved in the cycling of Atg9 between peripheral structures and the PAS [59]. Atg9 is a self-associating integral membrane protein that localizes to peripheral (i.e., non-PAS) punctate structures, and cycles to and from the PAS. Atg9 is the only integral membrane protein that is absolutely required for the initial stages of autophagy and the Cvt pathway, and is therefore thought to play a role in trafficking membrane from a donor source(s) to the PAS [60]. Atg9 accumulates at the PAS in atg1Δ, atg2Δ and atg18Δ mutant strains, indicating a role for these proteins in the retrograde (i.e., from the PAS to the peripheral sites) transport of Atg9.
The third group of core Atg proteins is the Ubl protein system. There are two separate yet related conjugation systems that are needed for autophagy. The first is the Atg8 conjugation system. Atg8 is conjugated to phosphatidylethanolamine (PE) and associates with the phagophore and autophagosome [43]. Atg8 expression is increased under starvation conditions [38,61], and this increase is implicated in regulating the size of the autophagosome; when the expression level of Atg8 is artificially decreased in starvation conditions, the size of the autophagosome is smaller compared to the wild-type phenotype [62]. Atg8 contains a C-terminal arginine residue that is removed by the cysteine protease Atg4 [38]. Atg8 is then conjugated to PE via the ultimate glycine residue through the actions of an E1 ubiquitin activating enzyme homolog, Atg7, and an E2 ubiquitin conjugating enzyme analog, Atg3 [63,64]. Atg8–PE is initially located on the inner and outer membranes of the phagophore. Upon completion of the autophagosome or Cvt vesicle, Atg8 is removed particularly from the PE in the outer membrane by a second Atg4-dependent cleavage [38]. The remaining Atg8–PE on the inner membrane is delivered to the vacuole as part of the autophagic (or Cvt) body and is degraded. The association of Atg8 with the inner membrane of the phagophore may play a role in cargo tethering in the Cvt pathway; Atg8 binds the prApe1 receptor Atg19, which may allow the sequestering membrane to enwrap the cargo [64].
The second conjugation system consists of the Atg12–Atg5 complex. Atg12 is conjugated to Atg5 by the formation of an irreversible isopeptide bond between a C-terminal glycine residue of Atg12 and a specific lysine residue of Atg5 [42]. Similar to the Atg8 conjugation system, Atg12 is conjugated to Atg5 through the actions of Atg7, and a different E2-like enzyme, Atg10 [63,64]. The Atg12–Atg5 complex associates with the multimeric protein Atg16; Atg5 binds non-covalently with the N terminus of Atg16 [65,66]. Atg16 is responsible for targeting the multimeric complex to the PAS [65]. The Atg12–Atg5-Atg16 complex appears to play some role in Atg8 conjugation to PE in vivo. Atg12–Atg5-Atg16 may act as an E3 enzyme by interacting with Atg3 and enhancing its E2-like activity, and Atg16 appears to dictate in part the location of Atg8 conjugation [67–69].
The last group of core proteins consists of those involved in the final stages of autophagy. At present, there are only two Atg proteins in this group, Atg15 and Atg22. Atg15 is a putative lipase that is needed for breakdown of the Cvt and autophagic bodies [70,71], whereas Atg22 is an amino acid permease in the vacuole membrane [72]. These components are critical for some types of autophagy such as starvation-induced nonselective autophagy; the cell cannot survive without degradation of the autophagosome cargo and release of the breakdown products back into the cytosol for reuse.
One question that remains to be answered is how the core machinery is able to switch from creating the smaller, and selective Cvt vesicles to the larger, nonselective autophagsomes upon nutrient starvation. It was first hypothesized that the phosphorylation state of Atg13 may act as the molecular switch to turn off the Cvt pathway and turn on autophagy since Atg13 is hyperphosphorylated during nutrient rich conditions and is rapidly dephosphorylated upon starvation [50]. However, the Cvt pathway and autophagy are not mutually exclusive processes, and therefore the molecular switch must be more complicated than just the dephosphorylation of Atg13 [73]. Another candidate for the molecular switch is the Atg17-Atg29-Atg31 ternary complex, which plays a role along with Atg1 and Atg13 in the nucleation of the PAS under starvation conditions. Cheong et al. (2005) show that in the atg17Δ mutant pexophagy is completely blocked and autophagy is partially defective while the Cvt pathway is unaffected, suggesting a role for Atg17 in controlling the magnitude of the autophagic response [74]. The Atg17-Atg29-Atg31 autophagy-specific complex could regulate the size of the autophagosome without affecting the Cvt vesicle. In addition, during nutrient rich conditions, Atg11, which is not needed for nonselective autophagy, is responsible for the organization of the PAS [53]. This suggests that the molecular switch between the two pathways is related to the actions of the Atg1-Atg13-Atg17(-Atg29-Atg31) starvation complex versus the Cvt pathway-specific protein Atg11. More research needs to be done to elucidate the mechanics of this proposed switching complex.
Specificity in the Cvt Pathway
Precursor Ape1 is preferentially targeted to the vacuole in both the Cvt pathway and autophagy, suggesting a specific targeting mechanism. A receptor for prApe1 was proposed when it was determined that prApe1 transport to the vacuole by the Cvt pathway is specific and saturable [75]. Two groups simultaneously discovered that Atg19 (originally Cvt19) has all of the characteristics needed to be a receptor for prApe1 in Cvt transport [75,76]. The protein was first identified in a genome wide yeast two-hybrid screen initiated to identify protein-protein interactions between full-length open reading frames predicted from the S. cerevisiae genome sequence [77]. Further characterization of the protein revealed that Atg19 is needed for the stabilization of prApe1 binding to the Cvt vesicle membrane, and that in atg19Δ cells prApe1 maturation is inhibited while autophagy is not affected [75]. In addition, Atg19 binds to prApe1 in a propeptide-dependent manner, suggesting that the propeptide region is responsible for the recognition of prApe1 by the Cvt pathway machinery [75]. Finally, Atg19 is a peripheral membrane protein that localizes to the PAS, and its half-life is consistent with that of prApe1 maturation. Atg19 has an expression level stoichiometric with prApe1 and it is delivered to the vacuole along with prApe1 [75]. Consistent with the Cvt pathway transporting both prApe1 and Ams1 to the vacuole, Atg19 is required for Ams1 vacuolar localization [28]. The binding domains for prApe1 and Ams1 on Atg19 are separate and therefore Atg19 is capable of delivering both prApe1 and Ams1 to the vacuole in the same Cvt vesicle [77]. The receptor-mediated Cvt pathway is illustrated in figure 3.
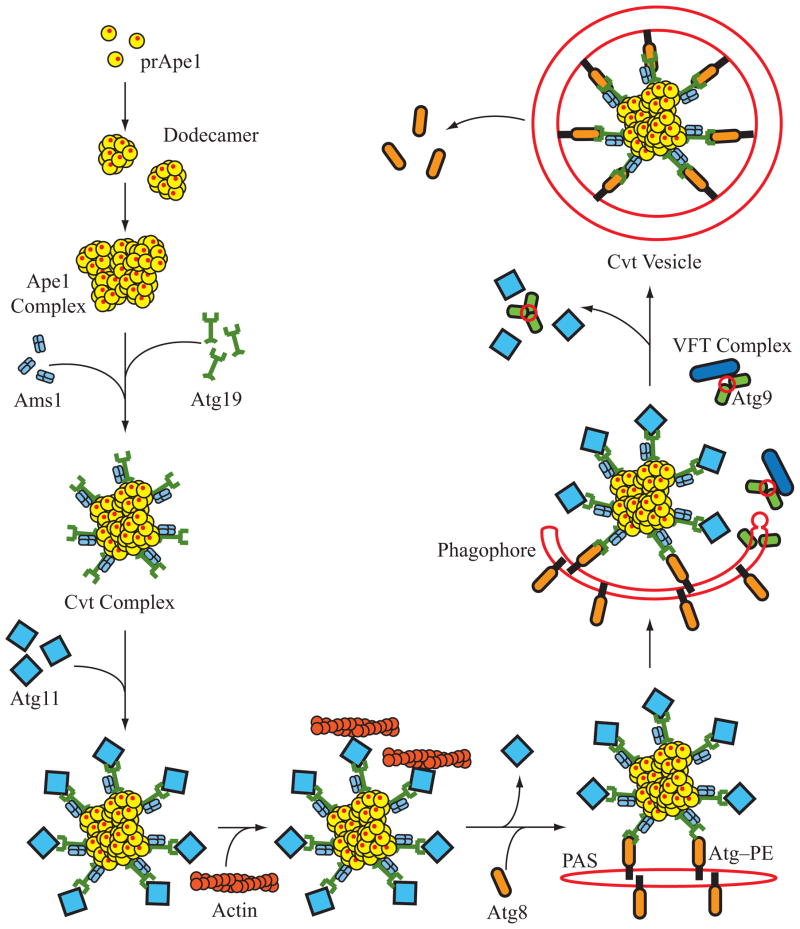
Figure 3. Cvt vesicle formation.
Precursor Ape1 is a proenzyme that is synthesized in the cytosol, and rapidly oligomerizes to form a homododecamer. The dodecamer further organizes into a higher order structure termed the Ape1 complex. The receptor protein, Atg19, then binds to the Ape1 complex forming the Cvt complex. The Ams1 oligomer is also able to bind Atg19 and can be incorporated into the same Cvt complex. Atg11 binds to Atg9 and transports the Cvt complex to the PAS. At the PAS the Cvt complex binds to the expanding phagophore membrane through an interaction between Atg19 and Atg8–PE. Transport of the Cvt complex to the PAS and the expansion of the phagophore require the actin cytoskeleton and the VFT complex, respectively; delivery of membrane to the expanding vesicle likely involves cycling of Atg9. The phagophore membrane expands around the Cvt complex (excluding bulk cytoplasm) forming the Cvt vesicle.
The prApe1-Atg19 complex (defined as the Cvt complex) is transported to the PAS through a mechanism that is dependent on Atg11 [78]. Atg11 specifically recognizes and binds a C-terminal domain of Atg19 [78], and this interaction is independent of the cargo proteins [79]; immunoprecipitation experiments show that Atg11 and Atg19 coprecipitate both in wild-type and ape1Δ cells [79]. The interaction between Atg11 and Atg19 is critical for the proper localization of the Cvt complex to the PAS, because cells lacking Atg11 show mislocalization of prApe1 and Atg19, as well as a defect in prApe1 maturation [78]. In addition, Atg11 localization is dependent upon both Atg19 and prApe1, suggesting that Atg11 associates with the Cvt complex prior to PAS localization [78,79]. These findings indicate that Atg11 is involved in the transport of the Cvt complex to the PAS. Proper localization of the Cvt complex to the PAS is also dependent upon the Vps53 (VFT) tethering complex [80]. The tethering complex is required for Cvt vesicle formation but not for autophagy, suggesting that the membrane source for Cvt vesicle and autophagosome formation may be, at least in part, different. The VFT complex is composed of four components, Vps51, Vps52, Vps53 and Vps54. This complex works in conjunction with Vps45, and the Q-SNAREs Tlg1 and Tlg2, to mediate Cvt vesicle formation [80]. In cells lacking components of the VFT complex, Atg19 and prApe1 are no longer transported to the PAS, but the defect in prApe1 maturation is reversed upon starvation [80]. Subsequent studies further suggest that the VFT complex plays a role in Cvt vesicle formation by facilitating the transport of Atg9 to the PAS [81].
A role for the actin cytoskeleton has also been elucidated in this process. Actin and the actin-binding complex Arp2/3 are required for the Cvt pathway. Cells treated with the actin-disrupting drug latrunculin A show a defect in prApe1 maturation, whereas the drug has no affect on autophagy, suggesting that the actin cytoskeleton is only critical for the Cvt pathway [82]. Further analysis by fluorescence microscopy in cell lines with specific actin mutants, suggests that the actin cytoskeleton is required for the localization of the Cvt complex and Atg9 to the PAS; however, disruption of actin does not affect the ability of the Cvt complex to bind to a membrane [82]. It is hypothesized that the actin cytoskeleton acts as a track to guide cargos to the PAS, and that Atg11 is the protein that binds the cargo to actin, whereas the Arp2/3 complex could act as a motor that drives the Cvt complex to the PAS [83].
The localization of the Cvt receptor complex to the PAS is essential for the proper organization of the PAS and Cvt vesicle formation. Without any component of the Cvt complex (and Atg11), other Atg proteins do not efficiently localize to the PAS [54]. Atg11 is the critical component for proper PAS organization during vegetative growth conditions, and the expression level of Atg11 correlates directly with the capacity of the Cvt pathway [84]. Though the C terminus of Atg11 is involved in binding to Atg19, there are other regions in Atg11 that are involved in forming complexes with other Atg proteins including its own self-interaction [79]. For example, Atg11 interacts with Atg9 to allow the delivery of the latter to the PAS [85], and Atg11 interacts with the Atg1-Atg13 kinase complex [79]. Likely as a result of its scaffold properties, the overexpression of Atg11 causes an increase in the amount of Atg8 and Atg9 recruited to the PAS. This results in the formation of more Cvt vesicles during nutrient rich conditions [84]. All of this evidence indicates a critical role for Atg11 in PAS organization and the formation of Cvt vesicles.
Atg19 interacts with Atg11 to transport the Cvt complex to the PAS, once at the PAS Atg19 interacts with Atg8. Atg8 plays two roles in the Cvt pathway; it is responsible for regulating vesicle formation, and is helps mediate the sorting of selective cargo by acting as a tether. The interaction with Atg19 is essential for Atg8’s role in tethering the Cvt complex to the phagophore [84]. The Cvt pathway is blocked in atg8Δ cells at the step of vesicle formation; precursor Ape1 accumulates and is protease accessible, but can still associate with membranes in the atg8Δ mutant [52]. Ho et al. was able to isolate these two functions in the sequence of Atg8. They determined that the residues Arg28, Tyr49 and Leu50 are involved in both vesicle formation and cargo selection, whereas Lys48 and Leu55 are only involved in vesicle formation [86]. It is thought that the binding of Atg19 to Atg8 is the anchor that forces the membrane to expand exclusively around the Cvt complex [54].
Atg11 is not required for nonselective autophagy, and hence has been termed Cvt-specific; however, Atg11 is also required for other types of selective autophagy. Other Atg proteins that are not required for nonselective autophagy include Atg20, Atg21, and Atg24. These latter proteins are phosphoinositide-binding proteins. Atg20 and Atg24 bind PtdIns(3)P through a phox homology (PX) domain and are dependent upon Atg14 and the PtdIns3K complex for proper localization [87]. Mutations in the PX domain of either protein prevent their localization at the PAS and partially block prApe1 maturation. Mutations in the PX domains of both proteins result in a complete block of the Cvt pathway. This suggests that the interaction between Atg20 and Atg24 offers partial complementation for a mutation in one of these two components [87]. Atg21 is similar to Atg18 in that it binds PtdIns(3)P, and is required for Cvt vesicle formation [58]. In atg21Δ cells, not only do Atg8 and Atg5 fail to localize to the PAS, there is also an observable decrease in Atg8 lipidation [58]. These proteins provide specificity for the role of the PtdIns3K complex in the Cvt pathway, and presumably other types of selective autophagy.
The Cvt Pathway and Selective Autophagy Require Specificty Factors
Selective autophagy is mediated by a cargo receptor and a specificity factor (adaptor) that together connect the cargo to the core autophagy machinery. As discussed previously, the cargo receptor and the specificity factor for the Cvt pathway are Atg19 and Atg11, respectively. Atg19 acts in two capacities; it first binds the cargo and transports it to the PAS through an interaction with Atg11, and second, it interacts with Atg8, a component of the core autophagy machinery, to aid in the formation of the Cvt vesicle. Atg19 interacts with Atg8 through a WXXL motif [88]. The WXXL motif has an extended β conformation and forms an intermolecular parallel β-sheet with β2 of Atg8. The WXXL motif of Atg19 is followed by acidic residues, which, with the motif, are required for binding to Atg8 [88]. The binding pocket for the WXXL motif of Atg8 is crucial only for the Cvt pathway and not nonselective autophagy, indicating that this interaction is specific only for selective autophagy. The WXXL binding pocket of Atg8 is highly conserved in higher eukaryotes [88]. The mammalian adaptor protein p62/SQSTM1, which binds ubiquitin, also contains a WXXL motif that is responsible for binding to the mammalian homologue of Atg8, LC3. These findings suggest a possible fundamental mechanism responsible for selective autophagy. Along these lines, two additional selective autophagy receptors with WXXL motifs have recently been identified in mammalian cells, NBR1 and Nix. Similar to p62, NBR1 contains both a ubiquitin binding domain and the WXXL LC3 binding domain, also termed the LC3-interacting region (LIR). NBR1 is involved in the clearance of ubiquitinated aggregates [89], whereas Nix plays a role in the selective clearance of mitochondria by autophagy (mitophagy) during reticulocyte maturation [90,91].
In the yeast S. cerevisiae, mitophagy is mediated through the receptor protein Atg32. Atg32 was first identified in a genomic screen for yeast mutants defective in mitophagy and it is not required for nonselective autophagy or the Cvt pathway [92]. Atg32 is located in the outer membrane of the mitochondria with its N terminus in the cytosol and the C terminus in the intermembrane space. Similar to Atg19, Atg32 interacts with both Atg11 and Atg8 as confirmed by yeast two-hybrid and co-immunoprecipitation experiments. Atg32 interacts with Atg11 to recruit mitochondria to the PAS. Atg32 also contains a WXXL domain on its N terminus, which is responsible for binding to Atg8, and mutation of the WXXL domain blocks binding to Atg8, but not to Atg11 [93]. The binding of Atg32 to Atg8 is required for the complete sequestration of mitochondria by the phagophore [92,93]. These observations suggest that Atg32 is a key receptor protein needed for mitophagy.
The selective removal of peroxisomes by autophagy (pexophagy) has mainly been studied in methylotrophic yeast, including Pichia pastoris. The P. pastoris protein PpAtg30 has been identified as the receptor for pexophagy. It was originally discovered in a screen of a collection of micropexophagy mutants, and was identified as a pexophagy-specific mutant; it is not required for the Cvt pathway or nonselective autophagy. PpAtg30 overexpression is able to stimulate pexophagy even under non-pexophagy inducing conditions. PpAtg30 binds to peroxisomes by interacting with the peroxins, PpPex3 and PpPex14, and it transiently associates with the PAS during pexophagy. PpAtg30 binds to PpAtg11 and PpAtg17, connecting the peroxisome to the core autophagy machinery. However, PpAtg30 does not contain a WXXL domain, and therefore does not interact with PpAtg8 [94]. It will be interesting to see if future studies can determine if there is an intermediary protein that links PpAtg30 to PpAtg8.
A recurring theme is apparent in selective autophagy. The cargo must first be recognized by a receptor protein, which is Atg19 for the Cvt pathway, Atg32 for mitophagy, and PpAtg30 for pexophagy. The receptor protein must then be able to bind an adaptor protein that connects the cargo to the core autophagy machinery. In yeast, Atg11 (and PpAtg11) acts as the adaptor protein. It binds receptor proteins and uses actin to transport the cargo to the PAS, and is responsible for the organization of the PAS during selective autophagy. Once at the PAS, the cargo protein, at least in the case of the Cvt pathway and mitophagy, is able to bind Atg8 via the receptor. This mechanism appears to be conserved in higher eukaryotes through the WXXL binding domain.
Conclusions
The Cvt pathway is the best characterized type of selective autophagy and therefore stands as a model for how specific cargos are delivered to the vacuole by the autophagy machinery. The Cvt pathway has only been characterized in yeast, specifically S. cerevisiae and P. pastoris, and is not evolutionarily conserved [95]. Even in fungi, there are certain differences, in that P. pastoris uses the two proteins PpAtg26 and PpAtg28 [96]; S. cerevisiae Atg26 is not involved in autophagy, and there is no ortholog of PpAtg28. Despite all of this, the study of the Cvt pathway can be beneficial for identifying and understanding selective types of autophagy in higher eukaryotes.
The study of selective autophagy should provide important information for future research related to human diseases. For example, selective autophagy is implicated in the response to pathogen infection; certain pathogens, including the herpes simplex virus, can be selectively degraded by xenophagy [97]. This process recognizes invading pathogens and sequesters them within autophagosomes, indicating a role for selective autophagy in innate immunity [5,97]. Little is known about this process, but Thursten et al. (2009) show that in human cells the protein NDP52 may act as a receptor by recognizing ubiquitin-coated Salmonella, and recruiting LC3 to the bacteria [98]. Selective autophagy also plays a key role in the prevention of neurodegenerative disorders. Huntington, Alzheimer, Parkinson and Creutzfeldt-Jakob diseases, are the result of toxic neuropeptides that accumulate and form large protein aggregates [99]. Mouse models show that autophagy can prevent neurodegeneration by degrading aggregate-prone proteins before they can damage neurons. The degradation of these protein aggregates depends on p62, which may act similar to Atg19 as noted above [100,101]. The basis for specificity in the different types of selective autophagy requires further study, but certain similarities between the types of selective autophagy occurring in higher eukaryotes and the yeast Cvt pathway are readily apparent. These pathways require both a specificity factor/adaptor and a receptor. The receptor in many cases contains a WXXL or LIR domain that is able to bind Atg8/LC3, connecting the cargo to the core autophagy machinery. Future studies using the Cvt pathway as a model may be able to identify receptors and specificity factors for selective autophagy pathways in higher eukaryotes. Defects in selective autophagy can result in the accumulation of damaged proteins and organelles that are associated with various diseases. Thus, being able to manipulate selective types of autophagy may have therapeutic potential for a range of human pathologies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Sources Cited
- 1.Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ. Cell biology: regulated self-cannibalism. Nature. 2004;431:31–2. doi: 10.1038/431031a. [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 4.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa I, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–40. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 6.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Müller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 7.Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung HS, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–24. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 9.Ebato C, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–32. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka Y, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–6. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 11.Yuan J, Lipinski M, Degterev A. Diversity in the mechanisms of neuronal cell death. Neuron. 2003;40:401–13. doi: 10.1016/s0896-6273(03)00601-9. [DOI] [PubMed] [Google Scholar]
- 12.Massey A, Kiffin R, Cuervo AM. Pathophysiology of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2420–34. doi: 10.1016/j.biocel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 13.Wang C-W, Klionsky DJ. Microautophagy. In: Klionsky DJ, editor. Autophagy. Landes Bioscience; Georgetown, TX: 2004. pp. 107–114. [Google Scholar]
- 14.Deter RL, Baudhuin P, de Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967;35:C11–6. doi: 10.1083/jcb.35.2.c11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–7. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 16.Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 17.Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci. 1999;112:4079–87. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- 18.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–53. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraft C, Reggiori F, Peter M. Selective types of autophagy in yeast. Biochim Biophys Acta. 2009;1793:1404–12. doi: 10.1016/j.bbamcr.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–4. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 21.Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci U S A. 1996;93:12304–8. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trumbly RJ, Bradley G. Isolation and characterization of aminopeptidase mutants of Saccharomyces cerevisiae. J Bacteriol. 1983;156:36–48. doi: 10.1128/jb.156.1.36-48.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matile P, Cortat M, Wiemken A, Frey-Wyssling A. Isolation of glucanase-containing particles from budding Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1971;68:636–40. doi: 10.1073/pnas.68.3.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frey J, Röhm KH. Subcellular localization and levels of aminopeptidases and dipeptidase in Saccharomyces cerevisiae. Biochim Biophys Acta. 1978;527:31–41. doi: 10.1016/0005-2744(78)90253-x. [DOI] [PubMed] [Google Scholar]
- 25.Metz G, Röhm KH. Yeast aminopeptidase I. Chemical composition and catalytic properties. Biochim Biophys Acta. 1976;429:933–49. doi: 10.1016/0005-2744(76)90338-7. [DOI] [PubMed] [Google Scholar]
- 26.Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–99. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshihisa T, Anraku Y. A novel pathway of import of α-mannosidase, a marker enzyme of vacuolar membrane, in Saccharomyces cerevisiae. J Biol Chem. 1990;265:22418–25. [PubMed] [Google Scholar]
- 28.Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J Biol Chem. 2001;276:20491–8. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott SV, Klionsky DJ. In vitro reconstitution of cytoplasm to vacuole protein targeting in yeast. J Cell Biol. 1995;131:1727–35. doi: 10.1083/jcb.131.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oda MN, Scott SV, Hefner-Gravink A, Caffarelli AD, Klionsky DJ. Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J Cell Biol. 1996;132:999–1010. doi: 10.1083/jcb.132.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segui-Real B, Martinez M, Sandoval IV. Yeast aminopeptidase I is post-translationally sorted from the cytosol to the vacuole by a mechanism mediated by its bipartite N-terminal extension. EMBO J. 1995;14:5476–84. doi: 10.1002/j.1460-2075.1995.tb00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez E, Jimenez MA, Segui-Real B, Vandekerckhove J, Sandoval IV. Folding of the presequence of yeast pAPI into an amphipathic helix determines transport of the protein from the cytosol to the vacuole. J Mol Biol. 1997;267:1124–38. doi: 10.1006/jmbi.1997.0925. [DOI] [PubMed] [Google Scholar]
- 34.Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J, Scott SV, Oda MN, Klionsky DJ. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1997;137:609–18. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–95. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148:465–80. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirisako T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–76. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klionsky DJ, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–45. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 40.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–13. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–30. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizushima N, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–8. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 43.Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–92. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 44.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–50. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 45.Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–58. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–6. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 47.Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004;279:20663–71. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:4180–9. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192:207–13. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 50.Scott SV, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840–9. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 51.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–50. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–81. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J, et al. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–96. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–94. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 56.Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes--Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1527–39. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dove SK, et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004;23:1922–33. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stromhaug PE, Reggiori F, Guan J, Wang CW, Klionsky DJ. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell. 2004;15:3553–66. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–18. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 60.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 61.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–78. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 62.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–8. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mizushima N, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–46. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–96. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277:18619–25. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 67.Ichimura Y, Imamura Y, Emoto K, Umeda M, Noda T, Ohsumi Y. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J Biol Chem. 2004;279:40584–92. doi: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- 68.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 69.Sou YS, Tanida I, Komatsu M, Ueno T, Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J Biol Chem. 2006;281:3017–24. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- 70.Epple UD, Suriapranata I, Eskelinen EL, Thumm M. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J Bacteriol. 2001;183:5942–55. doi: 10.1128/JB.183.20.5942-5955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teter SA, Eggerton KP, Scott SV, Kim J, Fischer AM, Klionsky DJ. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J Biol Chem. 2001;276:2083–7. doi: 10.1074/jbc.C000739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell. 2006;17:5094–104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khalfan WA, Klionsky DJ. Molecular machinery required for autophagy and the cytoplasm to vacuole targeting (Cvt) pathway in S. cerevisiae. Curr Opin Cell Biol. 2002;14:468–75. doi: 10.1016/s0955-0674(02)00343-5. [DOI] [PubMed] [Google Scholar]
- 74.Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang CW, Klionsky DJ. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–53. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001;7:1131–41. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leber R, Silles E, Sandoval IV, Mazon MJ. Yol082p, a novel CVT protein involved in the selective targeting of aminopeptidase I to the yeast vacuole. J Biol Chem. 2001;276:29210–7. doi: 10.1074/jbc.M101438200. [DOI] [PubMed] [Google Scholar]
- 77.Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 78.Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–37. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reggiori F, Wang CW, Stromhaug PE, Shintani T, Klionsky DJ. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J Biol Chem. 2003;278:5009–20. doi: 10.1074/jbc.M210436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reggiori F, Klionsky DJ. Atg9 sorting from mitochondria is impaired in early secretion and VFT-complex mutants in Saccharomyces cerevisiae. J Cell Sci. 2006;119:2903–11. doi: 10.1242/jcs.03047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reggiori F, Monastyrska I, Shintani T, Klionsky DJ. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:5843–56. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Monastyrska I, Reggiori F, Klionsky DJ. Harpooning the Cvt complex to the phagophore assembly site. Autophagy. 2008;4:914–6. doi: 10.4161/auto.6657. [DOI] [PubMed] [Google Scholar]
- 84.Geng J, Klionsky DJ. Quantitative regulation of vesicle formation in yeast nonspecific autophagy. Autophagy. 2008;4:955–7. doi: 10.4161/auto.6791. [DOI] [PubMed] [Google Scholar]
- 85.Chang CY, Huang WP. Atg19 mediates a dual interaction cargo sorting mechanism in selective autophagy. Mol Biol Cell. 2007;18:919–29. doi: 10.1091/mbc.E06-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ho KH, Chang HE, Huang WP. Mutation at the cargo-receptor binding site of Atg8 also affects its general autophagy regulation function. Autophagy. 2009;5:461–71. doi: 10.4161/auto.5.4.7696. [DOI] [PubMed] [Google Scholar]
- 87.Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J Biol Chem. 2002;277:30198–207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Noda NN, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–8. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 89.Kirkin V, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–16. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 90.Schweers RL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–5. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Novak I, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 94.Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–76. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meijer WH, van der Klei IJ, Veenhuis M, Kiel JAKW. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–16. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- 96.Farre JC, Vidal J, Subramani S. A cytoplasm to vacuole targeting pathway in P. pastoris. Autophagy. 2007;3:230–4. doi: 10.4161/auto.3905. [DOI] [PubMed] [Google Scholar]
- 97.Tallóczy Z, Virgin HW, IV, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–9. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- 98.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–21. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 99.Landles C, Bates GP. Huntingtin and the molecular pathogenesis of Huntington’s disease. Fourth in molecular medicine review series. EMBO Rep. 2004;5:958–63. doi: 10.1038/sj.embor.7400250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]