Abstract
Background
Drug discovery is a complex and unpredictable endeavor with a high failure rate. Current trends in the pharmaceutical industry have exasperated these challenges and are contributing to the dramatic decline in productivity observed over the last decade. The industrialization of science by forcing the drug discovery process to adhere to assembly-line protocols is imposing unnecessary restrictions, such as short project time-lines. Recent advances in nuclear magnetic resonance are responding to these self-imposed limitations and are providing opportunities to increase the success rate of drug discovery.
Objective/Method
A review of recent advancements in NMR technology that have the potential of significantly impacting and benefiting the drug discovery process will be presented. These include fast NMR data collection protocols and high-throughput protein structure determination, rapid protein-ligand co-structure determination, lead discovery using fragment-based NMR affinity screens, NMR metabolomics to monitor in vivo efficacy and toxicity for lead compounds, and the identification of new therapeutic targets through the functional annotation of proteins by FAST-NMR.
Conclusion
NMR is a critical component of the drug discovery process, where the versatility of the technique enables it to continually expand and evolve its role. NMR is expected to maintain this growth over the next decade with advancements in automation, speed of structure calculation, in-cell imaging techniques, and the expansion of NMR amenable targets.
Keywords: NMR, Drug Discovery, Structural Biology, Fragment-Based Screening, Metabolomics
1. INTRODUCTION
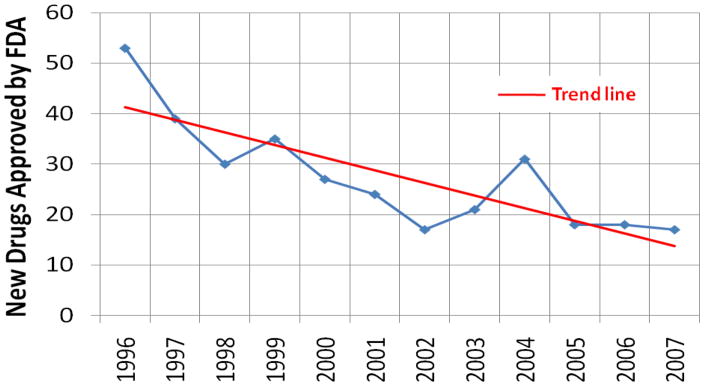
A troubling decline (Figure 1) has been observed in the productivity and creativity of the pharmaceutical industry over the last decade [1–5]. This is a complex issue and a number of factors are contributing to the observed reduction in new drugs approved by the FDA. One primary cause is the fact that the drug discovery process is a fundamentally challenging endeavor with a high failure rate and price tag. Estimates indicate that only one new chemical entity (NCE) out of 25 NCEs identified from active research projects will become a marketable drug. The corresponding success rate of clinical trials is only 11% [6, 7]. The development of the average drug costs > $800 million dollars [8], where current industrial trends to minimize these costs are contributing to the decline in new drugs [5].
Figure 1.
The number of new drugs approved by the U.S. Food and Drug Administration on a yearly basis. The straight line highlights the negative trend in drug approval rates.
Because of these high costs, pharmaceutical companies tend to focus their research efforts in therapeutic areas with potential profits exceeding a billion dollars a year [9]. These tend to be chronic diseases that require potentially life-time treatments for a significant percentage of the population, such as cardiovascular, oncology and central nervous system disorders. The end result is a concentrated effort in a very few research areas [10] leading to a high-level of competition, a limited number of new therapies and a number of redundant drugs [11] serving the same market [12]. Similar efforts to minimize costs also results in a negative impact on the discovery of new drugs.
The expanding trend of applying traditional business practices to the drug discovery process is misguided and having the unintended consequence of diminishing output and eliminating originality [2, 3, 13, 14]. Current methods being applied to reduce cost and increase efficiency include outsourcing [15, 16], metrics [17, 18], and limiting the duration of a research project [19]. This is despite the obvious fact that drug discovery is a complex, highly interdisciplinary process [20] and inherently unpredictable [21, 22]. Treating each component as simply an independent step in an assembly line protocol [23] clearly undermines the need for routine interactions and the exchange of ideas between each essential discipline [24]. In drug discovery, the whole is clearly greater than the sum of the parts where critical breakthroughs and insights come from the cross-fertilization of ideas and information between separate research groups. Outsourcing and metrics isolates these groups and places a high priority on inconsequential book-keeping: number of compounds synthesized, assays run, spectra collected and structures solved that creates the illusion of accomplishments [2]. Furthermore, placing artificial time-limits on research projects simply creates an endless cycle of identifying new therapeutic targets, generating chemical leads and rapidly abandoning a project when the typical challenges with bioavailability, stability, toxicity and efficacy are encountered. This focus on project lifetime evolved from the current drug discovery process that is based on high-throughput screening (HTS) and structure based drug design [25, 26].
HTS is an efficient and effective approach for identifying high-affinity binders to protein targets, but as recent reports have indicated it has not increased the number of successful drugs [27]. HTS chemical leads are not routinely translated into new drugs because a preponderance of inhibitors have undesirable modes of action [28, 29]. Misleading compounds that lack a confirmed correlation between functional activity and a direct binding interaction with the protein target routinely emerge from HTS [30]. These false leads are a significant factor in the delay or derailment of drug discovery projects, and they may contribute to clinical failures as well [28–32]. At the onset of a drug discovery program, it is clearly desirable to identify lead compounds that interact with the therapeutic target in a biologically relevant mechanism. In this manner, promising chemical leads can be quickly prioritized for more detailed evaluations, with an anticipated increase in the development of new drugs.
In the era where time is a critical consideration in the success of a drug discovery project, nuclear magnetic resonance (NMR) has become an integral component of the process [33–35]. The diversity of NMR enables the technique to be applied at multiple stages along the drug discovery pathway and is addressing some of these self-imposed challenges that are hindering progress in discovering new therapeutics [36]. NMR is being used for lead discovery and optimization, evaluating in vivo selectivity and efficacy, analyzing drug toxicity profiles and identifying new drug discovery targets. Recent advances in NMR technology enable NMR to rapidly determine protein and protein-ligand structures, to efficiently screen fragment-based libraries to identify biological relevant ligand interactions and identify new therapeutic targets, and to monitor changes in the metabolome from biofluids and cell lysates to explore in vivo drug activity. This review will discuss these recent advancements of NMR for drug discovery.
2. RAPID PROTEIN STRUCTURES
A high-resolution protein structure is a key requirement to evaluate the biological relevance of potential chemical leads identified from HTS. Validating that the compound binds specifically to a functional region of the protein structure dramatically increases the likelihood the compound can be evolved into a drug. Due to the increasingly limited time available for a research project to produce an NCE, obtaining a rapid protein structure during the early stages of the drug discovery project is essential. Despite the prevalent use of x-ray crystallography in the pharmaceutical industry for determining protein structures, NMR and x-ray should be viewed as complimentary techniques with limited redundancies [37, 38]. Recent statistical analysis indicates that ~20–40% of protein structures determined from structural genomics may be amenable to analysis by NMR, where a preponderance of protein structures were determined by NMR only or x-ray only. More importantly, NMR had an equal success rate for both prokaryote and eukaryote proteins [39], which is an important consideration for drug discovery.
The application of NMR has also been unnecessarily curtailed because of the general upper weight limitation of ≤ 25 kDa. Nevertheless, the average domain size for eukaryotic proteins is ~150 residues or ~17 kDa, which is within the MW range for NMR. Additionally, advancements in NMR methodology has significantly expanded this upper limit, where the NMR analysis of large MW complexes are becoming common place: 900kDa GroEL-GroES complex, 300-kDa cylindrical protease ClpP [40], 95 kDa homotrimeric complex of the acyltransferase protein [41], 82.4 kDa of malate synthase [42, 43], 69 kDa α1-proteinase inhibitor Pittsburgh-trypsin covalent complex [44], the 45.3 kDa catalytic domain of human BACE-1 [45], the 44 kDa nucleotide-binding domain [46], the 486 kDa TET2 aminopeptidase protein [47] and the 441 residue tau protein [48], among others. Obtaining these results required advanced labeling and NMR techniques that includes deuterium labeling [49], selective residue labeling [50], selective methyl labeling [51] and TROSY-based experiments [52]. This requires a robust expression system (e.g., E. coli or a cell-free system) to obtain milligram quantities of various labeled protein for multiple NMR samples [53–56]. Also, it is important to note that these studies did not yield high-resolution solution structures of the indicated large MW proteins. Generally, specific insights regarding the structure and dynamics of the proteins or complexes related to its biological function were obtained. This requires using existing x-ray structures. Chemical shift perturbations, residual dipolar coupling constants (RDCs) [57] and/or 13C-labeled methyl probes are typically used to model protein-protein complexes (from existing NMR or x-ray structures of each monomer), identify protein-ligand interactions or monitor the dynamics of domain or ligand interactions. Low-resolution structures may also be obtained from a minimal NOE dataset [58, 59] combined with other structural constraints such as RDCs [57] and pseudocontact shifts [60].
Similarly, significant improvements have been made to increase the throughput of NMR structure determination. This has occurred, in part, due to the Protein Structure Initiative (PSI) that has provided the incentive to develop the infrastructure and technology for high-throughput structure determination [61]. First, robotic systems have been developed for the automated production of protein samples for both NMR and x-ray [62]. Second, advancements in NMR pulse sequences and protocols for rapid data collection has shorten the time required to collect a complete set of NMR spectra for a protein structure. These methods include automated projection spectroscopy (APSY) [63], G-matrix Fourier transform NMR (GFT-NMR) [64, 65], high-resolution iterative frequency identification (HIFI-NMR) [66], projection-reconstruction NMR (PR-NMR) [67], and reduced-dimensionality NMR (RD-NMR) [68, 69]. The primary goal of these methods is to reduce the dimensionality of traditional multi-dimensional triple resonance experiments, and thus shorten the acquisition time, by joint sampling of multiple chemical shifts in a single indirect dimension. Generally, a series of low-dimensional NMR spectra are collected that are either linearly combined or used to reconstruct the desired higher-dimensionality spectrum. A general concern with reduced dimensionality experiments is the potential loss of information or resolution by projecting n indirect dimensions into a 2D spectrum. The encoding of multiple chemical shifts within a single resonance results in peak multiplicity with distinct phasing. As the size of the protein increases, peak overlap and the canceling of anti-phase peaks becomes an issue. Of course, reduced dimensionality experiments have been successfully applied to proteins with MW up to 20 kDa [64].
An alternative approach to reduce the acquisition time of multidimensional NMR experiments for protein structure determination is the application of sparse data collection or non-uniform sampling (NUS) [70–72]. The major factor that contributes to the long (~1–3 days) acquisition times for typical triple resonance experiments is the requirement to collect a uniform series of data points to properly represent the free induction decay (FID) along each indirect dimension of the multidimensional experiment. Even for a 3D experiment with modest resolution (512×128×32 complex points), the complete matrix of FIDS expands exponentially. The uniform sampling of each FID is necessary for the discrete fast Fourier transformation (FFT) algorithm to correctly convert the NMR data from the time-domain to the frequency domain for further analysis. But, acquiring the complete FID matrix overly defines the system and unnecessarily wastes instrument time and prolongs data collection. A simple solution is to randomly sample the FID matrix, usually at a >30% sampling rate, with a bias towards the early time points that have a higher-signal-to-noise [72]. This generally results in a 2- to 3-fold reduction in acquisition time along each indirect dimension. The incomplete FID matrix is then processed using non-FFT methods: filter diagonalization [73], maximum entropy techniques (MEM) [70, 74], multidimensional decomposition (MDD) [75, 76] or non-linear FT techniques [77–79]. The use of non-uniform sampling does introduce artifacts in the NMR spectra [71, 80], which includes ridges, aliasing artifacts, baseline artifacts and folding artifacts that may make it difficult to detect weak signals. The signal to artifact ratio increases as the square-root of the number of data points and artifacts are minimized using a random sampling scheme [72].
In addition to reducing the absolute number of FIDs that are collected, the overall experimental time can also be shortened by decreasing the time required to collect each individual FID [81]. The SOFAST-HMQC [82] pulse scheme permits the collection of a 2D 1H-15N HMQC spectrum in a few seconds by dramatically reducing the recycle time and allowing for high repetition rates. BEST-NMR (Band-selective Excitation Short-Transient) applies the same general concept within standard triple-resonance pulse sequences [81]. Rapid repetition rates are obtained by ensuring efficient T1 relaxation during the significantly shortened recycle delay (50–150 ms) relative to typical recycle delays of 0.75 to 1 second. A further decrease in experimental times could be gained by combining the BEST-NMR technique with non-uniform sampling or reduced dimensionality methods.
Third, the development of software for the semi-automated assignment of NMR spectra and the calculation of protein structures has greatly reduced the time required to generate a NMR protein structure. A number of software packages exist for the semi-automated assignment of protein NMR spectra (see [83] for a detailed review). These include: AutoAssign [84], BATCH [85], GARANT [86], DYNASSIGN [87], GANA [88], MATCH [89], and PISTACHIO [90], among others. Basically, theses programs implement a set of rules that mimic an expert’s approach to the analysis of spin-systems from a standard set of triple-resonance experiments [91]. The spin-systems are then joined by two or more overlapping Cαi, CαHi, C′i, Cβi and Cαi−1, CαHi−1, C′i−1, Cβi−1 chemical shifts. The primary challenge to the approach is due to incomplete data sets (missing peaks), noisy spectra (false peaks) and degeneracy (overlapping peaks). An expert deals with these problems through an iterative visual inspection of the NMR spectra. Conversely, an automated approach is either limited to peak lists or attempts to differentiate between a real peak and noise in the original spectra. Thus, the various software packages apply distinct approaches including exhaustive search algorithms, genetic algorithm, graph theory, heuristic algorithms neural networks, simulated annealing, system energy function or Monte Carlo algorithms in an attempt to overcome incomplete and noisy data. In general, these approaches tend to correctly assign backbone residues 70% to 100% of the time.
There is also a variety of software packages used to automate the calculation of a protein structure from NMR data (see [92] for a detailed review). This includes: AutoStructure [61, 93], ARIA [94], CLOUDS [95], CYANA [96], PASD [97], FLYA [98], and Rosetta [99], to name a few. One primary difference between these programs is the choice of the molecular mechanics/dynamics engine, XPLOR/CNS [100] or DYANA [101]. XPLOR/CNS refines the protein structure in distance space and tends to be significantly slower, but more accurate than DYANA that refines in torsion angle space. Again, the automated approach to refining a protein structure is basically achieved by incorporating rules to simulate the manual analysis of NMR structural data by an expert along with well-defined structural characteristics. For example, secondary structure elements α-helices, β-strands and turns have expected patterns of NOEs. Similarly, there are distinct regions of allowable backbone and side chain dihedral angles. Additionally, long-range contact or distance maps are generally self-consistent. The unambiguous assignment of a long-range NOE between non-sequential residues increases the likelihood that other NOEs between the same two residues or sequential neighbors of the two residues are observed.
Again, a primary challenge to the automated analysis of NMR spectra to generate a protein structure is incomplete and noisy data. Furthermore, the success of a structure determination is predicated on the completeness of the chemical shift assignments, since these assignments are required to correctly annotate the structural NOEs. It has been estimated that 90% completeness of chemical shift assignments are required for an accurate protein structure [102, 103]. Structure determination is an iterative process, where an initial structure is calculated based on a sub-set of unambiguous NOEs and other readily available structural constraints (torsion angles, chemical shifts, coupling constants, residual dipolar coupling (RDC) constants, etc). A consistency with the initial protein structure is then used to filter ambiguous NOEs. The success of the protocol is thus highly dependent on obtaining a correct fold for the initial structure, where either a significant lack of chemical shift assignments or unambiguous NOEs will result in failure [104]. To address these problems, ambiguous NOE methods [94, 96, 97], assignment-free methods [95], RDC-based methods [105, 106], and chemical shift-based methods (CS-ROSETTA) [107] are actively being developed. Protein structures determined solely by chemical shift information is particularly promising and exciting, since the chemical shift information is rapidly obtained and eliminates the need for additional NMR experiments to measure NOEs, coupling constants and RDCs. Protein structures calculated using CS-ROSETTA tend to yield a backbone root-mean squared deviation (rmsd) ≤ 2Å relative to original NMR structures using complete NOE datasets. CS-ROSETTA is currently limited to proteins < 130 residues. Similarly, assessments of protein structures emerging from structural genomics centers, which primarily utilize automated techniques, indicate a higher structure quality relative to NMR structures solved by traditional techniques [39, 108].
The availability of automated software and rapid NMR data collection schemes have resulted in a significant improvement in the throughput of NMR structure determination. This is illustrated by the >300 protein NMR structures generated as part of the Protein Structure Initiative and by a recent example (Figure 2) of eight NMR structures that required, on average, 1–2 weeks to complete each structure. The high-throughput generation of protein NMR structures is still generally restricted to proteins with MW < 30 kDa. But, expanding this limit beyond 50 kDa in the near future appears promising, especially since obtaining backbone chemical shift assignments have been demonstrated for a number of large MW proteins [42, 45, 46]. Obtaining a structure for high MW proteins may be achieved by expanding the CS-ROSETTA methodology to include additional structural constraints such as RDCs, pseudocontact shifts [60], minimal NOE constraints [59] and chemical crosslinking data [109].
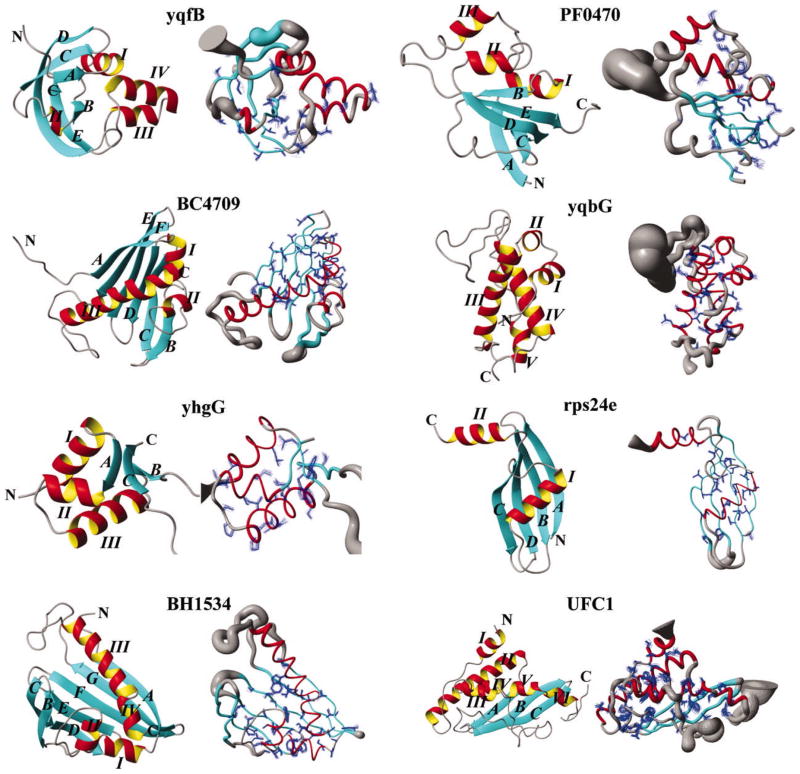
Figure 2.
High-quality NMR solution structures of Northeast Structural Genomics (NESG) consortium target proteins. A methodology for semiautomated data analysis was used to solve the eight NMR protein structures. NMR data collection was accomplished in ~1–9 days per structure and ~1–2 weeks of an expert’s time was required for semiautomated data analysis and structure calculation. For each structure, a ribbon drawing is shown on the left and a “sausage” representation of the backbone is shown on the right where the thickness reflects the precision achieved for the determination of the polypeptide backbone conformation. (Reprinted with permission from reference [64], Copyright 2005 by the National Academy of Sciences of the United States of America)
3. RAPID PROTEIN-LIGAND COMPLEX STRUCTURES
Obtaining a rapid protein structure provides an important foundation for a drug discovery project. It enables the validation of chemical leads through the determination of a protein-ligand co-structure. Confirmation that the compound binds in a biologically relevant region of the protein or induces a conformational change that affects the biological activity of the protein infers a viable lead compound. Again, obtaining this information rapidly is critical to the success of the drug discovery program especially since an iterative series of structures are required to optimize the chemical lead and a limited amount of time is available before the project is terminated. A number of recent techniques have been developed that enable NMR to rapidly determine a structure for a protein-ligand complex.
NMR chemical shift perturbations (CSPs) are routinely used to identify ligand bindings sites based on a clustering of residues on the proteins surface that incur CSPs. Generally, two-dimensional (2D) 1H-15N HSQC/HMQC or 2D 1H-13C HSQC/HMQC spectra are rapidly acquired in minutes (FHSQC) [110] or (SOFAST-HMQC) [82] for both the free protein and the protein-ligand complex. An overlay of the two spectra readily identifies the residues that incur CSPs, which are then simply mapped onto the protein’s surface. Thus, a ligand-binding site is obtained extremely rapidly and the process is automatable with the addition of robotic sample-changers or flow-probes to an NMR spectrometer [111]. It was recently demonstrated that a high-quality protein-ligand complex model can be obtained rapidly by combining traditional in situ ligand docking software with experimental NMR CSPs [112]. Specifically, the CSP identified ligand binding site is used to define a three-dimensional (3D) search-grid for AutoDock 4, which is the highest-cited ligand docking software package [113, 114]. AutoDock searches the 3D grid using a Lamarckian genetic algorithm to explore different translational, rotational, and torsional orientations for the ligand and estimates a free energy of binding. AutoDock is optimized for ligand-binding energetics and includes numerous terms related to dispersion/repulsion, directional hydrogen bonding, electrostatics, desolvation, and conformational energy. Typically, 120 different ligand conformations are calculated within the 3D search-grid, where the program AutoDockFilter (ADF) is used to select the best conformers based on a consistency with the experimental CSPs. ADF uses an NMR energy function based on the magnitude of the CSPs and the proximity of the ligand to amino-acid residues with a CSP. Simply, the ligand is expected to be closer to amino acids residues that incurred large CSPs. On average, the approach takes ~ 40 minutes to obtain a protein-ligand co-structure, where the resulting structure exhibits an average rmsd of 1.17 ± 0.74 Å relative to the original x-ray structure. Figure 3A illustrates the high similarity between the CSP-guided docking of thymidine 3′,5′-bisphosphate to S. aureus nuclease relative to the original x-ray structure (PDB-ID: 1SNC) [115].
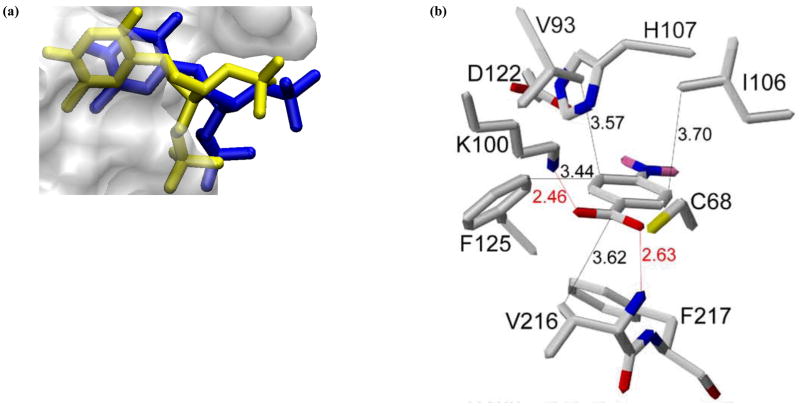
Figure 3.
(a) Superposition of the CSP-guided docking with ADF filtering conformers (blue) with the original x-ray structures (yellow) for S. aureus nuclease complexed to thymidine 3′,5′-bisphosphate (PDB-ID: 1SNC) (Reprinted with permission from reference [112], Copyright 2008 by American Chemical Society). (b) Expanded view of the HADDOCK model of human NAT1 bound to its substrate PABA. Carbon, nitrogen, oxygen, sulfur, and hydrogen atoms are colored white, blue, red, yellow, and pink, respectively. The lengths of several hydrophobic contacts are provided and the hydrogen bonds to the carboxylic acid group of PABA are indicated in red. (Reprinted with permission from reference [118], Copyright 2006 by Elsevier Ltd.)
Similarly, the program HADDOCK takes advantage of this relationship between CSPs and ligand binding to generate biomolecular complexes (protein-protein, protein-DNA, protein-RNA) [116]. HADDOCK utilizes ambiguous interaction constraints (AIR) [117] to refine a biomolecular complex using a three stage refinement: (i) a rigid body docking and energy minimization, (ii) a semirigid simulated annealing in torsional angle space and (iii) a final refinement with explicit solvent. Basically, HADDOCK defines an ambiguous intermolecular distance (≤ 3Å) between all sets of residues that incurred a CSP from molecule A to molecule B in the complex. HADDOCK has recently been applied to the docking of small molecular-weight compounds to proteins [64]. A clear advantage of the HADDOCK approach is the fact that the resulting co-structure is a result of a direct refinement against the experimental CSPs. But, HADDOCK uses XPLOR/CNS [100], which is optimized for protein refinement and dynamics, making it challenging to properly parameterize each new compound that is docked. Also, HADDOCK applies a more robust, multistage minimization and dynamics protocol resulting in a significantly longer computational time (~ 2 days) compared to AutoDock/ADF. Figure 3B illustrates the application of HADDOCK for the determination of the structure of human arylamine N-acetyltransferase (NAT) complexed with p-aminobenzoic acid (PABA) [118]. The clear advantages of the CSP approaches to determining a protein-ligand model is the speed, ease and relative simplicity of the techniques. But, these modeled structures should not be viewed as a replacement for traditional high-resolution protein-ligand complexes obtained by either NMR or x-ray crystallography for drug discovery. Instead, these CSP modeled structures provide a rapid approach to evaluate potential drug discovery targets before pursuing the more time-consuming and resource intensive experimental structures. Importantly, the CSP methods are restricted to proteins that do not undergo significant conformational changes upon ligand-binding. This is especially valid if only the apo-structure of the protein is available for modeling, since either a static version of the protein structure or a structure with limited side-chain mobility is used for ligand docking.
In addition to AutoDock/ADF and HADDOCK, other NMR approaches have been described to shorten the time-frame to determine a protein ligand co-structure. These methods are, in general, significantly slower, more complex to execute (requiring a variety of experiments and/or labeled samples), or limited in scope (requiring multiple known binders or unique site-specific labeling). But, the relative resolutions of some of these structures are increased compared to CSP-based structures. Again, these techniques are typically limited to protein structures that do not undergo significant conformational changes upon ligand binding since a static protein structure is used to model the protein-ligand complex. These alternative methods include NOE based protein-ligand models [119, 120], paramagnetic shifts [121], differential chemical shift perturbations [122], SOS-NMR [123], NMR-SOLVE [124], and NMR-DOC [125]. The NOE based protein-ligand model utilizes a refined NMR apo-protein structure and a minimal NMR dataset to assign intermolecular NOEs. Recent techniques minimize the need to re-assign the protein NMR spectra in the complex [126]. These NOEs are simply added to the complete set of structural constraints used to solve the apo-structure in order to solve the complex structure. The differential chemical shift approach uses a series of structurally similar ligands known to bind the protein. The differences in structure and CSPs are expected to correlate, which then identifies each ligand’s orientation within the similar binding pocket. The NMR-DOC approach is essentially identical, but simplifies the protein NMR spectra by using a series of residue-specific 13C-methly labeled protein samples. The NMR-SOLVE method is also very similar, it uses a reference ligand (natural cofactor) to identify chemical shifts associate with the ligand binding site. This information is then used to map the location and orientation of novel compounds.
The paramagnetic shifts and SOS-NMR methods also utilize selective labeling to identify the structure of the protein ligand complex. The SOS-NMR method uses a series of residue specific labeling to identify which residues are proximal to the ligand similar in concept to chemical shift perturbations. Every residue type except one is deuterium labeled. If a saturation transfer difference is still observed, the unlabeled residue is proximal to the ligand. The process is repeated for each relevant amino-acid. The paramagnetic shifts approach requires a site-specific lanthanide label proximal to the ligand binding site. The lanthanide binding site is identified from an x-ray crystal structure and Δχ tensors are determined from chemical shift changes in 2D 1H-15N HSQC spectra for the protein complexed to paramagnetic and diamagnetic lanthanides. Pseudocontact shifts (PCSs) are then measured for the ligand in the presence of the protein-lanthanide complex from 1D 1H and 13C NMR spectra. The PCSs are used to position the 1H and 13C nuclei from the ligand relative to the Δχ tensors to dock the ligand.
4. FRAGMENT-BASED LIGAND SCREENS
Traditional HTS is routinely used in the pharmaceutical industry to screen hundreds of thousands to millions of compounds against a therapeutic protein target [26]. But, HTS has an extremely high failure rate. Only 0.1 to 0.5 percent of the compounds in the screening library will be “active” in a particular screen [127]. A significant percentage of the best “hits”, highest observed inhibition, are false positives [30]. These compounds generate an HTS response through a number of undesirable mechanisms, such as: protein aggregation, protein denaturation, protein precipitation, micelle formation, chemical modification of the protein, non-specific binding, promiscuous binders or interfering with other reagents of the assay [28–31, 128]. These problematic chemical leads inevitably results in a significant amount of wasted resources and time.
Intrinsically linked to the success of HTS is the composition of the screening library. Obviously, the quality of “hits” identified from an HTS assay is dependent on the quality of compounds present in the library [32]. The accumulation of corporate chemical libraries occurs over years if not decades where the composition of the library is inherently biased to previous drug discovery projects. The analysis of typical large, random chemical libraries compared to the collection of known drugs identified a number of problems associated with corporate libraries while identifying a set of structural properties or Lipinski’s “rule of 5” that are exhibited by known drugs [129, 130]. Lipinski’s “rule of 5” is a general predictor for compound solubility and permeability that is related to bioactivity and bioavailability [131]. In contrast, compounds in chemical libraries have a tendency to trend toward higher molecular weight, higher lipophilicity and/or higher H-bond properties resulting in either poor solubility or permeability.
An outcome of these studies has been an extensive effort to design or reconfigure chemical libraries based on the Lipinski’s “rule of 5” or similar predictors of “drugability” [32, 132–135]. Concurrent with drug-like features are additional approaches to build in structural diversity in the design of chemical libraries [134, 136–140]. But, estimates indicate that approximately 1010 to 1050 compounds would be needed to properly cover structural space [141]. Regardless of the size of the chemical library, the number of compounds that can be practically screened represents an infinitesimally small fraction of the total number of potential compounds (1060) [142]. Concurrent with any effort to increase the size of the library, will be a proportional increase in the total number of troublesome false leads. From a practical perspective, it is not possible to follow-up all the hits identified from an HTS assay. Since false positives tend to over-populate the best inhibitors identified from an HTS assay, more effort is spent chasing after false leads [143]. Another practical consideration is that the integrity of individual compounds used in the HTS screen may be suspect because of issues associated with long-term storage of compounds in conditions amenable to HTS [144, 145]. Given these challenges, the decline in the productivity of the pharmaceutical industry has been attributed, in part, to the heavy reliance of HTS in the drug discovery process [146]. Fragment-based ligand screens provide an alternative to HTS and address a number of these problems [147]. NMR is the primary method being used to screen fragment-based libraries [148].
4.1 Library Design
Chemical libraries used for fragment-based screening are significantly different from the typical library used in a traditional HTS assay. This is because the goals are fundamentally different. In an HTS assay, the aim is to identify 3–5 chemical classes with the best binding affinity (KD ≤ μM) to the therapeutic target. These compounds go through an iterative series of chemical modifications to maximize binding affinity. Unfortunately, this process generally results in an increase in hydrophobicity and molecular-weight and a decrease in solubility [149]. Both factors are detrimental for evolving the lead compound into a drug. Critical absorption, distribution, metabolism, and excretion (ADME) and pharmacokinetic (PK) [150, 151] issues are typically dealt with at a later point in the drug discovery process [152, 153]. Conversely, the goal of a fragment-based screen is to identify a number of weak binders (KD ~μM to mM) that bind in separate but proximal sub-regions of a functionally relevant ligand binding site. In effect, the ligand-binding site is chemically mapped with low molecular-weight compound fragments (≤ 200–300 Da) [154, 155]. Chemical linking and optimization of the individual fragments generally results in a substantial improvement in binding affinity, where KDs in the nM range are commonly observed [156]. Importantly, an additional goal of the fragment-based approach is to maintain drug-like ADME and PK characteristics. This is a fundamental consideration in the design of the fragment-based screening library.
A number of reviews have discussed in detail the underlying theory in the construction of a fragment-based chemical library [154, 155, 157–159]. Basically, the objective is to identify small molecular-weight compounds (≤ 200–300 Da) that correspond to fragments of known drugs, adhere to the Lipinski “Rule of 5” and exhibit high aqueous solubility. A comparable approach is using a fragment-based library composed of biologically active compounds [160]. The relatively small libraries are usually composed of hundreds to thousands of compounds. Maintaining a chemical library of small molecular-weight compounds has a number of advantages that address common problems associated with HTS. First, it maximizes ligand efficiency (LE), the number of nonhydrogen atoms relative to free energy of binding [161]. The estimated maximum affinity per atom is ~1.5 kcal mol−1, which is approached by known drugs and fragments. Conversely, typical HTS leads exhibit an LE of ~ 0.3 kcal mol−1 [159]. Second, fragment-based libraries have a 10–1,000 times higher hit rate relative to traditional HTS corporate libraries [154]. Third, fragment-based libraries more efficiently cover structural space. This occurs because the number of low molecular-weight compounds is significantly reduced. Instead of a potential 1060 compounds that a HTS library is compared against, there are only ~ 14 million compounds with molecular-weights below 160 Da [162]. Thus, even a few thousand compounds represent a significant percentage of the possible number of compounds for a fragment-based library, which cannot be reasonably approached with higher-molecular weight HTS libraries. Additionally, since fragments are commonly linked to generate chemical leads, a fragment-based screen is actually examining a combinatorial number of compounds relative to the actual size of the library that is experimentally tested [163]. Fourth, smaller chemical libraries are significantly easier to handle, store, replenish and maintain quality control. Finally and most importantly, fragment-based libraries generate better-quality leads with a higher success rate [164]. This results from the smaller size of the fragments and the associated “drug-like” features. The smaller structures permit a greater flexibility to “grow” the fragments to improve binding affinities before inducing detrimental physiochemical characteristics that is common with HTS leads.
4.2 NMR Ligand Screening Methodologies
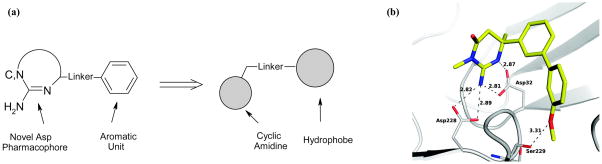
High-throughput NMR (HTS-NMR) ligand affinity screens cannot compete with the efficiency of traditional HTS that routinely assay millions of compounds in a few weeks [165]. Nevertheless, HTS-NMR is perfectible amenable to screening a fragment-based library composed of hundreds to thousands of compounds [148, 164, 166]. The value of NMR for HTS is its uniquely capability of providing direct evidence for binding between the ligand and protein target while subsequently being able to identify the binding site and determining a co-structure of the protein-ligand complex [91, 167–170]. An important advantage of NMR is the versatility of the technique to monitor protein-ligand interactions through a variety of physical measurements. Observation of a binding event may occur through changes in line-width and/or peak intensity (T1 and T2 relaxation changes) [171, 172], change in the measured diffusion coefficient for the ligand [173–175], chemical shift perturbations for either the ligand or protein [176–178], induced transferred NOE (trNOE) for the ligand [179, 180], a saturation transfer difference (STD) between either the protein or bulk solvent to the ligand [181, 182], or the appearance of new NOEs and/or intermolecular NOEs between the ligand and protein [167, 183]. Correspondingly, a variety of different NMR-based screens have been described (Table 1). The various HTS-NMR methods have a range of distinct advantages that include: (i) eliminating a need for isotopically labeled proteins, (ii) reducing the amount of protein required for the screen, (iii) increasing the throughput of the assay, (iv) identifying the ligand-binding site, (v) determining a protein-ligand co-structure, and (vi) eliminating the protein MW limitation. NMR fragment-based screens have resulted in a number of chemical leads that have advanced to clinical studies [164]. A recent example is the discovery of novel β-secretase inhibitors [184]. Cyclic amidine with an isocytosine core fragments were identified from an NMR screen as a lead hit. The further optimization of these fragments is illustrated in Figure 4a, which lead to a unique dihydroisocytosine inhibitor with a cellular activity of 470 nM (Figure 4b).
TABLE 1.
| Screening Technique | Method of Detecting Ligand Binding | Labeled Protein? | Protein-Ligand Co-structure? | Limited by Protein MW? | Ref. |
|---|---|---|---|---|---|
| 3-FABS | Chemical shift changes, requires fluorinated ligands | No | No | No | [252] |
| Affinity NMR | Change in translational diffusion | No | No | No | [174] |
| AIDA-NMR | Line-broadening change (T2) due to protein-protein complex formation, labeled protein or Trp reporter in ligand binding site | Yes/No | Yes/No | Yes | [253, 254] |
| FAXS | Line-broadening change (T2) due to ligand competition, requires fluorinated ligands | No | No | No | [255] |
| MS/NMR | Retention on size-exclusion column & chemical shift changes | Yes | Yes | Yes | [256] |
| Multi-Step NMR | Line-broadening change (T2) & chemical shift changes | Yes | Yes | Yes | [237] |
| NOE pumping | Transfer nuclear Overhauser effect (NOE) | No | No | No | [183] |
| RAMPED-UP NMR | Chemical shift changes, screening multiple proteins | Yes | No | Yes | [257] |
| SAR by NMR | Chemical shift changes | Yes | Yes | Yes | [258] |
| SLAPSTIC | Line-broadening change (T2) due to protein spin label | Yes | No | No | [259] |
| STD NMR | Saturation transfer difference from protein | No | No | No | [181] |
| WaterLOGSY | Saturation transfer difference from solvent | No | No | No | [182] |
Figure 4.
(a) General fragment-based scheme using a novel cyclic amidine-based aspartyl protease pharmacophore to generate BACE-1 inhibitors. (b) BACE-1 inhibitor complex. (Reprinted with permission from reference [184], Copyright 2007 by American Chemical Society)
5. METABOLOMICS
The discovery of a compound with a high-affinity to a therapeutic target does not necessarily lead to a marketable drug. Chemical leads routinely fail in pre-clinical and clinical trials because of problems with bioavailability, efficacy and toxicity [6, 7]. Typically, in vivo assays and animal studies attempt to identify these problems prior to proceeding with clinical trials. But, animal studies also have significant practical limitations [185–187]. They are fundamentally low-throughput, requiring kilogram quantities of the lead compound and weeks to months to complete a single study. Furthermore, there is typically little SAR or feed-back when a negative result is encountered, since such a small number of compounds are routinely evaluated. NMR-based metabolomics is providing drug-discovery with a filter to identify in vivo activity, selectivity and toxicity problems before proceeding with animal trials. Furthermore, NMR metabolomics techniques can be used as part of an animal study to identify and evaluate the mechanisms of drug induced toxicity. Similarly, mass spectrometry is also routinely used to monitor the metabolome [188, 189] and is complimentary to NMR [190]. MS is significantly more sensitive than NMR, but requires the inclusion of separation techniques because of the low molecular-weight diversity of metabolites and is limited to detecting metabolites that readily ionize. Conversely, NMR typically only observes the most abundant metabolites, but requires minimal sample preparation and can readily identify each metabolite from the multiple distinct peaks in its NMR spectra. Correspondingly, a number of techniques have been proposed that combine NMR and MS data for the analysis of metabolomic samples [190–195]. The application of NMR metabolomics in drug discovery has been recently reviewed [196–198] and will only be briefly summarized here.
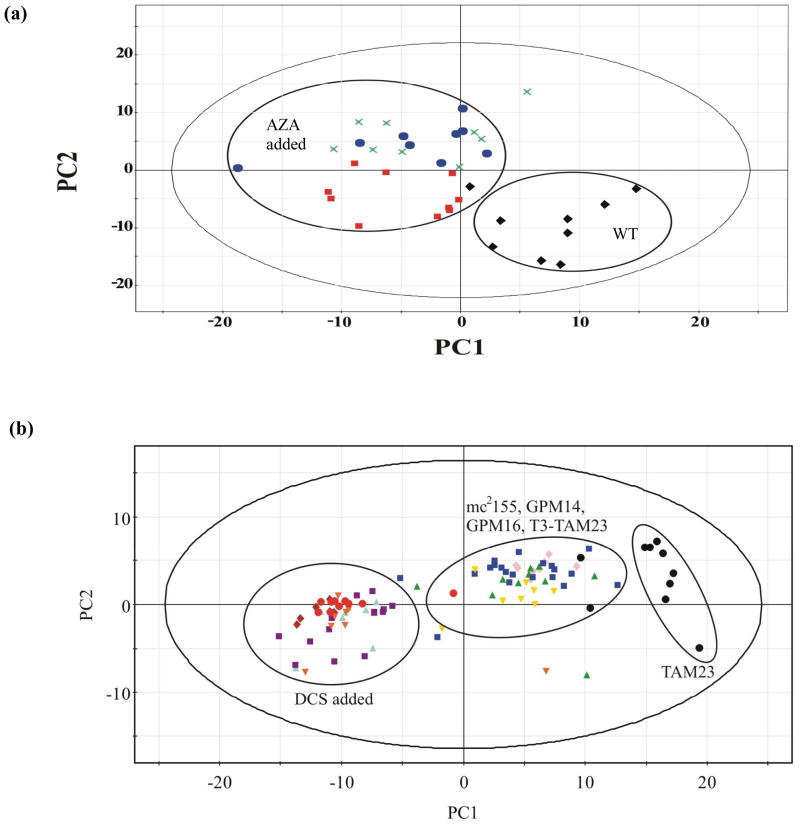
Basically, a 1D 1H NMR spectrum of a lysed cell captures its metabolic state. The metabolome directly measures the biological activity of the proteome since relative concentrations of various metabolites are dependent on enzymatic activity. Peak intensities in an NMR spectrum reflect both the abundance and the presence of specific metabolites and the corresponding activity of an enzyme. Environmental stress and drug activity will perturb protein activity resulting in changes in the metabolome that can be monitored by NMR. Principal component analysis (PCA) is a well-established statistical technique that is commonly used to analyze 1D 1H NMR spectra to monitor these perturbations in the metabolome [199]. The multivariate NMR spectrum is converted to a single point in PC-space that is usually represented by a 2D scores plot. Similar NMR spectra will cluster together in a 2D scores plot. The differential NMR metabolomics approach [200, 201] uses comparative clustering patterns in a 2D scores plot to monitor the in vivo activity and selectivity of a chemical lead. Simply, four different cell cultures are prepared with ten replicates each for a total of forty NMR samples. The four cultures correspond to: (i) wild-type cells, (ii) wild-type cells treated with the chemical lead, (iii) mutant cells where the protein target of the chemical lead has been inactivated, and (iv) mutant cells treated with the chemical lead. The metabolome for the wild-type cells and mutant cells will be different because of the inactivated protein resulting in distinct clustering in a 2D scores plot. The relative clustering of the wild-type cells and mutant cells treated with the chemical lead will determine the compound’s in vivo activity and selectivity. Specifically, if the wild-type cells treated with the chemical lead cluster together with the mutant cells with and without the addition of the chemical lead, this would then indicate that the chemical lead is active and selective in vivo. Inhibiting the cellular activity of the therapeutic target either chemically or genetically results in the same change in the metabolome. Different clustering patterns occur in the 2D scores plot if the chemical lead is inactive, inhibits multiple proteins or inhibits a different protein from the expected protein target [201]. Any of these outcomes would clearly indicate a problem, suggesting that other compounds with a positive response from the differential NMR metabolomics method should be prioritized for follow-up animal studies. The method has been successfully applied to demonstrate the selective activity of 8-azaxanthine against urate oxidase in Aspergillus nidulans (Figure 5A) and the promiscuous activity of D-cycloserine in Mycobacterium smegmatis (Figure 5B).
Figure 5.
(a) Analysis of the in vivo activity of 8-azaxanthine (AZA) in A. nidulans targeting urate oxidase. The PCA scores plot comparing A. nidulans inactive urate oxidase mutant (uaZ14) ( ), wild-type with AZA (
), wild-type with AZA ( ), uaZ14 mutant with AZA (
), uaZ14 mutant with AZA ( ), and wild-type cells (◆). Results clearly demonstrate the selective activity (see Figure 7b) of AZA (Reprinted with permission from reference [201], Copyright 2006 by American Chemical Society). (b) Analysis of the in vivo activity of D-cycloserine (DCS) in mycobacteria targeting alanine racemase. PCA scores plot comparing wild-type (mc2155) (
), and wild-type cells (◆). Results clearly demonstrate the selective activity (see Figure 7b) of AZA (Reprinted with permission from reference [201], Copyright 2006 by American Chemical Society). (b) Analysis of the in vivo activity of D-cycloserine (DCS) in mycobacteria targeting alanine racemase. PCA scores plot comparing wild-type (mc2155) ( ), inactive D-alanine racemase mutant (TAM23) (●), DCS resistant mutants (GPM14 (
), inactive D-alanine racemase mutant (TAM23) (●), DCS resistant mutants (GPM14 ( ), GPM16 (
), GPM16 ( )), restored D-alanine racemase activity mutant (TAM23 pTAMU3) (
)), restored D-alanine racemase activity mutant (TAM23 pTAMU3) ( ) mc2155 with DCS (
) mc2155 with DCS ( ), and TAM23 with DCS (
), and TAM23 with DCS ( ), GPM14 with DCS (
), GPM14 with DCS ( ), GPM16 with DCS (
), GPM16 with DCS ( ), and TAM23 pTAMU3 with DCS (
), and TAM23 pTAMU3 with DCS ( ). The results clearly demonstrate the active, non-selective inhibition of DCS (see Figure 7c). The secondary target of DCS is predicted to be D-alanine-D-alanine ligase (Reprinted with permission from reference [200], Copyright 2006 by American Chemical Society).
). The results clearly demonstrate the active, non-selective inhibition of DCS (see Figure 7c). The secondary target of DCS is predicted to be D-alanine-D-alanine ligase (Reprinted with permission from reference [200], Copyright 2006 by American Chemical Society).
The differential NMR metabolomics method is limited to disease states that can be model by a cell system. Also, the technique requires generating a viable mutant cell line where the activity of the target protein has been eliminated. This may be challenging if the protein is critical or essential to the cell’s viability. In these cases, a number of alternate strategies can be employed instead of simply generating a knock-out mutant. These include: a mutant protein with only diminished activity, supplementing the cell media or growth conditions to compensate for the inactive protein, treating the cells with sub-lethal dosages of siRNA or a known inhibitor of the target protein. The differential NMR metabolomics method is also predicated on the assumption that inactivating the target protein will perturb the metabolome in a manner that is detectable by NMR. It is plausible that eliminating the biological activity of a particular protein will not disturb the cellular metabolome or it may affect the relative flux of metabolites in a concentration range that is too low to be detectable by NMR.
NMR metabolomics can also be used to monitor body fluids (blood, urine, saliva, etc) to identify drug induced toxicity in animal trials. This can be achieved by the identification of metabolites known to be associated with toxicity, such as trimethylamine N-oxide and renal dysfunction or lipids and phospholipidosis [202–205]. Alternatively, a comparative analysis with compounds known to induce toxicity with other compounds with no known side-effect can be used to generate a predictive model for unknown compounds based on the correlated changes in the metabolome and NMR spectrum [202, 206]. Again, the method assumes that the metabolites associated with a toxic event are in a concentration range observable by NMR and present in the body fluid being examined. Thus, a lack of a change in the metabolite composition and concentration in a body fluid would only eliminate toxicities previously demonstrated to be observed by NMR for compounds with known toxic side-effects. Additionally, the metabolome is not completely defined and a reference spectrum for every observable metabolite is not currently available. Therefore, identifying every metabolite that is perturbed by treatment with a drug candidate may not be readily obtained. Nevertheless, there is an ongoing effort to catalog the NMR observable metabolites and there are a number of NMR metabolomic databases currently available [207–209].
NMR-based metabolomics have two important contributions to the current drug discovery protocol. First, the differential NMR metabolomics method could serve as a filter before proceeding with in vivo animal trials with lead candidates. Only compounds that proved to be active and selective for the desired protein target would continue on for animal testing. Other compounds would be eliminated or proceed through further rounds of chemical optimization. Changes in the structure that do not affect binding affinity, but are correlated with preferred activity in the differential NMR metabolomics screen could be pursued. Second, NMR-based metabolomics would augment animal studies. Initially, in vivo animal models are primarily focused on efficacy. The efficiency of these assays would be greatly improved by simultaneously analyzing animal biofluids by NMR for toxicity. Animal-based studies are typically time intensive assays requiring weeks to complete and usually result in sacrificing the animals. Furthermore, a detailed autopsy may be required to analyze the efficacy of the drug and identify any potential toxicity issues. NMR-based metabolomics may effectively identify a toxicity problem early enough in the study to stop the test, saving valuable time and resources. Alternatively, the lack of any observable toxicity problem by NMR in combination with an overall positive outcome for a particular lead candidate in an animal assay would support proceeding with NCE and clinical trials.
6. FUNCTIONAL ANNOTATIONS AND NEW THERAPEUTIC TARGETS
To date, 916 genomes have been completely sequenced with an additional 3,454 in progress resulting in ~6.5 million protein sequences [210–214]. Sequence similarity techniques may provide functional information for, at most, 50% of these proteins [215–217]. Valuable therapeutic targets are inevitably hidden within these vast numbers of unannotated proteins, providing an indispensable wealth of information for developing novel drugs [218–220]. Assigning a function is an essential first step for determining the potential utility of an unannotated protein in treating human disease; but this is an extremely challenging and time-consuming endeavor. Drug discovery cannot simply focus on the crucial or essential proteins since non-essential proteins play important roles in antibiotic resistance, cancer and tumor differentiation and metastasis, and the long-term development of Alzheimer, cardiovascular disease, diabetes and Parkinson disease. NMR is also making a significant contribution to the functional assignment of these unannotated proteins to identify new drug discovery targets.
The Functional Annotation Screening Technology using NMR (FAST-NMR) [221, 222] combines the NMR methodology described above in a single assay to assign a function through similarities in functional epitopes or ligand binding sites. This is based on the premise that amino-acid residues associated with active-sites are evolutionary stable relative to the remainder of the protein’s sequence [223–225]. Essential to this understanding is the knowledge that a protein’s active-site has been optimized by nature to interact with a unique and specific set of targets. Protein surfaces are exquisitely selective and only bind ligands at very specific locations [226–229]. Binding promiscuity is inherently detrimental to the overall biological process, which is evident by the high specificity of interactions that have been well-documented in numerous metabolic and signaling pathways [230–232]. This understanding is also an essential aspect of drug discovery and supports the observed rationale that high-affinity and selective compounds targeting a specific protein can be developed and used therapeutically [233–236].
FAST-NMR combines fragment-based ligand affinity screens with rapid determination of protein-ligand co-structures and bioinformatics. Specifically, a chemical library containing only biologically active compounds (co-factors, drugs, metabolites, inhibitors, substrates, etc) [160] are screened by NMR using a tiered approach [237]. Potential ligands are rapidly identified by a 1D 1H line-broadening experiment. These hits are then confirmed by chemical shift perturbations in a 2D 1H-15N HSQC spectrum, where the CSPs cluster together to identify a specific ligand binding site. The CSPs are then used to quickly determine a protein-ligand co-structure [112] that provides the input for the CPASS (Comparison of Protein Active-Site Structures) software and database [238]. CPASS compares the experimentally determined ligand-defined binding site against a database of ~35,000 unique ligand binding sites identified from the Protein database (PDB). A similarity in the sequence and structural characteristics of the ligand-defined binding site from the unannotated protein with a ligand-binding site for a protein of known function is then used to leverage a functional assignment.
FAST-NMR and CPASS were used to assign a function to hypothetical proteins SAV1430 from S. aureus [222] and PA1324 from Pseudomonas aeruginosa [239]. This analysis suggests SAV1430 is similar to an SH2 domain and may function as part of a multi-protein complex within the [Fe-S] cluster assembly network. SAV1430 may exhibit activity comparable to NifU or may regulate NifU activity. PA1324 may be involved in the binding and transport of sugars or polysaccharides as part of the peptidoglycan matrix required for biofilm formation. FAST-NMR has also been used to identify an evolutionary relationship between the bacterial type III secretion system and eukaryotic apoptosis [240]. This is based, in part, on the similarity in ligand-binding sites between PrgI (forms the needle complex in the type III secretion system) and Bcl-xL (involved in eukaryotic apoptosis).
7. EXPERT OPINION
The quantity of new drugs emerging from the pharmaceutical industry has declined dramatically. A number of self-inflicted factors have contributed to this decrease in productivity that has resulted from a need to decrease cost and, ironically, from a need to increase efficiency. Traditional business practices that include outsourcing, metrics and short timelines have been implemented into the drug discovery process. Scientific discovery is effectively being forced to adhere to an assembly-line protocol [23], isolating disciplines, diminishing creativity and negatively affecting morale [5]. This is antithesis to a successful drug discovery effort, which is fundamentally interdisciplinary, complex and unpredictable. Resolving these serious problems will require a major paradigm shift in the culture of the pharmaceutical industry, nevertheless evolving drug discovery technologies are providing avenues to improve the success rate [164] within these self-imposed limitations.
In this context, recent developments in NMR are greatly expanding its value to the drug discovery process. Specifically, NMR is a valuable and complimentary technique to x-ray for the high-throughput structure determinations of protein and protein-ligand complexes. It has been recently demonstrated that a significant percentage (20–40%) of protein structures are only solvable by NMR. Also, rapid (30–45 minute) protein-ligand co-structures are routinely obtainable by NMR as part of an iterative drug design program. This speed is essential given the short timelines imposed on a drug discovery project. Additionally, fragment-based NMR affinity screens are providing a more efficient and productive alternative to traditional HTS for lead generation. A fundamental advantage of NMR affinity screens is the direct knowledge of the chemical lead’s mechanism of action. This occurs because an NMR affinity screen identifies a specific binding interaction between the ligand and protein target, where the ligand binding site can be correlated with the functionally relevant regions of the protein. Furthermore, NMR-based metabolomics can assisted in the most challenging and problematic stage of the drug discovery process, turning a high-affinity ligand into a drug. This requires verification of in vivo efficacy with a corresponding lack of toxic side-effects and acceptable ADME profiles. Animal models are the primary source of this information, but are extremely costly and time consuming significantly limiting the number of leads that can be practically evaluated. Differential NMR metabolomics provides a simple, cheap and fast approach to filter-out problematic compounds and prioritize leads that demonstrate cellular activity and specificity while confirming the in vivo target(s) of the compound. Similar, NMR metabolomics can be incorporated as part of the animal trials to monitor body fluids and ascertain possible toxic side-effects of chemical leads. NMR is also being used to functionally annotate the proteome and identify novel drug discovery targets.
Despite the obvious important contributions of NMR to the drug discovery process, the pharmaceutical industry has reduced or completely eliminated biomolecular NMR groups over the last number of years. Again, this short-sighted decision has been primarily driven by cost and mergers, by the incorrect perspective that NMR is a redundant, but costlier and limited version of x-ray crystallography, and by the large financial investment in HTS robotics along with the perspective that weak-binding fragments are not viable drug leads. The pharmaceutical industry, like other businesses, is cyclical and responds to short-term needs instead of long-term goals. Thus, I believe the reduction in biomolecular NMR has bottom-out and this trend will begin to reverse. This will be driven, in part, by the growing acceptance and success of fragment-based ligand screens. Furthermore the achievements from structural genomics and the NIH Protein Structure Initiative along with the new NMR metabolomics technologies will further stimulate a renewed interest in biomolecular NMR by the pharmaceutical industry.
Finally, our basic understanding of the mechanism for generating a protein-ligand complex is undergoing a dramatic shift from the current induced-fit model or two-state model [241]. The recent application of relaxation dispersion NMR spectroscopy [242, 243] and RDCs [244] has supported the selected-fit model of ligand binding, where protein structures exist as an ensemble of conformations that includes sampling a higher-energy ligand bound form in the apo-state. The role of both protein and ligand dynamics [245–247] have been recognized as important factors in drug discovery, but have been rarely utilized [248]. Clearly, structure-focused drug discovery efforts will greatly benefit from the inclusion of a detailed NMR analysis [249] of the flexibility-function relationship. The growing recognition of the role dynamics plays in ligand binding is also expected to contribute to a revitalize interest and an improved appreciation by the pharmaceutical industry of the vital role for NMR in drug discovery.
A reversal in the decade-long decrease in the productivity of the pharmaceutical industry will obviously require more than the just the simple adoption of these recent advances in NMR. A major philosophical shift in the current approach to drug discovery is necessary. One potential avenue is for the pharmaceutical industry to move beyond the target-focused approach to drug discovery [1] and to adopt system biology techniques [250]. Basically, chemical genetics [251] is used to identify desirable phenotypic responses in cell-based disease models followed by the identification of the molecular target of the chemical probes. Once the target is identified and verified, traditional drug optimization techniques would be applied in an iterative fashion using positive feed-back from the original cell-based assays. NMR-based metabolomics would play an integral component of chemical genetics as an approach to classify and differentiate between different cellular phenotypes. NMR ligand affinity screens and NMR structural biology techniques would continue to play important roles in the drug optimization stage of the process. Thus, the diversity and flexibility of NMR will enable the technique to continually expand its contribution to drug discovery as the pharmaceutical industry responds to business pressures and progresses beyond current drug discovery paradigms.
Abbreviations
- 1D
one-dimensional
- 2D
two-dimensional
- 3D
three-dimensional
- ADF
AutoDockFilter
- ADME
absorption, distribution, metabolism, and excretion
- AIR
ambiguous interaction constraints
- APSY
automated projection spectroscopy
- AZA
8-azaxanthine
- BACE-1
β-secretase
- CDA
chenodeoxycholic acid
- cL-BABP
chicken liver bile acid binding protein
- CPASS
comparison of protein active-site structures
- CSPs
chemical shift perturbations
- DCS
D-cycloserine
- FAST-NMR
functional annotation screening technology using NMR
- FDA
US food and drug administration
- GFT-NMR
G-matrix Fourier transform NMR
- HIFI-NMR
high-resolution iterative frequency identification
- HMQC
heteronuclear multiple quantum coherence
- HSQC
heteronuclear single quantum coherence
- HTS
high-throughput screening
- HTS-NMR
high-throughput NMR
- LE
ligand efficiency
- mut
mutant
- MW
molecular weight
- MS
mass spectrometry
- NCE
new chemical entity
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- PC
principal component
- PCA
principal component analysis
- PCSs
pseudocontact shifts
- PDB
Protein database
- PK
pharmacokinetic
- PR-NMR
projection-reconstruction NMR
- PSI
Protein Structure Initiative
- RDC
residual dipolar coupling
- RD-NMR
reduced-dimensionality NMR
- RMSD
root-mead square deviation
- SAR
structure-activity relationship
- STD
saturation transfer difference
- trNOE
transferred NOE
- wt
wild-type
References
- 1.Sams-Dodd F. Target-based drug discovery: is something wrong? Drug Discovery Today. 2005;10(2):139–47. doi: 10.1016/S1359-6446(04)03316-1. [DOI] [PubMed] [Google Scholar]
- 2.Garnier J-P. Rebuilding the R&D engine in big pharma. Harv Bus Rev. 2008;86(5):68–70. 2–6, 128. [PubMed] [Google Scholar]
- 3.Weisbach JA, Moos WH. Diagnosing the decline of major pharmaceutical research laboratories: a prescription for drug companies. Drug Dev Res. 1995;34(3):243–59. [Google Scholar]
- 4.Horrobin DF. Realism in drug discovery-could Cassandra be right? Nat Biotechnol. 2001;19(12):1099–100. doi: 10.1038/nbt1201-1099. [DOI] [PubMed] [Google Scholar]
- 5.Cuatrecasas P. Drug discovery in jeopardy. J Clin Invest. 2006;116(11):2837–42. doi: 10.1172/JCI29999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kola I, Landis J. Opinion: Can the pharmaceutical industry reduce attrition rates? Nature Reviews Drug Discovery. 2004;3(8):711–6. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 7.Caldwell GW, Ritchie DM, Masucci JA, Hageman W, Yan Z. The new pre-preclinical paradigm: compound optimization in early and late phase drug discovery. Current Topics in Medicinal Chemistry (Hilversum, Netherlands) 2001;1(5):353–66. doi: 10.2174/1568026013394949. [DOI] [PubMed] [Google Scholar]
- 8*.Dickson M, Gagnon JP. Key factors in the rising cost of new drug discovery and development. Nat Rev Drug Discovery . 2004;3(5):417–29. doi: 10.1038/nrd1382. Provides a detailed analysis of the costs associated with the various stages of the drug discovery process. [DOI] [PubMed] [Google Scholar]
- 9.Karlberg JPE. Trends in disease focus of drug development. Nat Rev Drug Discovery. 2008;7(8):639–40. doi: 10.1038/nrd2618. [DOI] [PubMed] [Google Scholar]
- 10.Owens J. Big pharma slims down to bolster productivity. Nat Rev Drug Discovery. 2007;6(3):173–4. doi: 10.1038/nrd2284. [DOI] [PubMed] [Google Scholar]
- 11.Wermuth CG. Similarity in drugs: reflections on analogue design. Drug Discovery Today. 2006;11(7 & 8):348–54. doi: 10.1016/j.drudis.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Booth B, Zemmel R. Opinion: Quest for the best. Nat Rev Drug Discovery. 2003;2(10):838–41. doi: 10.1038/nrd1203. [DOI] [PubMed] [Google Scholar]
- 13.Zhong X, Moseley GB., III Mission possible: managing innovation in drug discovery. Nat Biotechnol. 2007;25(8):945–6. doi: 10.1038/nbt0807-945. [DOI] [PubMed] [Google Scholar]
- 14.Booth B, Zemmel R. Prospects for productivity. Nat Rev Drug Discovery. 2004;3(5):451–6. doi: 10.1038/nrd1384. [DOI] [PubMed] [Google Scholar]
- 15.Cavalla D. The extended pharmaceutical enterprise. Drug Discov Today. 2003;8(6):267–74. doi: 10.1016/s1359-6446(03)02634-5. [DOI] [PubMed] [Google Scholar]
- 16.Frantz S. Chemistry outsourcing going global. Nat Rev Drug Discovery. 2006;5(5):362–3. doi: 10.1038/nrd2049. [DOI] [PubMed] [Google Scholar]
- 17*.Ullman F, Boutellier R. Drug discovery: are productivity metrics inhibiting motivation and creativity? Drug Discovery Today. 2008;13(21/22):997–1001. doi: 10.1016/j.drudis.2008.06.015. An analysis of the impact of applying the standard business practice of metrics to the pharmaceutical industry. Scientists from over 50 companies were interviewed regarding their employer’s assessment protocol and the effect on their motivation and initiative. [DOI] [PubMed] [Google Scholar]
- 18.Ullman F, Boutellier R. A case study of lean drug discovery: from project driven research to innovation studios and process factories. Drug Discovery Today. 2008;13(11/12):543–50. doi: 10.1016/j.drudis.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 19**.Mackay M, Street SDA, McCall JM. Risk reduction in drug discovery and development. Curr Top Med Chem (Sharjah, United Arab Emirates) . 2005;5(11):1087–90. doi: 10.2174/156802605774297065. Illustrates a major trend and problem in the pharmaceutical industry. One approach to reduce cost is to reduce risk, which is being accomplished by shortening project timelines. Accelerated decision-making is being used to terminate projects at very early stages. [DOI] [PubMed] [Google Scholar]
- 20.Lewis CT, O’Connor PM. Discovering novel anticancer drugs: practical aspects and recent advances. Methods Mol Biol (Totowa, NJ, U S) 2003;223 doi: 10.1385/1-59259-329-1:425. [DOI] [PubMed] [Google Scholar]; Tumor Suppressor Genes. 2:425–63. [Google Scholar]
- 21.Senn S. Some statistical issues in project prioritization in the pharmaceutical industry. Stat Med. 1996;15(24):2689–702. doi: 10.1002/(SICI)1097-0258(19961230)15:24<2689::AID-SIM503>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 22.Schmid Esther F, Smith Dennis A. Is pharmaceutical R&D just a game of chance or can strategy make a difference? Drug Discov Today. 2004;9(1):18–26. doi: 10.1016/S1359-6446(04)02951-4. [DOI] [PubMed] [Google Scholar]
- 23*.Handen Jeffrey S. The industrialization of drug discovery. Drug Discov Today . 2002;7(2):83–5. doi: 10.1016/s1359-6446(01)02099-2. An editorial that articulates the current industry-wide philosophy to industrialize the science of drug discovery. [DOI] [PubMed] [Google Scholar]
- 24.Pichler FB, Turner SJ. The power and pitfalls of outsourcing. Nat Biotechnol. 2007;25(10):1093–6. doi: 10.1038/nbt1007-1093. [DOI] [PubMed] [Google Scholar]
- 25.Adam M. What to expect from rational drug design. Expert Opin Drug Discovery. 2007;2(6):773–6. doi: 10.1517/17460441.2.6.773. [DOI] [PubMed] [Google Scholar]
- 26.Sams-Dodd F. Drug discovery: selecting the optimal approach. Drug Discovery Today. 2006;11(9 & 10):465–72. doi: 10.1016/j.drudis.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 27.Lahana R. How many leads from HTS? Drug Discovery Today. 1999;4(10):447–8. doi: 10.1016/s1359-6446(99)01393-8. [DOI] [PubMed] [Google Scholar]
- 28*.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A Common Mechanism Underlying Promiscuous Inhibitors from Virtual and High-Throughput Screening. Journal of Medicinal Chemistry . 2002;45(8):1712–22. doi: 10.1021/jm010533y. Demonstrates that high-throughput screens lack information on the mechanism of action of potential lead compounds that results in an abundance of false positives. [DOI] [PubMed] [Google Scholar]
- 29.McGovern SL, Helfand BT, Feng B, Shoichet BK. A Specific Mechanism of Nonspecific Inhibition. Journal of Medicinal Chemistry. 2003;46(20):4265–72. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 30.Rishton GM. Reactive compounds and in vitro false positives in HTS. Drug Discovery Today. 1997;2(9):382–4. [Google Scholar]
- 31.Seidler J, McGovern SL, Doman TN, Shoichet BK. Identification and Prediction of Promiscuous Aggregating Inhibitors among Known Drugs. Journal of Medicinal Chemistry. 2003;46(21):4477–86. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- 32.Kubinyi H. Opinion: Drug research: myths, hype and reality. Nature Reviews Drug Discovery. 2003;2(8):665–8. doi: 10.1038/nrd1156. [DOI] [PubMed] [Google Scholar]
- 33.Pellecchia M, Bertini I, Cowburn D, Dalvit C, Giralt E, Jahnke W, et al. Perspectives on NMR in drug discovery: a technique comes of age. Nat Rev Drug Discovery. 2008;7(9):738–45. doi: 10.1038/nrd2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis B, Hubbard J. Applications of NMR in structure-based drug discovery. Struct-Based Drug Discovery. 2006:97–141. [Google Scholar]
- 35.Zartler ER, Shapiro MJ. Protein NMR-based screening in drug discovery. Curr Pharm Des. 2006;12(31):3963–72. doi: 10.2174/138161206778743619. [DOI] [PubMed] [Google Scholar]
- 36.Jahnke W. Perspectives of biomolecular NMR in drug discovery: the blessing and curse of versatility. J Biomol NMR. 2007;39(2):87–90. doi: 10.1007/s10858-007-9183-5. [DOI] [PubMed] [Google Scholar]
- 37*.Snyder DA, Chen Y, Denissova NG, Acton T, Aramini JM, Ciano M, et al. Comparisons of NMR Spectral Quality and Success in Crystallization Demonstrate that NMR and X-ray Crystallography Are Complementary Methods for Small Protein Structure Determination. Journal of the American Chemical Society . 2005;127(47):16505–11. doi: 10.1021/ja053564h. Demonstrates, along with the companion manuscript by Yee et al. (2005) [38], that the determination of a significant number of protein structures (20–40%) are only achievable using NMR. [DOI] [PubMed] [Google Scholar]
- 38.Yee AA, Savchenko A, Ignachenko A, Lukin J, Xu X, Skarina T, et al. NMR and X-ray Crystallography, Complementary Tools in Structural Proteomics of Small Proteins. Journal of the American Chemical Society. 2005;127(47):16512–7. doi: 10.1021/ja053565+. [DOI] [PubMed] [Google Scholar]
- 39.Montelione GT, Arrowsmith C, Girvin ME, Kennedy MA, Markley JL, Powers R, et al. Unique opportunities for NMR methods in structural genomics. J Struct Funct Genomics. 2009;10(2):101–6. doi: 10.1007/s10969-009-9064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sprangers R, Gribun A, Hwang PM, Houry WA, Kay LE. Quantitative NMR spectroscopy of supramolecular complexes: Dynamic side pores in ClpP are important for product release. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(46):16678–83. doi: 10.1073/pnas.0507370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain NU, Wyckoff TJO, Raetz CRH, Prestegard JH. Rapid Analysis of Large Protein-Protein Complexes Using NMR-derived Orientational Constraints: The 95kDa Complex of LpxA with Acyl Carrier Protein. Journal of Molecular Biology. 2004;343(5):1379–89. doi: 10.1016/j.jmb.2004.08.103. [DOI] [PubMed] [Google Scholar]
- 42.Tugarinov V, Muhandiram R, Ayed A, Kay LE. Four-Dimensional NMR Spectroscopy of a 723-Residue Protein: Chemical Shift Assignments and Secondary Structure of Malate Synthase G. Journal of the American Chemical Society. 2002;124(34):10025–35. doi: 10.1021/ja0205636. [DOI] [PubMed] [Google Scholar]
- 43.Tugarinov V, Kay LE. Quantitative 13C and 2H NMR Relaxation Studies of the 723-Residue Enzyme Malate Synthase G Reveal a Dynamic Binding Interface. Biochemistry. 2005;44(49):15970–7. doi: 10.1021/bi0519809. [DOI] [PubMed] [Google Scholar]
- 44.Peterson FC, Gettins PGW. Insight into the mechanism of serpin-proteinase inhibition from 2D [1H-15N] NMR studies of the 69 kDa a1-proteinase inhibitor Pittsburgh-trypsin covalent complex. Biochemistry. 2001;40(21):6284–92. doi: 10.1021/bi010100x. [DOI] [PubMed] [Google Scholar]
- 45.Liu D, Wang Y-S, Gesell JJ, Wilson E, Beyer BM, Wyss DF. Backbone resonance assignments of the 45.3 kDa catalytic domain of human BACE1. Journal of Biomolecular NMR. 2004;29(3):425–6. doi: 10.1023/B:JNMR.0000032509.81283.d3. [DOI] [PubMed] [Google Scholar]
- 46.Revington M, Zuiderweg ERP. Letter to the editor: TROSY-driven NMR backbone assignments of the 381-residue nucleotide-binding domain of the Thermus thermophilus DnaK molecular chaperone. Journal of Biomolecular NMR. 2004;30(1):113–4. doi: 10.1023/B:JNMR.0000042961.48233.f9. [DOI] [PubMed] [Google Scholar]
- 47.Amero C, Schanda P, Dura MA, Ayala I, Marion D, Franzetti B, et al. Fast Two-Dimensional NMR Spectroscopy of High Molecular Weight Protein Assemblies. J Am Chem Soc. 2009;131(10):3448–9. doi: 10.1021/ja809880p. [DOI] [PubMed] [Google Scholar]
- 48.Mukrasch MD, Bibow S, Korukottu J, Jeganathan S, Biernat J, Griesinger C, et al. Structural polymorphism of 441-residue tau at single residue resolution. PLoS Biol. 2009;7(2):399–414. doi: 10.1371/journal.pbio.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.LeMaster DM. Deuterium labeling in NMR structural analysis of larger proteins. Q Rev Biophys. 1990;23(2):133–74. doi: 10.1017/s0033583500005527. [DOI] [PubMed] [Google Scholar]
- 50.Kigawa T, Muto Y, Yokoyama S. Cell-free synthesis and amino acid-selective stable isotope labeling of proteins for NMR analysis. J Biomol NMR. 1995;6(2):129–34. doi: 10.1007/BF00211776. [DOI] [PubMed] [Google Scholar]
- 51.Goto NK, Kay LE. New developments in isotope labeling strategies for protein solution NMR spectroscopy. Curr Opin Struct Biol. 2000;10(5):585–92. doi: 10.1016/s0959-440x(00)00135-4. [DOI] [PubMed] [Google Scholar]
- 52.Zhu G, Yao X. TROSY-based NMR experiments for NMR studies of large biomolecules. Prog Nucl Magn Reson Spectrosc. 2008;52(1):49–68. [Google Scholar]
- 53.Keppetipola S, Kudlicki W, Nguyen BD, Meng X, Donovan KJ, Shaka AJ. From Gene to HSQC in under Five Hours: High-Throughput NMR Proteomics. J Am Chem Soc. 2006;128(14):4508–9. doi: 10.1021/ja0580791. [DOI] [PubMed] [Google Scholar]
- 54.Vinarov DA, Markley JL. High-throughput automated platform for nuclear magnetic resonance-based structural proteomics. Expert Rev Proteomics. 2005;2(1):49–55. doi: 10.1586/14789450.2.1.49. [DOI] [PubMed] [Google Scholar]
- 55.Folkers GE, van Buuren BNM, Kaptein R. Expression screening, protein purification and NMR analysis of human protein domains for structural genomics. J Struct Funct Genomics. 2004;5(1–2):119–31. doi: 10.1023/B:JSFG.0000029200.66197.0c. [DOI] [PubMed] [Google Scholar]
- 56.Ozawa K, Headlam MJ, Schaeffer PM, Henderson BR, Dixon NE, Otting G. Optimization of an Escherichia coli system for cell-free synthesis of selectively 15N-labelled proteins for rapid analysis by NMR spectroscopy. Eur J Biochem. 2004;271(20):4084–93. doi: 10.1111/j.1432-1033.2004.04346.x. [DOI] [PubMed] [Google Scholar]
- 57.de Alba E, Tjandra N. Residual dipolar couplings in protein structure determination. Methods Mol Biol (Totowa, NJ, U S) 2004;278:89–106. doi: 10.1385/1-59259-809-9:089. Protein NMR Techniques. [DOI] [PubMed] [Google Scholar]
- 58.Clore GM, Starich MR, Bewley CA, Cai M, Kuszewski J. Impact of Residual Dipolar Couplings on the Accuracy of NMR Structures Determined from a Minimal Number of NOE Restraints. J Am Chem Soc. 1999;121(27):6513–4. [Google Scholar]
- 59.Huang X, Moy F, Powers R. Evaluation of the Utility of NMR Structures Determined from Minimal NOE-Based Restraints for Structure-Based Drug Design, Using MMP-1 as an Example. Biochemistry. 2000;39(44):13365–75. doi: 10.1021/bi001658s. [DOI] [PubMed] [Google Scholar]
- 60.Gaponenko V, Sarma SP, Altieri AS, Horita DA, Li J, Byrd RA. Improving the Accuracy of NMR Structures of Large Proteins Using Pseudocontact Shifts as Long-Range Restraints. J Biomol NMR. 2004;28(3):205–12. doi: 10.1023/B:JNMR.0000013706.09264.36. [DOI] [PubMed] [Google Scholar]
- 61.Huang YJ, Moseley HNB, Baran MC, Arrowsmith C, Powers R, Tejero R, et al. An integrated platform for automated analysis of protein NMR structures. Methods in Enzymology. 2005;394:111–41. doi: 10.1016/S0076-6879(05)94005-6. Nuclear Magnetic Resonance of Biological Macromolecules, Part C. [DOI] [PubMed] [Google Scholar]
- 62.Acton TB, Gunsalus KC, Xiao R, Ma LC, Aramini J, Baran MC, et al. Robotic cloning and Protein Production Platform of the Northeast Structural Genomics Consortium. Methods in Enzymology. 2005;394:210–43. doi: 10.1016/S0076-6879(05)94008-1. Nuclear Magnetic Resonance of Biological Macromolecules, Part C. [DOI] [PubMed] [Google Scholar]
- 63.Hiller S, Fiorito F, Wuethrich K, Wider G. Automated projection spectroscopy (APSY) Proc Natl Acad Sci U S A. 2005;102(31):10876–81. doi: 10.1073/pnas.0504818102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64**.Liu G, Shen Y, Atreya HS, Parish D, Shao Y, Sukumaran DK, et al. NMR data collection and analysis protocol for high-throughput protein structure determination. Proc Natl Acad Sci U S A . 2005;102(30):10487–92. doi: 10.1073/pnas.0504338102. Demonstrates the recent advancements from structural genomics to rapidly determine protein NMR structures in a high-throughput fashion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65*.Atreya HS, Szyperski T. G-matrix Fourier transform NMR spectroscopy for complete protein resonance assignment. Proc Natl Acad Sci U S A . 2004;101(26):9642–7. doi: 10.1073/pnas.0403529101. Describes the G-matrix Fourier transform NMR approach that dramatically decreases NMR data acquisition time. GFT-NMR allows for a complete set of NMR spectra to assign a protein to be collected in ~1–2 days. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eghbalnia HR, Bahrami A, Tonelli M, Hallenga K, Markley JL. High-Resolution Iterative Frequency Identification for NMR as a General Strategy for Multidimensional Data Collection. J Am Chem Soc. 2005;127(36):12528–36. doi: 10.1021/ja052120i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coggins BE, Venters RA, Zhou P. Filtered Backprojection for the Reconstruction of a High-Resolution (4,2)D CH3-NH NOESY Spectrum on a 29 kDa Protein. J Am Chem Soc. 2005;127(33):11562–3. doi: 10.1021/ja053110k. [DOI] [PubMed] [Google Scholar]
- 68.Staykova DK, Fredriksson J, Bermel W, Billeter M. Assignment of protein NMR spectra based on projections, multi-way decomposition and a fast correlation approach. J Biomol NMR. 2008;42(2):87–97. doi: 10.1007/s10858-008-9265-z. [DOI] [PubMed] [Google Scholar]
- 69.Szyperski T, Yeh DC, Sukumaran DK, Moseley HNB, Montelione GT. Reduced-dimensionality NMR spectroscopy for high-throughput protein resonance assignment. Proc Natl Acad Sci U S A. 2002;99(12):8009–14. doi: 10.1073/pnas.122224599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rovnyak D, Frueh DP, Sastry M, Sun Z-YJ, Stern AS, Hoch JC, et al. Accelerated acquisition of high resolution triple-resonance spectra using non-uniform sampling and maximum entropy reconstruction. J Magn Reson. 2004;170(1):15–21. doi: 10.1016/j.jmr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 71.Mobli M, Stern AS, Hoch JC. Spectral reconstruction methods in fast NMR: Reduced dimensionality, random sampling and maximum entropy. J Magn Reson. 2006;182(1):96–105. doi: 10.1016/j.jmr.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 72.Hoch JC, Maciejewski MW, Filipovic B. Randomization improves sparse sampling in multidimensional NMR. J Magn Reson. 2008;193(2):317–20. doi: 10.1016/j.jmr.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mandelshtam VA, Taylor HS, Shaka AJ. Application of the filter diagonalization method to one- and two-dimensional NMR spectra. J Magn Reson. 1998;133(2):304–12. doi: 10.1006/jmre.1998.1476. [DOI] [PubMed] [Google Scholar]
- 74*.Schmieder P, Stern AS, Wagner G, Hoch JC. Improved resolution in triple-resonance spectra by nonlinear sampling in the constant-time domain. J Biomol NMR . 1994;4(4):483–90. doi: 10.1007/BF00156615. Demonstrates the feasibility of utilizing non-uniform data sampling to rapidly collect three- dimensional NMR spectra to solve a protein structure. [DOI] [PubMed] [Google Scholar]
- 75.Orekhov VY, Ibraghimov I, Billeter M. Optimizing resolution in multidimensional NMR by three-way decomposition. J Biomol NMR. 2003;27(2):165–73. doi: 10.1023/a:1024944720653. [DOI] [PubMed] [Google Scholar]
- 76.Orekhov VY, Ibraghimov IV, Billeter M. MUNIN: a new approach to multi-dimensional NMR spectra interpretation. J Biomol NMR. 2001;20(1):49–60. doi: 10.1023/a:1011234126930. [DOI] [PubMed] [Google Scholar]
- 77.Kazimierczuk K, Zawadzka A, Kozminski W. Optimization of random time domain sampling in multidimensional NMR. J Magn Reson. 2008;192(1):123–30. doi: 10.1016/j.jmr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 78.Kazimierczuk K, Zawadzka A, Kozminski W, Zhukov I. Random sampling of evolution time space and Fourier transform processing. J Biomol NMR. 2006;36(3):157–68. doi: 10.1007/s10858-006-9077-y. [DOI] [PubMed] [Google Scholar]
- 79.Kazimierczuk K, Kozminski W, Zhukov I. Two-dimensional Fourier transform of arbitrarily sampled NMR data sets. J Magn Reson. 2006;179(2):323–8. doi: 10.1016/j.jmr.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 80.Kazimierczuk K, Zawadzka A, Kozminski W, Zhukov I. Lineshapes and artifacts in Multidimensional Fourier Transform of arbitrary sampled NMR data sets. J Magn Reson. 2007;188(2):344–56. doi: 10.1016/j.jmr.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 81.Schanda P, Van Melckebeke H, Brutscher B. Speeding Up Three-Dimensional Protein NMR Experiments to a Few Minutes. J Am Chem Soc. 2006;128(28):9042–3. doi: 10.1021/ja062025p. [DOI] [PubMed] [Google Scholar]
- 82*.Schanda P, Kupce E, Brutscher B. SOFAST-HMQC Experiments for Recording Two- dimensional Heteronuclear Correlation Spectra of Proteins within a Few Seconds. J Biomol NMR . 2005;33(4):199–211. doi: 10.1007/s10858-005-4425-x. Extremely rapid acquisition of 2D 1H-15N HSQC spectra that is extremely beneficial to improving the throughput of NMR fragment-based screens. [DOI] [PubMed] [Google Scholar]
- 83.Moseley HNB, Monleon D, Montelione GT. Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Methods Enzymol. 2001;339:91–108. doi: 10.1016/s0076-6879(01)39311-4. Nuclear Magnetic Resonance of Biological Macromolecules, Part B. [DOI] [PubMed] [Google Scholar]
- 84.Zimmerman DE, Kulikowski CA, Huang Y, Feng W, Tashiro M, Shimotakahara S, et al. Automated analysis of protein NMR assignments using methods from artificial intelligence. Journal of Molecular Biology. 1997;269(4):592–610. doi: 10.1006/jmbi.1997.1052. [DOI] [PubMed] [Google Scholar]
- 85.Lescop E, Brutscher B. Highly automated protein backbone resonance assignment within a few hours: The ‘BATCH’ strategy and software package. J Biomol NMR. 2009;44(1):43–57. doi: 10.1007/s10858-009-9314-2. [DOI] [PubMed] [Google Scholar]
- 86.Bartels C, Billeter M, Guentert P, Wuethrich K. Automated sequence-specific NMR assignment of homologous proteins using the program GARANT. J Biomol NMR. 1996;7(3):207–13. doi: 10.1007/BF00202037. [DOI] [PubMed] [Google Scholar]
- 87.Schmucki R, Yokoyama S, Guentert P. Automated assignment of NMR chemical shifts using peak-particle dynamics simulation with the DYNASSIGN algorithm. J Biomol NMR. 2009;43(2):97–109. doi: 10.1007/s10858-008-9291-x. [DOI] [PubMed] [Google Scholar]
- 88.Lin H-N, Wu K-P, Chang J-M, Sung T-Y, Hsu W-L. GANA - a genetic algorithm for NMR backbone resonance assignment. Nucleic Acids Res. 2005;33(14):4593–601. doi: 10.1093/nar/gki768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Volk J, Herrmann T, Wuthrich K. Automated sequence-specific protein NMR assignment using the memetic algorithm MATCH. J Biomol NMR. 2008;41(3):127–38. doi: 10.1007/s10858-008-9243-5. [DOI] [PubMed] [Google Scholar]
- 90.Eghbalnia HR, Bahrami A, Wang L, Assadi A, Markley JL. Probabilistic Identification of Spin Systems and their Assignments including Coil-Helix Inference as Output (PISTACHIO) J Biomol NMR. 2005;32(3):219–33. doi: 10.1007/s10858-005-7944-6. [DOI] [PubMed] [Google Scholar]
- 91.Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Quarterly Reviews of Biophysics. 2000 Feb;33(1):29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- 92.Guentert P. Automated structure determination from NMR spectra. Eur Biophys J. 2009;38(2):129–43. doi: 10.1007/s00249-008-0367-z. [DOI] [PubMed] [Google Scholar]
- 93.Huang YJ, Tejero R, Powers R, Montelione GT. A topology-constrained distance network algorithm for protein structure determination from NOESY data. Proteins: Structure, Function, and Bioinformatics. 2006;62(3):587–603. doi: 10.1002/prot.20820. [DOI] [PubMed] [Google Scholar]
- 94.Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, Nilges M. ARIA2: Automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23(3):381–2. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 95.Grishaev A, Llinas M. Protein structure elucidation from minimal NMR data: The CLOUDS approach. Methods Enzymol. 2005;394:261–95. doi: 10.1016/S0076-6879(05)94010-X. Nuclear Magnetic Resonance of Biological Macromolecules, Part C. [DOI] [PubMed] [Google Scholar]
- 96.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol (Totowa, NJ, U S) 2004;278:353–78. doi: 10.1385/1-59259-809-9:353. Protein NMR Techniques. [DOI] [PubMed] [Google Scholar]
- 97.Kuszewski J, Schwieters CD, Garrett DS, Byrd RA, Tjandra N, Clore GM. Completely automated, highly error-tolerant macromolecular structure determination from multidimensional nuclear overhauser enhancement spectra and chemical shift assignments. J Am Chem Soc. 2004;126(20):6258–73. doi: 10.1021/ja049786h. [DOI] [PubMed] [Google Scholar]
- 98.Lopez-Mendez B, Guentert P. Automated Protein Structure Determination from NMR Spectra. J Am Chem Soc. 2006;128(40):13112–22. doi: 10.1021/ja061136l. [DOI] [PubMed] [Google Scholar]
- 99.Rohl CA. Protein structure estimation from minimal restraints using Rosetta. Methods Enzymol. 2005;394:244–60. doi: 10.1016/S0076-6879(05)94009-3. Nuclear Magnetic Resonance of Biological Macromolecules, Part C. [DOI] [PubMed] [Google Scholar]
- 100.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor-NIH for NMR molecular structure determination. Prog Nucl Magn Reson Spectrosc. 2006;48(1):47–62. [Google Scholar]
- 101.Mal TK, Bagby S, Ikura M. Protein structure calculation from NMR data. Methods Mol Biol (Totowa, NJ, U S) 2002;173 doi: 10.1385/1-59259-184-1:267. [DOI] [PubMed] [Google Scholar]; Calcium-Binding Protein Protocols. 2:267–83. [Google Scholar]
- 102.Jee J, Guentert P. Influence of the completeness of chemical shift assignments on NMR structures obtained with automated NOE assignment. J Struct Funct Genomics. 2003;4(2–3):179–89. doi: 10.1023/a:1026122726574. [DOI] [PubMed] [Google Scholar]
- 103.Herrmann T, Gntert P, Wthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319(1):209–27. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 104.Guntert P. Automated NMR protein structure calculation. Prog Nucl Magn Reson Spectrosc. 2003;43(3–4):105–25. [Google Scholar]
- 105.Delaglio F, Kontaxis G, Bax A. Protein Structure Determination Using Molecular Fragment Replacement and NMR Dipolar Couplings. J Am Chem Soc. 2000;122(9):2142–3. [Google Scholar]
- 106.Prestegard JH, Mayer KL, Valafar H, Benison GC. Determination of protein backbone structures from residual dipolar couplings. Methods Enzymol. 2005;394:175–209. doi: 10.1016/S0076-6879(05)94007-X. Nuclear Magnetic Resonance of Biological Macromolecules, Part C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107**.Shen Y, Vernon R, Baker D, Bax A. De novo protein structure generation from incomplete chemical shift assignments. J Biomol NMR . 2009;43(2):63–78. doi: 10.1007/s10858-008-9288-5. Describes the CS-ROSETTA protocol to determine a protein structure strictly from chemical shift assignments. Has the potential to significantly revolutionize NMR structural biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins Struct, Funct, Bioinf. 2007;66(4):778–95. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 109.Chakravarti B, Lewis SJ, Chakravarti DN, Raval A. Three dimensional structures of proteins and protein complexes from chemical cross-linking and mass spectrometry: a biochemical and computational overview. Curr Proteomics. 2006;3(1):1–21. [Google Scholar]
- 110.Mori S, Abeygunawardana C, Johnson MON, van Zijl PCM. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson, Ser B. 1995;108(1):94–8. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 111.Haner RL, Llanos W, Mueller L. Small Volume Flow Probe for Automated Direct-Injection NMR Analysis: Design and Performance. J Magn Reson. 2000;143(1):69–78. doi: 10.1006/jmre.1999.1983. [DOI] [PubMed] [Google Scholar]
- 112*.Stark J, Powers R. Rapid Protein-Ligand Costructures Using Chemical Shift Perturbations. Journal of the American Chemical Society . 2008;130(2):535–45. doi: 10.1021/ja0737974. Chemical shift perturbations are used to guide and filter an AutoDock simulation to rapidly determine protein-ligand models. [DOI] [PubMed] [Google Scholar]
- 113.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of Computational Chemistry. 1998;19(14):1639–62. [Google Scholar]
- 114.Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007 Apr 30;28(6):1145–52. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 115.Loll PJ, Lattman EE. The crystal structure of the ternary complex of staphylococcal nuclease, calcium, and the inhibitor pdTp, refined at 1.65 Ã…. Proteins Struct, Funct, Genet. 1989;5(3):183–201. doi: 10.1002/prot.340050302. [DOI] [PubMed] [Google Scholar]
- 116**.Dominguez C, Boelens R, Bonvin AMJJ. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc . 2003;125(7):1731–7. doi: 10.1021/ja026939x. HADDOCK uses chemical shift perturbations in combination with ambiguous interaction constraints to quickly and efficently model protein-protein complexes. [DOI] [PubMed] [Google Scholar]
- 117.Nilges M. Ambiguous distance data in the calculation of NMR structures. Folding Des. 1997;2(4):S53–S7. doi: 10.1016/s1359-0278(97)00064-3. [DOI] [PubMed] [Google Scholar]
- 118.Zhang N, Liu LH, Liu F, et al. NMR-based model reveals the structural determinants of mammalian arylamine N-acetyltransferase substrate specificity. J Mol Biol. 2006;363(1):188–200. doi: 10.1016/j.jmb.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 119.Moy FJ, Chanda PK, Chen JM, Cosmi S, Edris W, Skotnicki JS, et al. NMR solution structure of the catalytic fragment of human fibroblast collagenase complexed with a sulfonamide derivative of a hydroxamic acid compound. Biochemistry. 1999 Jun 1;38(22):7085–96. doi: 10.1021/bi982576v. [DOI] [PubMed] [Google Scholar]
- 120.Chen JM, Nelson FC, Levin JI, Mobilio D, Moy FJ, Nilakantan R, et al. Structure-Based Design of a Novel, Potent, and Selective Inhibitor for MMP-13 Utilizing NMR Spectroscopy and Computer-Aided Molecular Design. Journal of the American Chemical Society. 2000;122(40):9648–54. [Google Scholar]
- 121.John M, Pintacuda G, Park AY, Dixon NE, Otting G. Structure Determination of Protein-Ligand Complexes by Transferred Paramagnetic Shifts. J Am Chem Soc. 2006;128(39):12910–6. doi: 10.1021/ja063584z. [DOI] [PubMed] [Google Scholar]
- 122.Medek A, Hajduk PJ, Mack J, Fesik SW. The Use of Differential Chemical Shifts for Determining the Binding Site Location and Orientation of Protein-Bound Ligands. Journal of the American Chemical Society. 2000;122(6):1241–2. [Google Scholar]
- 123.Hajduk PJ, Mack JC, Olejniczak ET, Park C, Dandliker PJ, Beutel BA. SOS-NMR: A Saturation Transfer NMR-Based Method for Determining the Structures of Protein- Ligand Complexes. J Am Chem Soc. 2004;126(8):2390–8. doi: 10.1021/ja039480v. [DOI] [PubMed] [Google Scholar]
- 124.Sem DS, Yu L, Coutts SM, Jack R. Object-oriented approach to drug design enabled by NMR SOLVE: first real-time structural tool for characterizing protein-ligand interactions. J Cell Biochem. 2001;(Suppl 37):99–105. doi: 10.1002/jcb.10070. [DOI] [PubMed] [Google Scholar]
- 125.Pellecchia M, Meininger D, Dong Q, Chang E, Jack R, Sem DS. NMR-based structural characterization of large protein-ligand interactions. J Biomol NMR. 2002;22(2):165–73. doi: 10.1023/a:1014256707875. [DOI] [PubMed] [Google Scholar]
- 126.Metzler WJ, Claus BL, McDonnell PA, Johnson SR, Goldfarb V, Davis ME, et al. Application of protein-ligand NOE matching to the rapid evaluation of fragment binding poses. Fragm-Based Drug Discovery. 2008:99–133. [Google Scholar]
- 127.Dove A, Marshall A. Drug screening-beyond the bottleneck. Nature Biotechnology. 1999;17(9):859–63. doi: 10.1038/12845. [DOI] [PubMed] [Google Scholar]
- 128.Goode DR, Totten RK, Heeres JT, Hergenrother PJ. Identification of Promiscuous Small Molecule Activators in High-Throughput Enzyme Activation Screens. J Med Chem. 2008;51(8):2346–9. doi: 10.1021/jm701583b. [DOI] [PubMed] [Google Scholar]
- 129.Lipinski CA. Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods. 2001;44(1):235–49. doi: 10.1016/s1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 130.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Delivery Rev. 1997;23(1–3):3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 131.Andrews CW, Bennett L, Yu LX. Predicting human oral bioavailability of a compound: development of a novel quantitative structure-bioavailability relationship. Pharmaceutical Research. 2000;17(6):639–44. doi: 10.1023/a:1007556711109. [DOI] [PubMed] [Google Scholar]
- 132.Viswanadhan VN, Balan C, Hulme C, Cheetham JC, Sun Y. Knowledge-based approaches in the design and selection of compound libraries for drug discovery. Current Opinion in Drug Discovery & Development. 2002;5(3):400–6. [PubMed] [Google Scholar]
- 133.Matter H, Baringhaus K-H, Naumann T, Klabunde T, Pirard B. Computational approaches towards the rational design of drug-like compound libraries. Combinatorial Chemistry and High Throughput Screening. 2001;4(6):453–75. doi: 10.2174/1386207013330896. [DOI] [PubMed] [Google Scholar]
- 134.Xu J, Stevenson J. Drug-like Index: A New Approach To Measure Drug-like Compounds and Their Diversity. J Chem Inf Comput Sci. 2000;40(5):1177–87. doi: 10.1021/ci000026+. [DOI] [PubMed] [Google Scholar]
- 135.Jelic D, Mesar V, Basic I, Nadramija D, Verbanac D. Novel approach to drug discovery in PLIVA - establishing of HTS unit and unique compounds library. PharmaChem. 2003;2(6):64–7. [Google Scholar]
- 136.Xue L, Bajorath J. Molecular descriptors in chemoinformatics, computational combinatorial chemistry, and virtual screening. Comb Chem High Throughput Screening. 2000;3(5):363–72. doi: 10.2174/1386207003331454. [DOI] [PubMed] [Google Scholar]
- 137.Lewis RA, Pickett SD, Clark DE. Computer-aided molecular diversity analysis and combinatorial library design. Rev Comput Chem. 2000;16:1–51. [Google Scholar]
- 138.Willett P. Chemoinformatics - similarity and diversity in chemical libraries. Curr Opin Biotechnol. 2000;11(1):85–8. doi: 10.1016/s0958-1669(99)00059-2. [DOI] [PubMed] [Google Scholar]
- 139.Spellmeyer DC, Grootenhuis PDJ. Recent developments in molecular diversity. Computational approaches to combinatorial chemistry. Annu Rep Med Chem. 1999;34:287–96. [Google Scholar]
- 140.Gorse D, Lahana R. Functional diversity of compound libraries. Curr Opin Chem Biol. 2000;4(3):287–94. doi: 10.1016/s1367-5931(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 141.Villar HO, Koehler RT. Comments on the design of chemical libraries for screening. Molecular diversity. 2000;5(1):13–24. doi: 10.1023/a:1011326914800. [DOI] [PubMed] [Google Scholar]
- 142.Bohacek RS, McMartin C, Guida WC. The art and practice of structure-based drug design: a molecular modeling perspective. Medicinal Research Reviews. 1996;16(1):3–50. doi: 10.1002/(SICI)1098-1128(199601)16:1<3::AID-MED1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 143.Mestres J, Veeneman GH. Identification of “Latent Hits” in Compound Screening Collections. J Med Chem. 2003;46(16):3441–4. doi: 10.1021/jm034078c. [DOI] [PubMed] [Google Scholar]
- 144.Kozikowski BA, Burt TM, Tirey DA, Williams LE, Kuzmak BR, Stanton DT, et al. The Effect of Freeze/Thaw Cycles on the Stability of Compounds in DMSO. Journal of Biomolecular Screening. 2003;8(2):210–5. doi: 10.1177/1087057103252618. [DOI] [PubMed] [Google Scholar]
- 145.Kozikowski BA, Burt TM, Tirey DA, Williams LE, Kuzmak BR, Stanton DT, et al. The Effect of Room-Temperature Storage on the Stability of Compounds in DMSO. Journal of Biomolecular Screening. 2003;8(2):205–9. doi: 10.1177/1087057103252617. [DOI] [PubMed] [Google Scholar]
- 146.Posner BA. High-throughput screening-driven lead discovery: Meeting the challenges of finding new therapeutics. Curr Opin Drug Discovery Dev. 2005;8(4):487–94. [PubMed] [Google Scholar]
- 147.Jahnke W. Fragment-based approaches. Compr Med Chem II. 2006;3:939–57. [Google Scholar]
- 148.Schultz J. Practical aspects of using NMR in fragment-based screening. Fragm-Based Drug Discovery. 2008:63–98. [Google Scholar]
- 149*.Gribbon P, Sewing A. High-throughput drug discovery: what can we expect from HTS? Drug Discov Today . 2005;10(1):17–22. doi: 10.1016/S1359-6446(04)03275-1. An analysis of the Pfizer corporate library and the impact on screening results. The analysis indicates a continual trend toward higher molecular-weight and low solubility compounds that result in an increase in apparent inhibition and false leads. [DOI] [PubMed] [Google Scholar]
- 150.Schmitt W, Willmann S. Physiology-based pharmacokinetic modeling: ready to be used. Drug Discovery Today Technol. 2004;1(4):449–56. doi: 10.1016/j.ddtec.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 151.Wienkers LC, Heath TG. Predicting in vivo drug interactions from in vitro drug discovery data. Nat Rev Drug Discovery. 2005;4(10):825–33. doi: 10.1038/nrd1851. [DOI] [PubMed] [Google Scholar]
- 152.Zimmerman RJ. How and when to apply absorption, distribution, metabolism, excretion, and toxicity. Compr Med Chem II. 2006;2:559–72. [Google Scholar]
- 153.Korfmacher W. Strategies and techniques for higher throughput ADME/PK assays. High- Throughput Anal Pharm Ind. 2009:205–31. [Google Scholar]
- 154.Schuffenhauer A, Ruedisser S, Marzinzik AL, Jahnke W, Blommers M, Selzer P, et al. Library design for fragment based screening. Curr Top Med Chem (Sharjah, United Arab Emirates) 2005;5(8):751–62. doi: 10.2174/1568026054637700. [DOI] [PubMed] [Google Scholar]
- 155.Erlanson DA. Fragment-based lead discovery: A chemical update. Curr Opin Biotechnol. 2006;17(6):643–52. doi: 10.1016/j.copbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 156.Jencks WP. On the attribution and additivity of binding energies. Proc Natl Acad Sci U S A. 1981;78(7):4046–50. doi: 10.1073/pnas.78.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fischer M, Hubbard RE. Fragment-based ligand discovery. Mol Interventions. 2009;9(1):22–30. doi: 10.1124/mi.9.1.7. [DOI] [PubMed] [Google Scholar]
- 158.Congreve M, Murray CW, Carr R, Rees DC. Fragment-based lead discovery. Annu Rep Med Chem. 2007;42:431–48. [Google Scholar]
- 159.Siegal G, Ab E, Schultz J. Integration of fragment screening and library design. Drug Discovery Today. 2007;12(23&24):1032–9. doi: 10.1016/j.drudis.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 160.Mercier KA, Germer K, Powers R. Design and characterization of a functional library for NMR screening against novel protein targets. Combinatorial Chemistry & High Throughput Screening. 2006;9(7):515–34. doi: 10.2174/138620706777935342. [DOI] [PubMed] [Google Scholar]
- 161*.Hopkins Andrew L, Groom Colin R, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discov Today . 2004;9(10):430–1. doi: 10.1016/S1359-6446(04)03069-7. Describes the concept of ligand efficiency, a major paradigm shift in the philosophy of drug discovery that supports the expanding role of fragment-based screens. Binding energy is interpreted on a per atom basis instead of a per molecule. [DOI] [PubMed] [Google Scholar]
- 162.Fink T, Bruggesser H, Reymond J-L. Virtual exploration of the small-molecule chemical universe below 160 D. Angew Chem, Int Ed. 2005;44(10):1504–8. doi: 10.1002/anie.200462457. [DOI] [PubMed] [Google Scholar]
- 163.Hajduk PJ. SAR by NMR: putting the pieces together. Mol Interventions. 2006;6(5):266–72. doi: 10.1124/mi.6.5.8. [DOI] [PubMed] [Google Scholar]
- 164**.Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat Rev Drug Discovery . 2007;6(3):211–9. doi: 10.1038/nrd2220. A review describing the benefits and continuing growth of NMR fragment-based screening. Of particular note are the documented successes of fragment-based screening and the impact the procedure is having on the drug discovery business. [DOI] [PubMed] [Google Scholar]
- 165.Mayr LM, Fuerst P. The future of high-throughput screening. J Biomol Screening. 2008;13(6):443–8. doi: 10.1177/1087057108319644. [DOI] [PubMed] [Google Scholar]
- 166.Schade M. Fragment-based lead discovery by NMR. Front Drug Des Discovery. 2007;3:105–19. [Google Scholar]
- 167.Clore GM, Gronenborn AM. Structures of larger proteins, protein-ligand and protein-DNA complexes by multidimensional heteronuclear NMR. Protein Science. 1994;3:372–90. doi: 10.1002/pro.5560030302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cooke RM. Protein NMR extends into new fields of structural biology. Curr Opin Chem Biol. 1997;1(3):359–64. doi: 10.1016/s1367-5931(97)80074-9. [DOI] [PubMed] [Google Scholar]
- 169.Kay LE. NMR methods for the study of protein structure and dynamics. Biochem Cell Biol. 1997;75(1):1–15. doi: 10.1139/o97-023. [DOI] [PubMed] [Google Scholar]
- 170.Roberts GCK. Applications of NMR in drug discovery. Drug Discovery Today. 2000;5(6):230–40. doi: 10.1016/s1359-6446(00)01479-3. [DOI] [PubMed] [Google Scholar]
- 171.Rossi C, Donati A, Sansoni MR. Nuclear magnetic resonance as a tool for the identification of specific DNA-ligand interaction. Chem Phys Lett. 1992;189(3):278–80. [Google Scholar]
- 172.Hajduk PJ, Olejniczak ET, Fesik SW. One-Dimensional Relaxation- and Diffusion-Edited NMR Methods for Screening Compounds That Bind to Macromolecules. J Am Chem Soc. 1997;119(50):12257–61. [Google Scholar]
- 173.Lin M, Shapiro MJ, Wareing JR. Screening Mixtures by Affinity NMR. J Org Chem. 1997;62(25):8930–1. [Google Scholar]
- 174.Lin M, Shapiro MJ, Wareing JR. Diffusion-Edited NMR-Affinity NMR for Direct Observation of Molecular Interactions. J Am Chem Soc. 1997;119(22):5249–50. [Google Scholar]
- 175.Waldeck AR, Kuchel PW, Lennon AJ, Chapman BE. NMR diffusion measurements to characterize membrane transport and solute binding. Prog Nucl Magn Reson Spectrosc. 1997;30(1–2):39–68. [Google Scholar]
- 176*.Wüthrich K. NMR of proteins and nucleic acids. New York: John Wiley & Sons, Inc; 1986. The now classic book by the Noble prize winning author that describes the fundamental concepts behind the application of NMR to solve protein and nucleic acid structures. [Google Scholar]
- 177.Shirakawa M, Lee SJ, Takimoto M, Matsuo H, Akutsu H, Kyogoku Y. Interaction of the l-cro repressor protein with operator DNA fragments monitored as to amide proton magnetic resonances. J Mol Struct. 1991;242:355–66. [Google Scholar]
- 178.Otting G. Experimental NMR techniques for studies of protein-ligand interactions. Curr Opin Struct Biol. 1993;3(5):760–8. [Google Scholar]
- 179.Ni F. Recent developments in transferred NOE methods. Prog Nucl Magn Reson Spectrosc. 1994;26(6):517–606. [Google Scholar]
- 180.Vogtherr M, Peters T. Application of NMR Based Binding Assays to Identify Key Hydroxy Groups for Intermolecular Recognition. J Am Chem Soc. 2000;122(25):6093–9. [Google Scholar]
- 181*.Mayer M, Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew Chem, Int Ed . 1999;38(12):1784–8. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. 1D 1H NMR STD experiments introduced ligand-detected approaches to NMR high-throughput ligand affinity screens. [DOI] [PubMed] [Google Scholar]
- 182.Dalvit C, Pevarello P, Tato M, Veronesi M, Vulpetti A, Sundstrom M. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J Biomol NMR. 2000;18(1):65–8. doi: 10.1023/a:1008354229396. [DOI] [PubMed] [Google Scholar]
- 183.Chen A, Shapiro MJ. NOE Pumping: A Novel NMR Technique for Identification of Compounds with Binding Affinity to Macromolecules. J Am Chem Soc. 1998;120(39):10258–9. [Google Scholar]
- 184.Edwards PD, Albert JS, Sylvester M, Aharony D, Andisik D, Callaghan O, et al. Application of fragment-based lead generation to the discovery of novel, cyclic amidine Î2-secretase inhibitors with nanomolar potency, cellular activity, and high ligand efficiency. J Med Chem. 2007;50(24):5912–25. doi: 10.1021/jm070829p. [DOI] [PubMed] [Google Scholar]
- 185.Dixon AK, Fisch HU. Animal models and ethological strategies for early drug-testing in humans. Neurosci Biobehav Rev. 1998;23(2):345–58. doi: 10.1016/s0149-7634(98)00036-0. [DOI] [PubMed] [Google Scholar]
- 186.Preziosi P. Predictive values of animal versus clinical testing in drug evaluation, problems and difficulties. Adv Clin Pharmacol. 1977;13:41–69. (Probl. Clin. Pharmacol. Ther. Res.: Phase 1, Proc. Int. Symp. Clin. Pharmacol., 14th) [Google Scholar]
- 187.Fox TM. Strains and species variations in pharmacological responses. Symp Ser Immunobiol Stand. 1967;5:133–48. [Google Scholar]
- 188.Halket JM, Waterman D, Przyborowska AM, Patel RKP, Fraser PD, Bramley PM. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. Journal of Experimental Botany. 2005;56(410):219–43. doi: 10.1093/jxb/eri069. [DOI] [PubMed] [Google Scholar]
- 189.Wilson ID, Plumb R, Granger J, Major H, Williams R, Lenz EM. HPLC-MS-based methods for the study of metabonomics. Journal of Chromatography, B: Analytical Technologies in the Biomedical and Life Sciences. 2005;817(1):67–76. doi: 10.1016/j.jchromb.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 190.Pan Z, Raftery D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Analytical and Bioanalytical Chemistry. 2007;387(2):525–7. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- 191.Chen H, Pan Z, Talaty N, Raftery D, Cooks RG. Combining desorption electrospray ionization mass spectrometry and nuclear magnetic resonance for differential metabolomics without sample preparation. Rapid Communications in Mass Spectrometry. 2006;20(10):1577–84. doi: 10.1002/rcm.2474. [DOI] [PubMed] [Google Scholar]
- 192.Crockford DJ, Holmes E, Lindon JC, Plumb RS, Zirah S, Bruce SJ, et al. Statistical Heterospectroscopy, an Approach to the Integrated Analysis of NMR and UPLC-MS Data Sets: Application in Metabonomic Toxicology Studies. Analytical Chemistry. 2006;78(2):363–71. doi: 10.1021/ac051444m. [DOI] [PubMed] [Google Scholar]
- 193.Crockford DJ, Maher AD, Ahmadi KR, Barrett A, Plumb RS, Wilson ID, et al. 1H NMR and UPLC-MSE Statistical Heterospectroscopy: Characterization of Drug Metabolites (Xenometabolome) in Epidemiological Studies. Analytical Chemistry (Washington, DC, United States) 2008;80(18):6835–44. doi: 10.1021/ac801075m. [DOI] [PubMed] [Google Scholar]
- 194.Dumas M-E, Canlet C, Debrauwer L, Martin P, Paris A. Selection of biomarkers by a multivariate statistical processing of composite metabonomic data sets using multiple factor analysis. Journal of Proteome Research. 2005;4(5):1485–92. doi: 10.1021/pr050056y. [DOI] [PubMed] [Google Scholar]
- 195.Pan Z, Gu H, Talaty N, Chen H, Shanaiah N, Hainline BE, et al. Principal component analysis of urine metabolites detected by NMR and DESI-MS in patients with inborn errors of metabolism. Analytical and Bioanalytical Chemistry. 2007;387(2):539–49. doi: 10.1007/s00216-006-0546-7. [DOI] [PubMed] [Google Scholar]
- 196.Powers R. NMR metabolomics and drug discovery. Magnetic Resonance in Chemistry. 2009 doi: 10.1002/mrc.2461. Published online(Jun 5 2009) [DOI] [PubMed] [Google Scholar]
- 197.Robertson DG, Reily MD, Baker JD. Metabonomics in Pharmaceutical Discovery and Development. J Proteome Res. 2007;6(2):526–39. doi: 10.1021/pr060535c. [DOI] [PubMed] [Google Scholar]
- 198.Wishart DS. Applications of metabolomics in drug discovery and development. Drugs R&D. 2008;9(5):307–22. doi: 10.2165/00126839-200809050-00002. [DOI] [PubMed] [Google Scholar]
- 199*.Stoyanova R, Brown TR. NMR spectral quantitation by principal component analysis. NMR in biomedicine. 2001 Jun;14(4):271–7. doi: 10.1002/nbm.700. Provides an introduction to the application of the principal component analysis statistical technique for the analysis of NMR metabolomics data. [DOI] [PubMed] [Google Scholar]
- 200.Halouska S, Chacon O, Fenton RJ, Zinniel DK, Barletta RG, Powers R. Use of NMR Metabolomics To Analyze the Targets of D-Cycloserine in Mycobacteria: Role of D- Alanine Racemase. Journal of Proteome Research. 2007;6(12):4608–14. doi: 10.1021/pr0704332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201**.Forgue P, Halouska S, Werth M, Xu K, Harris S, Powers R. NMR Metabolic Profiling of Aspergillus nidulans to Monitor Drug and Protein Activity. Journal of Proteome Research . 2006;5(8):1916–23. doi: 10.1021/pr060114v. Demonstrates the differential NMR metabolomics method to easily and rapidly ascertain in vivo drug activity, specificity and toxicity by monitoring drug induced changes in the metabolome of lysed cells using 1D 1H NMR and principal component analysis. [DOI] [PubMed] [Google Scholar]
- 202.Lienemann K, Ploetz T, Pestel S. NMR-based urine analysis in rats: Prediction of proximal tubule kidney toxicity and phospholipidosis. J Pharmacol Toxicol Methods. 2008;58(1):41–9. doi: 10.1016/j.vascn.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 203.Clarke CJ, Haselden JN. Metabolic profiling as a tool for understanding mechanisms of toxicity. Toxicol Pathol. 2008;36(1):140–7. doi: 10.1177/0192623307310947. [DOI] [PubMed] [Google Scholar]
- 204.Park J-C, Hong Y-S, Kim YJ, Yang J-Y, Kim E-Y, Kwack SJ, et al. A Metabonomic Study on the Biochemical Effects of Doxorubicin in Rats Using 1H-NMR Spectroscopy. J Toxicol Environ Health, Part A. 2009;72(6):374–84. doi: 10.1080/15287390802647195. [DOI] [PubMed] [Google Scholar]
- 205.Shi C, Wu C-q, Cao A-m, Sheng H-Z, Yan X-z, Liao M-y. NMR-spectroscopy-based metabonomic approach to the analysis of Bay41-4109, a novel anti-HBV compound, induced hepatotoxicity in rats. Toxicol Lett. 2007;173(3):161–7. doi: 10.1016/j.toxlet.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 206.Dieterle F, Schlotterbeck G, Ross A, Niederhauser U, Senn H. Application of Metabonomics in a Compound Ranking Study in Early Drug Development Revealing Drug-Induced Excretion of Choline into Urine. Chem Res Toxicol. 2006;19(9):1175–81. doi: 10.1021/tx060094b. [DOI] [PubMed] [Google Scholar]
- 207*.Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, et al. Metabolite identification via the Madison Metabolomics Consortium Database. Nature Biotechnology . 2008;26(2):162–4. doi: 10.1038/nbt0208-162. A valuable metabolomics database ( http://mmcd.nmrfam.wisc.edu/) that contains 1D and 2D NMR reference spectra for known metabolites and tools to assist in the analysis of complexe metabolome mixtures. [DOI] [PubMed] [Google Scholar]
- 208.Robinette SL, Zhang F, Bruschweiler-Li L, Bruschweiler R. Web Server Based Complex Mixture Analysis by NMR. Analytical Chemistry (Washington, DC, United States) 2008;80(10):3606–11. doi: 10.1021/ac702530t. [DOI] [PubMed] [Google Scholar]
- 209.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, et al. HMDB: the Human Metabolome Database. Nucleic Acids Research. 2007;35(Database Iss):D521–D6. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Frishman D, Mokrejs M, Kosykh D, Kastenmuller G, Kolesov G, Zubrzycki I, et al. The PEDANT genome database. Nucleic Acids Research. 2003;31(1):207–11. doi: 10.1093/nar/gkg005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Mewes HW, Frishman D, Gueldener U, Mannhaupt G, Mayer K, Mokrejs M, et al. MIPS: a database for genomes and protein sequences. Nucleic Acids Research. 2002;30(1):31–4. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, et al. PANTHER: A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Research. 2003;31(1):334–41. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Bernal A, Ear U, Kyrpides N. Genomes OnLine Database (GOLD): a monitor of genome projects world-wide. Nucleic Acids Research. 2001;29(1):126–7. doi: 10.1093/nar/29.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Black MT, Hodgson J. Novel target sites in bacteria for overcoming antibiotic resistance. Advanced Drug Delivery Reviews. 2005;57(10):1528–38. doi: 10.1016/j.addr.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 215.Jensen LJ, Gupta R, Staerfeldt HH, Brunak S. Prediction of human protein function according to gene ontology categories. Bioinformatics. 2003;19(5):635–42. doi: 10.1093/bioinformatics/btg036. [DOI] [PubMed] [Google Scholar]
- 216.Pouliot Y, Gao J, Su QJ, Liu GG, Ling XB. DIAN: a novel algorithm for genome ontological classification. Genome Research. 2001;11(10):1766–79. doi: 10.1101/gr.183301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Brenner SE. Target selection for structural genomics. Nat Struct Biol. 2000;7(Suppl):967–9. doi: 10.1038/80747. [DOI] [PubMed] [Google Scholar]
- 218.Buysse JM. The role of genomics in antibacterial target discovery. Current Medicinal Chemistry. 2001;8(14):1713–26. doi: 10.2174/0929867013371699. [DOI] [PubMed] [Google Scholar]
- 219.Kramer R, Cohen D. Functional genomics to new drug targets. Nature Reviews Drug Discovery. 2004;3(11):965–72. doi: 10.1038/nrd1552. [DOI] [PubMed] [Google Scholar]
- 220.Roses AD, St Jean PL, Ehm MG. Use of whole-genome association scans in disease gene identification, drug discovery and development. IDrugs. 2007;10(11):797–804. [PubMed] [Google Scholar]
- 221.Powers R, Copeland J, Mercier K. Application of FAST-NMR in Drug Discovery. Drug Discovery Today. 2008;13(3–4):172–9. doi: 10.1016/j.drudis.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222**.Mercier KA, Baran M, Ramanathan V, Revesz P, Xiao R, Montelione GT, et al. FAST-NMR: Functional Annotation Screening Technology Using NMR Spectroscopy. Journal of the American Chemical Society . 2006;128(47):15292–9. doi: 10.1021/ja0651759. NMR fragment-based ligand affinity screens, rapid protein-ligand co-structure determination and bioinformatic tools that compare similarities in active-sites are combined to assign a function to novel, unannotated proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Livingstone CD, Barton GJ. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. CABIOS, Computer Applications in the Biosciences. 1993;9(6):745–56. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- 224.Mirny LA, Shakhnovich EI. Universally Conserved Positions in Protein Folds: Reading Evolutionary Signals about Stability, Folding Kinetics and Function. Journal of Molecular Biology. 1999;291(1):177–96. doi: 10.1006/jmbi.1999.2911. [DOI] [PubMed] [Google Scholar]
- 225.Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annual Review of Biochemistry. 2001;70:209–46. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 226.Hajduk PJ, Huth JR, Fesik SW. Druggability Indices for Protein Targets Derived from NMR-Based Screening Data. Journal of Medicinal Chemistry. 2005;48(7):2518–25. doi: 10.1021/jm049131r. [DOI] [PubMed] [Google Scholar]
- 227.English AC, Groom CR, Hubbard RE. Experimental and computational mapping of the binding surface of a crystalline protein. Protein Engineering. 2001;14(1):47–59. doi: 10.1093/protein/14.1.47. [DOI] [PubMed] [Google Scholar]
- 228.Allen KN, Bellamacina CR, Ding X, Jeffery CJ, Mattos C, Petsko GA, et al. An Experimental Approach to Mapping the Binding Surfaces of Crystalline Proteins. Journal of Physical Chemistry. 1996;100(7):2605–11. [Google Scholar]
- 229.Liepinsh E, Otting G. Organic solvents identify specific ligand-binding sites on protein surfaces. Nature Biotechnology. 1997;15(3):264–8. doi: 10.1038/nbt0397-264. [DOI] [PubMed] [Google Scholar]
- 230.Takai-Igarashi T, Kaminuma T. A pathway finding system for the cell signaling networks database. In Silico Biology. 1999;1(3):129–46. [PubMed] [Google Scholar]
- 231.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Research. 2004;32(Database):D277–D80. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Krieger CJ, Zhang P, Mueller LA, Wang A, Paley S, Arnaud M, et al. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Research. 2004;32(Database):D438–D42. doi: 10.1093/nar/gkh100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Gubernator K, Boehm HJ. Examples of active areas of structure based-design. Methods Princ Med Chem. 1998;6:15–36. [Google Scholar]
- 234.Kubinyi H. Structure-based design of enzyme inhibitors and receptor ligands. Curr Opin Drug Discovery Dev. 1998;1(1):4–15. [PubMed] [Google Scholar]
- 235.Sintchak MD, Nimmesgern E. The structure of inosine 5′-monophosphate dehydrogenase and the design of novel inhibitors. Immunopharmacology. 2000;47(2–3):163–84. doi: 10.1016/s0162-3109(00)00193-4. [DOI] [PubMed] [Google Scholar]
- 236.Mihelich ED, Schevitz RW. Structure-based design of a new class of anti-inflammatory drugs: secretory phospholipase A2 inhibitors, SPI. Biochimica et Biophysica Acta, Molecular and Cell Biology of Lipids. 1999;1441(2–3):223–8. doi: 10.1016/s1388-1981(99)00157-2. [DOI] [PubMed] [Google Scholar]
- 237.Mercier KA, Shortridge M, Powers R. A Multi-Step NMR Screen for the Identification and Evaluation of Chemical Leads for Drug Discovery. Combinatorial Chemistry & High Throughput Screening. 2009;12(3):285–95. doi: 10.2174/138620709787581738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Powers R, Copeland JC, Germer K, Mercier KA, Ramanathan V, Revesz P. Comparison of Protein Active Site Structures for Functional Annotation of Proteins and Drug Design. PROTEINS: Struct Funct Bioinformatics. 2006;65(1):124–35. doi: 10.1002/prot.21092. [DOI] [PubMed] [Google Scholar]
- 239.Mercier Kelly A, Cort John R, Kennedy Michael A, Lockert Erin E, Ni S, Shortridge Matthew D, et al. Structure and function of Pseudomonas aeruginosa protein PA1324 (21–170) Protein Sci. 2009;18(3):606–18. doi: 10.1002/pro.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Shortridge MD, Powers R. Evidence for an Evolutionary Relationship between the Bacterial Type III Secretion System and Eukaryotic Apoptosis. PLoS ONE. 2009 doi: 10.1371/journal.pone.0007442. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lee GM, Craik CS. Trapping Moving Targets with Small Molecules. Science (Washington, DC, U S) 2009;324(5924):213–5. doi: 10.1126/science.1169378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242**.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science (Washington, DC, U S) . 2006;313(5793):1638–42. doi: 10.1126/science.1130258. An exceptional manuscript that experimental defines the range of conformational changes for the DHFR catalytic cycle. The results support the selected-fit model of ligand binding. [DOI] [PubMed] [Google Scholar]
- 243**.Mauldin RV, Carroll MJ, Lee AL. Dynamic Dysfunction in Dihydrofolate Reductase Results from Antifolate Drug Binding: Modulation of Dynamics within a Structural State. Structure (Cambridge, MA, U S) . 2009;17(3):386–94. doi: 10.1016/j.str.2009.01.005. An important companion paper to the manuscript by Boehr et al. (2006) that illustrates the important role that dynamics plays in enzyme activity. The results demonstrate that an inhibitor may also function by disrupting a functionally vital dynamic process. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244**.Lange OF, Lakomek N-A, Fares C, Schroeder GF, Walter KFA, Becker S, et al. Recognition Dynamics Up to Microseconds Revealed from an RDC-Derived Ubiquitin Ensemble in Solution. Science (Washington, DC, U S) . 2008;320(5882):1471–5. doi: 10.1126/science.1157092. Another critical paper in redefining the mechanism of protein-ligand binding and further supporting the selection model. An ensemble of ubiquitin structures determined from RDC data comprises all observed ubiquitin structural changes resulting from complex formation. [DOI] [PubMed] [Google Scholar]
- 245.Teague SJ. Implications of protein flexibility for drug discovery. Nat Rev Drug Discovery. 2003;2(7):527–41. doi: 10.1038/nrd1129. [DOI] [PubMed] [Google Scholar]
- 246.Moy FJ, Chanda PK, Chen J, Cosmi S, Edris W, Levin JI, et al. Impact of Mobility on Structure-Based Drug Design for the MMPs. Journal of the American Chemical Society. 2002;124(43):12658–9. doi: 10.1021/ja027391x. [DOI] [PubMed] [Google Scholar]
- 247.Zintsmaster JS, Wilson BD, Peng JW. Dynamics of Ligand Binding from 13C NMR Relaxation Dispersion at Natural Abundance. J Am Chem Soc. 2008;130(43):14060–1. doi: 10.1021/ja805839y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Peng JW. Communication Breakdown: Protein Dynamics and Drug Design. Structure (Cambridge, MA, U S) 2009;17(3):319–20. doi: 10.1016/j.str.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 249.Palmer AG., III NMR characterization of the dynamics of biomacromolecules. Chem Rev (Washington, DC, U S) 2004;104(8):3623–40. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- 250*.Terstappen GC, Schluepen C, Raggiaschi R, Gaviraghi G. Target deconvolution strategies in drug discovery. Nat Rev Drug Discovery . 2007;6(11):891–903. doi: 10.1038/nrd2410. A collection of analytical techniques for identifying molecular targets obtained from a chemical genetics screening effort are illustrated. The manuscript demonstrates methodologies the pharmaceutical industry could readily adopt to change the current drug discovery paradigm from target-focused to a system biology approach. [DOI] [PubMed] [Google Scholar]
- 251.Strausberg RL, Schreiber SL. From Knowing to Controlling: A Path from Genomics to Drugs Using Small Molecule Probes. Science (Washington, DC, U S) 2003;300(5617):294–5. doi: 10.1126/science.1083395. [DOI] [PubMed] [Google Scholar]
- 252.Dalvit C, Ardini E, Fogliatto GP, Mongelli N, Veronesi M. Reliable high-throughput functional screening with 3-FABS. Drug Discovery Today. 2004;9(14):595–602. doi: 10.1016/S1359-6446(04)03161-7. [DOI] [PubMed] [Google Scholar]
- 253.Bista M, Kowalska K, Janczyk W, Doemling A, Holak TA. Robust NMR Screening for Lead Compounds Using Tryptophan-Containing Proteins. J Am Chem Soc. 2009;131(22):7500–1. doi: 10.1021/ja901863h. [DOI] [PubMed] [Google Scholar]
- 254.D’Silva L, Ozdowy P, Krajewski M, Rothweiler U, Singh M, Holak TA. Monitoring the Effects of Antagonists on Protein-Protein Interactions with NMR Spectroscopy. J Am Chem Soc. 2005;127(38):13220–6. doi: 10.1021/ja052143x. [DOI] [PubMed] [Google Scholar]
- 255.Dalvit C, Fagerness PE, Hadden DTA, Sarver RW, Stockman BJ. Fluorine-NMR experiments for high-throughput screening: Theoretical aspects, practical considerations, and range of applicability. J Am Chem Soc. 2003;125(25):7696–703. doi: 10.1021/ja034646d. [DOI] [PubMed] [Google Scholar]
- 256.Moy FJ, Haraki K, Mobilio D, Walker G, Tabei K, Tong H, et al. MS/NMR: A Structure-Based Approach for Discovering Protein Ligands and for Drug Design by Coupling Size Exclusion Chromatography, Mass Spectrometry, and Nuclear Magnetic Resonance Spectroscopy. Analytical Chemistry. 2001;73(3):571–81. doi: 10.1021/ac0006270. [DOI] [PubMed] [Google Scholar]
- 257.Zartler ER, Hanson J, Jones BE, Kline AD, Martin G, Mo H, et al. RAMPED-UP NMR: Multiplexed NMR-Based Screening for Drug Discovery. J Am Chem Soc. 2003;125(36):10941–6. doi: 10.1021/ja0348593. [DOI] [PubMed] [Google Scholar]
- 258**.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science (Washington, D C) . 1996;274(5292):1531–4. doi: 10.1126/science.274.5292.1531. Seminal paper that demonstrates the practicality of using NMR for ligand-affinity screens and initiated the fragment-based drug discovery trend. [DOI] [PubMed] [Google Scholar]
- 259.Jahnke W, Ruedisser S, Zurini M. Spin label enhanced NMR screening. J Am Chem Soc. 2001;123(13):3149–50. doi: 10.1021/ja005836g. [DOI] [PubMed] [Google Scholar]