Abstract
Aims
Geography-based genetic differentials operating on entire biochemical pathways may reflect different adaptive evolutionary processes that separated populations may have undergone. They may also influence treatment outcome for a variety of drugs – an emerging and important area of study. This research article leverages the International HapMap Consortium data to identify pathway components that differ in genotype frequency for four populations: individuals of Northern European descent from the USA (CEU), individuals from West Africa (YRI), Japan (JPT) and China (CHB).
Materials & methods
By identifying loci with fixed or large frequency differences (δ = 1) between paired population samples (CEU vs YRI, CEU vs CHB, CEU vs JPT, YRI vs CHB, YRI vs JPT and CHB vs JPT), and reconstructing the physiological functions of genes at these loci, we report a list of pathways affected by natural selection during human evolution.
Results
Of the 3.7 million HapMap SNPs, 463 loci (which mapped to 38 genes) were fixed (δ = 1) in at least one population pair. These private loci included four nonsynonymous coding SNPs: rs4536103 (NEUROG3), rs1385699 (EDA2R), rs11946338 (ARHGAP24) and rs4422842 (CACNA1B). A total of four additional genes demonstrated evidence of recent positive selection: three genes in European subjects (IER5L, NPNT and SESTD1) and a single gene in Asian subjects (EXOC6B).
Discussion
Gene ontology and pathway analyses suggest that cellular differentiation, apoptosis and activation of the NF-κB transcription factor vary between populations in genomic regions of fixed (private) SNPs identified in this study. Variability in these pathways may provide important clues into the mechanisms of human adaptation to different environments. An improved understanding of their variability may also help to explain race-specific differences in the treatment outcomes observed for a variety of modern drugs.
Keywords: gene ontology, genomic diversity, HapMap, Ingenuity Pathways Analysis, IPA, pathway, private SNPs, selection
Genomic landscapes are not uniform across chromosomes and they demonstrate strong hetero geneity in humans across the different ethnic groups [1]. These differences could contribute to some of the observed variance in disease susceptibility, onset and treatment outcome. SNPs cataloged within the HapMap resource [101] capture a significant representative fraction of common human sequence variability across different genomic regions, and this resource can be used to test for the presence of population structure related to geography and, presumably, ancestry. Variability between genomic regions can reflect natural selection, gene conversion and mutation [2]; conversely, genome-wide average variability reflects patterns of geographic subdivision and the size of breeding populations, factors associated with gene-frequency evolution [3]. Both provide tantalizing clues to possible selection pressures experienced by ancestral populations [4,5].
HapMap data have provided critical evidence in support of recent positive selection, or selection in favor of new alleles, for genes influencing the pathogenicity of infectious agents (e.g., malaria), genes involved in nutrient metabolizing pathways (e.g., disaccharides and fatty acids) and genes associated with pigmentation differences [6,7]. These genes confer advantages at different geographic and climatic conditions, as well as at different latitudes. Analyses to identify such population-specific effects rely on the fact that, under strong positive selection, an allele may rise to high frequency so rapidly that associations extend for substantial distances along chromosomes, primarily because there has not been sufficient time for significant recombination [8,9].
On a worldwide scale, human populations show large phenotypic variability, particularly for skin color, face and body shape, susceptibility to pathogens and prevalence of disease [10]. However, most of the genetic variation in humans is found within populations rather than among populations or geographic regions [11-13]. Still, many studies have focused on traits or loci showing geographically restricted distribution, or on loci showing drastic allele frequency differences between two regions. These particular cases can indeed reveal important information about local selective pressures or about the demographic histories of different populations [14].
SNPs that are fixed in only one population sample but absent in others are considered ‘private SNPs’ [15]. Populations whose genetic makeup was shaped through thousands of generations in distinct, relatively fixed environments were suddenly exposed to an entirely new world and unfamiliar environment. This introduction to a drastically different environment, composed of distinct pathogens as well as diverse cultural and social influences, may have provided opportunities for an individual’s private SNPs to exist and rapidly adapt and adjust. Investigation of SNPs with major frequency differences among populations may yield valuable insight into pathways governing responses to environmental pathogens, and other functional effects of pathophysiologic or pharmacogenomic interest. With the growing emphasis on genome-wide association studies using high-density genome-wide SNP arrays, and the recent accumulation of large and publicly available datasets, there exists an increasing need for finer resolution within and between population structure to identify significant genomic regions with genetic influence on disease risk [16,17].
Our overall goals were to characterize and determine the regional and chromosomal distribution of HapMap private SNP loci and associated genes among four human populations, to identify enriched biological functions/processes or pathways associated with these SNPs, and to evaluate the ecological and adaptational implications of these loci and genes. Our approach included the application of genomic information available within functional annotation databases (e.g., gene ontology [GO] and pathways), and bioinformatics tools such as Ingenuity Pathways Analysis (IPA), PupaSuite, function analysis and selection tool for SNPs (FastSNP), Integrated haplotype score (iHS), Haplotter and Onto-Express. By enhancing our understanding of genomic regions of extreme frequency differences, this study may provide more insight into genetic variation influencing human disease, and affect our adaptation of ‘personalized’, DNA-based treatment strategies [18].
Materials & methods
Data mining & processing
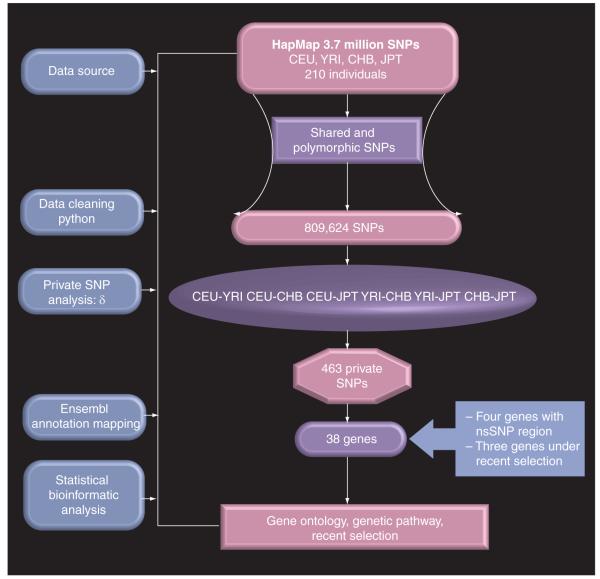
We downloaded the HapMap SNP data [101] for 210 unrelated samples in the four HapMap populations (60 centre d’etude du polymorphisme humain [CEPH] North Americans with European ancestry [CEU]; 60 Yorubans from Ibadan [YRI], Nigeria; 45 Japanese from Tokyo [JPT]; and 45 Han Chinese from Beijing [CHB]). These data represent a complete HapMap Phase II dataset available for each of the representative populations. Two criteria were used to filter the SNPs included in the analysis. First, the SNP should be shared by at least two populations. Second, all filtered SNPs were required to show polymorphism in one or more HapMap population(s) for over 90% of the samples in each population. A computer program using Python™ (Python Software Foundation, NH, USA) [102] was written to export and pre-process SNP genotype information from the databases. There are approximately 3.7 million SNPs in the HapMap data release [19]. Genotypes were summarized for each population. For each dataset, the number of alleles per locus (SNP) was coded to a string of numbers to obtain a full design matrix of alleles (the cells give the number of copies of each major allele for each individual: 0, 1 or 2). Figure 1 depicts our approach to private SNP mining and analysis. The numbers of SNP markers used are shown in Table 1.
Figure 1. Schematic presentation of private SNP mining and analysis strategy.
There are approximately 3.7 million SNPs in the HapMap data release. Genotypes were summarized for each population. For each dataset, the number of alleles per locus (SNP) was coded to a string of numbers to obtain a full design matrix of alleles (the cells give the number of copies of each major allele for each individual: 0, 1 or 2). Two criteria were used to filter the SNPs included in the analysis: first, the SNP should be shared by at least two populations; second, all filtered SNPs were required to show polymorphism in one or more HapMap population(s) for over 90% of the samples in each population. From the total of approximately 3.7 million SNPs in the HapMap data release, only 809,624 SNPs were polymorphic across the population and were eligible for analysis.
CEU: North Americans with European ancestry; CHB: Han Chinese from Beijing; JPT: Japanese from Tokyo; nsSNP: Nonsynonymous SNP; YRI: Yorubans from Ibadan, Nigeria.
Table 1.
Chromosome length and number of SNP markers across the genome in all four populations of the HapMap Phase II dataset*.
| Chromosome | Length (Mb) |
Number of SNP markers in each population |
SNPs genotyped in at least two populations |
SNPs genotyped in at least two populations and polymorphic |
|||
|---|---|---|---|---|---|---|---|
| CEU | CHB | JPT | YRI | ||||
| 1 | 247.2 | 296743 | 300845 | 300845 | 295045 | 283235 | 54622 |
| 2 | 243.0 | 317542 | 318502 | 318502 | 310060 | 302700 | 61155 |
| 3 | 199.5 | 246274 | 246743 | 246743 | 241275 | 231525 | 59955 |
| 4 | 191.3 | 235794 | 236037 | 236037 | 229979 | 221158 | 43242 |
| 5 | 180.9 | 240113 | 240886 | 240886 | 234787 | 227811 | 63441 |
| 6 | 170.9 | 261167 | 265375 | 265375 | 258668 | 249426 | 62085 |
| 7 | 158.8 | 206003 | 206785 | 206785 | 201730 | 193685 | 45227 |
| 8 | 146.3 | 207474 | 211166 | 211166 | 206488 | 196972 | 41581 |
| 9 | 140.3 | 176336 | 178339 | 178339 | 175111 | 166855 | 32255 |
| 10 | 135.4 | 203991 | 206390 | 206390 | 202537 | 194722 | 41097 |
| 11 | 134.5 | 198333 | 199773 | 199773 | 193186 | 187845 | 42856 |
| 12 | 132.3 | 186292 | 187515 | 187515 | 185020 | 173541 | 35841 |
| 13 | 114.1 | 151413 | 153859 | 153859 | 150960 | 144852 | 33299 |
| 14 | 106.4 | 119915 | 120682 | 120682 | 117364 | 113919 | 36834 |
| 15 | 100.3 | 104116 | 104725 | 104725 | 101682 | 98797 | 26962 |
| 16 | 88.8 | 106660 | 106754 | 106754 | 103910 | 100674 | 25828 |
| 17 | 78.8 | 86957 | 86902 | 86902 | 84923 | 82403 | 20534 |
| 18 | 76.1 | 115675 | 116564 | 116564 | 114373 | 109595 | 19203 |
| 19 | 63.8 | 54237 | 54387 | 54387 | 53003 | 51115 | 12658 |
| 20 | 62.4 | 116276 | 116309 | 116309 | 114079 | 111080 | 26591 |
| 21 | 46.9 | 48217 | 50053 | 50053 | 48541 | 45466 | 6948 |
| 22 | 49.7 | 52986 | 54881 | 54881 | 54008 | 50584 | 8300 |
| X | 154.9 | 106780 | 107710 | 107710 | 106026 | 102744 | 9110 |
| Y | 57.8 | 69 | 65 | 65 | 63 | 49 | 0 |
| Total | 3080.4 | 3839363 | 3871247 | 3871247 | 3782818 | 3640753 | 809624 |
Release no. 22 on NCBI B36 assembly, SNP database b126.
CEU: North Americans with European ancestry; CHB: Han Chinese from Beijing; JPT: Japanese from Tokyo; NCBI: National Center for Biotechnology Information; YRI: Yorubans from Ibadan.
Allele frequency difference estimation
Private loci and various levels of marker informativeness were quantified as a measure of ancestry among populations, using the allele frequency difference termed ‘δ procedure’. Marker informativeness for ancestry is defined as the absolute value of the difference of the frequency of a particular allele observed for two ancestral populations. For example, if we let p11 represent the frequency of a reference allele in the first parental population and p21 represent the frequency of the same allele in the second parental population, then the δ value is given by δ = ∣p11 – p21∣. A marker with δ = 1 provides perfect information regarding its ancestry, whereas a marker with δ = 0 carries no information for ancestry [20].
Gene ontology analysis
Onto-Express [103] was used to identify any enriched GO [104] terms in the genes that mapped to private SNPs using functional profiles of GO terms for biological function, biological process and cellular components. A false discovery rate (FDR) of 20% (corrected p-value) after Benjamini–Hochberg (BH) correction [21] was used for significance in these enrichment analyses, allowing us to distinguish between significant biological processes/function and random events.
Pathways & network analyses
A dataset containing gene identifiers was uploaded into the IPA 7.0 (Ingenuity Systems, CA, USA) to map and generate putative networks based on the manually curated knowledge database of pathway interactions extracted from the literature. The network was generated by the input genes, called focus genes, using both direct and indirect relationships/connectivity. These networks were ranked by scores that measured the probability that the genes were included in the network by chance alone. Networks with scores of three or more represent genes not being generated by random chance [22]. The overlapping networks were merged to produce the largest possible network, such that the number of biological relationships to be examined was maximized. The significance threshold for Fisher’s exact test to determine the probability that each biological function and/or disease assigned to that network is due to chance alone was 0.05 or less. Canonical pathways associated with input genes were elucidated with a statistical significance value.
PupaSuite & FastSNP functional analyses
We used PupaSuite [105] to search for and retrieve SNPs with potential phenotypic effects, SNPs that could affect conserved regions identified by alignment with the mouse genome, exonic splicing silencers (ESS) that often contribute to alternative splicing, predicted transcription factor binding sites (TFBS) and changes in amino acids in the encoded proteins. PupaSuite uses the complete set of SNPs cataloged in version 44 of Ensembl, which includes SNP database (dbSNP) 126 genotype data and Sanger-caller Celera SNPs, collectively accounting for over 11 million SNPs. To explore potential functions of the SNPs, we used the FastSNP program [106] that analyzes SNP functions based on up-to-date information extracted from 11 external bioinformatic databases at query time [23].
Recent positive selection analysis
The iHS, which measures the possibility that a gene has undergone recent positive selection, was developed to detect evidence of recent positive selection at a locus. This approach is based on the differential levels of linkage disequilibrium surrounding a positively selected allele compared with the background allele at the same position [6]. We used the Haplotter-calculated iHS [107] to measure if the 38 genes associated with differentially fixed SNPs had undergone recent positive selection among CEU, CHB, JPT and YRI. Using an empirical significance threshold of 0.05, the proportion of SNPs with an ∣iHS∣ greater than 2 for each bin of 50 neighboring SNPs was generated by Haplotter [6]. Haplotter is a web application used to check and display results if a particular gene has been a target of recent positive selection. Large positive and negative values of iHS indicate unusually long haplotypes carrying the ancestral and derived allele, respectively.
Bootstrap analysis
To confirm that the distribution of private SNPs has not been influenced by stochastic effects, bootstrap analysis was employed [24]. All SNPs (809,624) that initially went into the analysis were randomly permuted for each population (CEU, YRI, CHB and JPT) to generate 1000 independent replicates (i.e., 1000 unique datasets). We then calculated the δ value as measured previously with: δ = ∣p11 – p21∣. The SNPs with δ values of 1 (private SNPs) of the randomly permuted datasets between populations were then statistically compared with that of the private SNPs generated from the original dataset. The δ value between the private SNPs and the bootstrapped data were statistically different (p ≤ 0.01), indicating that none of these private SNPs were identified by random chance or by stochastic variation that could affect the robustness of our conclusions.
Results
Allele frequency characterization & racial variation
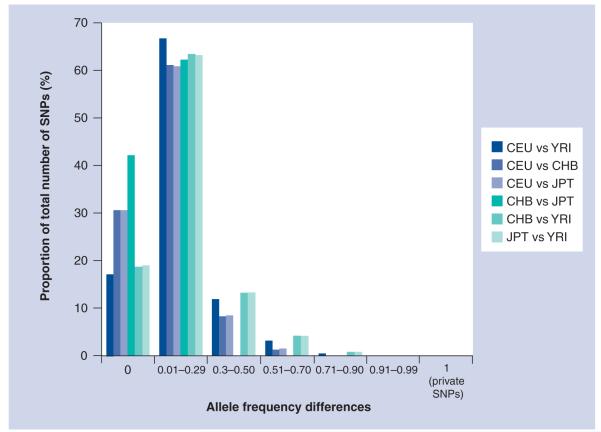
Of the total HapMap SNPs where allele frequencies were available for YRI, CEU, CHB and JPT, we used allele frequency difference (δ) to extract monomorphic SNPs and SNPs with various levels of polymorphism (including 100% informative SNPs between populations). Figure 2 illustrates the distribution of allele frequencies between populations. Of the Phase II HapMap SNPs, 17% were 100% noninformative for ancestry between CEU and YRI, 40% were between CHB and JPT, and 19% for CHB/JPT and YRI. Closely related populations such as CHB and JPT are similar across the genome and only approximately 0.04% of SNPs display δ values of 0.3–0.5, while 8–13% of the SNPs for the same δ range exist between other populations. Interpopulation differences across the 3.7 million Phase II HapMap SNPs demonstrate that 30, 29, 34, 163 and 207 HapMap loci have fixed allele differences between YRI and CEU, CEU and CHB, CEU and JPT, CHB and YRI, and JPT and YRI population groups, respectively.
Figure 2. HapMap SNP allele frequency differences (δ) between HapMap populations.
CEPH: Centre d’etude du polymorphisme humain; CEU: North Americans with European ancestry; CHB: Han Chinese from Beijing; JPT: Japanese from Tokyo; YRI: Yorubans from Ibadan, Nigeria.
In general, the genome-wide SNP allele frequency data showed that most SNP markers do not vary significantly among the major continental populations. Of note, despite allele frequency differences between CHB and JPT populations, none of the 3.7 million sites showed a fixed difference between these two populations (no private SNPs); that is, there was no diallelic SNP wherein one allele was fixed in the CHB population and the other allele was fixed in the JPT population.
Chromosomes harboring private SNPs
The density and distribution of SNPs and genes vary among chromosomes. Across the genome, a total of 463 private SNPs or fixed loci were identified in the HapMap population. Of the total 463 private SNPs, the highest proportion of private loci were observed on the X chromosome (68%), followed by chromosome 2 (9%), chromosome 7 (5%), chromosome 4 (4%) and chromosome 20 (3%). Several chromosomes, including chromosomes 1, 5, 6, 11, 13, 14, 15, 16, 17, 19 and Y, did not have any fixed or private SNPs across these populations (Table 2). The higher number of private SNPs in the X chromosome compared with autosomes might indicate that demographic history of populations affects genetic variation on sex chromosomes in a different way from the genetic variation of autosomes [25].
Table 2.
List of genes mapped to 463 private SNPs (δ = 1) among populations including mouse (Mus musculus) conserved regions.
| Chromosome | Gene ID | No. of SNPs |
Ensembl transcript ID | Location in Ensembl gene |
Conserved region in mouse |
|---|---|---|---|---|---|
| 2 | EXOC6B * | 61 | ENST00000272427 | Intronic | 72642587–72643868 |
| 2 | SESTD1 * | 1 | ENST00000335289 | Intronic | – |
| 3 | CNTN4 | 2 | ENST00000397461 | Intronic | – |
| 3 | MYRIP | 2 | ENST00000302541 | Intronic | – |
| 4 | ARHGAP24 ‡ | 2 | ENST00000380408 | Coding | 86614613–86616183 |
| 4 | PRDM5 | 1 | ENST00000264808 | Intronic | – |
| 4 | LPHN3 | 3 | ENST00000355061 | Intronic | 62606685–62607480 |
| 4 | NDST3 | 2 | ENST00000296499 | Intronic | – |
| 4 | NPNT * | 2 | ENST00000379987 | Intronic | 107094932–107098649 |
| 7 | SDK1 | 3 | ENST00000389530 | Intronic | – |
| 7 | PSCD3 | 2 | ENST00000396741 | Intronic | – |
| 7 | GRM8 | 3 | ENST00000341617 | Intronic | – |
| 8 | DLC1 | 2 | ENST00000276297 | Intronic | 13346713–13348002 |
| 9 | IER5L * | 2 | ENST00000372491 | 5′ upstream | 130977175–130981687 |
| 9 | Q6ZW12 | 1 | ENST00000372490 | Intronic | – |
| 9 | XPA | 2 | ENST00000375128 | Intronic | – |
| 9 | FREQ | 1 | ENSG00000107130 | Intronic | – |
| 9 | Q6ZR86 | 1 | ENST00000371390 | 3′ UTR | 139896075–139897503 |
| 9 | CACNA1B ‡ | 6 | ENST00000277549 | Coding | – |
| 10 | NEUROG3 ‡ | 1 | ENST00000242462 | Coding | 71002005–71004012 |
| 12 | IFNG | 1 | ENST00000229135 | 3′ UTR | 66834494–66840406 |
| 18 | CDH2 | 1 | ENST00000269141 | Intronic | – |
| 20 | DOK5 | 12 | ENST00000262593 | Intronic | 52687136–52687323 |
| 20 | HNRPA3P2 | 3 | ENST00000373803 | Intronic | 34802883–34804166 |
| 21 | DSCAM | 2 | ENST00000400454 | Intronic | 40968804–40969534 |
| 22 | FLJ27365 | 3 | ENST00000396006 | 3′ downstream | 44881459–44883328 |
| X | USP11 | 4 | ENST00000218348 | Intronic | 46984113–46987132 |
| X | ARHGEF9 | 3 | ENST00000374878 | Intronic | 62880250–62882786 |
| X | KIAA1166 | 1 | ENST00000337990 | Intronic | – |
| X | VSIG4 | 1 | ENST00000374737 | Intronic | 65163682–65165088 |
| X | HEPH | 9 | ENST00000374727 | Intronic | 65305279–65305291 |
| X | EDA2R ‡ | 9 | ENST00000396050 | Coding | 65735687–65736483 |
| X | AR | 28 | ENST00000374690 | Intronic | 66704619–66705600 |
| X | KIAA2022 | 2 | ENST00000055682 | Intronic | 73905011–73905786 |
| X | ABCB7 | 3 | ENST00000253577 | Intronic | – |
| X | IL1RAPL2 | 2 | ENST00000344799 | Intronic | – |
| X | PAK3 | 4 | ENST00000360648 | Intronic | 110304872–110305030 |
| X | ARHGAP6 | 4 | ENSG00000047648 | Intronic | – |
Under recent selection.
Contain at least one nonsynonymous coding SNP.
Across each population and genome, CEU and YRI have 119 and 212 private SNPs, respectively, whereas CHB and JPT share all of the SNPs. Using private SNPs as a descriptor, CEU and CHB populations were best discriminated from the CEU and JPT populations by chromosome 7. The YRI population was best separated from CEU and CHB/JPT by private SNPs that are on the X chromosome. Few SNPs that are private in two populations were demonstrated to be private in other populations as well. For example, SNP rs2132498 is fixed in CHB (δ = 1) but not in CEU and YRI (δ = 0). SNPs rs2132498 and rs6979384 are fixed in JPT but not in CEU and YRI (δ = 0). SNP rs7772008 is fixed in CEU, CHB and JPT population but not in YRI.
Chromosome regions with fixed SNPs might indicate regions of strong selection or drift. Using the recent human genome assembly (NCBI build 36, March 2008) and gene-structure annotation from the Ensembl database (version 49, March 2008) [26], we mapped private SNPs using BIOMART [108] options of the Ensembl genome browser. There were 38 genes that mapped to private SNPs across the genome. The chromosomal distribution of these 38 genes is shown in Table 2. Similar to the chromosomal distribution of private SNPs, most genes are on the X chromosome.
Relevant gene networks & pathways
The GO annotation does not cover every aspect of biology. Furthermore, many GO classes are overlapping or redundant, owing to the hierarchical nature of the annotation terms. The use of network analyses for gene data, as an alternative to hierarchical cluster analysis, is particularly helpful for the prioritization of genes within pathways. Therefore, all 38 genes associated with the 463 private SNPs were also characterized using a network-based approach. Through application of IPA, five molecular and cellular functions were identified among these 38 genes (Table 3). The cell–cell signaling interaction had the highest p-value (p = 1.87 × 10−4–1.16 × 10−2) and included ten genes. The functions of these genes were found to be related to lipid metabolism and molecular transport.
Table 3.
IPA summary of associated networks, molecular and cellular functions, diseases and disorders and canonical pathways for the 38 genes mapped to private SNPs.
| IPA categories | Statistical measures | Associated gene(s) |
|---|---|---|
| Associated network functions | Network score * | |
| Immune and lymphatic system development and function, cardiovascular system development and function, cell death |
25 |
AR, CDH2, CNTN4, DOK5, EDA2R, FREQ, GRMB, IFNG,
MYRIP, NEUROG3, VSIG4 |
| Cancer, cellular growth and proliferation, reproductive system disease |
25 |
ABCB7, CACNA18, DSCAM, EXOC6B, HEPH, IER5L,
NPNT, PRDM5, PSCD3, SDK1, XPA |
| Cellular assembly and organization, nervous system development and function, cell morphology |
7 | ARHGAP6, ARHGEF9, DLC1, PAK3 |
| Molecular and cellular functions | p-value ‡ | |
| Cell–cell signaling and interaction | 1.87E-04–1.16E-02 |
AR, CDH2, IFNG, CACNA1B, FREQ, GRM8, CNTN4,
DSCAM, NPNT, PSCD3 |
| Cellular assembly and organization | 2.63E-04–1.16E-02 | ARHGEF9, CDH2, IFNG, PAK3, AR, XPA, CDH2 |
| Cell death | 6.18E-04–1.16E-02 | CDH2, IFNG, PRDM5, AR |
| Gene expression | 8.77E-04–1.16E-02 | AR, IFNG, XPA, USP11, |
| Cellular compromise | 1.07E-03–1.16E-02 | AR, IFNG, USP11, XPA, CDH2, SDK1, ARHGAP24 |
| Associated disease and disorders | p-value ‡ | |
| Nutritional disease | 4.70E-04–4.70E-04 | AR, XPA |
| Reproductive system disease | 6.18E-04–1.16E-02 | CDH2, IFNG, PRDM5, AR, DLC1 |
| Immunological disease | 7.14E-04–9.31E-03 | AR, IFNG, XPA |
| Hematological disease | 1.41E-03–9.31E-03 | IFNG, XPA, ABCB7, AR |
| Cancer | 2.34E-03–1.16E-02 | IFNG, PRDM5, PAK3, AR, CDH2, DLC1, NPNT, XPA |
| Top canonical pathways | p-value ‡ | |
| Interferon signaling | 6.57E-02 | IFNG |
| Protein ubiquitination pathway | 6.7E-02 | IGNG, USP11 |
| Leukocyte extravasation signaling | 6.96E-02 | ARHGAP6, DLC1 |
| Nucleotide excision repair pathway | 7.66E-02 | XPA |
| Glucocorticoid receptor signaling | 1.14E-01 | AR, IFNG, |
| Physiological system development and function | p-value ‡ | |
| Skeletal and muscular system development and function |
1.87E-04–1.16E-02 | AR, CDH2, IFNG |
| Tissue morphology | 1.07E-03–1.16E-02 | CDH2, IFNG, NEUROG3, AR |
| Cardiovascular system development and function | 2.34E-03–1.16E-02 | CACNA1B, ARHGAP24, CDH2, IFNG |
| Connective tissue development and function | 2.34E-03–1.16E-02 | IFNG, AR, ARHGAP24, |
| Hair and skin development and function | 2.34E-03–1.16E-02 | CDH2, IFNG |
| Digestive system development and function | 2.34E-03–2.34E-03 | IFNG |
Networks with scores ≥3 have a 99.9% confidence of not being generated by random chance.
The IPA computes p-values of statistically significant findings by comparing the number of molecules of interest relative to the total number of occurrences of these molecules in all functional/pathway annotations stored in the Pathways Knowledge Base (Fisher’s exact test with p-value adjusted using the Benjamini–Hochberg multiple testing correction).
IPA: Ingenuity Pathways Analysis.
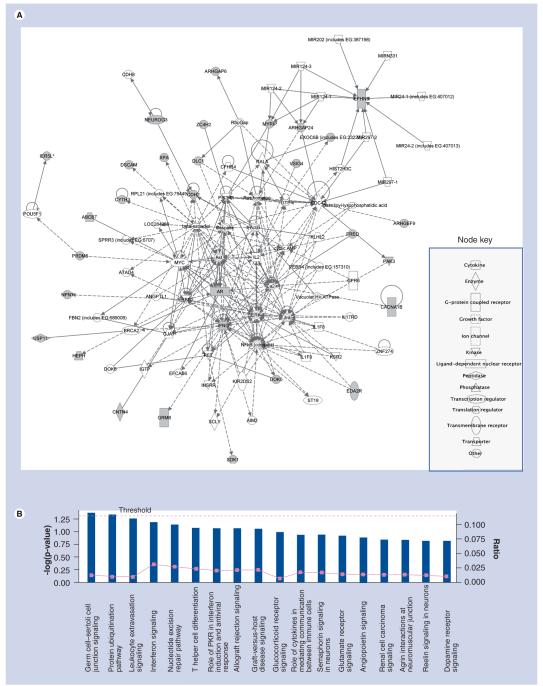
The 38 genes of interest were also overlaid onto a global molecular network developed from literature-reported connectivity that is recorded in the Ingenuity Pathways Knowledge Base (Figure 3A & Box 1). Unfilled nodes are genes that are identified by IPA as being part of the networks, but are not our focus genes. The biological relationship between two nodes is represented in Figure 3A as an ‘edge’. Solid lines indicate a direct interaction whereas dashed lines indicate an indirect interaction. We used IPA to characterize the enrichment of specific pathway components into functionally differentiated gene groups [27]. IPA canonical pathways enriched in the 38 genes associated with private SNPs (p < 0.05) were cell junction signaling and the protein ubiquitination pathway (Figure 3B & Box 1). These most enriched IPA pathways were known to play essential roles in development. The ubiquitination pathway [28] was also found by to be enriched in the differential genes between CEU and YRI.
Figure 3. IPA network and pathway analyses for 38 genes mapped to 463 private SNPs (see right).
(A) IPA network for 38 genes mapped to 463 private SNPs. Genes with shaded nodes are focused genes in our analysis, the others are generated through the network analysis from the Pathways Knowledge Base [112]. Edges are displayed with labels that describe the nature of the relationship between the nodes. The lines between genes represent known interactions, with solid lines representing direct interactions and dashed lines representing indirect interactions. Nodes are displayed using various shapes that represent the functional class of the gene product. (B) The 38 genes linked to 463 private SNPs canonical pathways from IPA. The significance threshold, shown in yellow, represents a p-value of greater than 0.05. The first two sets of functions shown below represent a p-value of less than 0.01. Bars that are above the line indicate significant enrichment of a pathway.
IPA: Ingenuity Pathways Analysis.
Box 1. Summary note for Figures 3, 4 and 6.
The purpose of the Ingenuity Pathways Analysis (IPA) network generation algorithm is to find networks of highly connected focus genes. Genes of interest, that are uploaded by users and directly interact with other genes in the Ingenuity Pathways Knowledge Base (IPKB), are identified as focus genes. Focus genes serve as the ‘seeds’, or focal points, for generating networks. Networks are preferentially enriched for focus genes with the most extensive interactions, and for which the interactions are specific among the other genes in the network and exist in the IPKB. Additional nonfocus genes from the IPKB are then recruited and added to the growing networks. Networks are scored for the likelihood of finding the focus gene(s) in that given network. The higher the score, the lower the probability that you would find the focus gene(s) you see in a given network by random chance. Highly-interconnected subnetworks are likely to represent those that are significantly enriched with genes with specific functional annotations [55]. Genes with no known molecular interactions with other genes will appear as a single node on a network, without connections to other genes or proteins. Canonical pathways associated with input genes of interest were elucidated with a statistical significance value.
Analysis of nonsynonymous SNPs
Since nonsynonymous SNPs (nsSNPS) are more likely to have phenotypic effects in populations that show interpopulation frequency differences [29,30], we searched the private SNP dataset for nsSNPs. Of the 463 SNPs, four SNPs (rs4536103, rs1385699, rs11946338 and rs4422842) were nonsynonymous. These four SNPs mapped to NEUROG3, EDA2R, ARHGAP24 and CACNA1B, respectively (Table 4). The nsSNPs positioned in three of these genes were found to have 0% variability within the YRI population (EDA2R, ARHGAP24 and CACNA1B). This is particularly noteworthy, since the YRI population is evolutionarily older and, in general, more genetically diverse. The reason that these three nonsynonymous coding SNPs across the three genes are highly conserved within the YRI population remains unclear, but it may be that these SNPs in these gene regions serve critical roles in development, and it is conceivable that their selection may be related to an undefined geographic or environmental advantage.
Table 4.
Nonsynonymous coding SNPs, allele frequency distribution, associated genes and their functional role.
| SNP ID | Gene symbol |
Gene name | Cytoband | AA change | Possible functional effect |
Population | Allele frequency |
Minor allele |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | G | C | T | ||||||||
| rs11946338 (A/G) | ARHGAP24 | Rho GTPase activating protein 24 |
4q21.3 | Serine → glycine | Modulator of angiogenesis (signal transduction) |
CHB | 0 | 1 | – | – | A |
| Ancestral allele: A | JPT | 0 | 1 | – | – | A | |||||
| Derived allele: G | YRI | 1 | 0 | – | – | G | |||||
| rs4422842 (C/G) | CACNA1B | Calcium channel, α1B subunit |
9q34.3 | Asparagine → lysine | Voltage-gated calcium channel activity (pain) |
CEU | – | 0 | 1 | – | G |
| Ancestral allele: G | |||||||||||
| Derived allele: C | YRI | – | 1 | 0 | – | C | |||||
| rs4536103 (A/G ) | NEUROG3 | Neurogenin 3 | 10q21.3 | Phenylalanine → serine | Polymorphism contributes to glucose intolerance |
CEU | 0 | 1 | – | – | A |
| Ancestral allele: G | CHB | 0.98 | 0.02 | – | – | G | |||||
| Derived allele: A | JPT | 1 | 0 | – | – | G | |||||
| YRI | 0.98 | 0.02 | – | – | G | ||||||
| rs1385699 (C/T) | EDA2R | Ectodysplasin A2 receptor |
Xq12 | Arginine → lysine | Tumor necrosis factor receptor (apoptosis) |
CEU | – | – | 0.23 | 0.78 | C |
| Ancestral allele: C | CHB | – | – | 0 | 1 | C | |||||
| Derived allele: T | JPT | – | – | 0 | 1 | C | |||||
| YRI | – | – | 1 | 0 | T | ||||||
AA: Amino acid; CEU: North Americans with European ancestry; CHB: Han Chinese from Beijing; JPT: Japanese from Tokyo; YRI: Yorubans from Ibadan, Nigeria.
EDA-A1 and EDA-A2 are two isoforms of ectodysplasin that are encoded by the anhidrotic ectodermal dysplasia (EDA) gene. Genetic variability in the EDA ligand has been associated with loss of hair, sweat glands and teeth [31]. The nsSNP rs1385699, identified within the EDA2 receptor gene, EDA2R, is fixed in both Asian populations, where an R57K substitution in EDA2R has derived allele (T) frequencies of 100% (Table 4) [9]. The EDA2R gene product is involved in the positive regulation of NF-κB transcription factor activity, specifically within the hair follicle, tumor necrosis factor receptor activity, embryonic development and apoptosis [32]. ARHGAP24, on the other hand, is located in the cytoplasm and at cell junctions, where it functions as an important modulator of angiogenesis [33]. ARHGAP24 is a negative regulator of Rho family GTPases, and genetic variation in this gene may therefore also influence actin remodeling, cell polarity, cell migration, differentiation and development [104]. The ARHGAP24 gene also has derived allele (G) frequencies of 100% in CHB and JPT populations. No information exists for ARHGAP24 within the CEU population.
Conversely, the CACNA1B gene has derived allele (C) frequencies of 100% in the CEU, and no information exists for this gene within CHB and JPT populations. CACNA1B is located in the plasma membrane, within a multimeric voltage-gated calcium channel complex [34]. CACNA1B has been proposed as a candidate gene involved in the pathogenetic mechanism underlying several of the neurodevelopmental abnormalities seen within the context of Down Syndrome [35].
The fourth nonsynonymous private SNP is located within Neurogenin 3 (NEUROG3). This gene encodes a member of the subfamily of basic helix-loop-helix (bHLH) transcription factors involved in the differentiation of endocrine progenitor cells in the adult pancreas where it may contribute to the development of diabetes mellitus in some races [36]. NEUROG3 has derived allele (A) frequencies of 100% in JPT, 98% in CHB and YRI, and 0% in the CEU population. The NEUROG3 variant is notable because it is highly differentiated between the CEU and the rest of the populations (YRI, CHB and JPT).
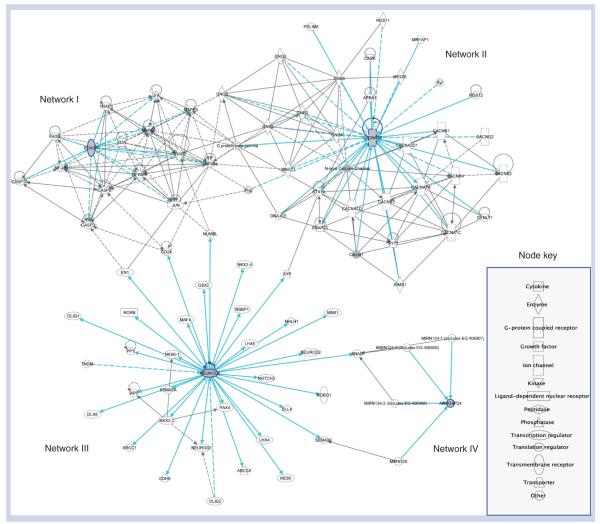
We looked further at the gene network among the four genes mapped to the four nsSNPs (NEUROG3, EDA2R, ARHGAP24 and CACNA1B) using IPA network analysis, and found an independent (nonoverlapping) and disconnected subgene networks for each gene, indicating that these genes are involved in independent biological activities and have no functional commonalities (Figure 4 & Box 1). An IPA summary of associated networks, molecular and cellular functions, diseases and disorders, and canonical pathways for the four genes mapped to nsSNPs are presented in Table 5. The four genes associated with the nsSNPs were further characterized using GO analysis [104]. Data were categorized based on three independent GO terms: biological process, molecular function and cellular component. Similar to the gene network analysis, these genes do not share GO annotation function, process and component terms (data not shown). The reason could be that these genes are involved in diverse and unrelated biological activities that are specific to each geographic region.
Figure 4. Four networks merged and centered around the four genes (NEUROG3, EDA2R, CACNA1B and ARHGAP24) mapped to private nsSNPs.
The light blue line connection indicates focus genes of interest. No overlapping networks exist between ARHGAP24 and the other three focused genes. Edges are displayed with labels that describe the nature of the relationship between the nodes. The lines between genes represent known interactions, with solid lines representing direct interactions and dashed lines representing indirect interactions. Nodes are displayed using various shapes that represent the functional class of the gene product.
Table 5.
An IPA summary of associated networks, molecular and cellular functions, diseases and disorders and canonical pathways for the four genes mapped to nsSNPs.
| IPA categories | Statistical measures | Associated gene(s) |
|---|---|---|
| Associated network functions | Network score * | |
| Cancer, cell death, endocrine system disorder | 3 | EDA2R |
| Cellular development, endocrine system development and function, tissue morphology | 2 | NEUROG3 |
| Cell signaling, molecular transport, vitamin and mineral metabolism | 2 | CACNA1B |
| Molecular and cellular functions | p-value ‡ | |
| Cellular assembly and organization | 1.75E-03–3.77E-02 | ARHGAP24 |
| Cellular compromise | 1.75E-03–7.70E-03 | ARHGAP24 |
| Cell signaling | 2.45E-03–2.67E-02 | CACNA1B |
| Molecular transport | 2.45E-03–3.39E-02 | CACNA1B |
| Vitamin and mineral metabolism | 2.55E-03–2.67E-02 | CACNA1B |
| Associated disease and disorders | p-value ‡ | |
| Cardiovascular disease | 2.10E-03–1.50E-02 | CACNA1B |
| Organisimal injury and abnormality | 2.10E-03–4.86E-02 | CACNA1B |
| Neurological disorder | 3.50E-03–1.40E-02 | CACNA1B |
| Cancer | 1.05E-02–1.47E-02 | NEUROG3 |
| Reproductive system disease | 1.05E-02–1.47E-02 | NEUROG3 |
| Top canonical pathways | p-value | Gene(s) |
| Huntington’s disease signaling | 1.14E+00 | CACNA1B |
| Physiological system development and function | p-value ‡ | |
| Cardiovascular system development and function | 3.51E-04–4.65E-02 | CACNA1B, ARHGAP24 |
| Endocrine system development and function | 3.51E-04–2.09E-02 | NEUROG3 |
| Organ morphology | 3.51E-04–1.47E-02 | CACNA1B |
| Tissue morphology | 3.51E-04–2.09E-02 | NEUROG3 |
| Cellular assembly and organization | 1.75E-03–3.77E-02 | ARHGAP24 |
| Cellular compromise | 1.75E-03–7.7E-03 | ARHGAP24 |
| Connective tissue development and function | 1.75E-03–1.75E-03 | ARHGAP24 |
| Cardiovascular disease | 2.1E-03–1.5E-02 | CACNA1B |
| Organismal injury and abnormalities | 2.1E-03–4.86E-02 | CACNA1B |
| Cell signaling | 2.45E-03–2.67E-02 | CACNA1B |
| Molecular transport | 2.45E-03–3.39E-02 | CACNA1B |
| Nervous system development and function | 2.45E-03–1.78E-02 | CACNA1B, NEUROG3 |
| Vitamin and mineral metabolism | 2.45E-03–2.67E-02 | CACNA1B |
| Neurological disease | 3.5E-03–1.4E-02 | CACNA1B |
| Tissue development | 5.6E-03–1.74E-02 | ARHGAP24 |
| Cancer | 1.05E-02–1.47E-02 | NEUROG3 |
| Reproductive system disease | 1.05E-02–1.47E-02 | NEUROG3 |
| Small molecule biochemistry | 1.53E-02–1.53E-02 | CACNA1B |
| Cellular growth and proliferation | 4.48E-02–4.48E-02 | ARHGAP24 |
Networks with scores ≥3 have a 99.9% confidence of not being generated by random chance.
The IPA computes p-values of statistically significant findings by comparing the number of molecules of interest relative to the total number of occurrences of these molecules in all functional/pathway annotations stored in the Pathways Knowledge Base (Fisher’s exact test with p-value adjusted using the Benjamini–Hochberg multiple testing correction).
IPA: Ingenuity Pathways Analysis; nsSNP: nonsynonymous SNP.
Influence of selection
Using the recent selection model [6] of the web application Haplotter, three genes in the European sample showed strong evidence for recent selection (empirical p < 0.05) and association with cell development and differentiation in the European sample (Table 6). The first gene, NPNT (also named POEM), is on chromosome 4 and has an important role in cell morphology and tissue development [37]. The NPTN gene is implicated to be involved in embryonic development and development and function of various tissues, such as kidney, bone, muscles and endocrine organs [38]. An enhanced expression of NPNT is known to regenerate tubular epithelium and could be a useful tissue and urine biomarker for both the development and evolution of nephrotoxic acute renal injury [39]. The second gene, SESTD1 (on chromosome 2), is enriched in membrane-containing cell fractions and has been implicated in vesicle trafficking. The third gene, IER5L (on chromosome 9), is related to cellular development and embryonic development (Figure 5).
Table 6.
Investigation of recent selection from 38 genes mapped to 463 private SNPs among CEU, YRI and ASN (CHB and JPT) populations.
| Gene Symbol |
Gene name | Cytoband | CEU* | YRI* | ASN* | Population under selection |
|---|---|---|---|---|---|---|
| ABCB7 | ATP-binding cassette, subfamily B, member 7 | Xq13 3 | N/A | N/A | N/A | – |
| AR | Androgen receptor isoform 1 | Xq12 | N/A | N/A | N/A | – |
| ARHGAP24 | Rho GTPase activating protein 24 isoform 1 | 4q21.23 | 0.62904 | 0.756717 | 0.124424 | – |
| ARHGAP6 | Rho GTPase activating protein 6 isoform 4 | Xp22.2 | N/A | N/A | N/A | – |
| ARHGEF9 | Cdc42 guanine exchange factor 9 | Xq11.1 | N/A | N/A | N/A | – |
| CACNA1B | Calcium channel, voltage-dependent, L type | 9q34.3 | 0.18775 | 0.165134 | 0.138348 | – |
| CDH2 | Cadherin 2, type 1 preproprotein | 18q12.1 | 0.78591 | 0.133875 | 0.755444 | – |
| CNTN4 | Contactin 4 isoform a precursor | 3p26.3 | 0.44594 | 0.31137 | 0.500523 | – |
| DLC1 | Deleted in liver cancer 1 isoform 2 | 8p22 | 0.2933 | 0.354531 | 0.499215 | – |
| DOK5 | DOK5 protein isoform b | 20q13.2 | 0.23201 | 0.209368 | 0.420436 | – |
| DSCAM | Down syndrome cell-adhesion molecule isoform | 21q22.2 | 0.23242 | 0.273062 | 0.358825 | – |
| EDA2R | X-linked ectodysplasin receptor | Xq12 | N/A | N/A | N/A | – |
| EXOC6B | Exocyst complex component 6B | 2p13.3 | 0.21042 | 0.05593 | 0.03392 | ASN |
| FLJ27365 | FLJ27365 protein | 22q13.31 | 0.08474 | 0.148687 | 0.271252 | – |
| FREQ | Frequenin homolog | 9q34.11 | 0.38686 | 0.148687 | 0.138348 | – |
| GRM8 | Glutamate receptor, metabotropic 8 precursor | 7q31.33 | 0.38418 | 0.20804 | 0.124215 | – |
| HEPH | Hephaestin isoform a | Xq12 | N/A | N/A | N/A | – |
| HNRPA3P2 | Heterogeneous nuclear ribonucleoprotein A3 pseudogene 2 |
20q11.23 | N/A | N/A | N/A | – |
| IER5L | Immediate early response 5-like protein | 9q34.11 | 0.03883 | 0.54597 | 0.503402 | CEU |
| IFNG | Interferon γ | 12q15 | 0.78705 | 0.468536 | 0.755444 | – |
| IL1RAPL2 | Interleukin 1 receptor accessory protein-like 2 | Xq22.3 | N/A | N/A | N/A | – |
| KIAA1166 | Hepatocellular carcinoma-associated antigen 127 | Xq11.1 | N/A | N/A | N/A | – |
| KIAA2022 | Hypothetical protein LOC340533 | Xq13.3 | N/A | N/A | N/A | – |
| LPHN3 | Latrophilin 3 precursor | 4q13.1 | 0.2934 | 0.638727 | 0.269315 | – |
| MYRIP | Myosin VIIA and Rab interacting protein | 3p22.1 | 0.52788 | 0.643222 | 0.097309 | – |
| NDST3 | N-deacetylase/N-sulfotransferase (heparan) | 4q26 | 0.2588 | 0.761058 | 0.503402 | – |
| NEUROG3 | Neurogenin 3 | 10q21.3 | N/A | N/A | N/A | – |
| NPNT | Nephronectin | 4q24 | 0.04131 | 0.273572 | 0.240473 | CEU |
| PAK3 | Protein activated kinase 3 | Xq22.3 | N/A | N/A | N/A | – |
| PRDM5 | PR domain containing 5 | 4q27 | 0.16844 | 0.886403 | 0.070456 | – |
| PSCD3 | Pleckstrin homology, Sec7 and coiled/coil | 7p22.1 | 0.38686 | 0.761058 | 0.755444 | – |
| Q6ZR86 | Hypothetical LOC100133077 | 9q34 | N/A | N/A | N/A | – |
| Q6ZW12 | Immediate early response 5-like | 9q34 | N/A | N/A | N/A | – |
| SDK1 | Sidekick homolog 1 | 7p22.2 | 0.52241 | 0.353867 | 0.114374 | – |
| SESTD1 | SEC14 and spectrin domains 1 | 2q31.2 | 0.03269 | 0.273572 | 0.105737 | CEU |
| USP11 | Ubiquitin-specific protease 11 | Xp11.3 | N/A | N/A | N/A | – |
| VSIG4 | V-set and immunoglobulin domain containing 4 | Xq12 | N/A | N/A | N/A | – |
| XPA | Xeroderma pigmentosum, complementation group A | 9q22 33 | 0.38686 | 0.13459 | 0.755444 | – |
Genes in bold are genes under recent selection in the specified (bold) population.
Empirical p-value reported by Haplotter [6].
ASN: Asian sample consisting of the CHB and JPT populations; CEU: North Americans with European ancestry; CHB: Han Chinese from Beijing; JPT: Japanese from Tokyo; YRI: Yorubans from Ibadan, Nigeria.
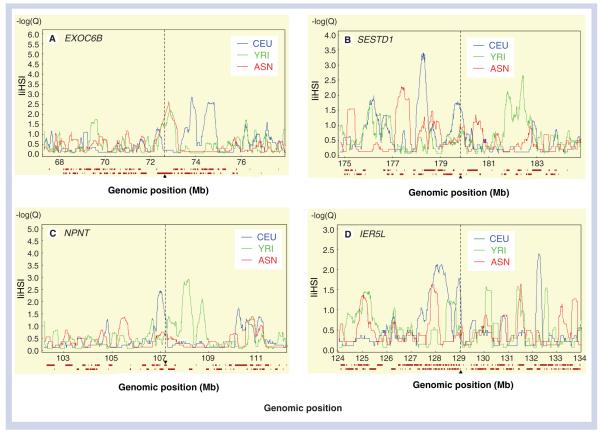
Figure 5. Genes under recent positive selection.
(A) EXOC6B chromosome 2 (72314768:72964925) in ASN (red). (B) SESTD1 chromosome 2 (179799037:179882102). (C) NPNT chromosome 4 (107174209:107250432). (D) IER5L Chr 9 [129018502:129020080] in CEU population (blue).
X-axis is genomic position (Hapmap release 22, dbSNP b126). Y-axis is ∣iHS∣ score. The ∣iHS∣ cut-off for selection is 2. The target gene is marked by a vertical dashed black bar. The 100-kb flanking regions are also shown. The horizontal bars displayed under each panel of the graphic display represent genes present in the region.
ASN: Asian sample made up of the Han Chinese and Japanese populations; CEU: North Americans with European ancestry; ∣iHS∣: Integrated haplotype score; YRI: Yorubans from Ibadan, Nigeria.
In the Asian samples ([ASN] made up of the CHB and JPT populations), we found evidence of selection for the EXOC6B gene (on chromosome 2), which is involved in protein transport and vesicle docking during exocytosis (Figure 5). Two other genes (MYRIP and PRDM5) showed suggestive evidence of recent selection. MYRIP is involved in Rab GTPase binding, apoptosis, retinal melanosomes, carcinogenesis and intracellular protein transport [40], whereas PRDM5 is a zinc finger protein belonging to the tumor suppressor protein family [41]. Both genes also showed suggestive recent selection in the African sample.
The presence of recent selection in the ASN and CEU populations is not surprising given that these two populations may have migrated from Africa and adapted to new environments approximately 50,000–100,000 years ago [42]. The small number of genes (three out of 38) with recent selection in this study suggests that divergence among populations appears to be affected primarily by genetic drift and is, to a lesser degree, due to positive selection.
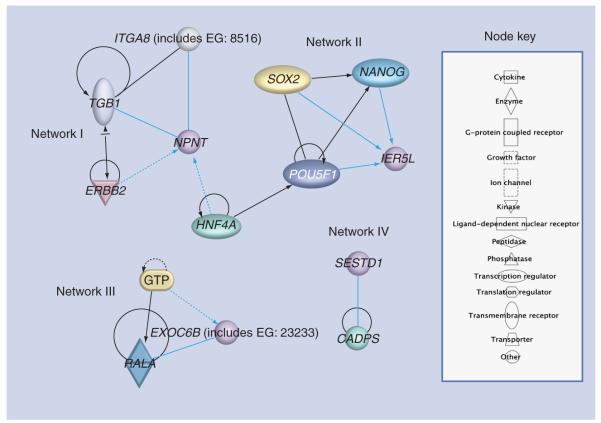
Lastly, we ran the IPA to identify major pathways and networks for genes under recent selection (Figure 6 & Box 1). IPA analysis revealed that these four genes are involved in embryonic, organ and tissue development, and cell-to-cell signaling and interaction (Table 7). Genes from the ASN population (EXOC6B) have independent pathways, whereas the two genes (IER5L and NPNT) that show evidence for recent selection in the CEU population were indirectly linked in their network systems. These genes were also not enriched by shared GO terms (data not shown). The reason could be that these genes that are fixed in the different populations might have been evolved in separate pathways that involve completely different biological activities specific to each geographic region.
Figure 6. Four networks centered around genes that were under recent selection (NPNT, IER5L, EXOC6B and SESTD1).
Nonoverlapping networks were generated among the four genes except between NPNT and IER5L.
The light blue line connection indicates focus genes of interest. The lines between genes represent known interactions, with solid lines representing direct interactions and dashed lines representing indirect interactions.
Table 7.
An IPA summary of associated networks, molecular and cellular functions, diseases and disorders and canonical pathways for the four genes under recent selection.
| IPA categories | Statistical measures |
Associated gene(s) |
|---|---|---|
| Associated network functions | Network score * | |
| Drug metabolism, small molecule biochemistry, molecular transport | 4 | SESTD1 |
| Infection mechanism, lipid metabolism, small molecule biochemistry | 3 | EXOC6B |
| Cellular function and maintenance, cellular growth and proliferation, embryonic development |
3 | IER5L |
| Tissue development, cell morphology, cell-to-cell signaling and interaction | 3 | NPNT |
| Molecular and cellular functions | p-value ‡ | |
| Cell-to-cell signaling and interaction | 1.14E-02–1.14E-02 | NPNT |
| Cellular assembly and organization | 1.14E-02–1.14E-02 | NPNT |
| Associated disease and disorders | p-value | |
| Developmental disorder | 2.80E-03–2.80E-03 | NPNT |
| Renal and urological disease | 2.80E-03–2.80E-03 | NPNT |
| Gastrointestinal disease | 2.33E-02–3.57E-02 | SESTD1, EXOC6B |
| Hepatic system disease | 2.33E-02–2.33E-02 | SESTD1 |
| Physiological system development and function | p-value | |
| Embryonic development | 2.16E-04–7.10E-03 | NPNT |
| Organ morphology | 2.80E-03–2.80E-03 | NPNT |
| Tissue development | 6.03E-03–1.70E-02 | NPNT |
Networks with scores ≥3 have a 99.9% confidence of not being generated by random chance.
The IPA computes p-values of statistically significant findings by comparing the number of molecules of interest relative to the total number of occurrences of these molecules in all functional/pathway annotations stored in the Pathways Knowledge Base (Fisher’s exact test with p-value adjusted using the Benjamini–Hochberg multiple testing correction).
IPA: Ingenuity Pathways Analysis.
Discussion
The goal of this study was to characterize the regional and chromosomal distribution of private SNPs among human populations using 3.7 million HapMap SNPs genotyped in four racial populations: CEU, CHB, JPT and YRI. We report 463 genomic regions containing SNPs with maximal allele frequency differences (100%) between populations. These SNPs may be appropriate to identify the chromosomal regions showing significant local differences in ancestry that are associated with co-descendent phenotypic traits. These SNPs may also help elucidate the genetic basis of interethnic differences in rates of complex diseases through approaches such as genomic ancestry mapping [43].
The recent approval by the US FDA of a combination of isosorbide dinitrate and hydralazine for the treatment of congestive heart failure primarily in self-identified black patients suggests that race can (and sometimes should) be included in treatment decisions, particularly if the inclusion of such information leads to the optimization of a patient’s care [109]. Hence, a detailed examination of population SNP allele-frequency difference may be important in our effort to develop implementation strategies for personalized medicine across populations of different and potentially mixed ethnic origin. Our data revealing very few non-random private SNPs across the human genome within racial populations indicate that knowing one’s ancestry with a high level of accuracy may be extremely challenging [44,45]. We observed 0.001% of the entire HapMap data that were fixed in only one population sample (private SNPs). The vast majority of the SNPs were present in all four populations, which suggests that they have been present since before humans emerged from Africa. Most private SNPs (77%) were found in the African sample. This was anticipated since it had been previously reported that African populations harbor more unique polymorphic alleles than non-African populations [46].
Our analyses demonstrated that the SNP databases in their current status might have some limitations for studies of complex disorders, especially in different ethnic groups, as a result of an incomplete or uneven representation of SNPs along the genome [47]. In critically evaluating our results, it is important to note that our analyses, and hence interpretations, are subject to several limitations. First, many of our analyses relied on data derived from available databases with contents that are, and will continue to be for some time, in a state of change. Therefore, our results represent a snapshot based on currently available data, and ultimately, when the human genome annotation becomes more stable, it will be important to verify these results. Second, the SNP allele frequencies were determined using relatively small sample sizes (see Methods & materials) and preclude any definite conclusion regarding the complete absence of a particular allele in any given population. Moreover, the fundamental theorem under pinning HapMap is the common disease/common variance (CD/CV) hypothesis [48]. How much information we can capture from rare variants is not clear [49]. Several studies have discussed the similarities between human populations in terms of genetic constituents, and hence, a large sample size may enable the detection of small differences in rare outcomes. The analytical caveats associated with each database, such as how surrogates are YRI or CEU to each ancestral population, and how much of the data (e.g., in HapMap) is transferable to the diverse populations in Africa where there is extreme adaptive variation along the various countries is also debatable.
While the synonymous SNPs (neutrally evolving variants) reflect evolutionary time more accurately than the other SNP types [50], the nsSNPs could affected selection through altering protein structure and function. In the present study, of the total 463 private SNPs, only four SNPs were nonsynonymous: rs4536103 (NEUROG3), rs1385699 (EDA2R), rs11946338 (ARHGAP24) and rs4422842 (CACNA1B). Ontology and pathway-based analyses revealed that the expression of these genes impacts cellu lar differentiation, apoptosis, and activation of the NF-κB transcription factor.
The differences in SNP allele frequencies or complete replacement of one allele by others could reflect population-specific selective pressures including diet or other environmental signals. Three genes in European subjects (IER5L, NPNT and SESTD1) and a single gene in Asian subjects (EXOC6B) have been under recent positive selection. Therefore, based on the current data, the majority of the genes do not show evidence for recent positive selection suggesting that other mechanisms (e.g., genetic drift) could be responsible for their variation. Zhang and Li reported that human SNPs showed no frequent positive selection across the genome [51]. Studies in other organisms have also suggested that divergence among populations is primarily affected by drift and, to a lesser degree, by directional selection [52]. A recent study using the HapMap genotypic data demonstrated that negative selection has globally reduced human population differentiation at amino acid-altering mutations, particularly in disease-related genes, while positive selection has ensured the regional adaptation of human populations by increasing population differentiation in gene regions [53].
Since the HapMap genotypic data may not be able to capture all genetic variations among these populations [54], we cannot rule out the possibility that we could have missed some signals of selection because of the untyped and undiscovered SNPs. The results from the deep resequencing projects, such as the SeattleSNPs Project [110] and the National Institute of Environmental Health Sciences (NIEHS) Environmental Genome Project [111], will therefore likely improve our power to identify more differentially fixed genes.
In conclusion, we report the presence of SNPs with extreme allele frequency differences among human populations that could potentially indicate the existence of variants or loci that could potentially be linked to phenotypic variation. Genes neighboring the private SNPs implicate specific biological processes, molecular functions, cellular components and signaling pathways. Although the relevant phenotypes in human studies are generally not known, such loci should be of particular interest in mapping studies of complex traits. Our results can thus provide unique insights into the evolutionary history of variation in gene–environment–race interaction. Follow-up studies and an improved understanding of these genes may help to explain race-specific treatment outcome, as ancient genes are exposed to modern drugs.
Executive summary.
Of 3.7 million HapMap SNPs, 463 loci (which mapped to 38 genes) were fixed (δ = 1).
A total of four of these genes contained private nonsynonymous SNPs: NEUROG3, EDA2R, ARHGAP24 and CACNA1B.
A total of four additional genes showed evidence of recent positive selection: IER5L, NPNT and SESTD1 in European subjects, and EXOC6B in Asian subjects.
Gene ontology and network analyses helped to reconstruct potential and enriched biological functions and processes or pathways associated with these private loci.
Acknowledgments
Financial & competing interests disclosure This work was in part supported by Grant HL74168 (MO), 1R01DK080007 (RAW) and U19AI070235 (TMB).
Footnotes
Ethical conduct of research The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4:E1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paabo S. The mosaic that is our genome. Nature. 2003;421:409–412. doi: 10.1038/nature01400. ■■ Report about the heterogeneity of the genome.
- 3.Slatkin M. Linkage disequilibrium – understanding the evolutionary past and mapping the medical future. Nat. Rev. Genet. 2008;9:477–485. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas S, Akey JM. Genomic insights into positive selection. Trends Genet. 2006;22:437–446. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 5.De La Vega FM, Isaac H, Collins A, et al. The linkage disequilibrium maps of three human chromosomes across four populations reflect their demographic history and a common underlying recombination pattern. Genome Res. 2005;15:454–462. doi: 10.1101/gr.3241705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:E72. doi: 10.1371/journal.pbio.0040072. ■■ Relevant to analyzing positive selection.
- 7.Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. Recent and ongoing selection in the human genome. Nat. Rev. Genet. 2007;8:857–868. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabeti PC, Reich DE, Higgins JM, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 9.Sabeti PC, Varilly P, Fry B, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. ■ Describes positive selection in human populations in greater depth.
- 10.Lewontin R. Human Diversity. Scientific American Library; NY, USA: 1995. [Google Scholar]
- 11.Lewontin R. The apportionment of human diversity. Evol. Biology. 1972;6:381–398. [Google Scholar]
- 12.Barbujani G, Magagni A, Minch E, Cavalli-Sforza LL. An apportionment of human DNA diversity. Proc. Natl Acad. Sci. USA. 1997;94:4516–4519. doi: 10.1073/pnas.94.9.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 14.Balaresque PL, Ballereau SJ, Jobling MA. Challenges in human genetic diversity: demographic history and adaptation. Hum. Mol. Genet. 2007;16(Spec 2):R134–R139. doi: 10.1093/hmg/ddm242. [DOI] [PubMed] [Google Scholar]
- 15.Hinds DA, Stuve LL, Nilsen GB, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. ■ Discusses private SNPs in human population.
- 16.Plagnol V, Wall JD. Possible ancestral structure in human populations. PLoS Genet. 2006;2:E105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoehe MR, Timmermann B, Lehrach H. Human inter-individual DNA sequence variation in candidate genes, drug targets, the importance of haplotypes and pharmacogenomics. Curr. Pharm. Biotechnol. 2003;4:351–378. doi: 10.2174/1389201033377300. [DOI] [PubMed] [Google Scholar]
- 19.International HapMap Consortium. Frazer KA, Ballinger DG, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenberg NA, Li LM, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. Am. J. Hum. Genet. 2003;73:1402–1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false-discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 22.Raponi M, Belly RT, Karp JE, et al. Microarray analysis reveals genetic pathways modulated by tipifarnib in acute myeloid leukemia. BMC Cancer. 2004;4:56. doi: 10.1186/1471-2407-4-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan HY, Chiou JJ, Tseng WH, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–W641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Efron B, Tibshirani R. Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Stat. Sci. 1986;1:54–77. [Google Scholar]
- 25.Keinan A, Mullikin JC, Patterson N, Reich D. Accelerated genetic drift on chromosome X during the human dispersal out of Africa. Nat. Genet. 2009;41:66–70. doi: 10.1038/ng.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flicek P, Aken BL, Beal K, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganter B, Giroux CN. Emerging applications of network and pathway analysis in drug discovery and development. Curr. Opin. Drug Discov. Devel. 2008;11:86–94. [PubMed] [Google Scholar]
- 28.Storey JD, Madeoy J, Strout JL, et al. Gene-expression variation within and among human populations. Am. J. Hum. Genet. 2007;80:502–509. doi: 10.1086/512017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes AL, Packer B, Welch R, et al. Effects of natural selection on interpopulation divergence at polymorphic sites in human protein-coding loci. Genetics. 2005;170:1181–1187. doi: 10.1534/genetics.104.037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myles S, Tang K, Somel M, et al. Identification and analysis of genomic regions with large between-population differentiation in humans. Ann. Hum. Genet. 2008;72:99–110. doi: 10.1111/j.1469-1809.2007.00390.x. [DOI] [PubMed] [Google Scholar]
- 31.Monreal AW, Zonana J, Ferguson B. Identification of a new splice form of the EDA1 gene permits detection of nearly all X-linked hypohidrotic ectodermal dysplasia mutations. Am. J. Hum. Genet. 1998;63:380–389. doi: 10.1086/301984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan M, Wang LC, Hymowitz SG, et al. Two-amino acid molecular switch in an epithelial morphogen that regulates binding to two distinct receptors. Science. 2000;290:523–527. doi: 10.1126/science.290.5491.523. [DOI] [PubMed] [Google Scholar]
- 33.Katoh M, Katoh M. Identification and characterization of ARHGAP24 and ARHGAP25 genes in silico. Int. J. Mol. Med. 2004;14:333–338. [PubMed] [Google Scholar]
- 34.Williams ME, Brust PF, Feldman DH, et al. Structure and functional expression of an ω-conotoxin-sensitive human N-type calcium channel. Science. 1992;257:389–395. doi: 10.1126/science.1321501. [DOI] [PubMed] [Google Scholar]
- 35.Lubec G, Sohn SY. RNA microarray analysis of channels and transporters in normal and fetal Down syndrome (trisomy 21) brain. J. Neural. Transm. 2003;(Suppl):215–224. doi: 10.1007/978-3-7091-6721-2_19. [DOI] [PubMed] [Google Scholar]
- 36.Gradwohl G, Dierich A, LeMeur M, Guillemot F. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl Acad. Sci. USA. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morimura N, Tezuka Y, Watanabe N, et al. Molecular cloning of POEM: a novel adhesion molecule that interacts with α8β1 integrin. J. Biol. Chem. 2001;276:42172–42181. doi: 10.1074/jbc.M103216200. [DOI] [PubMed] [Google Scholar]
- 38.Huang JT, Lee V. Identification and characterization of a novel human nephronectin gene in silico. Int. J. Mol. Med. 2005;15:719–724. [PubMed] [Google Scholar]
- 39.Cheng CW, Ka SM, Yang SM, et al. Nephronectin expression in nephrotoxic acute tubular necrosis. Nephrol. Dial. Transplant. 2008;23:101–109. doi: 10.1093/ndt/gfm496. [DOI] [PubMed] [Google Scholar]
- 40.El-Amraoui A, Schonn JS, Kussel-Andermann P, et al. MyRIP, a novel Rab effector, enables myosin VIIa recruitment to retinal melanosomes. EMBO Rep. 2002;3:463–470. doi: 10.1093/embo-reports/kvf090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duan Z, Person RE, Lee HH, et al. Epigenetic regulation of protein-coding and microRNA genes by the Gfi1-interacting tumor suppressor PRDM5. Mol. Cell. Biol. 2007;27:6889–6902. doi: 10.1128/MCB.00762-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tishkoff SA, Reed FA, Friedlaender FR, et al. The genetic structure and history of Africans and African–Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. ■■ Comprehensive report about African population and migration ‘out of Africa’.
- 43.Redden DT, Divers J, Vaughan LK, et al. Regional admixture mapping and structured association testing: conceptual unification and an extensible general linear model. PLoS Genet. 2006;2:E137. doi: 10.1371/journal.pgen.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolnick DA, Fullwiley D, Duster T, et al. Genetics. The science and business of genetic ancestry testing. Science. 2007;318:399–400. doi: 10.1126/science.1150098. [DOI] [PubMed] [Google Scholar]
- 45.McKeigue PM. Mapping genes underlying ethnic differences in disease risk by linkage disequilibrium in recently admixed populations. Am. J. Hum. Genet. 1997;60:188–196. [PMC free article] [PubMed] [Google Scholar]
- 46.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 47.Dvornyk V, Long JR, Xiong DH, et al. Current limitations of SNP data from the public domain for studies of complex disorders: a test for ten candidate genes for obesity and osteoporosis. BMC Genet. 2004;5:4. doi: 10.1186/1471-2156-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 49.Barnes MR. Navigating the HapMap. Brief Bioinform. 2006;7:211–224. doi: 10.1093/bib/bbl021. ■ Describes the HapMap and its limitations.
- 50.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge University Press; Cambridge, UK: 1983. [Google Scholar]
- 51.Zhang L, Li WH. Human SNPs reveal no evidence of frequent positive selection. Mol. Biol. Evol. 2005;22:2504–2507. doi: 10.1093/molbev/msi240. [DOI] [PubMed] [Google Scholar]
- 52.Whitehead A, Crawford DL. Variation within and among species in gene expression: raw material for evolution. Mol. Ecol. 2006;15:1197–1211. doi: 10.1111/j.1365-294X.2006.02868.x. [DOI] [PubMed] [Google Scholar]
- 53.Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat. Genet. 2008;40:340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- 54.Tantoso E, Yang Y, Li KB. How well do HapMap SNPs capture the untyped SNPs? BMC Genomics. 2006;7:238. doi: 10.1186/1471-2164-7-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc. Natl Acad. Sci. USA. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.HapMap [Accessed 13 March 2007]; Release no. 22, on NCBI B36 assembly, dbSNP b126 www.hapmap.org.
- 102.PYTHON www.python.org.
- 103.Onto-Express http://vortex.cs.wayne.edu.
- 104.Gene Ontology www.geneontology.org.
- 105.PupaSuite http://pupasuite.bioinfo.cipf.es.
- 106.FastSNP http://fastsnp.ibms.sinica.edu.tw.
- 107.iHS and Haplotter http://hg-wen.uchicago.edu.
- 108.Ensembl www.ensembl.org.
- 109.US FDA FDA approves BiDil heart failure drug for black patients. 2005 www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/2005/ucm108445.htm.
- 110.Seattle SNPs Variation discovery resource. http://pga.mbt.washington.edu/
- 111.The National Institute of Environmental Health Science www.niehs.nih.gov/
- 112.Ingenuity Pathways Analysis (IPA) The Ingenuity Knowledge Base. www.ingenuity.com/products/pathways_knowledge.html.