Abstract
Numerous studies have shown that neuronal lipids are highly susceptible to oxidative stress including in those brain areas directly involved in the neurodegenerative process of Alzheimer’s disease (AD). Lipid peroxidation directly damages membranes and also generates a number of secondary biologically active products (toxic aldehydes)that are capable of easily attacking lipids, proteins, and DNA. Accumulating evidence has demonstrated regionally increased brain lipid peroxidation in patients with AD; however, extensive studies on specific targets of lipid peroxidation-induced damage are still missing. The present study represents a further step in understanding the relationship between oxidative modification of protein and neuronal death associated with AD. We used a proteomics approach to determine specific targets of lipid peroxidation in AD brain, both in hippocampus and inferior parietal lobule, by coupling immunochemical detection of 4-hydroxynonenal-bound proteins with 2-D polyacrylamide gel electrophoresis and MS analysis. We identified 4-hydroxynonenal-bound proteins in the hippocampus and inferior parietal lobule brain regions of subjects with AD. The identified proteins play different biological functions including energy metabolism, antioxidant system, and structural proteins, thus impairing multiple molecular pathways. Our results provide further evidence for the role of lipid peroxidation in the pathogenesis of AD.
Keywords: 4-Hydroxynonenal, Alzheimer’s disease, Lipid peroxidation, Protein oxidation, Redox proteomics
1 Introduction
Alzheimer’s disease (AD) is the most common neurodegenerative disorder and the leading cause of dementia in the elderly. Although the initiating events are still unknown, it is clear that AD results from the combination of genetic risk factors with different epigenetic events. Besides the pathological hallmarks of the disease, which include the accumulation of protein deposits in the brain as extracellular neuritic plaques containing amyloid β-peptide (Aβ) and as intraneuronal neurofibrillary tangles, AD brains exhibit evidence of ROS-mediated injury [1, 2]. The oxidative stress hypothesis postulates that accumulation of ROS during aging results in the damage to major components of cells including DNA, RNA, lipids, and proteins. Furthermore, it has been suggested that in the initial stages of the development of AD, Aβ deposition and hyperphosphorylated tau function as compensatory responses to ensure that neuronal cells do not succumb to oxidative damage [3]. However, during AD progression, other events still unknown contribute to exacerbate increasing oxidative stress conditions that ultimately result in the pro-oxidant activity of Aβ [4]. Postmortem analysis of AD brain showed elevated markers of oxidative stress including protein 3-nitrotyrosine, protein carbonyls, lipid oxidation products, and oxidized DNA bases [5–9].
Lipid peroxidation is one of the major sources of oxidative stress in the brain due to its high oxygen utilization, low level of antioxidants, and high level of polyunsaturated fatty acids – a substrate for lipid peroxidation. This complex process involves the interaction of oxygen-derived free radicals with polyunsaturated fatty acids, resulting in a variety of highly reactive electrophilic aldehydes that are capable of facile covalent binding to proteins by forming adducts with cysteine, lysine, or histidine residues. Among the aldehydes formed, malondialdehyde, 4-hydroxynonenal (HNE), and acrolein represent the major products of lipid peroxidation.
The HNE-protein adducts are detected in the brain of patients with AD [10], and HNE is considered to play a crucial role in oxidative injury of biomolecules [11]. HNE is one of the most abundant and toxic aldehydes generated through ROS-mediated peroxidation of lipids, and it is a highly reactive electrophile. This alkenal has been shown to form Michael adducts with the sulfhydryl group of Cys residues, the imidazole group of His residues, and the ε-amino group of Lys and Arg residues on a large number of proteins [12]. HNE has also been shown to react with the sulfhydryl groups of lipoic acid moieties on proteins [13]. Modification of proteins by HNE has the potential to have serious detrimental effects in a cell because of the modification of amino acids and the potential to form cross links in proteins [14]. Since protein oxidation generally leads to its dysfunction, the identification of oxidatively modified proteins, i. e. HNE-modification, provides insights into the biochemical and pathological alterations involved in AD pathogenesis.
Numerous studies have demonstrated increased levels of lipid peroxidation in the brain of patients with AD compared with age-matched controls [15]; however, extensive studies on specific targets of lipid peroxidation-induced damage are still missing. Recently, we showed increased levels of oxidative damage in the brain of subjects with mild cognitive impairment (MCI), an early form of AD [16–19]. To investigate the role of lipid peroxidation in the pathogenesis of AD, we used a redox proteomic approach to identify specific HNE-bound proteins in inferior parietal lobule (IPL) and hippocampus (HP) specimens obtained from short postmortem interval (PMI) AD subjects and corresponding controls.
2 Material and methods
2.1 Control and AD brains
For the present study, IPL samples and HP samples were obtained at autopsy from six late-stage AD subjects and six age- and sex-matched controls. The Rapid Autopsy Program of the University of Kentucky Clinical Center (UK ADC) provided autopsy samples with average PMIs of 2.1 h for AD subjects and 2.9 h for control subjects (Table 1). This short PMI offers a distinct advantage for proteomics analysis, since postmortem changes in brain, a common problem in many studies in AD, are minimal. All AD subjects displayed progressive intellectual decline and met NINCDS-ADRDA Workgroup criteria for the clinical diagnosis of probable AD. Hematoxylin-eosin and modified Bielschowsky staining and 10-D-5 and α-synuclein immunohistochemistry were used on multiple neocortical, hippocampal, entorhinal, amygdala, brainstem, and cerebellum sections for diagnosis. Some patients were also diagnosed with AD plus dementia with Lewy bodies, but the results of this study showed no difference between AD patients with or without the presence of Lewy bodies. Control subjects underwent annual mental status testing and semi-annual physical and neurological examinations as a part of the UK ADC normal volunteer longitudinal aging study and did not have a history of dementia or other neurological disorders. All control subjects had test scores in the normal range. Neuropathologic evaluation of control brains revealed only age-associated gross and histopathologic alterations.
Table 1.
Demographic characteristics of AD and control subjects
| Sample (n = 6) |
Age (years) |
Gender (M/F) |
Postmortem interval (h) |
|---|---|---|---|
| Control | 85.8 ± 4.1 | 4/2 | 2.9 ± 0.23 |
| Late-stage AD | 84.5 ± 5.2 | 4/2 | 2.1 ± 0.47 |
2.2 Sample preparation
Brain samples were minced and suspended in 10 mM HEPES buffer (pH 7.4) containing 137 mM NaCl, 4.6 mM KCl, 1.1 mM KH2PO4, 0.1 mM EDTA, and 0.6 mM MgSO4 as well as proteinase inhibitors: leupeptin (0.5 mg/mL), pepstatin (0.7 µg/mL), type II S soybean trypsin inhibitor (0.5 µg/mL), and PMSF (40 µg/mL). Homogenates were centrifuged at 14000g for 10 min to remove debris. Protein concentration in the supernatant was determined by the BCA method (Pierce, Rockford, IL, USA).
2.3 2-D gel electrophoresis
Samples (200 µg) from IPL and from HP were prepared as previously described [19]. Proteins (200 µg) were precipitated by the addition of ice-cold 100% trichloroacetic acid to obtain a final concentration of 15% trichloroacetic acid and placed on ice for 10 min to allow the precipitation of proteins. Precipitates were centrifuged at 15 800 × g for 2 min. The pellets were washed with 0.5 mL of 1: 1 v/v ethanol/ethyl acetate solution. The samples were then dissolved with 185 µL of rehydration buffer (8 M urea, 2 M thiourea, 20 mM DTT 2.0% w/v CHAPS, 0.2% Biolytes, and bromophenol blue).
For first-dimension electrophoresis, 200 µL of the sample solution was applied to a ReadyStrip™ IPG strip pH 3–10 (Bio-Rad, Hercules, CA, USA). The strips were previously soaked in the sample solution for 1 h to allow the uptake of proteins. The strip was then actively rehydrated in protean IEF cell (Bio-Rad) for 16 h at 50 V. The IEF was performed at 300 V for 2 h linearly; 500 V for 2 h linearly; 1000 V for 2 h linearly, 8000 V for 8 h linearly, and 8000 V for 10 h rapidly. All the processes above were carried out at 22°C. The focused IEF strips were stored at −0°C until second-dimension electrophoresis was performed.
For second-dimension electrophoresis, thawed strips were equilibrated for 10 min in 50 mM Tris-HCl (pH 6.8) containing 6 M urea, 1% w/v SDS, 30% v/v glycerol, and 0.5% DTT and then re-equilibrated for 15 min in the same buffer containing 4.5% iodacetamide in place of DTT. Linear Gradient (8–16%) Precast Criterion Tris-HCl gels (Bio-Rad) were used to perform second-dimension electrophoresis. Precision Protein™ Standards (Bio-Rad) were run along with the sample at 200 V for 65 min.
After electrophoresis, the gels were incubated in fixing solution (7% acetic acid and 10% methanol) for 20 min. Approximately 40 mL of SYPRO Ruby Gel Stain (Bio-Rad) was used to stain the gels for 2 h, on a gently continuous rocker. The gels were placed in deionized water overnight for destaining.
2.4 Western blotting
The same amount of protein samples (200 µg) was used for 2-D-immunoblotting analysis, and the electrophoresis was carried out as described above. The proteins from the second-dimension electrophoresis gels were transferred to nitrocellulose membranes (Bio-Rad) using a Transblot-Blot SD Semi-Dry Transfer Cell (Bio-Rad) at 15 V for 2 h. HNE-protein adducts were detected on the nitrocellulose paper using a primary rabbit antibody (Intergen) specific for HNE-bound protein (1: 100), followed by a secondary goat anti-rabbit IgG antibody coupled to alkaline phosphatase (Sigma, St Louis, MO, USA). The resultant stain was developed using BCIP/NBT solution (SigmaFast tablets; Sigma).
2.5 Image analysis
Images from SYPRO Ruby-stained gels (n = 6 controls and n = 6 AD), used to measure protein content, were obtained with a UV transilluminator (λex = 470 nm, λem = 618 nm; Molecular Dynamics, Sunnyvale, CA, USA). The 12 nitrocellulose blots were scanned and saved in TIF format using a Scanjet 3300C (Hewlett Packard). PDQuest 2-D Analysis Software (Bio-Rad) was used for matching and analysis of visualized protein spots among differential gels and membranes to compare protein and HNE immunoreactivity content between AD IPL and AD HP samples and respective age-matched controls. Powerful automatching algorithms quickly and accurately match gels or blots and sophisticated statistical analysis tools identify experimentally significant spots. The principles of measuring intensity values by 2-D Analysis Software were similar to those of densitometric measurement. The average mode of background subtraction was used to normalize intensity values, which represents the amount of protein (total protein on gel and HNE-bound protein on the membrane) per spot. After completion of spot matching, the normalized intensity of each protein spot from individual gels (or membranes) was compared between groups using statistical analysis. Statistical significance was assessed by a two-tailed Student’s t-test. p values <0.05 were considered significant for comparison between control (age-matched subjects) and experimental data (AD subjects). This is the method of statistical analysis most appropriate for proteomic analysis of small number of protein spots in contrast to statistical approach used for large analysis common in gene micro-array studies [20].
2.6 Trypsin digestion
In-gel digestion on selected gel spots was performed according to Thongboonkerd et al. [21]. The selected protein spots were excised with a clean blade and transferred into clean microcentrifuge tubes. The protein spots were then washed with 0.1M ammonium bicarbonate (NH4HCO3) at room temperature for 15 min. ACN was added to the gel pieces and incubated at room temperature for 15 min. The solvent was removed, and the gel pieces were dried in a flow hood. The protein spots were incubated with 20 µL of 20 mM DTT in 0.1 NH4HCO3 at 56°C for 45 min. The DTT solution was then removed and replaced with 20 µL of 55 mM iodoacetamide in 0.1 NH4HCO3. The solution was incubated at room temperature in the dark for 30 min. The iodoacetamide was removed and replaced with 0.2 mL of 50 mM NH4HCO3 and incubated at room temperature for 15 min. ACN (200 µL) was added, and after a 15-min incubation, the solvent was removed and the gel spots were dried in a flow hood for 30 min. The gel pieces were rehydrated with 20 ng/µL methylated trypsin (Promega, Madison, WI, USA) in 50 mM NH4HCO3 with the minimal volume to cover the gel pieces. The gel pieces were chopped into smaller pieces and incubated at 37°C overnight in shaking incubator.
2.7 MS
All mass spectra reported in this study were acquired at the Department of Pharmacology in the University of Louisville School of Medicine. A Spec 2E MALDI TOF (MALDI-time of flight) mass spectrometer operated in the reflectron mode was used to generate peptide mass fingerprints. Peptides resulting from in-gel digestion with trypsin were analyzed on a 384 position, 600 µm AnchorChip™ Target (Bruker Daltonics, Bremen, Germany) and prepared according to AnchorChip recommendations (AnchorChip Technology, Rev. 2, Bruker Daltonics). Briefly, 1 µL of digest was mixed with 1 µL of CHCA (0.3 mg/mL in ethanol: acetone, 2: 1 ratio) directly on the target and allowed to dry at room temperature. The sample spot was washed with 1 µL of a 1% TFA solution for approximately 60 s. The TFA droplet was gently blown off the sample spot with compressed air. The resulting diffuse sample spot was recrystallized (refocused) using 1 µL of a solution of ethanol: acetone: 0.1% TFA (6: 3: 1 ratio). Reported spectra are a summation of 100 laser shots. External calibration of the mass axis was used for acquisition and internal calibration using either trypsin autolysis ions or matrix clusters and was applied postacquisition for accurate mass determination.
The MALDI spectra used for protein identification from tryptic fragments were searched against the NCBI protein databases using the MASCOT search engine (http://www.matrixscience.com). Peptide mass fingerprinting used the assumption that peptides are monoisotopic, oxidized at methionine residues, and carbamidomethylated at cysteine residues [8, 22, 23]. Up to one missed cleavage, 50 ppm measurement tolerance, oxidation at methionine (variable modification) and carbamidomethyl cysteine (fixed modification) were considered. Identifications were accepted when the probability-based MOWSE protein score was significant according to MASCOT [24].
Probability-based MOWSE scores were estimated by comparison of search results against estimated random match population and were reported as −10*LOG10(p), where p is the absolute probability of random protein identification. The criteria for positive identification of proteins were set as follows: (i) at least four matching peptide masses, (ii) 50 ppm or better mass accuracy, and (iii) Mr and pI of identified proteins should match estimated values obtained from images analysis.
2.8 Immunoprecipitation
To confirm that correct identifications of the proteins were determined by MS, the identity of MnSOD, whose MOWSE score was less than 60, was validated using immunochemical methods. Control or AD samples (200 µg) were first precleared by incubation with protein A-agarose (Amersham Pharmacia Biotech, Piscataway, NJ, USA) for 1 h at 4°C. Samples were then incubated overnight with the relevant antibody (MnSOD antibody) followed by 1 h of incubation with protein A-agarose, then washed three times with buffer B (50 mM Tris-HCl pH 8.0, 150 mM NaCl, and 1% NP40). Proteins were resolved by SDS-PAGE followed by immunoblotting on a nitrocellulose membrane (Bio-Rad). MnSOD was detected by the alkaline phosphate-linked secondary antibody and BCIP/NBT solution (Sigma).
2.9 Enzyme assay
2.9.1 ATP synthase
Mitochondrial ATP synthase activity was measured spectrophotometrically at 340 nm by coupling the production of ADP to the oxidation of NADPH via the pyruvate kinase and lactate dehydrogenase reaction (coupled assay) as described [25]. The reaction mixture (0.2 mL final volume) contained: 100 mM Tris (pH 8.0), 4 mM Mg-ATP, 2 mM MgCl2, 50 mM KCl, 0.2 mM EDTA, 0.23 mM NADH, 1 mM phosphoenolpyruvate, 1.4 unit of pyruvate kinase, 1.4 unit of lactate dehydrogenase, and about 25–50 µg of proteins (brain homogenates), and was assayed at 30°C. The assay was carried out in a microtiter plate reader (Bio-Tek Instrument, Winooski, VT, USA). Specific activity was also calculated for ATP synthase in AD and control IPL.
2.9.2 Aconitase activity
According to the recent literature, decreased aconitase enzyme activity is a sensitive and specific indicator of oxidative damage during aging, Parkinson’s, and other neurodegenerative diseases [26]. Because of its role in cellular energy production, aconitase function is well positioned to serve as an important marker of biological decline.
Aconitase activity in brain homogenates (25–50 µg of protein) was assayed immediately after thawing by following absorbance at 340 nm at 22°C in a 1.0-mL reaction mixture containing 50 mM Tris-HC1, pH 7.4, 30 mM sodium citrate, 0.6 mM MnC12, 0.2 mM NADP+, and 1–2 units/mL of isocitrate dehydrogenase. One milliunit of aconitase activity catalyzed the formation of 1 nmol of isocitrate/min. Specific activity was calculated for aconitase in AD and control HP.
2.9.3 MnSOD (SOD2) activity
SOD2 activity was measured spectrophotmetrically at 550 nm as the inhibition of cytochrome c by superoxide radical. Superoxide was produced by converting xanthine to uric acid via xanthine oxidase. A reaction “cocktail” consisting of 216 mM potassium phosphate buffer (pH 7.8), 10.7 mM EDTA, 1.1 mM cytochrome c solution, 0.108 mM xanthine solution was added to brain homogenates (25–50 µg of protein). To this cocktail, xanthine oxidase enzyme was added. The reaction (final volume of 0.2 mL) was monitored at 25°C.
2.10 Statistics
Two-tailed, Student’s t-tests were used to analyze differences in protein levels between AD and control subjects. A p-value of less than 0.05 was considered statistically significant. The significance of the change in HNE-modification of specific proteins in the proteomics study also was evaluated by Student’s t-tests. A value of p<0.05 was considered statistically significant. The HNE level of each spot was normalized to the protein level of the corresponding spot on the gel. According to Maurer and Peters [20], generalized statistical tests applicable to proteomics data are unavailable, in contrast to microarray data. Because of the small number of proteins identified in this study, compared with the large quantities of genes that are analyzed by microarrays, Student’s t-tests were used for analysis.
3 Results
3.1 IPL proteomics studies
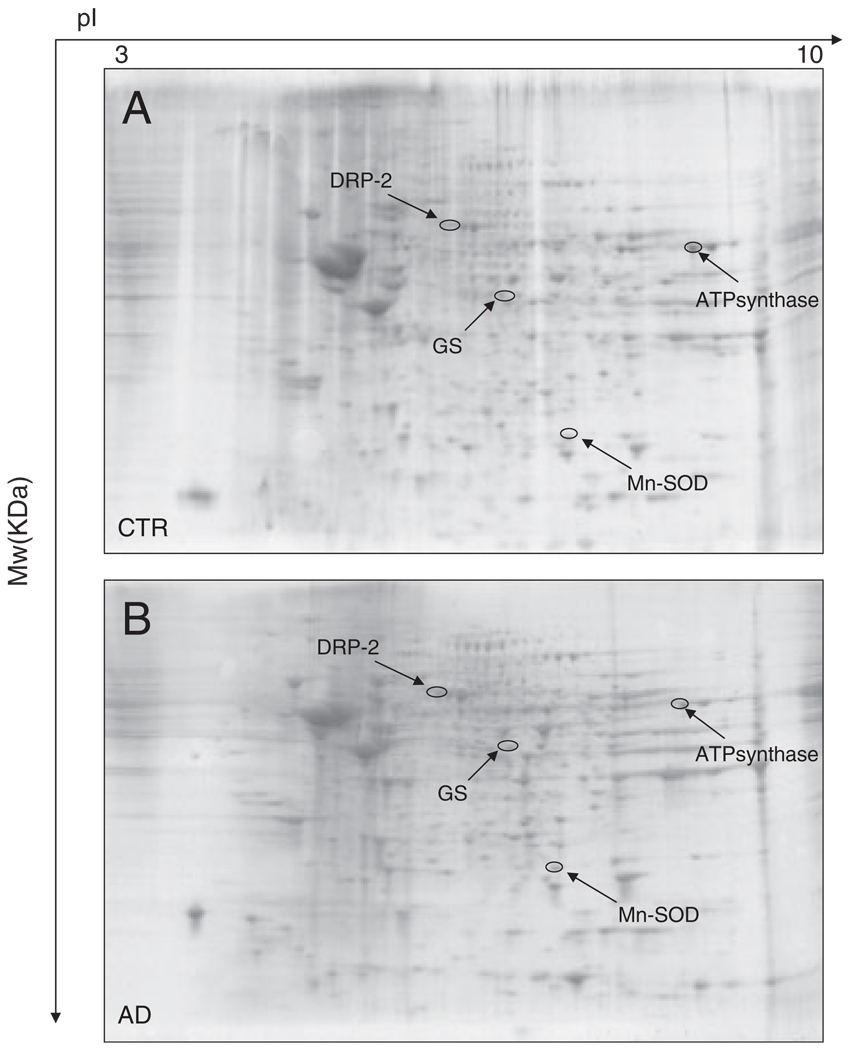
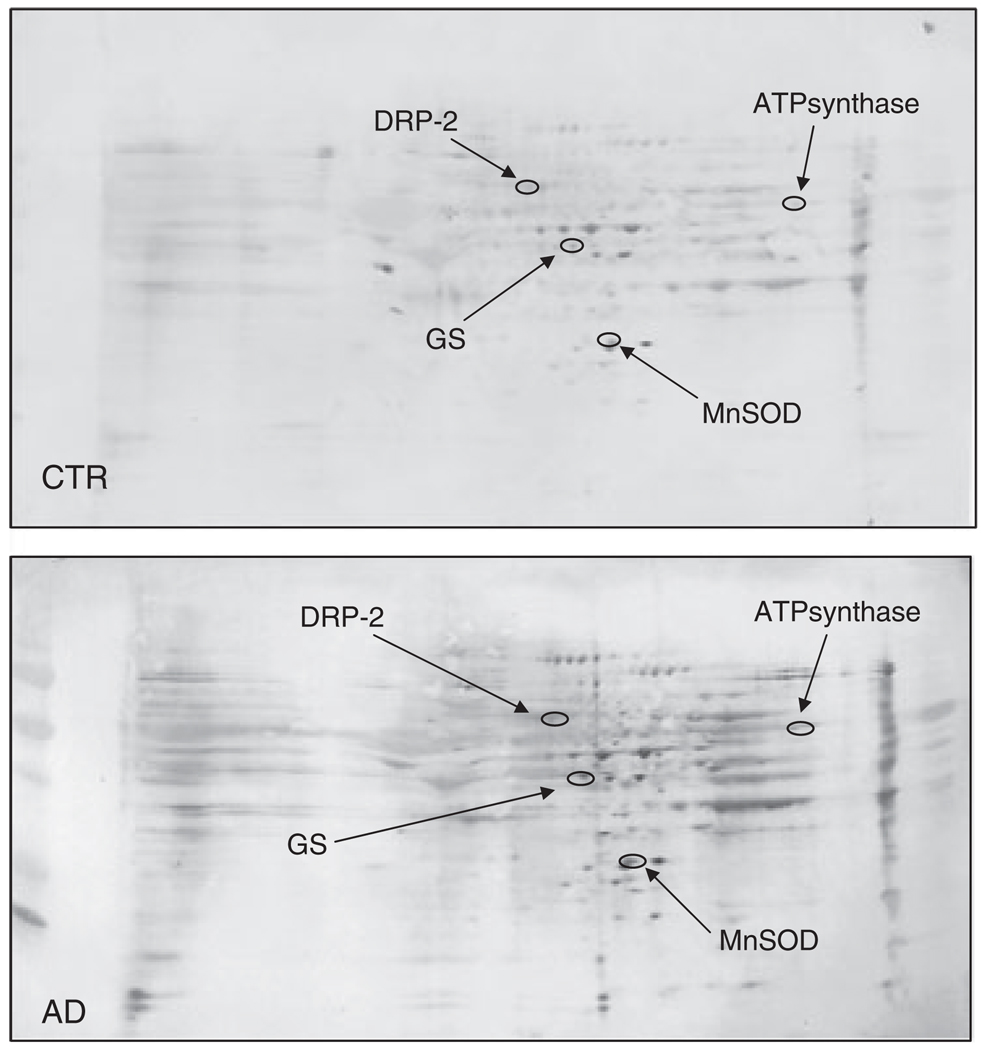
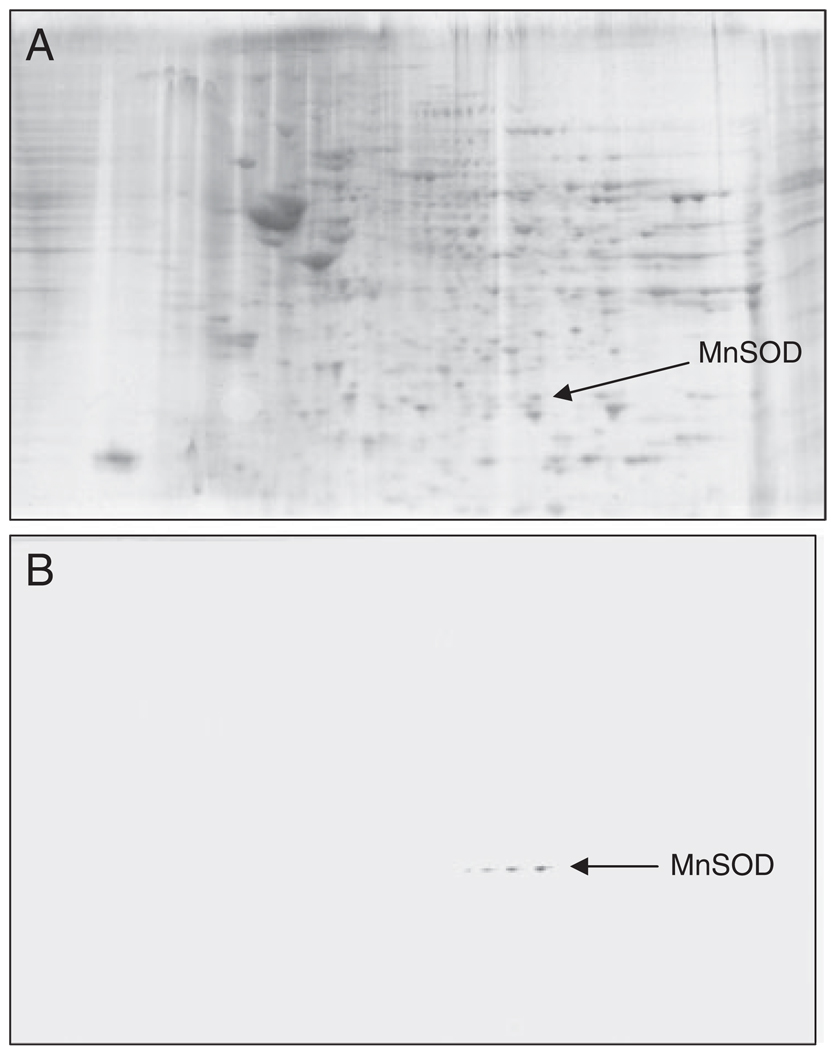
Western blot and subsequent immunochemical detection of protein-bound HNE allowed identification of HNE-modified proteins in the IPL and HP isolated from postmortem brains of AD subjects in comparison with their age-matched controls. These data confirm previous studies from our laboratory and from others that showed increased levels of lipid peroxidation in AD brain compared with control brain [10, 15, 27, 37]. We used a parallel approach to quantify the protein levels by SYPRO Ruby staining and the extent of HNE binding by immunochemistry (Fig. 1 and Fig. 2). SYPRO Ruby fluorescent stain achieves a linear and sensitive staining of gel slabs, and immunoblotting with HNE antibody allows specific detection of HNE adducts in the IPL and HP of AD subjects in comparison with their age-matched controls. The specific HNE-bound levels were determined by dividing the HNE level of a protein spot on the blotted membrane by the protein level of its corresponding protein spot on the gel. Figure 1 shows representative 2-D-electrophoresis gels of IPL from AD brain and control brain (CTR) after SYPRO Ruby staining. Figure 2 shows representative 2-D Western blots from AD IPL and control IPL (CTR). Comparing the densitometric intensities of individual spots, we determined that four proteins were statistically significantly modified by HNE in IPL of AD subjects when compared with those of controls. The HNE-modified proteins identified in the IPL of AD subjects were as follows: dihydropyrimidinase-related protein 2 (DRP-2), glutamine synthase, ATP synthase, and MnSOD (Table 2). Although the MOWSE score for MnSOD was less than 60, the correct identification of this protein was validated by immunochemistry (Fig. 3). The presence of four spots with the same Mw and different pI likely is due to fragmentation of MnSOD with post-traslational modification [28, 29]. This result also indicates that identification of proteins by MS is equivalent to those by immunochemistry, thus providing confidence in the redox proteomics identification of the other HNE-modified proteins reported in this study.
Figure 1.
Representative SYPRO Ruby 2-D gels of the IPL from control (A) or AD (B) brain. Proteins (200 µg) were separated on immobilized pH 3–10 IPG strips followed by separation on an 8–16% gradient SDS-PAGE gels.
Figure 2.
2-D HNE immunoblots from AD and control subjects (IPL). Positions of the four identified proteins are shown on the blots. Relative changes in HNE immunoreactivity, after normalization of the immunostaining intensities to the protein content, were significant for four proteins. See text.
Table 2.
Summary of the brain proteins identified by redox proteomics that have elevated HNE bound in AD subjects when comparing these proteins with controls
| HNE-modified proteins |
Brain region |
Expression level (AD versus CTR) |
Fold increase (AD versus CTR) |
Peptides matched |
% Coverage |
pl, Mr (kDa) |
MOWSE score |
p-Value |
|---|---|---|---|---|---|---|---|---|
| ATP synthase | IPL | ↓ | 2.56 ± 0.54 | 13/18 | 32 | 9.21, 59.7 | 165 | <0.04 |
| Glutamine synthase | IPL | ↔ | 1.89 ± 0.31 | 5/9 | 13 | 6.43, 42.6 | 69 | <0.02 |
| MnSOD | IPL | ↑ | 1.70 ± 0.37 | 8/20 | 21 | 6.97, 21.7 | 58 | <0.02 |
| Dihydropyrminidase | IPL | ↔ | 3.23 ± 0.53 | 10/42 | 28 | 5.95, 62.7 | 77 | <0.05 |
| like-2 | ||||||||
| α-Enolase | HP | ↑ | 3.90 ± 0.44 | 9/21 | 35 | 6.99, 47 | 119 | <0.002 |
| Aconitase | HP | ↔ | 1.85 ± 0.26 | 9/20 | 27 | 7.36, 86.1 | 60 | < 0.02 |
| Aldolase | HP | ↑ | 2.78 ± 0.3 | 6/20 | 30 | 6.46, 39.6 | 68 | < 0.04 |
| Peroxiredoxin 6 | HP | ↔ | 4.20 ± 0.5 | 9/27 | 46 | 6.00, 25.1 | 109 | < 0.05 |
| α-Tubulin | HP | ↔ | 2.89 ± 0.3 | 11/19 | 29 | 4.9, 50.1 | 130 | < 0.02 |
For each protein, the HNE immunoreactivity/protein expression values were averaged (n = 6) and expressed as fold-increase compared to control ± SEM. Also shown are the differences in protein levels of the identified proteins compared with controls (↑, increased levels; ↔, unchanged levels; ↓, decreased levels). The p-value listed is the statistical significance of elevated HNE-bound proteins relative to control (see text). IPL, inferior parietal lobule; HP, hippocampus.
Figure 3.
Validation of MnSOD identified by MS using immunoprecipitation and Western blot analysis: (A) represents gel of total HP protein, a box is drawn around the MnSOD, which is one of the HNE-modified proteins. (B) Represents blot probed with HNE antibody. n = 3.
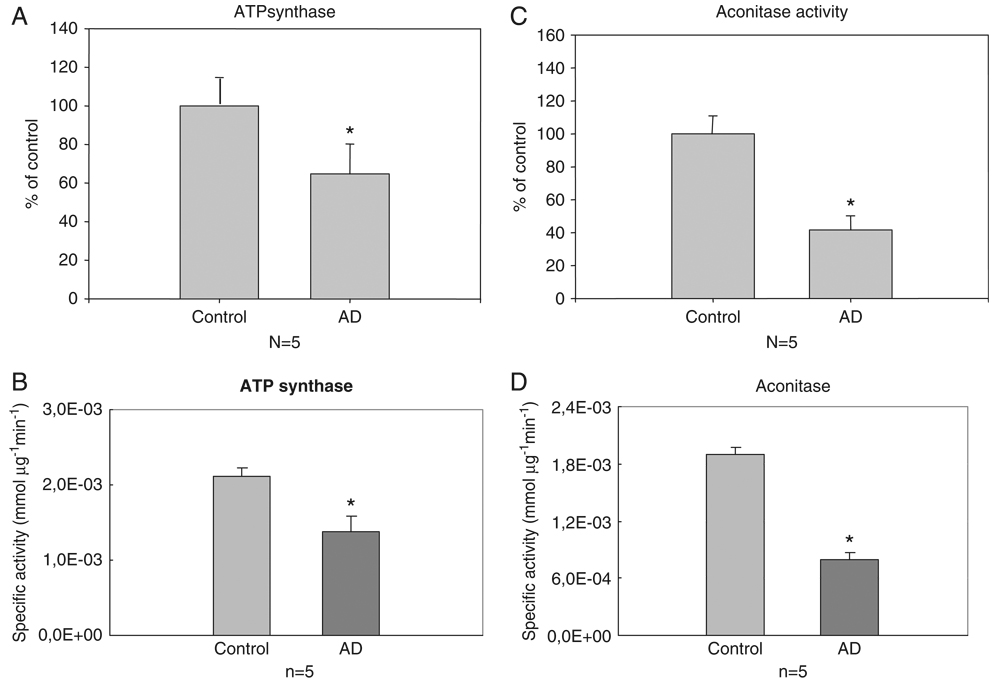
Since oxidatively modified proteins generally have diminished activity [30, 31], we measured the enzyme activities of ATP synthase and MnSOD. ATP synthase was not only oxidatively modified, but also oxidatively inhibited, demonstrated by the decreased activity (48%) in IPL of AD subjects when compared with controls (Fig. 6A and B). We also found decreased protein levels of this enzyme in AD samples compared with controls (Table 2), suggesting that impairment of ATP synthase results from both down-regulation of its protein levels together with its oxidative modification. In contrast, MnSOD enzyme activity did not show any significant change between AD and control samples (data not shown), although we found increased protein levels of this enzyme in AD samples compared with controls (Table 2).
Figure 6.
(A) Activity of ATP synthase in AD IPL samples compared with controls. The activity of ATP synthase is significantly decreased in AD samples. Bars represent mean ± SE *p<0.05; n = 5 for each group. (B) Specific activity of ATP synthase in AD IPL versus control. Bars represent mean ± SE *p<0.05; n = 5 for each group. (C) Activity of aconitase in AD samples compared with controls (HP). The activity of aconitase is significantly decreased in AD samples. Bars represent mean ± SE *p<0.05; n = 5 for each group. (D) Specific activity of aconitase in AD HP versus control. Bars represent mean ± SE *p<0.05; n = 5 for each group.
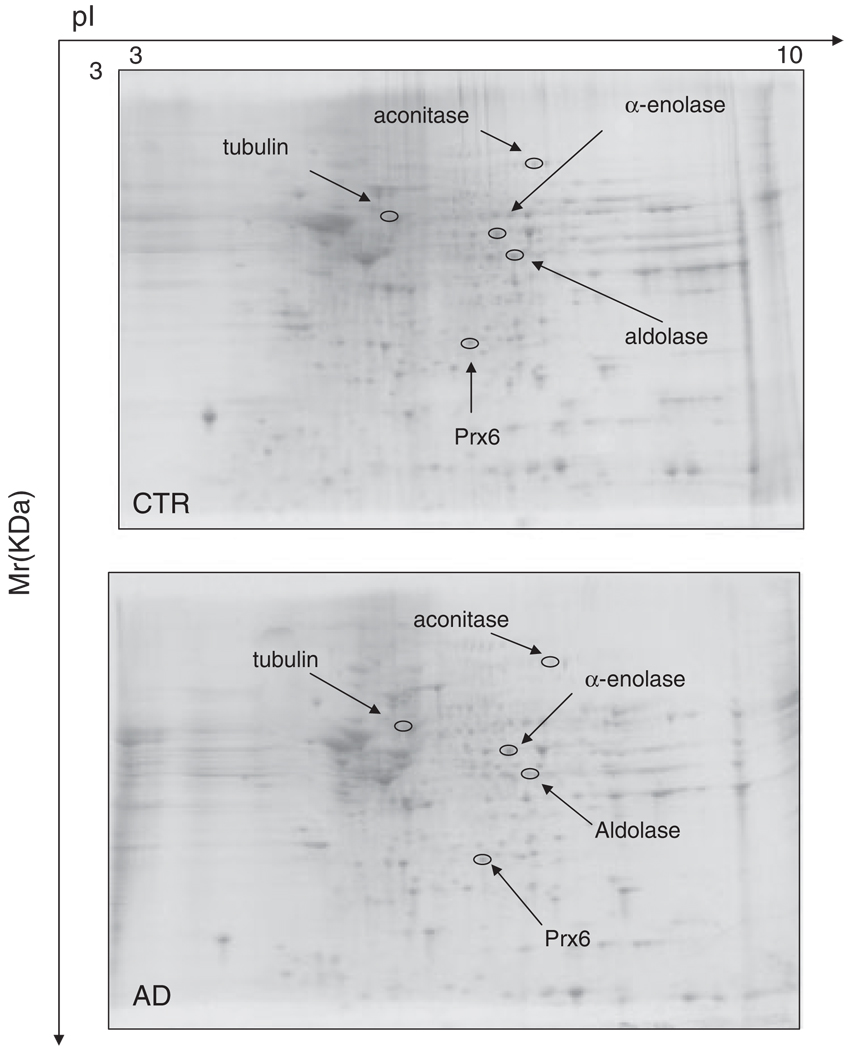
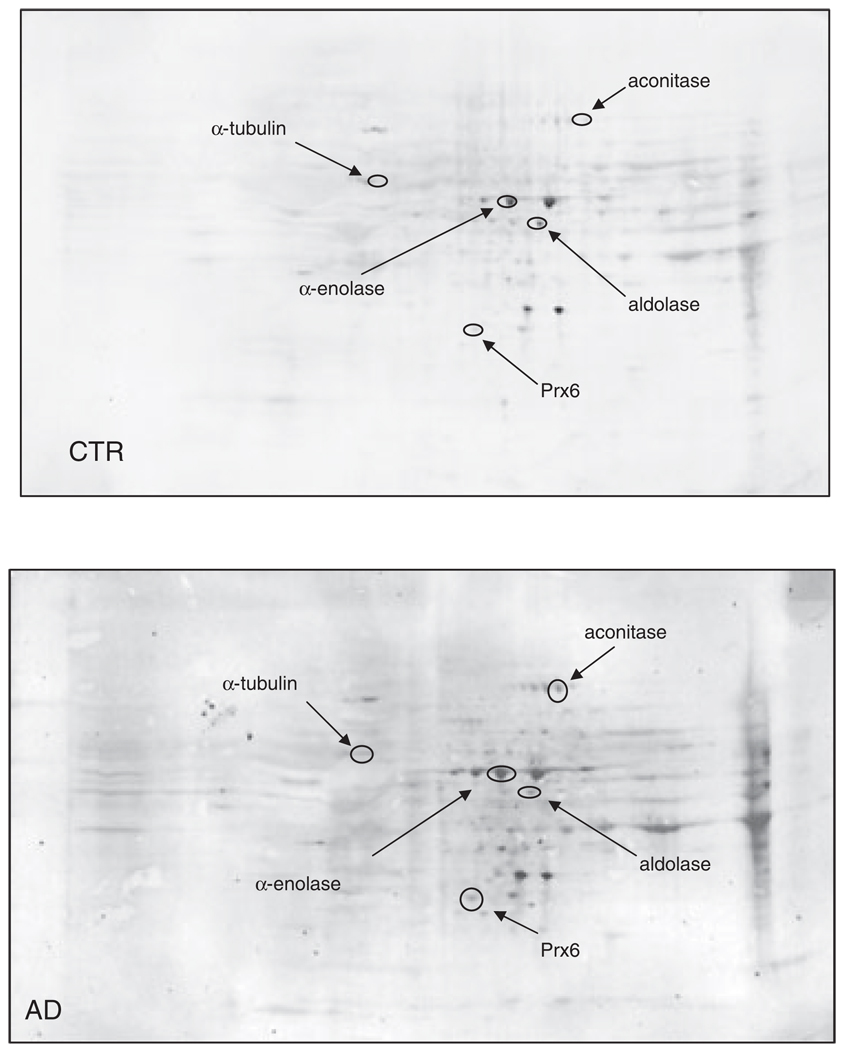
3.2 HP proteomics studies
Figure 4 illustrates representative 2-D-electrophoresis gels of hippocampal proteins from control subjects and AD subjects after SYPRO Ruby staining, while Fig. 5 shows representative 2-D Western blots of hippocampal proteins for control and AD. The statistically significant HNE-modified hippocampal proteins identified in AD compared with control were a follows: α-enolase, aldolase, peroxiredoxin 6 (Prx6), aconitase, and α-tubulin. Table 2 shows the proteins that were successfully identified by MS along with the peptides matched, percentage coverage, pI, migration rate values, protein levels, and the increase of HNE levels, indexed as fold-increase from control (fold-increase of oxidative modification).
Figure 4.
Representative SYPRO Ruby 2-D gels of the HP from control (CTR) or AD brain. Proteins (200 µg) were separated on immobilized pH 3–10 IPG strips followed by separation on an 8–16% gradient SDS-PAGE gels.
Figure 5.
2-D HNE immunoblots from AD and control subjects (HP). Positions of the five identified proteins are shown on the blots. Relative changes in HNE immunoreactivity, after normalization of the immunostaining intensities to the protein content, were significant for five proteins. See text.
We also found that the enzymatic activity of aconitase was significantly decreased (50%) in HP of AD subjects compared with controls (Fig. 6C and D). These data are consistent with the above results obtained on ATP synthase activity in IPL samples of AD subjects compared with controls and are consonant with the notion that oxidative modification of proteins generally leads to their dysfunction. In agreement with the results in AD in this current study, previous investigations from our laboratory found decreased enzyme activity of oxidatively modified α-enolase in brain of subjects with MCI [32].
4 Discussion
Numerous studies have demonstrated increased lipid peroxidation in the brain of patients with AD compared with age-matched controls [10, 15, 33–37]. Lipid peroxidation causes structural membrane damage and produces diffusible secondary bioactive aldehydes including HNE, malondialdehyde, and acrolein, all of which are increased in several brain regions of late-stage AD subjects [38–40]. There are numerous findings supporting an important role of HNE in the development of AD. Accordingly, an elevated level of protein-bound HNE in brain from subjects with MCI was observed [17, 41]. Based on these notions, we aimed to identify specific proteins oxidatively modified by HNE in AD IPL and HP.
The proteins that were found to be statistically significantly HNE-modified in AD HP include the following: α-enolase, aldolase, Prx6, aconitase, and α-tubulin. In IPL, we found ATP synthase a chain, glutamine synthase, DRP-2, and MnSOD to be statistically significantly HNE-modified in AD compared with control subjetcs. These proteins are involved in the regulation of a number of important cellular functions including: energy metabolism, cellular signalling, antioxidant, and detoxification, in addition to regulating structural functions of brain cells (Table 3). Interestingly, previous studies from our laboratory demonstrated that some of these proteins were oxidatively modified as indexed by increased carbonyl levels or by increased nitration in AD [23, 42, 43] or MCI brain [19, 32, 44] (Table 3).
Table 3.
Functionalities of identified HNE-modified proteins in AD HP and IPL in comparison with previous studies from our laboratory that showed oxidative modification of selected proteins
| Functions | Proteins involved | Brain region (oxidative modification) | Pathology (AD or MCI) |
|---|---|---|---|
| Energy or mitochondrial dysfunction | α-Enolase | IPL, HP (nitrated and carbonylated) | MCI and AD |
| IPL (HNE-modified) | MCI | ||
| Aconitase | HP (HNE-modified) | AD | |
| ATP synthase | IPL (HNE-modified) | AD | |
| HP (HNE-modified) | MCI | ||
| HP (nitrated) | AD | ||
| Aldolase | |||
| Antioxidant | Peroxiredoxin 6 | IPL (nitrated) | MCI |
| HP (HNE-modified) | AD | ||
| MnSOD | IPL (HNE-modified) | AD | |
| Structural dysfunction | Dihydropyrminidase like-2 | IPL (HNE-modified) | AD |
| HP (carbonylated and nitrated) | AD | ||
| α-Tubulin | HP (HNE-modified) | AD | |
| Excitoxicity | Glutamine synthase | IPL, HP (carbonylated) | AD |
| HP (carbonylated) | MCI |
4.1 Energy dysfunction: α-enolase, aldolase, aconitase, and ATP synthase
One of the most consistent alterations in AD is a reduced brain glucose utilization [45]. Most of the evidence suggesting an impairment of cerebral energy metabolism has come from in vivo studies of glucose consumption using positron emission tomography [45–47]. Besides the reduction of metabolic rate for glucose in AD brain, decreased concentrations of GLUT1 and GLUT3 glucose transporters [48] and decreased activities of some of the enzymes involved in glucose metabolism have also been reported [49, 50]. These biochemical alterations are consistent with the identification of aldolase and α-enolase as HNE-modified proteins using redox proteomics, since each of these proteins is involved in glucose metabolism.
Impairment of energy metabolism, which culminates in a reduced ATP production, also results from the oxidation – HNE-modification – of aconitase and ATP synthase. Aconitase, a key Krebs cycle enzyme, contains a Fe-S cluster and is highly sensitive to oxidative stress, which can inhibit its activity. It has been shown that aconitase activity is decreased in both lymphocytes and purified mitochondria from AD patients compared with controls [51]. This is the first report to show that reduced enzyme activity of aconitase in AD brain could result from its modification by HNE and sheds light on the role of lipid peroxidation in protein dysfunction in this disorder. Loss of ATP has pronounced effects on the maintenance of ionic gradients, activity of pumps and channels, and increases neurotoxicity [52].
Mitochondria dysfunction is further supported by our finding of ATP synthase oxidative modification and by the parallel reduction of its enzyme activity. ATP synthase has been shown to be highly sensitive to oxidative damage, and our data, in addition to previous studies, suggest that the function of ATP synthase is significantly altered in AD brain [42]. Recent studies from our laboratory reported increased nitration of ATP synthase á chain in the HP of AD patients [42] and HNE-modification in the IPL of MCI subjects [19]. Interestingly, the decrease of aconitase and ATP synthase activities in brain from subjects with AD is markedly duplicated in subjects with MCI [19, 53], arguably a prodromal stage of AD. The oxidation of ATP synthase leads to the inactivation of this mitochondrial complex, resulting in possible electron leakage and parallel ROS production. Taken together, these findings confirm and extend the notion that the impairment of energy metabolism plays a pivotal role in the pathogenesis and progression of AD [9, 54].
4.2 Antioxidant system dysfunction: Prx6 and MnSOD
Prx6 was found to be HNE-modified in HP of AD subjetcs compared with controls. Proxiredoxins play an important role in regulating cellular processes by decreasing the levels of oxidants or by removing toxic compounds that are generated in the cell. Peroxiredoxins are a ubiquitously expressed family of thiol peroxidases that reduce hydrogen peroxide, peroxynitrite, and hydroperoxides using a highly conserved cysteine and employing thioredoxin reducing equivalents [55]. Conceivably, the presence of catalytic cysteines makes them highly susceptible to oxidative damage. Recent studies from our laboratory have shown increased levels of nitrated peroxiredon 6 in the IPL of MCI subjects [44], confirming the potential involvement of this enzyme in the pathogenesis and progression of AD. Clearly, oxidative dysfunction of a key antioxidant enzyme would contribute to the elevated oxidative environment of AD.
To our knowledge, this is the first study to show oxidative modification of a key antioxidant enzyme, MnSOD, in AD brain. MnSOD (SOD2), the main superoxide scavenger in the mithocondria, is enriched around amyloid plaques [56] and brain microvessels [57] in hAPP transgenic mice. MnSOD is excessively nitrated in brain and neurons from human double mutant APP/PS-1 knock-in mice [58, 59]. Increased expression of antioxidant enzymes, such as superoxide dismutases, and their co-localization to senile plaques and dystrophic neurites have established a firm association between free-radical-mediated injury and AD neuropathology. While several studies have confirmed these findings, there is conflicting information regarding the alteration of enzyme activity. It has been shown that increased expression of SOD2 in the HP of AD patients functions as a compensatory mechanism from free-radical oxidative damage [60]. We confirmed this observation, and we observed that SOD2 enzyme activity was almost unchanged in AD IPL with respect to controls. Based on our result and that from others, we hypothesise that in AD there is an attempt to counteract increased ROS by increasing the expression of MnSOD, but the increased expression of SOD2 is not accompanied (coupled) by increased enzyme activity due to its oxidative modification. Thus, oxidative modification of antioxidant defence systems, such as Prx6 and SOD2, compromises the ability of neuronal cells to countercat increasing oxidative damage and likely contributes to the neurodegenerative process involved in AD.
4.3 Structural dysfunction: DRP-2, α-tubulin
One of the characteristic features of AD is synaptic loss. A number of studies reported β-actin and DRP-2 as down-regulated and oxidatively modified in AD brain [8, 43, 61]. Here, we show for the first time elevated levels of HNE-bound α-tubulin in AD brain compared with control brain. Previous studies have shown that tubulin transcription was reduced in neurons containing neurofibrillary tangles with a parallel reduction in the number of micro-tubules [62]. Accordingly, our results suggest a possible link between oxidative damage and microtubule disassembly through the tubulin modification by increased levels of HNE. Our finding confirmed previous studies from others [63] and supports the hypothesis that HNE-adduction to tubulin is a primary mechanism involved in the HNE-induced loss of the microtubule network.
DRP-2 plays an important function in maintaining interneuronal communication, neuronal repair, and regulates dendritic length [64]. We and others have proposed that oxidative post-translational modification of neuronal cytoskeletal protein is one potential link between pathogenic processes initiated by Aβ accumulation and the cytoskeletal abnormalities that are closely related to neuron dysfunction and degeneration in AD [2, 63]. Oxidation of these proteins could lead to loss of membrane integrity, activation of cellular events that may lead to apoptosis, loss of interneuronal connections, neuronal repair, and shortened dendritic lengths as observed in AD. Such an alteration would lead to decreased intraneuronal connections, which is important in a memory disorder.
4.4 Excitotoxicity: Glutamine synthase
Oxidative stress is intimately linked to glutamate neurotoxicity, known as “excitotoxicity,” which results in a cascade of events leading to neural cell death. The glutamate transporter EAAT2 and glutamine synthase regulate the extraneuronal levels of glutamate and neurotransmission. EAAT2 (Glt-1) is oxidatively modified by HNE in AD brain, and Aβ (1–42) also leads to HNE binding to EAAT2 [10]. Glutamine synthase is oxidatively modified by carbonylation in AD brain as determined by previous proteomics studies from our laboratory [23]. Glutamine synthase is localized in astroglial cells, and it protects neurons by converting the potential neurotoxic glutamate and ammonia into glutamine. Glutamine synthase enzymatic activity has been shown to be decreased in AD brain [9, 65, 66], which together with decreased EAAT2 activity increases the probability of exacerbated excitotoxic death in AD brain. Reduced glutamine synthase activity in AD brain was localized to areas with increased oxidation products mostly present in the vicinity of amyloid plaques [9]. Our proteomics results further support the hypothesis that loss of activity of glutamine synthase is associated with its oxidative modification, either by carbonylation or by HNE-modification.
In conclusion, our redox proteomics study led to the identification of specifically HNE-bound proteins in the HP and IPL of AD postmortem brains. We have demonstrated a regional-specific oxidative damage in the IPL and HP of AD brain, thus confirming that both these regions are highly vulnerable to oxidative damage, as it is shown by pathological hallmarks. Previous studies from our laboratory demonstrated that some of these proteins were oxidatively modified by increased carbonyl levels or by increased nitration in AD [23, 42, 43] or MCI brain [19, 32, 44] (Table 3). Taken together, these findings indicate that these proteins – ATP synthase, glutamine synthase, DRP-2, and α-enolase – are selectively impaired by multiple oxidative modifications and therefore are consequently dysfunctional. The identification of “target” proteins provides evidence to the importance of oxidative damage in the progression of AD and potentially establishes a link between oxidative stress-induced protein modification and neurodegeneration. Our results provide insight into the sequelae of lipid peroxidation in the pathogenesis of this dementing disorder and further support the notion that impaired energy metabolism, synaptic loss, and exitotoxicity are crucially involved in the complex mechanisms of AD brain pathology.
Acknowledgments
The authors thank the University of Kentucky ADC Clinical and Neuropathology Cores for providing the brain specimens used for this study. This research was supported in part by grants from NIH [AG-10836; AG-05119; AG-029839].
Abbreviations
- AD
Alzheimer’s disease
- Aβ
amyloid β-peptide
- DRP-2
dihydropyrimidinase-related protein 2
- HNE
4-hydroxynonenal
- HP
hippocampus
- IPL
inferior parietal lobule
- MCI
mild cognitive impairment
- PMI
postmortem interval
- Prx6
peroxiredoxin 6
Footnotes
The authors have declared no conflict of interest.
References
- 1.Markesbery WR. Oxidative stress hypothesis in Alzheimer’s disease. Free Radic. Biol. Med. 1997;23:134–147. doi: 10.1016/s0891-5849(96)00629-6. [DOI] [PubMed] [Google Scholar]
- 2.Butterfield DA, Reed T, Newman SF, Sultana R. Roles of amyloid beta-peptide-associated oxidative stress and brain protein modifications in the pathogenesis of Alzheimer’s disease and mild cognitive impairment. Free Radic. Biol. Med. 2007;43:658–677. doi: 10.1016/j.freeradbiomed.2007.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreira PI, Zhu X, Liu Q, Honda K, et al. Compensatory responses induced by oxidative stress in Alzheimer disease. Biol. Res. 2006;39:7–13. doi: 10.4067/s0716-97602006000100002. [DOI] [PubMed] [Google Scholar]
- 4.Gandy S. The role of cerebral amyloid beta accumulation in common forms of Alzheimer disease. J. Clin. Invest. 2005;115:1121–1129. doi: 10.1172/JCI25100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butterfield DA. Proteomics: a new approach to investigate oxidative stress in Alzheimer’s disease brain. Brain Res. 2004;1000:1–7. doi: 10.1016/j.brainres.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Aksenov MY, Aksenova MV, Butterfield DA, Geddes JW, et al. Protein oxidation in the brain in Alzheimer’s disease. Neuroscience. 2001;103:373–383. doi: 10.1016/s0306-4522(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 7.Montine TJ, Neely MD, Quinn JF, Beal MF, et al. Lipid peroxidation in aging brain and Alzheimer’s disease. Free Radic. Biol. Med. 2002;33:620–626. doi: 10.1016/s0891-5849(02)00807-9. [DOI] [PubMed] [Google Scholar]
- 8.Castegna A, Thongboonkerd V, Klein JB, Lynn B, et al. Proteomic identification of nitrated proteins in Alzheimer’s disease brain. J. Neurochem. 2003;85:1394–1401. doi: 10.1046/j.1471-4159.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 9.Hensley K, Hall N, Subramaniam R, Cole P, et al. Brain regional correspondence between Alzheimer’s disease histopathology and biomarkers of protein oxidation. J. Neurochem. 1995;65:2146–2156. doi: 10.1046/j.1471-4159.1995.65052146.x. [DOI] [PubMed] [Google Scholar]
- 10.Lauderback CM, Hackett JM, Huang FF, Keller JN, et al. The glial glutamate transporter, GLT-1, is oxidatively modified by 4-hydroxy-2-nonenal in the Alzheimer’s disease brain: the role of Abeta1–42. J. Neurochem. 2001;78:413–416. doi: 10.1046/j.1471-4159.2001.00451.x. [DOI] [PubMed] [Google Scholar]
- 11.Liu Q, Raina AK, Smith MA, Sayre LM, et al. Hydroxynonenal, toxic carbonyls, and Alzheimer disease. Mol. Aspects Med. 2003;24:305–313. doi: 10.1016/s0098-2997(03)00025-6. [DOI] [PubMed] [Google Scholar]
- 12.Uchida K, Toyokuni S, Nishikawa K, Kawakishi S, et al. Michael addition-type 4-hydroxy-2-nonenal adducts in modified low-density lipoproteins: markers for atherosclerosis. Biochemistry. 1994;33:12487–12494. doi: 10.1021/bi00207a016. [DOI] [PubMed] [Google Scholar]
- 13.Humphries KM, Szweda LI. Selective inactivation of alpha-ketoglutarate dehydrogenase and pyruvate dehydrogenase: reaction of lipoic acid with 4-hydroxy-2-nonenal. Biochemistry. 1998;37:15835–15841. doi: 10.1021/bi981512h. [DOI] [PubMed] [Google Scholar]
- 14.Cohn JA, Tsai L, Friguet B, Szweda LI. Chemical characterization of a protein-4-hydroxy-2-nonenal crosslink: immunochemical detection in mitochondria exposed to oxidative stress. Arch. Biochem. Biophys. 1996;328:158–164. doi: 10.1006/abbi.1996.0156. [DOI] [PubMed] [Google Scholar]
- 15.Sultana R, Perluigi M, Butterfield DA. Protein oxidation and lipid peroxidation in brain of subjects with Alzheimer’s disease: insights into mechanism of neurodegeneration from redox proteomics. Antioxid. Redox Signal. 2006;8:2021–2037. doi: 10.1089/ars.2006.8.2021. [DOI] [PubMed] [Google Scholar]
- 16.Butterfield DA, Reed TT, Perluigi M, De Marco C, et al. Elevated levels of 3-nitrotyrosine in brain from subjects with amnestic mild cognitive impairment: implications for the role of nitration in the progression of Alzheimer’s disease. Brain Res. 2007;1148:243–248. doi: 10.1016/j.brainres.2007.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butterfield DA, Reed T, Perluigi M, De Marco C, et al. Elevated protein-bound levels of the lipid peroxidation product, 4-hydroxy-2-nonenal, in brain from persons with mild cognitive impairment. Neurosci. Lett. 2006;397:170–173. doi: 10.1016/j.neulet.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 18.Keller JN, Schmitt FA, Scheff SW, Ding Q, et al. Evidence of increased oxidative damage in subjects with mild cognitive impairment. Neurology. 2005;64:1152–1156. doi: 10.1212/01.WNL.0000156156.13641.BA. [DOI] [PubMed] [Google Scholar]
- 19.Reed T, Perluigi M, Sultana R, Pierce WM, et al. Redox proteomic identification of 4-hydroxy-2-nonenal-modified brain proteins in amnestic mild cognitive impairment: insight into the role of lipid peroxidation in the progression and pathogenesis of Alzheimer’s disease. Neurobiol. Dis. 2008;30:107–120. doi: 10.1016/j.nbd.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Maurer HH, Peters FT. Toward high-throughput drug screening using mass spectrometry. Ther. Drug Monit. 2005;27:686–688. doi: 10.1097/01.ftd.0000180224.19384.f0. [DOI] [PubMed] [Google Scholar]
- 21.Thongboonkerd V, McLeish KR, Arthur JM, Klein JB. Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. Kidney Int. 2002;62:1461–1469. doi: 10.1111/j.1523-1755.2002.kid565.x. [DOI] [PubMed] [Google Scholar]
- 22.Butterfield DA, Boyd-Kimball D, Castegna A. Proteomics in Alzheimer’s disease: insights into potential mechanisms of neurodegeneration. J. Neurochem. 2003;86:1313–1327. doi: 10.1046/j.1471-4159.2003.01948.x. [DOI] [PubMed] [Google Scholar]
- 23.Castegna A, Aksenov M, Aksenova M, Thongboonkerd V, et al. Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. Part I: creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic. Biol. Med. 2002;33:562–571. doi: 10.1016/s0891-5849(02)00914-0. [DOI] [PubMed] [Google Scholar]
- 24.Pappin DJ. Peptide mass fingerprinting using MALDI-TOF mass spectrometry. Methods Mol. Biol. 2003;211:211–219. doi: 10.1385/1-59259-342-9:211. [DOI] [PubMed] [Google Scholar]
- 25.Zheng J, Ramirez VD. Rapid inhibition of rat brain mitochondrial proton F0F1-ATPase activity by estrogens: comparison with Na+, K+-ATPase of porcine cortex. Eur. J. Pharmacol. 1999;368:95–102. doi: 10.1016/s0014-2999(99)00012-6. [DOI] [PubMed] [Google Scholar]
- 26.Yan LJ, Levine RL, Sohal RS. Oxidative damage during aging targets mitochondrial aconitase. Proc. Natl. Acad. Sci. USA. 1997;94:11168–11172. doi: 10.1073/pnas.94.21.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pratico D, Lee VMY, Trojanowski JQ, Rokach J, et al. Increased F2-isoprostanes in Alzheimer’s disease: evidence for enhanced lipid peroxidation in vivo. FASEB J. 1998;12:1777–1783. doi: 10.1096/fasebj.12.15.1777. [DOI] [PubMed] [Google Scholar]
- 28.Vohradsky J, Branny P, Li XM, Thompson CJ. Effect of protein degradation on spot M(r) distribution in 2-D gels – a case study of proteolysis during development of Streptomyces coelicolor cultures. Proteomics. 2008;8:2371–2375. doi: 10.1002/pmic.200701075. [DOI] [PubMed] [Google Scholar]
- 29.Halligan BD, Ruotti V, Jin W, Laffoon S, et al. ProMoST (Protein Modification Screening Tool): a web-based tool for mapping protein modifications on two-dimensional gels. Nucl. Acids Res. 2004;32:W638–W644. doi: 10.1093/nar/gkh356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aksenova MV, Aksenov MY, Carney JM, Butterfield DA. Protein oxidation and enzyme activity decline in old brown Norway rats are reduced by dietary restriction. Mech. Ageing Dev. 1998;100:157–168. doi: 10.1016/s0047-6374(97)00133-4. [DOI] [PubMed] [Google Scholar]
- 31.Butterfield DA, Stadtman ER. Protein oxidation processes in aging brain. Adv. Cell Aging Gerontol. 1997;2:161–191. [Google Scholar]
- 32.Butterfield DA, Poon HF, St Clair D, Keller JN, et al. Redox proteomics identification of oxidatively modified hippocampal proteins in mild cognitive impairment: insights into the development of Alzheimer’s disease. Neurobiol. Dis. 2006;22:223–232. doi: 10.1016/j.nbd.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi T, Shishido N, Nakayama K, Nunomura A, et al. Lipid peroxidation and 4-hydroxy-2-nonenal formation by copper ion bound to amyloid-beta peptide. Free Radic. Biol. Med. 2007;43:1552–1559. doi: 10.1016/j.freeradbiomed.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Murray IV, Liu L, Komatsu H, Uryu K, et al. Membrane-mediated amyloidogenesis and the promotion of oxidative lipid damage by amyloid beta proteins. J. Biol. Chem. 2007;282:9335–9345. doi: 10.1074/jbc.M608589200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montine TJ, Morrow JD. Fatty acid oxidation in the pathogenesis of Alzheimer’s disease. Am. J. Pathol. 2005;166:1283–1289. doi: 10.1016/S0002-9440(10)62347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pamplona R, Dalfo E, Ayala V, Bellmunt MJ, et al. Proteins in human brain cortex are modified by oxidation, glycoxidation, and lipoxidation. Effects of Alzheimer disease and identification of lipoxidation targets. J. Biol. Chem. 2005;280:21522–21530. doi: 10.1074/jbc.M502255200. [DOI] [PubMed] [Google Scholar]
- 37.Montine KS, Quinn JF, Zhang J, Fessel JP, et al. Isoprostanes and related products of lipid peroxidation in neurodegenerative diseases. Chem. Phys. Lipids. 2004;128:117–124. doi: 10.1016/j.chemphyslip.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 38.Zarkovic K. 4-Hydroxynonenal and neurodegenerative diseases. Mol. Aspects Med. 2003;24:293–303. doi: 10.1016/s0098-2997(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 39.Picklo MJ, Sr, Montine TJ. Mitochondrial effects of lipid-derived neurotoxins. J. Alzheimers Dis. 2007;12:185–193. doi: 10.3233/jad-2007-12209. [DOI] [PubMed] [Google Scholar]
- 40.Butterfield DA, Castegna A, Lauderback CM, Drake J. Evidence that amyloid beta-peptide-induced lipid peroxidation and its sequelae in Alzheimer’s disease brain contribute to neuronal death. Neurobiol. Aging. 2002;23:655–664. doi: 10.1016/s0197-4580(01)00340-2. [DOI] [PubMed] [Google Scholar]
- 41.Williams TI, Lynn BC, Markesbery WR, Lovell MA. Increased levels of 4-hydroxynonenal and acrolein, neurotoxic markers of lipid peroxidation, in the brain in Mild Cognitive Impairment and early Alzheimer’s disease. Neurobiol. Aging. 2006;27:1094–1099. doi: 10.1016/j.neurobiolaging.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 42.Sultana R, Poon HF, Cai J, Pierce WM, et al. Identification of nitrated proteins in Alzheimer’s disease brain using a redox proteomics approach. Neurobiol. Dis. 2006;22:76–87. doi: 10.1016/j.nbd.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 43.Sultana R, Boyd-Kimball D, Poon HF, Cai J, et al. Redox proteomics identification of oxidized proteins in Alzheimer’s disease hippocampus and cerebellum: an approach to understand pathological and biochemical alterations in AD. Neurobiol. Aging. 2006;27:1564–1576. doi: 10.1016/j.neurobiolaging.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 44.Sultana R, Reed T, Perluigi M, Coccia R, et al. Proteomic identification of nitrated brain proteins in amnestic mild cognitive impairment: a regional study. J. Cell Mol. Med. 2007;77:839–851. doi: 10.1111/j.1582-4934.2007.00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pietrini P, Alexander GE, Furey ML, Hampel H, et al. The neurometabolic landscape of cognitive decline: in vivo studies with positron emission tomography in Alzheimer’s disease. Int. J. Psychophysiol. 2000;37:87–98. doi: 10.1016/s0167-8760(00)00097-0. [DOI] [PubMed] [Google Scholar]
- 46.Frackowiak RS, Pozzilli C, Legg NJ, Du Boulay GH, et al. Regional cerebral oxygen supply and utilization in dementia. A clinical and physiological study with oxygen-15 and positron tomography. Brain. 1981;104:753–778. doi: 10.1093/brain/104.4.753. [DOI] [PubMed] [Google Scholar]
- 47.Ishii K, Sasaki M, Kitagaki H, Yamaji S, et al. Reduction of cerebellar glucose metabolism in advanced Alzheimer’s disease. J. Nucl. Med. 1997;38:925–928. [PubMed] [Google Scholar]
- 48.Vannucci SJ, Maher F, Koehler E, Simpson IA. Altered expression of GLUT-1 and GLUT-3 glucose transporters in neurohypophysis of water-deprived or diabetic rats. Am. J. Physiol. 1994;267:E605–E611. doi: 10.1152/ajpendo.1994.267.4.E605. [DOI] [PubMed] [Google Scholar]
- 49.Hoyer S. Risk factors for Alzheimer’s disease during aging. Impacts of glucose/energy metabolism. J. Neural Transm. Suppl. 1998;54:187–194. doi: 10.1007/978-3-7091-7508-8_18. [DOI] [PubMed] [Google Scholar]
- 50.Schubert D. Glucose metabolism and Alzheimer’s disease. Ageing Res. Rev. 2005;4:240–257. doi: 10.1016/j.arr.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 51.Polidori MC, Griffiths HR, Mariani E, Mecocci P. Hallmarks of protein oxidative damage in neurodegenerative diseases: focus on Alzheimer’s disease. Amino Acids. 2007;32:553–559. doi: 10.1007/s00726-006-0431-x. [DOI] [PubMed] [Google Scholar]
- 52.Del Rio P, Montiel T, Chagoya V, Massieu L. Exacerbation of excitotoxic neuronal death induced during mitochondrial inhibition in vivo: relation to energy imbalance or ATP depletion? Neuroscience. 2007;146:1561–1570. doi: 10.1016/j.neuroscience.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 53.Mecocci P. Oxidative stress in mild cognitive impairment and Alzheimer disease: a continuum. J. Alzheimers Dis. 2004;6:159–163. doi: 10.3233/jad-2004-6207. [DOI] [PubMed] [Google Scholar]
- 54.Poon HF, Joshi G, Sultana R, Farr SA, et al. Antisense directed at the Abeta region of APP decreases brain oxidative markers in aged senescence accelerated mice. Brain Res. 2004;1018:86–96. doi: 10.1016/j.brainres.2004.05.048. [DOI] [PubMed] [Google Scholar]
- 55.Rouhier N, Jacquot JP. The plant multigenic family of thiol peroxidases. Free Radic. Biol. Med. 2005;38:1413–1421. doi: 10.1016/j.freeradbiomed.2004.07.037. [DOI] [PubMed] [Google Scholar]
- 56.Blanchard V, Moussaoui S, Czech C, Touchet N, et al. Time sequence of maturation of dystrophic neurites associated with Abeta deposits in APP/PS1 transgenic mice. Exp. Neurol. 2003;184:247–263. doi: 10.1016/s0014-4886(03)00252-8. [DOI] [PubMed] [Google Scholar]
- 57.Tong XK, Nicolakakis N, Kocharyan A, Hamel E. Vascular remodeling versus amyloid beta-induced oxidative stress in the cerebrovascular dysfunctions associated with Alzheimer’s disease. J. Neurosci. 2005;25:11165–11174. doi: 10.1523/JNEUROSCI.4031-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anantharaman M, Tangpong J, Keller JN, Murphy MP, et al. Beta-amyloid mediated nitraiton of manganese super-oxide dismutase: implication for oxidative stress in a APPNLH/NLH X PS-1P264L/P264L double knock-in mouse model of Alzheimer’s disease. Am. J. Pathol. 2006;165:1608–1616. doi: 10.2353/ajpath.2006.051223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sompol P, Ittarat W, Tangpong J, Chen Y, et al. A neuronal model of Alzheimer’s disease: an insight into the mechanisms of oxidative stress-mediated mitochondrial injury. Neuroscience. 2008;153:120–130. doi: 10.1016/j.neuroscience.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marcus DL, Strafaci JA, Freedman ML. Differential neuronal expression of manganese superoxide dismutase in Alzheimer’s disease. Med. Sci. Monit. 2006;12:BR8–BR14. [PubMed] [Google Scholar]
- 61.Butterfield DA, Gnjec A, Poon HF, Castegna A, et al. Redox proteomics identification of oxidatively modified brain proteins in inherited Alzheimer’s disease: an initial assessment. J. Alzheimers Dis. 2006;10:391–397. doi: 10.3233/jad-2006-10407. [DOI] [PubMed] [Google Scholar]
- 62.Brion JP, Flament-Durand J. Distribution and expression of the alpha-tubulin mRNA in the hippocampus and the temporal cortex in Alzheimer’s disease. Pathol. Res. Pract. 1995;191:490–498. doi: 10.1016/s0344-0338(11)80867-8. [DOI] [PubMed] [Google Scholar]
- 63.Neely MD, Boutte A, Milatovic D, Montine TJ. Mechanisms of 4-hydroxynonenal-induced neuronal microtubule dysfunction. Brain Res. 2005;1037:90–98. doi: 10.1016/j.brainres.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 64.Hamajima N, Matsuda K, Sakata S, Tamaki N, et al. A novel gene family defined by human dihydropyrimidinase and three related proteins with differential tissue distribution. Gene. 1996;180:157–163. doi: 10.1016/s0378-1119(96)00445-3. [DOI] [PubMed] [Google Scholar]
- 65.Butterfield DA, Hensley K, Cole P, Subramaniam R, et al. Oxidatively induced structural alteration of glutamine synthetase assessed by analysis of spin label incorporation kinetics: relevance to Alzheimer’s disease. J. Neurochem. 1997;68:2451–2457. doi: 10.1046/j.1471-4159.1997.68062451.x. [DOI] [PubMed] [Google Scholar]
- 66.Le Prince G, Delaere P, Fages C, Lefrancois T, et al. Glutamine synthetase (GS) expression is reduced in senile dementia of the Alzheimer type. Neurochem. Res. 1995;20:859–862. doi: 10.1007/BF00969698. [DOI] [PubMed] [Google Scholar]