Abstract
From the structures of isolated protein complexes to the molecular dynamics of whole cells, neutron methods can achieve a resolution in complex systems that is inaccessible to other techniques. Biology is fortunate in that it is rich in water and hydrogen, and this allows us to exploit the differential sensitivity of neutrons to this element and its major isotope, deuterium. Furthermore, neutrons exhibit wave properties that allow us to use them in similar ways to light, X-rays and electrons. This review aims to explain the basics of biological neutron science to encourage its greater use in solving difficult problems in the life sciences.
Keywords: small-angle neutron scattering, reflectivity, deuteration, neutrons, diffraction
1. Introduction
This article is written by a biochemist who only lately has developed an interest in neutrons. An article of this type necessarily reflects personal influences, the types of experiments we have tried, the facilities that we have used and the successes and failures we have experienced, all of which play a part in deciding what the neophyte neutron scatterer should know. Thus, any omissions, unwarranted inclusions and downright misconceptions are entirely my own fault and should not reflect upon the excellent people who have helped me get this far in the wonderful world of the neutron. If I succeed in stimulating your interest in these methods, then the excellent reference book Neutron Scattering in Biology should be your next step (Fitter et al. 2006).
2. Why neutrons?
The first question to ask is why the neutron has any use in biological science. To most of us, it is simply a core nuclear particle that is involved in the nuclear chain reaction and, therefore, its usefulness to biology is not immediately obvious. One way to illustrate this is to compare the neutron with the other tiny ‘particles’ we can shoot at biological molecules. We use them in order to overcome the fundamental human failing that we cannot see the molecules that many of us spend our working lives studying. In these lives, the first tool we mostly use is the photon, which we might take for granted.
3. Uses of photons
While we cannot see the proteins, we can see green chlorophyll containing light harvesting complexes in plants and red haemoglobin in blood. This use of colours means that, already with the naked eye, we can use wavelength differences to distinguish different proteins. Later, perhaps in the teaching laboratory, the absorption spectrometer allows us to use ultraviolet and infrared wavelengths we cannot see. Furthermore, the use of instrumentation introduces quantitation so that we can determine concentrations by measuring the number of photons absorbed. Such simple approaches show the use of two important and easily measurable variables, wavelength and intensity, which are important in many research fields.
If we shine visible light at a solution of red blood cells, we get two separate bits of information that use both of these variables. By measuring the transmitted intensity through the sample, we observe a wavelength-dependent transmission that could be used to estimate the haemoglobin concentration. Thus, we choose a wavelength, measure the number of photons that make it through the sample compared with the number that set out and we have a measure of concentration. In reality, this is unlikely to work for red blood cell suspensions as the red blood cells are larger than the wavelength of the light. This means that there will be significant Mie scattering of light owing to the large size of the cells and the sample will be turbid. If the solution is solubilized by detergents and clarified by centrifugation, then a more reasonable absorption spectrum can be collected. This trivial problem nevertheless reveals something else about particles and molecules; that scattered particles can provide information on molecular size.
Another useful property of visible light in measurement is interference. One example that we all recognize are the colours that a thin film displays on the surface of water, e.g. diesel oil on a roadside puddle. The colours vary depending upon the thickness of the film and the angle at which we view the surface. This is due to the extra distance that some photons have to travel through the oil film compared with those reflected at the top. If this extra distance is some multiples of the wavelength, then that colour predominates owing to constructive interference. Thus, if we wished, we could use visible light to try and determine film thicknesses down to 1 µm or less by such a reflectivity approach. The reflection occurs in this case owing to the difference in refractive index between the air, oil and water (Fresnel reflection).
4. Use of X-rays
As our measurement precision is related to the wavelength, it loosely follows that in order to probe distances in very thin layers, we need very short wavelengths and X-rays are the obvious tools. This is because modern synchrotrons produce very intense X-rays and even laboratory-based generators can satisfy many research requirements (Kim et al. 2005). X-ray photons have a wavelength of approximately 1 Å (0.1 nm) and are scattered by the electrons around the atoms. This allows very thin layers from a few angstroms upwards to be measured. X-rays are very penetrating, and their refractive index differences are very small. Nevertheless, for our purposes, the same basic principles apply, and as X-ray refractive index is related to electron density, the layers can still be resolved as differences in structure.
The greatest use of X-rays in structural biology is in crystallography where X-ray diffraction data have enabled us to obtain high-resolution models of structures as large as viruses but more commonly of individual proteins. Diffraction works like a multi-dimensional example of the thin film experiment with the specific repeating distances within crystals combining with the angle of incidence and the wave properties of light to produce interference effects. The X-ray sources are generally monochromatic (emit a single wavelength at a time) and instead of distributions of colour being scattered from the crystal, we obtain bright spots (reflections) where constructive interference happens for the chosen wavelength. A full set of these reflections from many angles for a crystal, combined with the required phase information, provides enough information on the electron density distribution to build the high-resolution protein structural models with which we are all familiar. Elsewhere, X-rays are also used in biology in similar ways to visible light, e.g. X-ray absorption spectroscopy is used to probe the structure of metalloproteins.
Reflectometry and diffraction studies are thus the results of scattering of X-rays from systems with varying degrees of long range order. In addition to the thin films and crystals mentioned above, diffraction from oriented fibres can also be used, most famously in the solving of the structure of DNA (Schindler 2008). A fibre diffraction experiment uses samples where rotation around the long axis of the molecule does not change the signal. Compared with three-dimensional crystallography, fibre diffraction data thus provide information in two dimensions, e.g. for a helix, the width and the repeat distance along the axis. These interference effects combine to amplify the data from relatively widely scattered X-rays (Forsyth & Parrot 2006). On the other hand, small-angle (0.1–10°) scattering is used in the case of molecules in a solution with no long range order. Here, the separate scattering centres within each molecule contribute to the interference pattern but, because the molecules are randomly distributed, the scattered X-ray pattern reduces to a single angle versus amplitude dimension.
Nevertheless, because the small-angle scattering data come from every orientation possible, hidden within them is the information to reconstruct the molecular size and shape in three dimensions. In simple terms, this is accomplished in two ways. Firstly, if the structure is completely unknown, ab initio (latin for ‘from first principles’) techniques can be used. These, initially, define the largest dimension in the structure and, then, use model-building approaches to build a structure that in simulations most closely reproduces the original scattering profile. Secondly, if the structure is a complex of small subunits whose high-resolution X-ray structures are known, the model building can use these as rigid body components. Thus, solutions of homogeneous molecules can provide size and shape data down to approximately 10 Å resolution (Neylon 2008).
5. Use of electrons
The electron microscope is the obvious use of electrons in biological structure determination. Here, the electrons are accelerated through a vacuum by an electric field. As with observing a sample using a light microscope, the pattern of electron scattering by a sample gives a picture of the distribution of the material and therefore an image. Electrons, being charged particles, interact strongly with matter and can be used to image very thin samples. An electric field corresponding to 100 kV gives an electron a speed of 0.55 times the speed of light and the matter wave theory, as defined by de Broglie, predicts that every particle has a wavelength that is inversely proportional to its momentum. As an electron has a significant mass (9.11 × 10−31 kg at rest) compared to a photon (approx. 0) it ends up with a much greater momentum and a much shorter wavelength (0.0039 nm at 100 kV) than the commonly used X-ray photons. Thus, very high resolutions are possible but unfortunately rarely achieved. Firstly because electron microscope optics (lenses) introduce aberrations and secondly, more importantly for biological samples, the mass of the electron also gives it a high kinetic energy which on absorption causes sample damage. Nevertheless images with resolutions of less than 10 Å are possible and the wave properties allow diffraction data to be collected from two-dimensional thin crystals. Combined with image data this has allowed high-resolution structures of membrane proteins to be solved (Breyton et al. 2002).
6. So where are the neutrons?
The simple answer is that neutrons can be employed in just about every technique described above because they have a particle/wave nature too (Fitter et al. 2006; Neylon 2008). Neutron sources are either nuclear reactors or spallation sources (in which neutrons are produced by hitting a target with high energy protons; Teixeira et al. 2008). In both cases the neutrons initially have much higher velocities and, thus, wavelengths shorter than the relevant distances to be measured in biological samples. Neutrons have a greater mass (1.67 × 10−27 kg) than electrons so the de Broglie calculation gives a velocity of only approximately 1 km s−1 for a useful wavelength of 5 Å. The fast neutrons are thus moderated by passing them through a cold medium (e.g. hydrogen/methane at 26 K) where they equilibrate with the kinetic energy of the medium. This is the origin of the term ‘cold neutrons’, which applies to those with wavelengths between approximately 2 and 20 Å, which are generally used for biological work. This increased mass and slower speed leads to the interesting result that the fall of neutrons owing to gravity must be taken into account along the beam length. Furthermore, neutrons have certain properties that increase the value of neutron experiments for biologists. These properties include the ability to detect hydrogen and differentiate it from its stable isotope deuterium, its weak interaction with matter making it penetrative and non-destructive, its useable wavelength range, the sensitivity to magnetic structures and a simple scattering process (Fitter et al. 2006). They also adopt two spin states that increase the resolution in some special applications (Majkrzak 1990).
The fundamental reason for most of these effects is that neutrons are uncharged species that interact only with nuclei. Being insensitive to the electron density, their interaction with hydrogen is unaffected by there being just a single electron, unlike X-ray methods where hydrogen is difficult to resolve. X-ray scattering is proportional to the number of electrons and hence the atomic number, thus heavy atoms scatter more than light ones (figure 1). Neutron scattering ‘strength’ is defined by the scattering length, which for simplicity we can envisage as the refractive index for neutrons. It depends upon complex features of nuclei and follows neither atomic number nor periodic table families. Thus, replacement by a different element in the same family may cause a large change in scattering length, e.g. sulphur (2.85 fm) and selenium (7.97 fm). However, the most useful contrast technique results from deuterium/hydrogen exchange owing to the size of the scattering length difference (1H = −3.74 fm and 2H = 6.67 fm), the large abundance of hydrogen in biological molecules and the generally small effect of such exchange upon function.
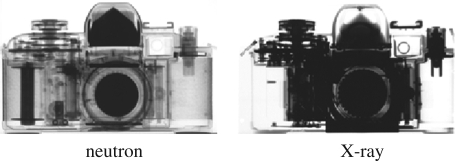
Figure 1.
Images of a camera taken with neutrons and X-rays. The plastic components are well resolved by neutrons owing to their hydrogen content while the metallic body is penetrated easily. The complementarity with X-ray methods is shown by their resolution of the higher atomic number, electron-rich metallic parts. Adapted from the Atomic Institute of the Austrian Universities, Vienna, Austria.
7. Small-angle neutron scattering in biology
The clearest example of this process is to imagine the small-angle neutron scattering (SANS) experiment for a protein dissolved in an aqueous buffer, typically about 200 µl of 2 mg ml−1 in a 1 mm pathlength cuvette. The neutron scattering by the sample is determined by the scattering length density (SLD), which is the sum of the scattering lengths of its components (nitrogen, carbon oxygen, hydrogen etc.) divided by its molecular volume. For example, H2O is −0.56 × 1010 cm−2 and D2O is 6.38 × 1010 cm−2. As, in proteins, hydrogen is a minor fraction of the total and the remaining elements (carbon, nitrogen) have positive scattering lengths, the SLD for hydrogenated polypeptides is roughly 2 × 10−4 nm−2. We, however, must also consider the exchangeable protons on the protein which will gradually be converted entirely to deuterium if the protein is dissolved in 100 per cent D2O, giving an SLD nearer to 3 × 10−4 nm−2. Backbone amide protons in hydrogen-bonded secondary structure will be the slowest and may not exchange at all on the time scales used. Thus, good scattering data will be achieved for a hydrogenated protein in both 100 per cent H2O and 100 per cent D2O. This sort of data could, however, be achieved more easily with small-angle X-ray scattering (SAXS) because X-ray sources are more intense. However, and this is crucial, the protein will have the same SLD as its solvent (the match point) at approximately 40–45% D2O and here it will not scatter neutrons. In figure 2, the same effect is produced by increasing the refractive index around glass and plastic objects by replacing water with glycerol. Owing to the reduced difference in refractive index between solid and liquid states, there is less scattering and the objects are nearly transparent. So neutrons really come into their own in complex mixtures. If we observe a protein complex composed of more than one species by SAXS or SANS, we can observe the total envelope. If, however, we selectively deuterate one component, we can use SANS to describe where that component is, and a good example is a recent paper on the neuroligin/neurexin complex (Comoletti et al. 2007). If the protein is a membrane protein it is possible to observe the protein and not the lipids or detergent. This is even possible without deuteration of either lipid or protein. For example, in a toxin–lipid complex, the hydrogenated lipid (1,2-dimyristoyl-sn-glycero-3-phosphatidylglycerol) match point was 14.5 per cent D2O and the protein (Colicin A) match point was 41 per cent (Jeanteur et al. 1994). Deuterated lipids make the system even more flexible such that individual lipid species can be observed separately, for example during phase separation (Pencer et al. 2005). Finally, nucleic acids, which are matched at 65 per cent D2O, can be seen separately from proteins as in a study of tRNAs in the deuterated ribosome (Nierhaus et al. 1998).
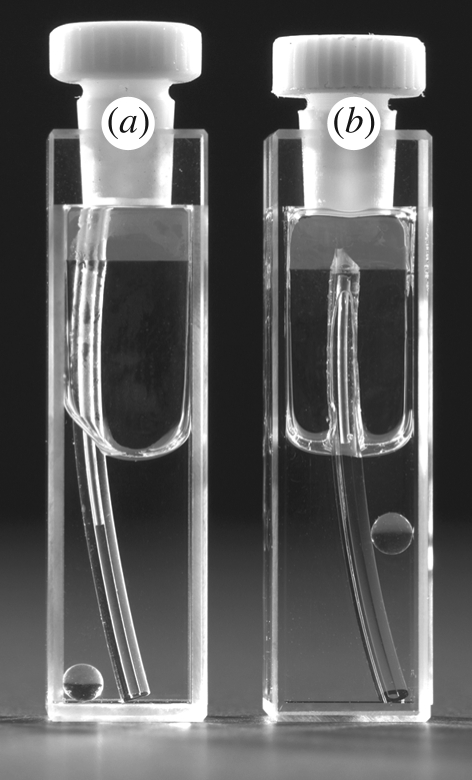
Figure 2.
Effect of solution refractive index (n) upon light scattering from solid objects. Cuvettes contain a section of plastic tubing and a glass bead. (a) In water (n = 1.333), the scattering of plastic tube (n = 1.55) and glass bead (1.52) is strong. (b) In 100 per cent glycerol (n = 1.47), which more closely matches both plastic and glass, light scatter is greatly reduced and objects blend with the background. If we were able to use a solvent with n = 1.55, the plastic would have been matched out completely. Based upon the original demonstration by Don Engelman (Kreuger & Wignal 2006).
8. Neutron diffraction in biology
These advantages are not just limited to SANS; neutrons can be used instead of X-rays in diffraction experiments. These experiments are challenging, particularly because larger crystals are needed and data acquisition times are significantly longer, but again the approach is well suited to clearly answer questions about complex biological systems (Fitter et al. 2006). Furthermore, new beamlines and instruments are continually improving the technology and making it more accessible (Blakeley et al. 2008). Thus, biologists need to be aware of their unique capabilities. One example is the distribution of detergent in membrane protein crystals. Although an essential component of the crystal in replacing the membrane lipids, only tightly bound detergents are observed by X-ray methods. Using non-deuterated detergents, which like the lipids mentioned above have a match point near to 14 per cent D2O, the detergent distributions in both the reaction centre and OmpF crystals were determined by a combination of X-ray and neutron diffraction methods (Roth et al. 1989; Pebay-Peyroula et al. 1995). Deuteration can extend the selectivity to particular species as in the case of bacteriorhodopsin (Weik et al. 1998) where specific interactions with glycolipids were resolved by feeding the Halobacterium salinarum cells with deuterated glucose. This resulted in specific labelling of the glycolipid head groups, and combined with the use of contrast matching with H2O/D2O, it enabled the resolution of two specific sites for glycolipid binding. In a similar way, the distribution of cholesterol in bilayers can be studied (Harroun et al. 2008). Away from membranes, the sensitivity of neutrons to hydrogen enables determination of proton roles in enzyme catalysis (Blakeley et al. 2008), unusual hydrogen bonds in proteins (Yamaguchi et al. 2009) and hydration patterns in protein structure (Niimura et al. 2006).
9. Neutron reflection in biology
As in the case of light reflection from the oil film on water, the neutron reflection (NR) technique is used for studying thin films either on water or on solid surfaces. In general, the biologically relevant experiments use specular NR, which gives high-resolution information about the distribution of molecules along the axis perpendicular to the surface. Thus, for a membrane, it will describe the cross-sectional distribution of lipids and proteins from one water interface to the other. Off-specular reflection methods give data about the distribution of material in the other axes and may develop to be more generally used for biology than they are now. The reflection is performed on very flat surfaces several square centimetres in area at very low angles, i.e. less than 5° to the sample plane. Surfaces include the air–water interface on a Langmuir trough, liquid–liquid interfaces, polished silicon and sputtered gold.
It is complementary to surface plasmon resonance, ellipsometry, Brewster angle microscopy, grazing incidence Fourier transform infrared spectroscopy (Terrettaz et al. 2002), atomic force microscopy (Cisneros et al. 2006), surface acoustic wave and other methods as in the study of amyloid proteins in membranes (Valincius et al. 2008). X-ray reflection is similar and has the usual advantages of high beam intensity and the option to do it in a laboratory rather than a centralized facility. Once again, and this should not now be a surprise, it is the lipid versus protein and deuterium versus hydrogen contrasts that make the trip to the neutron source worthwhile. One additional feature is that mixtures of D2O and H2O can be used to match the SLD of the underlying phase, thus air (8% D2O), silicon (38%) and gold (72%) matched water allows just the reflections from the thin layer to be viewed if that retains enough contrast. This has been used to observe lipid monolayers (Vaknin et al. 1991), their digestion by phospholipase (Wacklin et al. 2007) and their interactions with pulmonary surfactant (Fullagar et al. 2008). Tethering bilayers on a gold (Krueger et al. 2001; Holt et al. 2005; Le Brun et al. 2007) or silicon surface (Hughes et al. 2008) can help impose the orientation and asymmetry found in natural systems. Furthermore, this can exploit yet another feature of neutrons, which is their magnetic sensitivity. This property has been used extensively in the measurement of magnetic layers used in storage media etc. (Majkrzak 1990), and uses the fact that the two spin states of neutrons ‘see’ magnetized layers differently. Recently, we have used it to increase the resolution of NR experiments for biology. A magnetic layer (e.g. iron) is buried beneath a supported membrane and in a magnetic field displays two different SLDs for spin-up and spin-down neutrons. This also provides two separate sets of reflection data in which the biological sample is unchanged. This means that the datasets can be combined to provide a unique solution to the modelling of the biological layers. This provides a significant increase in resolution and reliability in NR experiments, allowing even very complex layers of lipids, membranes and antibodies to be determined (Le Brun et al. 2007; Holt et al. 2009).
10. Neutrons for measuring biological dynamics
The methods discussed so far use coherently and elastically scattered neutrons, which means that the phase and energy of the neutron are conserved after the scattering event; however, this can change when neutrons interact with the scattering material. Hydrogen has a very large incoherent scattering cross section, and so is the main origin of this type of scattering signal measured from biological samples. Whether this includes elastic or inelastic scatter, the resulting neutrons carry information on the dynamics and energy of protons both within the biomolecule or in the hydration layers (Zaccai 2000). This gives a unique set of measurements that can be used with techniques such as Raman and NMR spectroscopy to probe the dynamics of proteins, DNA, lipids and their hydration shells. Measurements can even be carried out on whole cells or organelles (Tehei et al. 2007; Jasnin et al. 2008). There is a range of instruments, such as time-of-flight, backscattering spectrometers and neutron spin-echo instruments, which can investigate dynamics and which have been described recently (Teixeira et al. 2008).
11. What next? how do i start?
To understand how such studies are done, it is of course best to consult the literature for examples close to your own interests, some of which may appear in this volume. There are very good websites such as Thomas (rkt.chem.ox.ac.uk) or Gilbert laboratories (www.strubi.ox.ac.uk/people/gilbert/neutrons.html), NIST-NCNR (www.ncnr.nist.gov) etc. Next, you should define the clear biological question that can be addressed and get advice whether you have a suitable system, e.g. amounts of protein, solubility, deuterated forms, to produce data from a neutron experiment. Now, but hopefully earlier, you must ensure that the same question cannot be solved by some much simpler approach. For example, some SANS-derived information such as radius of gyration and aggregation state is better collected by an analytical ultracentrifuge or SAXS (Solovyova et al. 2004), some features of layers may be profitably studied by dual-polarization interferometry, Brewster angle microscopy or ellipsometry (Armstrong et al. 2003) instead of reflectometry and some dynamics questions are better answered by NMR (Hecht et al. 2008). Having defined what you would like to do, you should approach a facility that has the ability to perform the measurements.
12. Getting beam time
Initial enquiries should lead you to an instrument scientist who can give you a clear opinion whether the project is suitable or not and how much material and time it might take. Using this advice, you need to apply for beam time, and usually there are calls at regular times each year with a corresponding deadline. The applications are peer reviewed, and as facility time is expensive and precious, you need to make the case that the experiment is interesting, feasible and needs the facility to answer it. Neutron time is expensive, so the value of the unique data obtainable from the experiment needs to be stressed. You also need to show that you can make the samples in the required quantities, that you have previously characterized them and that they are stable for the duration of your experiment. For example, it is unlikely that a proposal will get funded if the protein has not yet been purified in sufficient amounts. However, some facilities offer protein deuteration and purification as well, and this can be carried out before a specific date for beam time is allocated to the project. When successful, you may be allocated between a day and a week of beam time, but, with newer and faster sources, even shorter allocations may become commonplace. The date will be fixed and you have the responsibility to ensure that the samples are ready and delivered to the neutron source, so be aware of any shipping issues if you need to cross national borders. You will carry out the experiments with the beamline scientist, and in some cases your travel and lodging are paid as part of the grant. A basic training course in the local safety procedures has to be taken, and varies between a Web-based version completed before arrival and a full day on site before starting. A steeper learning curve follows as the computer software that controls the experiments is not standardized and often difficult to grasp for the first-time user. However, you should be able to run repetitive data collection runs without the instrument scientist present either because there is an automatic sample changer or because the same software routine can be run easily. The biological experiments mostly do not run as time-tabled, and flexibility is important, which can be helped by preparing extra samples. The laboratory facilities on site need to be understood: do they have pipettes and a pH meter, buffers and pure water? At most sites, biologists are still in the minority, so it is best not to assume that the basic equipment is always available. Nevertheless, biology is part of the field of soft-matter/large-scale structures that have been studied extensively at these sites, and many new instruments have been justified in part by their biological applications. Good biology is always welcomed.
13. Data analysis
When the experiments have been performed, the data need to be treated and in many cases fitted to models to provide a publishable result. You may wish to embark on learning the required procedures, and the fitting programs are mostly freely available. Courses are put on by some major facilities and are aimed at graduate students or early post-doc level. They are mostly oversubscribed, so applications need to be planned well in advance. The experimental data need to be put into a form suitable for further analysis, and this is often called data reduction. It is important to ensure that this is sorted out before leaving the site. If you have a single burning question that neutrons can solve for you and are unlikely to need them again, then you should collaborate to obtain the publishable data that you can include, hopefully with results from other experimental procedures, in the final paper (Valincius et al. 2008). This may be all you need, or it may pave the way to further forays into the neutron world. Whichever applies, the important thing is to recognize where neutrons can provide a unique answer and to have a go.
Acknowledgements
Our neutron work has been supported by the Wellcome Trust, BBSRC, Orla Protein Technologies Ltd. I am grateful to my many collaborators and especially Stephen Holt, Chuck Majkrzak, Sofian M. Daud, Anton Le Brun, Phil Callow, Peter Timmins and Alexandra Solovyova. The facilities at ISIS, ILL, NIST and HIFR have all kindly given us beam time. Finally, I thank Bob Thomas and Jeff Penfold for inspiration and getting me started.
Footnotes
One contribution of 13 to a Theme Supplement ‘Biological physics at large facilities’.
References
- Armstrong J., Salacinski H. J., Mu Q. S., Seifalian A. M., Peel L., Freeman N., Holt C. M., Lu J. R. 2003. Interfacial adsorption of fibrinogen and its inhibition by RGD peptide: a combined physical study. In Meeting on Biological Surfaces and Interfaces, pp. S2483–S2491. Castelvecchio Pascoli, Italy: IOP Publishing Ltd. [Google Scholar]
- Blakeley M. P., Langan P., Niimura N., Podjarny A. 2008. Neutron crystallography: opportunities, challenges, and limitations. Curr. Opin. Struct. Biol. 18, 593–600. ( 10.1016/j.sbi.2008.06.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyton C., Haase W., Rapoport T. A., Kuhlbrandt W., Collinson I. 2002. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature 418, 662–665. ( 10.1038/nature00827) [DOI] [PubMed] [Google Scholar]
- Cisneros D. A., Muller D. J., Daud S. M., Lakey J. H. 2006. An approach to prepare membrane proteins for single-molecule imaging. Angew. Chem. Int. Ed. 45, 3252–3256. ( 10.1002/anie.200504506) [DOI] [PubMed] [Google Scholar]
- Comoletti D., Grishaev A., Whitten A. E., Tsigelny I., Taylor P., Trewhella J. 2007. Synaptic arrangement of the neuroligin/beta-neurexin complex revealed by X-ray and neutron scattering. Structure 15, 693–705. ( 10.1016/j.str.2007.04.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter J., Gutberlet T., Katsaras K. (eds) 2006. Neutron scattering in biology: techniques and applications. Heidelberg, Germany: Springer. [Google Scholar]
- Forsyth V. T., Parrot I. M. 2006. High angle neutron fiber diffraction in the study of biological systems. In Neutron scattering in biology: techniques and applications (eds Fitter J., Gutberlet T., Katsaras K.), pp. 85–106. Heidelberg, Germany: Springer. [Google Scholar]
- Fullagar W. K., Holt S. A., Gentle I. R. 2008. Structure of SP-B/DPPC mixed films studied by neutron reflectometry. Biophys. J. 95, 4829–4836. ( 10.1529/biophysj.108.134395) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harroun T. A., Katsaras J., Wassall S. R. 2008. Cholesterol is found to reside in the center of a polyunsaturated lipid membrane. Biochemistry 47, 7090–7096. ( 10.1021/bi800123b) [DOI] [PubMed] [Google Scholar]
- Hecht O., Ridley H., Boetzel R., Lewin A., Cull N., Chalton D. A., Lakey J. H., Moore G. R. 2008. Self-recognition by an intrinsically disordered protein. FEBS Lett. 582, 2673–2677. ( 10.1016/j.febslet.2008.06.022) [DOI] [PubMed] [Google Scholar]
- Holt S. A., Lakey J. H., Daud S. M., Keegan N. 2005. Neutron reflectometry of membrane protein assemblies at the solid/liquid interface. In Australian Colloid and Interface Symposium (ACIS), pp. 674–677. Sydney, Australia: CSIRO Publishing. [Google Scholar]
- Holt S. A., Le Brun A. P., Majkrzak C. F., McGillivray D. J., Heinrich F., Lösche M., Lakey J. H. 2009. An ion channel containing model membrane: structural determination by magnetic contrast neutron reflectometry. Soft Matter 5, 2576–2586. ( 10.1039/b822411K) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. V., Howse J. R., Dabkowska A., Jones R. A. L., Lawrence M. J., Roser S. J. 2008. Floating lipid bilayers deposited on chemically grafted phosphatidylcholine surfaces. Langmuir 24, 1989–1999. ( 10.1021/la702050b) [DOI] [PubMed] [Google Scholar]
- Jasnin M., Moulin M., Haertlein M., Zaccai G., Tehei M. 2008. In vivo measurement of internal and global macromolecular motions in Escherichia coli. Biophys. J. 95, 857–864. ( 10.1529/biophysj.107.124420) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanteur D., Pattus F., Timmins P. A. 1994. Membrane-bound form of the pore-forming domain of Colicin-A—a neutron-scattering study. J. Mol. Biol. 235, 898–907. ( 10.1006/jmbi.1994.1047) [DOI] [PubMed] [Google Scholar]
- Kim K., Byun Y., Kim C., Kim T. C., Noh D. Y., Shin K. 2005. Combined study of X-ray reflectivity and atomic force microscopy on a surface-grafted phospholipid monolayer on a solid. J. Colloid Interface Sci. 284, 107–113. ( 10.1016/j.jcis.2004.09.068) [DOI] [PubMed] [Google Scholar]
- Kreuger J. K., Wignal G. D. 2006. Small angle scattering from biological molecules. In Neutron scattering in biology: techniques and applications (eds Fitter J., Gutberlet T., Katsaras K.), pp. 127–160. Heidelberg, Germany: Springer. [Google Scholar]
- Krueger S., Meuse C. W., Majkrzak C. F., Dura J. A., Berk N. F., Tarek M., Plant A. L. 2001. Investigation of hybrid bilayer membranes with neutron reflectometry: probing the interactions of melittin. Langmuir 17, 511–521. ( 10.1021/la001134t) [DOI] [Google Scholar]
- Le Brun A. P., Holt S. A., Shah D. S., Majkrzak C. F., Lakey J. H. 2007. Monitoring the assembly of antibody-binding membrane protein arrays using polarised neutron reflection. In Workshop on Proteins at Work 2007, pp. 639–645. Perugia, Italy: Springer. [DOI] [PubMed] [Google Scholar]
- Majkrzak C. F. 1990. Polarized neutron reflectometry. In Workshop on Methods of Analysis and Interpretation of Neutron Reflectivity Data, pp. 75–88. Argonne, IL: Elsevier Science. [Google Scholar]
- Neylon C. 2008. Small angle neutron and X-ray scattering in structural biology: recent examples from the literature. Eur. Biophys. J. Biophys. Lett. 37, 531–541. ( 10.1007/s00249-008-0259-2) [DOI] [PubMed] [Google Scholar]
- Nierhaus K. H., Wadzack J., Burkhardt N., Junemann R., Meerwinck W., Willumeit R., Stuhrmann H. B. 1998. Structure of the elongating ribosome: arrangement of the two tRNAs before and after translocation. Proc. Natl Acad. Sci. USA 95, 945–950. ( 10.1073/pnas.95.3.945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura N., Arai S., Kurihara K., Chatake T., Tanaka I., Bau R. 2006. Recent results on hydrogen and hydration in biology studied by neutron macromolecular crystallography. Cell. Mol. Life Sci. 63, 285–300. ( 10.1007/s00018-005-5418-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebay-Peyroula E., Garavito R. M., Rosenbusch J. P., Zulauf M., Timmins P. A. 1995. Detergent structure in tetragonal crystals of Ompf Porin. Structure 3, 1051–1059. ( 10.1016/S0969-2126(01)00241-6) [DOI] [PubMed] [Google Scholar]
- Pencer J., Mills T., Anghel V., Krueger S., Epand R. M., Katsaras J. 2005. Detection of submicron-sized raft-like domains in membranes by small-angle neutron scattering. Eur. Phys. J. E 18, 447–458. ( 10.1140/epje/e2005-00046-5) [DOI] [PubMed] [Google Scholar]
- Roth M., Lewitbentley A., Michel H., Deisenhofer J., Huber R., Oesterhelt D. 1989. Detergent structure in crystals of a bacterial photosynthetic reaction center. Nature 340, 659–662. ( 10.1038/340659a0) [DOI] [Google Scholar]
- Schindler S. 2008. Model, theory, and evidence in the discovery of the DNA structure. Br. J. Phil. Sci. 59, 619–658. ( 10.1093/bjps/axn030) [DOI] [Google Scholar]
- Solovyova A. S., Nollmann M., Mitchell T. J., Byron O. 2004. The solution structure and oligomerization behavior of two bacterial toxins: pneumolysin and perfringolysin O. Biophys. J. 87, 540–552. ( 10.1529/biophysj.104.039974) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehei M., et al. 2007. Neutron scattering reveals extremely slow cell water in a Dead Sea organism. Proc. Natl Acad. Sci. USA 104, 766–771. ( 10.1073/pnas.0601639104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira S. C. M., et al. 2008. New sources and instrumentation for neutrons in biology. Chem. Phys. 345, 133–151. ( 10.1016/j.chemphys.2008.02.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrettaz S., Ulrich W.-P., Vogel H., Hong Q., Dover L. G., Lakey J. H. 2002. Stable self-assembly of a protein engineering scaffold on gold surfaces. Protein Sci. 11, 1917–1925. ( 10.1110/ps.0206102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaknin D., Kjaer K., Alsnielsen J., Losche M. 1991. Structural properties of phosphatidylcholine in a monolayer at the air–water interface—neutron reflection study and reexamination of X-ray reflection measurements. Biophys. J. 59, 1325–1332. ( 10.1016/S0006-3495(91)82347-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valincius G., Heinrich F., Budvytyte R., Vanderah D. J., McGillivray D. J., Sokolov Y., Hall J. E., Losche M. 2008. Soluble amyloid beta-oligomers affect dielectric membrane properties by bilayer insertion and domain formation: implications for cell toxicity. Biophys. J. 95, 4845–4861. ( 10.1529/biophysj.108.130997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacklin H. P., Tiberg F., Fragneto G., Thomas R. K. 2007. Distribution of reaction products in phospholipase A(2) hydrolysis. Biochim. Biophys. Acta Biomembr. 1768, 1036–1049. ( 10.1016/j.bbamem.2006.10.020) [DOI] [PubMed] [Google Scholar]
- Weik M., Patzelt H., Zaccai G., Oesterhelt D. 1998. Localization of glycolipids in membranes by in vivo labeling and neutron diffraction. Mol. Cell 1, 411–419. ( 10.1016/S1097-2765(00)80041-6) [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Kamikubo H., Kurihara K., Kuroki R., Niimura N., Shimizu N., Yamazaki Y., Kataoka M. 2009. Low-barrier hydrogen bond in photoactive yellow protein. Proc. Natl Acad. Sci. USA 106, 440–444. ( 10.1073/pnas.0811882106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccai G. 2000. Biochemistry—how soft is a protein? A protein dynamics force constant measured by neutron scattering. Science 288, 1604–1607. ( 10.1126/science.288.5471.1604) [DOI] [PubMed] [Google Scholar]