Abstract
Fluorescence two-dimensional difference gel electrophoresis (DiGE) is rapidly becoming established as a powerful technique for the characterization of differences in protein expression levels between two or more conditions. In this review, we consider the application of DiGE—both minimal and saturation labelling—to biomaterials research, considering the challenges and rewards of this approach.
Keywords: fluorescence two-dimensional difference gel electrophoresis, two-dimensional gel, biomaterials, cell–material interactions, topography, scarce samples
1. Fluorescence two-dimensional difference gel electrophoresis: a useful tool for biomaterials research
The cellular proteome is the total complement of proteins expressed by a cell under a set of defined conditions. The composition and relative abundance of proteins in the proteome can change in response to a multitude of factors, which in a biomaterials context includes the interaction of cells with metals, polymers and chemically or topographically micro- or nanopatterned surfaces. In recent years, the development of fluorescence two-dimensional difference gel electrophoresis (DiGE) has offered additional scope for the evaluation of cell–material interactions at the protein level, allowing relative changes in protein abundance and post-translational modifications (PTMs) to be mapped across the proteome between the control and test states. This is advantageous because proteins are the functional effectors of cellular responses to biomaterials, and changes in protein expression or PTMs are likely to equate with changes in cellular state. This is not necessarily true of gene expression data, where the translational activity of each mRNA species in a microarray experiment is unknown, and transcript levels are subject to additional levels of control, including degradation (Wu et al. 2006) and sequestration (Liu et al. 2005) of transcripts by the miRNA machinery, before the protein level is reached. In biomaterials research, the use of DiGE affords the opportunity to investigate global changes in cellular protein profile in response to new materials, devices and implants, to identify responsive proteins of interest, and groups of proteins (‘biomarkers’) associated with the cell and tissue responses to different materials or constructs. This has the potential to give valuable insight into cell surface interfaces at the protein level, which should assist with the development of next-generation materials and implants with the capacity to tailor cellular responses to particular laboratory, industrial or therapeutic applications. Samples for DiGE could be derived from in vitro, in vivo and clinical sources. In contrast to most other fields, in biomaterials research, many conventional biochemical techniques (such as binding assays and immunoprecipitation) are very difficult to perform, owing to the typically small sample yield and often time-consuming and labour-intensive nature of sample generation from these sources. DiGE offers the capability for simultaneous screening of large numbers of proteins for changes in abundance between different conditions, combined with the ability to cope with very scarce samples. The analyses are readily multiplexable, allowing comparison between replicate samples, over time courses or between multiple conditions. These features should assist biomaterials researchers in maximizing the data output of their experiments, while minimizing the amount of sample required.
Prior to the advent of DiGE, evaluation of relative changes in protein abundance could be performed using traditional two-dimensional gel electrophoresis. In this technique, individual samples are initially separated by native charge in the first dimension, using isoelectric focusing (IEF), and partitioned by molecular mass in the second dimension using SDS–PAGE (Carrette et al. 2006). In the biomaterials field, this approach has been applied to investigate the response of monocyte-derived macrophages to polycarbonate–urethane (Dinnes et al. 2007) and dermal fibroblasts to titanium surfaces (Derhami et al. 2001). The technique suffers from gel-to-gel variation and poor reproducibility, however, as one sample is resolved per gel in the absence of an internal standard, and therefore a large number of gels are required to perform a robust pairwise comparison. In addition, relatively large quantities of protein are required for this approach (at least 50 µg for silver staining, the most sensitive non-fluorescent post-stain, and up to approximately 500 µg protein with colloidal Coomassie Blue staining, equivalent to two or three 75 cm3 tissue culture flasks of confluent adherent cells). Sufficient protein was generated in the aforementioned investigations by using relatively high cell densities (Derhami et al. 2001; Dinnes et al. 2007) and small-format gels (less protein required, but with lower resolution—fewer distinct spots—than large-format gels) (Derhami et al. 2001). These approaches are not always practicable, depending on the biomaterial system, because samples can be very scarce and high resolution is desirable.
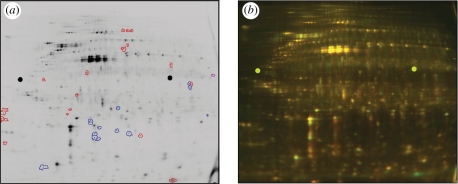
In DiGE, proteins extracted from a control extract are labelled with one CyDye (Cy3 or Cy5 conjugated; GE Healthcare),1 and proteins isolated from a test extract labelled with the other colour of CyDye fluorophore, which are size and charge matched. These labelled protein extracts are mixed and co-resolved (often with the addition of an internal standard, which can be labelled with Cy2) on large-format two-dimensional gels for analysis of expression changes in the resulting pattern of spots (‘spot maps’) (Unlu et al. 1997; Carrette et al. 2006). In comparison with two-dimensional gel electrophoresis, DiGE offered the advantage that multiple samples could be compared on a single gel (‘multiplexing’), and afforded the ability to stain control and test samples with different fluorescent dyes prior to electrophoresis. This advance alleviated issues of gel-to-gel comparison and decreased the number of gels required. The capability to include an internal standard, composed of an equal fraction of all the samples in an experiment, also improved inter-gel matching and facilitated normalization of matched spots in replicate samples on multiple gels. The crucial advantage of the DiGE approach for biomaterial applications is that the use of CyDyes to label proteins, in place of non-fluorescent post-stains, can give a large enhancement of sensitivity for protein detection (Shaw et al. 2003). This enables analysis of even very scarce protein samples, including small areas of laser-microdissected tissue (Greengauz-Roberts et al. 2005; Kondo & Hirohashi 2007), low cell numbers from topographical substrates (figure 1; see also Kantawong et al. 2009a, 2009b) and other material sources.
Figure 1.
DiGE gel of protein extracted from human fibroblasts (hTERT BJ-1) cultured at low density for 24 h on microgrooved (G; 5 µm deep × 25 µm pitch) versus planar (P) poly(dimethylsiloxane) (PDMS) substrates. At a 2.5-fold threshold, around 33 proteins appeared differentially regulated on this single gel. (For a multi-gel comparison, an internal standard should normally be included.) (a) Cy3 gel channel shown following software analysis with DeCyder v. 5.0. Blue, proteins upregulated in cells grown on G versus P; red, proteins downregulated in cells on G versus P. (b) Same gel as (a); green, Cy3 (extract from cells on P); red, Cy5 (extract from cells on G).
Current alternatives to DiGE for quantitative comparative proteomics are all gel-free approaches that involve chromatographic separation of peptides followed by mass spectrometry (MS; reviewed in Aebersold & Mann (2003)). The principal techniques include stable isotopic labelling by amino acids in cell culture (SILAC), an approach for metabolic isotopic labelling of proteins in cultured cells (Ong et al. 2002; Amanchy et al. 2005), isotope-coded affinity tag (ICAT) technology and the isobaric tags for relative and absolute quantification (iTRAQ) approach (Wu et al. 2005; Wiese et al. 2007). The latter approach was recently used to compare the response of osteoblasts to hydroxyapatite (HA) and HA reinforced with carbon nanotubes (Xu et al. 2008). Gel-free comparative approaches can be easier to perform than DiGE, but are generally difficult to multiplex, can be less quantitative and may not be well suited to the study of PTMs. In addition, iTRAQ can suffer from difficulties in isolating suitable ions for fragmentation (Wu et al. 2005). These methods are better suited to the investigation of certain groups of proteins, such as hydrophobic proteins, that are difficult to evaluate using two-dimensional gels. The above techniques are discussed more fully elsewhere (Wu et al. 2005; Mirza & Olivier 2008), and can be complementary to DiGE studies.
2. The DiGE workflow: considerations for biomaterial applications
2.1. Sample preparation from biomaterial sources
The method of protein extraction must be considered carefully. It is important to account for the source material and how it will be most effectively harvested while minimizing artefactual changes to the proteome. A detailed description of the optimization of a DiGE-compatible protein extraction procedure from a murine epithelial source, for example, can be found in Hannigan et al. (2007).
Cells can be directly lysed on their culture substratum, which is likely to result in the fewest artefacts at the cellular level, and cell scrapers can be used to assist collection. This approach can be costly in terms of reagents, depending on the volume required to cover the surfaces. It may also sample adherent proteins from the material surface, which could result in a significant background of adsorbed serum proteins, and mask lower abundance cellular proteins. If cell and extracellular matrix (ECM) analysis is desired, samples can be depleted of particularly high-abundance serum constituents, such as albumin, using affinity columns (Zolotarjova et al. 2005) or commercial kits. Alternatively, cells can be trypsinized from the surface prior to lysis to remove much of the serum and ECM. Although this may alter the proteome, if control and test samples are treated equally, the effects should not present large differential expression changes using DiGE. The surface chemistry of the material should also be taken into account. Surface chemistry and topography will almost certainly have an impact upon protein deposition upon the surface, with downstream effects at both the cellular and ECM levels, and control materials should be carefully chosen to minimize artificial differences arising in the experiment between samples. Such artefacts would be expected, for example, in the comparison of DiGE gels of protein from cells cultured on different heights of poly(caprolactone) microgrooves with extracts from an unsuitable control material of a different surface chemistry, such as planar poly(methylmethacrylate). Surface treatments, such as micro-contact printed surface patterns, should be checked to ensure that there is no downstream interference with DiGE (for example, if the chosen chemistry affects protein labelling). Protein coatings (e.g. pro-adhesive proteins such as collagen, poly-l-lysine) should also be chosen with care. The protein should be capable of adsorbing onto the surface (for example, by opposite charge interactions, such as poly-l-lysine (+) on quartz (−)), and its isoelectric point (pI) and molecular mass should be considered, to ensure that if any artificial spots result from the protein coating, they should appear in a part of the gel that would cause minimal disruption to the spot pattern in the region of interest. In the example of poly-l-lysine-coated quartz, the highly basic coating could result in spotting in the basic region of the gel, particularly if extraction is performed to harvest both cellular and ECM proteins. It should be possible to exclude spots derived from poly-l-lysine by using a first dimension IEF gel targeted to an acidic pH range, such as a pH 4–7 strip. Narrow pH-range IEF gels (known as ‘zoom’ gels) can also be useful to focus on and expand the resolution of a specific pH range of particular interest. This can assist with the detection of less abundant proteins and can be helpful in resolving particularly acidic or basic proteins, which are troublesome to resolve with DiGE, particularly in broad pH-range IEF gels.
In addition to in vitro studies, it is also possible to microdissect tissue samples into lysis buffer from histological sections (Kondo et al. 2003; Sitek et al. 2006; Kondo & Hirohashi 2007), which could provide a useful means of isolating areas of tissue of interest around an implanted biomaterial or device. If stained histological sections are to be used as the source material for DiGE, the tissue stains should be carefully chosen, as most stains affect protein recovery (Craven et al. 2002; Mouledous et al. 2002; Kirana et al. 2009), and some can interfere with saturation labelling (Kirana et al. 2009). Cleaning of toluidine blue-stained samples, to decrease the amount of stain remaining prior to IEF, improved sample yield and increased the number of spots that could be detected in DiGE gels (Kirana et al. 2009). The ability to remove toluidine blue from samples was also helpful for MS analysis (Li et al. 2004).
Once cells have been harvested, they are typically lysed using a DiGE compatible lysis buffer, which can be aided mechanically by processes such as sonication, freeze–thaw, homogenization and grinding. The lysis buffer normally contains chaotropes (urea, or urea with thiourea) and amphiphilic or non-ionic detergents (such as 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulphonate (CHAPS)) to denature and solubilize proteins. The composition of the buffers used (particularly the rehydration/sample, equilibration and lysis buffers) should be considered, and if necessary optimized for the sample type under investigation, while maintaining the optimum pH (see §2.2 for further detail) for the labelling regimen. It is important to reduce contamination from exogenous proteins, salts and chemical impurities during sample preparation and later gel processing for MS to ensure the best quality DiGE results. Keratin contamination (from hair and skin flakes) is particularly problematic for MS analysis. If the concentration of the sample of interest is likely to be lower than that of the contaminating keratin (for example, in the peptide mix extracted from a faint gel spot), the resulting spectrum can be dominated by keratin peaks, and give a stronger ‘hit’ (significance value) for keratin than for the protein of interest when an identification is sought from peptide mass databases.
Proteolytic degradation and unwanted protein modification can be circumvented using chaotropes, protease inhibitors, high-quality reagents and by ensuring the protein extracts remain adequately chilled (−20°C to 4°C, depending on the procedure) during preparation. Useful troubleshooting advice can be found in Tannu & Hemby (2006) and Carrette et al. (2006). Following lysis, proteins must be concentrated and contaminants removed as far as possible. Protein precipitation performs multiple useful functions and is applicable to many sample types. Precipitation can be used to enrich protein, inactivate proteases and remove salts, lipids and phenols from samples. Unfortunately, this approach can also lead to protein loss through protein insolubility following concentration or adhesion to the microfuge tube wall.
As the resolution of large-format two-dimensional gels is limited to approximately 2000–3000 protein spots (mostly highly abundant cellular components), if a subset of proteins is of particular interest (and especially if these are also low abundance), fractionation and enrichment procedures may also be required. After extraction, the protein concentration of the samples is usually determined using a detergent compatible quantification assay or by incorporating lysis buffer into the standard curve of an assay that is partially detergent incompatible. Sample losses during sample preparation and quantification must be accounted for when assessing the quantity of cellular material required. The biological replicates should be samples from independent sources (e.g. a number of metal discs each seeded with fibroblasts, or cells from identical implants used in a number of different patients) and subjected to the same conditions. A pilot study should be performed to estimate the number of biological replicates needed to obtain sufficient statistical power from the experiment, considering the expected magnitude of changes in expression level, and inherent biological ‘noise’ (biological variation between replicates), to anticipate whether greater or fewer replicates are necessary (discussed in Karp & Lilley (2007)). At the protein level, biological noise appears to be low in established cell lines and higher in inbred mice (Karp & Lilley 2007; Karp et al. 2007). It could be extrapolated that noise would be highest in outbred populations.
If sufficient material cannot be harvested from a single biomaterial source to perform DiGE, after cell seeding densities and the surface area of culture material have been optimized, it is possible to pool protein from a number of equivalent sources to generate a pooled biological replicate with enough protein. Note, however, that it is important to estimate the biological noise of the study system before this is undertaken, since subtle changes in response in particular subgroups may become masked by the overall variation in the system (discussed for microarray data in Zhang et al. (2007); Mary-Huard et al. (2007); Kendziorski et al. (2005)). If pooling has to be performed to generate sufficient material for a single sample, this should also be carried out for the other replicates, without overlapping between samples, to prevent statistical bias (discussed for pooling of mRNA samples in Kendziorski et al. (2005); Zhang et al. (2007); Mary-Huard et al. (2007)). Although such pooling can be suboptimal compared with the use of a single replicate per sample, it can assist in protein extraction from scarce samples, as protein pellets may become visible and more straightforward to process. It can also offset protein losses, and potentially enable repeats or confirmatory Western blotting to be performed with the same samples. If adequate sample material is available, a pooling strategy can still be useful for reducing the resources required for two-dimensional analysis. The relative merits of various pooled designs for DiGE were evaluated in Karp & Lilley (2009). This study suggested that pooling for ‘biological averaging’ of samples (analogous to the pooling for scarce isolates described above) could be particularly useful in applications with a higher level of biological variation between samples of the same type. The disadvantage of this experimental design is that the contributions from each individual sample in the pool would not be separable.
Another form of pooling is used to construct the internal standard, which should be present on every gel in an experiment to aid gel-to-gel matching and normalization of fluorescence intensity between gels, for more robust spot volume analysis. The internal standard is composed of an equal portion of each of the control and test replicates in the study, and should thus contain protein corresponding to every spot present in the gels. This is useful as it prevents protein spots being discounted when apparently absent in one set of samples relative to another, as these spots may be subject to the most dramatic differential regulation. If necessary, when the control and test samples are too scarce to constitute the pooled standard or preparative gel (a high-load gel used for MS identification of proteins), these can be made from a ‘related’ source to the samples under analysis. For cells grown on topographical substrates, for example, protein for the preparative gel would ideally be extracted from cells cultured on a planar piece of material of the same chemistry. If this is not feasible, however, the lysate can be prepared from cells on tissue culture-grade polystyrene. Similarly, the preparative extract for comparison of protein profiles from cells at different sites in a small implanted scaffold or device, or between different scaffolds, could be prepared from the surrounding tissue.
2.2. Minimal or saturation labelling?
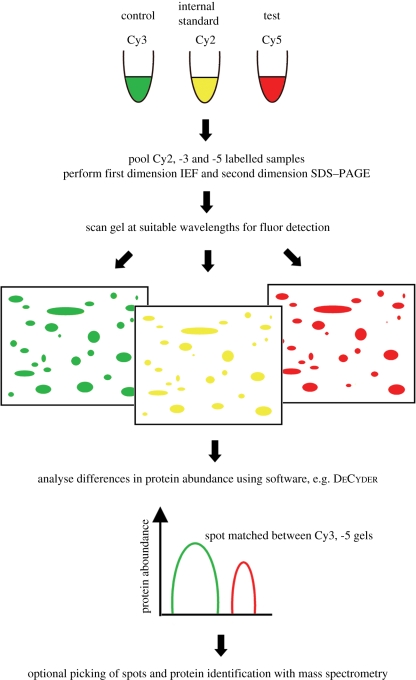
If the protein yield from the particular sample preparation regime is high enough to generate more than 100 µg protein per sample (50 µg for each control or test sample, with a further 50 µg to contribute to the internal standard), the most common form of DiGE, minimal labelling (DiGEmin), can be used. The minimal dyes have N-hydroxy succinimidyl ester chemistry and are used to label approximately 5 per cent of the lysine residues in proteins. The samples are labelled with Cy3 or Cy5 fluorophores, and the internal standard is labelled with Cy2. The workflow for DiGEmin can be seen in figure 2, and a step-by-step protocol for DiGEmin is available (Viswanathan et al. 2006). Minimal labelling is best suited to applications where greater cell mass can be generated, such as tissue samples, cells grown at high density (e.g. in bioreactor cultures), or on large areas of biomaterials. If protein identities are desired, a preparative gel (typically 300–500 µg protein load) can be post-stained with a fluorescent stain such as Sypro Orange, and matched to the analytical gels. Following software analysis of the spot patterns in the analytical gels, protein spots of interest can be manually or robotically excised and submitted for MS (§2.3).
Figure 2.
Basic workflow for DiGEmin (‘traditional’ experimental design). Cy2, -3 and -5 labelled samples are co-resolved on a single gel. The Cy2-labelled samples are used in spot normalization and matching between gels, and differences in protein abundance are quantitated using software. If desired, a picklist can be generated for manual or robotic spot-picking and analysis by MS.
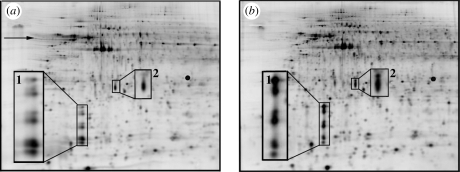
Saturation labelling (DiGEsat), which uses Cy3 and Cy5 fluorophores with maleimide chemistry, is used to label all cysteine residues within the extracted proteins. Unlike in minimal labelling, the protein samples are reduced prior to labelling. To ensure saturation is achieved, titration is required to find the optimal stoichiometry of the ratios of the reducing agent tris(2-carboxyethyl)phosphine (TCEP) : protein : dye for the particular sample (figure 3). This involves a qualitative assessment of the gels to determine which concentration results in the fewest artefacts in the resulting spot map. The experimental design recommended by the CyDye manufacturer uses Cy3 to label a pooled standard and the preparative gel, and Cy5 to label the pooled or test samples, because there is no Cy2-conjugated saturation dye available. Saturation labelling has greatly enhanced sensitivity over minimal labelling (Greengauz-Roberts et al. 2005), however, because all cysteines in a protein are labelled (rather than a small fraction of the lysine residues). This means that only 5 µg protein per biological replicate is required for analytical gels (with an additional 5 µg for the production of the internal standard), and substantially less material has been used in some studies (Shaw et al. 2003; Sitek et al. 2006; Kondo & Hirohashi 2007). This feature makes DiGE analysis accessible to even very low protein yield biomaterial applications. This is particularly important when sample area is restricted in academic-scale studies or when the study necessitates the use of a population of single cells without intercellular contact. An in-depth protocol for saturation labelling of microdissected samples is available (Kondo & Hirohashi 2007). The two-dye system requires twice as many gels as the minimal labelling approach, however, and the cysteine-reactive chemistry limits detection to proteins containing this less common residue. This restricts the number of proteins that can be detected (although approx. 96.3% of human proteins possess at least one cysteine residue (Sitek et al. 2005)), but this disadvantage is offset by the increased sensitivity of saturation labelling, and generally only becomes a concern if particular proteins of interest fail to be labelled owing to a lack of cysteine residues. For basic proteins, Kirana et al. (2009) found that saturation-labelled samples were less well resolved than unlabelled samples in pH 6-11 IEF gels, but reported preliminary data that this effect could be alleviated by additional sample clean-up following labelling.
Figure 3.
Part of a saturation labelling dye titration experiment using the same protein extract (from human fibroblasts grown to approx. 70% confluence on tissue culture-grade polystyrene) labelled with two different reducing agents and dye concentrations. (a) 1 nmol TCEP, 2 nmol dye; (b) 2 nmol TCEP, 4 nmol dye. Underlabelling issues are visible in (a) (horizontal streaking (arrow), spot doubling (inset 1), mass shifting in certain proteins (insets 1 and 2)), cf. optimal labelling stoichiometry shown in (b) (no visible artefacts).
Minimal and saturation staining approaches were found to have similar success rates for protein identification by MS (Greengauz-Roberts et al. 2005). Although saturation labelling affords increased sensitivity over minimal labelling for the detection of additional protein species, difficulties are likely to be encountered in identifying proteins from the smaller saturation-labelled spots because these spots will contain limiting quantities of protein. The success will be protein-specific. Current tandem MS instrumentation can generate robust fragmentation spectra from less than 1 fmol of starting material, but saturation labelling can result in the detection of spots that contain significantly less protein. Cy3 saturation-labelled peptides appear to be less efficiently detected by MS than unlabelled peptides, which was attributed to loss of these hydrophobic peptides by precipitation, and most labelled peptides failed to generate additional fragmentation data (Sitek et al. 2005). This should not affect the rate of identification for the majority of proteins, however, as there should still be a sufficient number of unlabelled peptides for successful MS. If identification fails, it may be possible to identify the spots by another type of MS, or de novo sequencing (§2.3), but even in the absence of a confirmed identity, these spots can still be valuable. For most applications, successful identification of the proteins of interest by MS is a prerequisite; however, the detection of patterns of differentially expressed proteins can be useful in the generation of biomarker maps. This could be used, for example, to study the protein profile of mouse tissue immediately surrounding the site of an implanted biomaterial, compared between animals where the implants were well versus poorly tolerated (or bioactive versus bioinert). This would enable the researcher to build up a map of the group of protein spots that were subject to differential regulation, and therefore likely to be important in determining the cellular reaction to the implant, even if these spots could not all be identified by MS. If multiple samples were collected over time, the resulting spot maps might be useful as a predictor of the outcome in a new animal exhibiting a particular protein profile prior to, and following, implantation.
Until recently, the minimal labelling approach was presumed to be the most robust of the DiGE variants, as the control, test and pooled internal standards are co-resolved on the same gel, reducing issues of gel-to-gel variation. This approach can lead to bias, however, through the matching of both samples to the pooled standard (Karp et al. 2007). The authors proposed that the experimental design used in saturation labelling (one sample—control or test—per gel, co-resolved with a pooled standard) should also be used for minimal labelling, which successfully removed the bias. This approach also negates any potential problems with preferential binding of one fluorophore over another to certain proteins, although this could be overcome by incorporating dye switches, or ‘fluor flips’, into the experimental design, where the fluor colour is alternated within a group of biological replicates. To prevent bias, fluor flips are most effectively applied to even numbers of samples. With four control samples and four test samples in a dye-flipped design, for example, two of each of the control and test samples would be labelled with Cy3 and two with Cy5. It is advisable not to mix replicate types (such as a combination of biological and technical replicates, i.e. different biological samples and gel repeats of the same sample) in a single experiment, as this confounds the statistical analysis (Karp et al. 2005).
If the amount of source material is insufficient for an unpooled minimal labelling study, but is enough for either a pooled minimal design with pooling of two or more source samples per biological replicate or an unpooled saturation design, it is prudent to consider the source material, available resources and expertise of the investigators. Assuming that the design with one sample per gel, together with a pooled standard, is adopted in either case, the saturation labelling approach should detect more protein spots, owing to its higher sensitivity, but it can be a more technically demanding technique for inexperienced users, owing to the difficulties in handling small samples, the narrow working range of pH conditions for labelling and the requirement for dye titration. If resources are restricted and the number of gels must be reduced while retaining a pooled standard, however, or the protein extracts have high inter-sample biological variability, the pooled three-dye minimal approach may be more readily applicable. Using either the two- or three-dye designs, it is possible to analyse an additional test sample during the same analysis, such as an equivalent extract from a different time point, or from a second topography or scaffold, if the samples can be compared with the same control. This provides a useful means of cross-comparing data from equivalent samples in a single experiment, such as spot maps from cells grown on a planar substrate compared with those from two different heights or types of topographic features with the same surface chemistry.
Some studies have elected to use a saturation labelling approach in the absence of an internal standard (Kantawong et al. 2009a, 2009b). This reduces the amount of protein required, lowers material costs and decreases labour required, and matching between the intra-gel sample pairs (control–test) can be more straightforward than the inter-gel matching required by the incorporation of an internal standard. For these reasons, this approach may be more accessible to a wider group of biomaterials scientists, but has the caveat that the data may be less robust. Statistical analysis can be more challenging for this approach, if the chosen gel analysis software is not equipped to facilitate this experimental design. Fluor flips should be included if this protocol is used. This design was also commonplace for minimal labelling studies, prior to the introduction of the Cy2 dye (Unlu et al. 1997; Zhou et al. 2002).
2.3. Identification of differentially expressed proteins and validation of results
Following software analysis, spots of interest, such as those subject to differential regulation, are usually submitted for identification by MS. A useful introduction to MS in the context of proteomics can be found in Aebersold & Mann (2003) and Steen & Mann (2004). For both minimal and saturation labelling studies, proteins should be picked from the high-load preparative gels for MS analysis. The reasons for this are that, in saturation labelling, the protein load is usually insufficient for identification of spots from the analytical gels, and in minimal labelling, although the protein load is higher, there is a shift between labelled and unlabelled proteins (as only approx. 3–5% of lysine residues are labelled), which could lead to failed or false identifications. In general, more abundant proteins are more readily and robustly identified, as there should be more peptide ions corresponding to these proteins detected in the mass spectrometer, which simplifies database identification. In general, large, intensely stained spots on DiGE gels correspond to abundant proteins, particularly if the spots are still detectable with a less sensitive post-stain such as colloidal Coomassie Blue, which interacts with proteins regardless of lysine or cysteine content. An exception to this could be cysteine-rich saturation labelled proteins, which might exhibit intense staining despite being lower abundance than suggested by the fluorescence intensity, but this could be assessed using a post-stain.
It is often useful to combine different types of MS, such as a high-throughput approach with a low-throughput, higher sensitivity approach, to maximize both the number of identifications and their relevance to the experimental question. The former approach, typified by Matrix-Assisted Laser Desorption Ionization/Time-of-Flight (MALDI/TOF/TOF) MS is useful for rapid identification of large numbers of spots, for example in attempting to assemble a comprehensive spot map of proteins detectable in the DiGE gel (whether or not the spots are differentially regulated). In contrast, when a particular subset of spots is of interest, a more focused analysis with the higher sensitivity approach, such as a liquid chromatography-based separation, followed by Electrospray Surface Ionization Quadrupole TOF/TOF (ESI-Q-TOF/TOF) is useful because chromatographic resolution separates and concentrates discrete peptides, and the more efficient fragmentation that can be achieved in a quadrupole mass spectrometer aids database matching. In combining two or more complementary MS techniques, the likelihood of generating a useful set of successful protein identifications is increased, and the identification of the same protein by two different types of MS is a useful means of confirming protein identifications. Gel fixation, storage and the optimal extraction of protein from gel plugs will also affect the success of protein identification. Kondo & Hirohashi (2007) acknowledged that considerable work may be required to optimize sample preparation for MS for each DiGE sample to maximize the number of successful identifications, and proposed an optimized protocol used in the identification of approximately 2500 saturation labelled gel spots.
Once the gel plugs have been digested to tryptic peptides, it can be helpful to split the sample into smaller aliquots, to ensure that there is sufficient peptide digest remaining to perform two complementary techniques, and, ideally, leave some remaining to allow for technical difficulties that would necessitate repetition of the MS. This is particularly important for biomaterials studies, where the initial samples may be scarce and difficult or time-consuming to collect and process for DiGE. Ideally, both MS and tandem MS (MS/MS) spectra should be collected, to increase the chance of successfully identifying the protein of interest and the confidence in its putative identification, allowing both peptide mass fingerprinting (mass-based analysis) and MS/MS analysis (includes sequence data derived from additional subpeptide fragmentation) to be performed. Tandem MS differs from MS in that an additional round of fragmentation of strong mass peaks (usually by collision-induced dissociation, where the peptides are broken into increasingly smaller pieces by repeated collision of the primary peptide ions with an inert gas) is induced following the initial ‘survey’ scan. Peptide masses are detected in the survey scan (known as MS), and strong mass peaks identified at this stage can be subsequently fragmented by MS/MS. Following this, suitable spectra are compared with a database of theoretical trypsin-derived peptides (such as Mascot, at http://www.matrixscience.com, which also has detailed guidelines for performing database searches) to generate hits (most probable protein identifications, together with significance values) for the sample.
It is useful to widen the initial database searches to include the species from which trypsin originated (e.g. if porcine trypsin is used to digest protein extracted from human samples, the initial taxonomic search criterion could specify ‘Mammalia (mammals)’, rather than the more restrictive ‘Homo sapiens (human)’). This serves as an internal control to ensure that both MS and identification have proceeded appropriately, as the trypsin should be readily identified from spectral peaks of autolysis, where the enzyme has digested itself. Later, if desired, the search can be directed specifically towards the species of interest (human, in the previous example), which will decrease the time required for searching the database, although it should be noted that contaminants derived from other species (such as bovine serum albumin) will be missed using the restricted taxonomic criterion. For all types of MS with trypsin-digested samples, the enzyme should be identified as trypsin, usually allowing up to one missed cleavage, and the parameters set to search for the appropriate charge of the ionized peptides generated by the particular type of mass spectrometer (e.g. 2+, 3+ product ions), with selection of the mass modifications that could be expected under the experimental conditions used. Common modifications include variable methionine oxidation and fixed cysteine carbamidomethylation, and in saturation labelling, a fixed modification of +672.85 Da is added by the Cy3 dye conjugate.
Confidence in a protein identity comes from a number of sources. The threshold for significance of peptide mass fingerprints is usually set to p < 0.05, but this can be adjusted manually. There may be more than one protein match assigned to the group of peptides if the particular peptides could have derived from more than one protein in the database. In this case, the more likely identification should have a molecular mass and pI comparable to the region of the gel from which the spots were excised. Confirmation of appropriate molecular mass and pI is also useful as a general check for the plausibility of a putative identity. In contrast, the peptide ion scores are important for the significance of MS/MS results. It must be confirmed that there are multiple highly scoring peptide ions contributing to the total score, indicative of a good match to the identified protein, rather than an aggregated score of many peptides that individually have low scores (for example, short, ill-matched or highly modified peptides, or those with several missed cleavages), which results in a cumulative increase in score. Multidimensional Protein Identification Technology (MudPIT) scoring can be used to remove spurious protein hits that have many low-scoring peptides, but this is more stringent and generally reserved for complex samples (where many proteins are present within a single digest, which should not be the case for protein spots extracted from DiGE gels), although an ion score cut-off filter can be applied to remove low-scoring peptides. In Mascot, the ions are ranked from 1 (best) to 10 (worst), according to the quality of the match. As mentioned, the same proposed identification revealed by two different MS techniques enhances confidence in the proposed protein identity.
One difficulty that is frequently encountered in DiGE experiments is the presence of multiple protein species within a single spot. This confounds the differential analysis, but can usually only be detected at the level of MS identification. These multi-protein spots generally have to be excluded, or taken as the combination of the constituent spots, unless the proteins can be analysed separately in a repeat experiment, or using another technique, such as Western blotting. It has been suggested that this can be improved by decreasing the protein load of the preparative gel somewhat, thus reducing the amount of contaminating substances present during IEF in incompletely cleaned samples, and so enhancing the resolution of neighbouring spots (Kondo & Hirohashi 2007), but ideally this should not be at the expense of maximizing the number of successful identifications. Another problem can be encountered when proteins fail to be identified by more than one type of MS. In this case, de novo sequencing, where software is used to generate protein sequence data from mass spectra (discussed in Steen & Mann (2004), used with two-dimensional gel electrophoresis in Tannu & Hemby (2007) and applied to DiGE samples in Oehlers et al. (2007)), is an option when the spectra are strong (assessed by the intensity of the unassigned peptides) but fail to match a protein entry in the database. This technique can also be used as a means of validating protein identities.
Differentially regulated proteins that are subsequently successfully identified by MS should ideally be validated by another technique, usually by Western blotting. It should be borne in mind, however, that some discrepancies between DiGE with MS analysis and Western blotting data have been reported in the literature (Masayo et al. 2009; Suehara et al. 2009). It is likely that such non-concordant data could result from differences in the form of the protein detected by DiGE/MS versus an antibody-based validation, since DiGE is naturally suited to resolving distinct PT modifications and protein isoforms, whereas Western blotting detects the overall signal from a protein species without differentiating between the subforms of the protein (unless isoform-specific antibodies are used, which would require prior knowledge of the particular PTM or isoform under study, and availability of a suitably specific antibody). Alternatives to Western blotting following one-dimensional SDS–PAGE could include the use of two-dimensional gels for confirmatory blotting against non-concordant protein species, or protein microarrays (reviewed in Templin et al. 2002). Antibody-based validation can suffer from sensitivity issues, for example, if the antibody fails to detect the small quantities of protein available, if there are issues due to batch variation of antibodies (Pozner-Moulis et al. 2007), or the blotting procedure does not provide accurate quantitative data over the required dynamic range. The latter problem has been observed with the use of β-actin as an internal control in Western blotting, as it failed to show a linear response to increasing protein loads (Dittmer & Dittmer 2006). The authors recommended using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a more reliable loading control. Since GAPDH levels can also fluctuate according to cellular state, however, an alternative means of inter-sample standardization has been proposed. This utilizes total protein staining to check loading across the whole sample, followed by fluorescent Western blotting, for quantitative analysis of individual proteins, and has been successfully applied to the validation of DiGE data (Zellner et al. 2008).
If blotting is performed efficiently, scarce biomaterial samples can be examined for several antigens, with minimal sample use. If sample and antibody titration is performed using protein extracted from a higher yielding source to the sample of interest (such as protein from cells grown in a tissue culture flask), the appropriate antibody dilution and minimum protein load per lane can be optimized for the successful detection of each antigen of interest, with low background, before the scarce samples are analysed under optimal conditions. Furthermore, by cutting the membranes with the transferred protein into smaller pieces, it may be possible to probe for multiple antigens on the same membrane, without cross-reaction between antibodies. With fluorescent Western blotting, this could also be achieved using multiple directly fluorophore-conjugated primary antibodies with spectrally distinct fluorophores or indirect conjugates (fluorescently labelled secondary detection antibodies) with primary antibodies raised in different species. These approaches should be useful for the conservation of the limited sample volume that would be anticipated in many biomaterial studies.
3. Current research: DiGE and biomaterials
Recent modifications of the traditional DiGE procedures have enabled an elegant examination of cell surface proteins with protein labelling performed prior to lysis (Mayrhofer et al. 2006), and comparison of the redox status of cellular proteins under different conditions using minimal labels and modified saturation dyes (Chan et al. 2005). Another modification, known as Blue Native-DiGE, has been employed to improve the resolution of hydrophobic proteins, and also gives some indication of the differences in native protein structures (discussed in Dani & Dencher 2008). Similarly, it has recently been possible to adapt a technique for improved two-dimensional electrophoretic display of hydrophobic proteins (described in Bridges et al. 2008) for minimal labelling DiGE (D. J. Bridges & R. J. S. Burchmore 2006, unpublished data). Such continual developments should enable flexibility for novel applications of the technique to the biomaterials field, such as the investigation of cell stress in response to materials with redox DiGE, and global analysis of the membrane and surface markers expressed over time by differentiating cells on structured substrata.
To date, DiGE has been employed successfully in the biomaterials field to investigate cell responses to nano- (Kantawong et al. 2009b) and microtopographical substrates (Kantawong et al. 2009a; fig. 1) using DiGEsat. Preliminary analysis suggests that approximately 33 proteins appeared differentially regulated between human fibroblasts grown on a planar poly(dimethylsiloxane) (PDMS) substrate and an equivalent area of PDMS, patterned with 5 µm deep × 25 µm pitch microgrooves (figure 1), at a threshold of 2.5-fold change.
These pioneering studies should pave the way for future investigations using DiGE in biomaterial applications, and the versatility of the technique gives flexibility for its adaptation to the study system of interest. The ability to probe differential protein expression patterns and PTMs has great potential to provide invaluable insight into the molecular biology of cell–material interactions, identify biomarkers for laboratory and clinical applications using biomaterials and inform future material design for the direction of cellular responses.
Acknowledgements
L.E.M. is funded by BBSRC Doctoral Training Grant BB/D526329/1. M.J.D. and R.B. are funded by the BBSRC and MRC, respectively. The authors would like to thank Ms F. Kantawong and Dr K. Jayawardena for their assistance and advice, and all in the Sir Henry Wellcome Functional Genomics Facility and the Centre for Cell Engineering for help and fruitful discussion.
Footnotes
One contribution to a Theme Supplement ‘NanoBioInterface: crossing borders’.
Two-dimensional electrophoresis, principles and methods (GE Healthcare). Available online at: http://www5.gelifesciences.com/aptrix/upp01077.nsf/Content/orderonline_handbooks.
References
- Aebersold R., Mann M. 2003. Mass spectrometry-based proteomics. Nature 422, 198–207 (doi:10.1038/nature01511) [DOI] [PubMed] [Google Scholar]
- Amanchy R., Kalume D. E., Pandey A. 2005. Stable isotope labeling with amino acids in cell culture (SILAC) for studying dynamics of protein abundance and posttranslational modifications. Sci. STKE 2005, pl2 (doi:10.1126/stke.2672005pl2) [DOI] [PubMed] [Google Scholar]
- Bridges D. J., Pitt A. R., Hanrahan O., Brennan K., Herzyk P., De Koning H. P., Burchmore R. J. S. 2008. Characterisation of the plasma membrane subproteome of bloodstream form Trypanosoma brucei. Proteomics 8, 83–99 (doi:10.1002/pmic.200700607) [DOI] [PubMed] [Google Scholar]
- Carrette O., Burkhard P. R., Sanchez J.-C., Hochstrasser D. F. 2006. State-of-the-art two-dimensional gel electrophoresis: a key tool of proteomics research. Nat. Protoc. 1, 812–823 (doi:10.1038/nprot.2006.104) [DOI] [PubMed] [Google Scholar]
- Chan H.-L., Gharbi S., Gaffney P. R., Cramer R., Waterfield M. D., Timms J. F. 2005. Proteomic analysis of redox- and ErbB2-dependent changes in mammary luminal epithelial cells using cysteine- and lysine-labelling two-dimensional difference gel electrophoresis. Proteomics 5, 2908–2926 (doi:10.1002/pmic.200401300) [DOI] [PubMed] [Google Scholar]
- Craven R. A., Totty N., Harnden P., Selby P. J., Banks R. E. 2002. Laser capture microdissection and two-dimensional polyacrylamide gel electrophoresis: evaluation of tissue preparation and sample limitations. Am. J. Pathol. 160, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani D., Dencher N. A. 2008. Native-DIGE: A new look at the mitochondrial membrane proteome. Biotechnol. J. 3, 817–822 (doi:10.1002/biot.200800030) [DOI] [PubMed] [Google Scholar]
- Derhami K., Zheng J., Li L., Wolfaardt J. F., Scott P. G. 2001. Proteomic analysis of human skin fibroblasts grown on titanium: novel approach to study molecular biocompatibility. J. Biomed. Mater. Res. 56, 234–244 (doi:10.1002/1097-4636(200108)56:2<234::AID-JBM1090>3.0.CO;2-#) [DOI] [PubMed] [Google Scholar]
- Dinnes D. L., Marcal H., Mahler S. M., Santerre J. P., Labow R. S. 2007. Material surfaces affect the protein expression patterns of human macrophages: a proteomics approach. J. Biomed. Mater. Res. A 80, 895–908 (doi:org/10.1002/jbm.a.30967) [DOI] [PubMed] [Google Scholar]
- Dittmer A., Dittmer J. R. 2006. beta-Actin is not a reliable loading control in Western blot analysis. Electrophoresis 27, 2844–2845 (doi:10.1002/elps.200500785) [DOI] [PubMed] [Google Scholar]
- Greengauz-Roberts O., Stoppler H., Nomura S., Yamaguchi H., Goldenring J. R., Podolsky R. H., Lee J. R., Dynan W. S. 2005. Saturation labeling with cysteine-reactive cyanine fluorescent dyes provides increased sensitivity for protein expression profiling of laser-microdissected clinical specimens. Proteomics 5, 1746–1757 (doi:10.1002/pmic.200401068) [DOI] [PubMed] [Google Scholar]
- Hannigan A., Burchmore R., Wilson J. B. 2007. The optimization of protocols for proteome difference gel electrophoresis (DiGE) analysis of preneoplastic skin. J. Proteome Res. 6, 3422–3432 (doi:10.1021/pr0606878) [DOI] [PubMed] [Google Scholar]
- Kantawong F., Burchmore R., Wilkinson C. D. W., Oreffo R. O. C., Dalby M. J. 2009a. Differential in-gel electrophoresis (DIGE) analysis of human bone marrow osteoprogenitor cell contact guidance. Acta Biomater. 5, 1137–1146 (doi:10.1016/j.actbio.2008.11.001) [DOI] [PubMed] [Google Scholar]
- Kantawong F., Burchmore R., Gadegaard N., Oreffo R. O. C., Dalby M. J. 2009b. Proteomic analysis of human osteoprogenitor response to disordered nanotopography. J. R. Soc. Interface 6, 1075–1086 (doi:10.1098/rsif.2008.0447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp N. A., Lilley K. S. 2007. Design and analysis issues in quantitative proteomics studies. Proteomics 7(Suppl. 1), 42–50 (doi:org/10.1002/pmic.200700683) [DOI] [PubMed] [Google Scholar]
- Karp N. A., Lilley K. S. 2009. Investigating sample pooling strategies for DIGE experiments to address biological variability. Proteomics 9, 388–397 (doi:10.1002/pmic.200800485) [DOI] [PubMed] [Google Scholar]
- Karp N. A., Spencer M., Lindsay H., O'Dell K., Lilley K. S. 2005. Impact of replicate types on proteomic expression analysis. J. Proteome Res. 4, 1867–1871 (doi:10.1021/pr050084g) [DOI] [PubMed] [Google Scholar]
- Karp N. A., Mccormick P. S., Russell M. R., Lilley K. S. 2007. Experimental and statistical considerations to avoid false conclusions in proteomics studies using differential in-gel electrophoresis. Mol. Cell. Proteomics 6, 1354–1364 (doi:10.1074/mcp.M600274-MCP200) [DOI] [PubMed] [Google Scholar]
- Kendziorski C., Irizarry R. A., Chen K. S., Haag J. D., Gould M. N. 2005. On the utility of pooling biological samples in microarray experiments. Proc. Natl Acad. Sci. USA 102, 4252–4257 (doi:10.1073/pnas.0500607102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirana C., Ward T., Jordan T. W., Rawson P., Royds J., Shi H. J., Stubbs R., Hood K. 2009. Compatibility of toluidine blue with laser microdissection and saturation labeling DIGE. Proteomics 9, 485–490 (doi:10.1002/pmic.200800197) [DOI] [PubMed] [Google Scholar]
- Kondo T., Hirohashi S. 2007. Application of highly sensitive fluorescent dyes (CyDye DIGE Fluor saturation dyes) to laser microdissection and two-dimensional difference gel electrophoresis (2D-DIGE) for cancer proteomics. Nat. Protoc. 1, 2940–2956 (doi:10.1038/nprot.2006.421) [DOI] [PubMed] [Google Scholar]
- Kondo T., Seike M., Mori Y., Fujii K., Yamada T., Hirohashi S. 2003. Application of sensitive fluorescent dyes in linkage of laser microdissection and two-dimensional gel electrophoresis as a cancer proteomic study tool. Proteomics 3, 1758–1766 (doi:10.1002/pmic.200300531) [DOI] [PubMed] [Google Scholar]
- Li C., et al. 2004. Accurate qualitative and quantitative proteomic analysis of clinical hepatocellular carcinoma using laser capture microdissection coupled with isotope-coded affinity tag and two-dimensional liquid chromatography mass spectrometry. Mol. Cell. Proteomics 3, 399–409 (doi:org/10.1074/mcp.M300133-MCP200) [DOI] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M. A., Hannon G. J., Parker R. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7, 719–723 (doi:10.1038/ncb1274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mary-Huard T., Daudin J.-J., Baccini M., Biggeri A., Bar-Hen A. 2007. Biases induced by pooling samples in microarray experiments. Bioinformatics 23, i313–i318 (doi:10.1093/bioinformatics/btm182) [DOI] [PubMed] [Google Scholar]
- Masayo Y., Kiyonaga F., Koji K., Setsuo H., Tadashi K. 2009. The Proteomic Profile of pancreatic cancer cell lines corresponding to carcinogenesis and metastasis. J. Proteomics Bioinform. 2, 1–18(doi:org/10.4172/jpb.1000057) [Google Scholar]
- Mayrhofer C., Krieger S., Allmaier G., Kerjaschki D. 2006. DIGE compatible labelling of surface proteins on vital cells in vitro and in vivo. Proteomics 6, 579–585 (doi:10.1002/pmic.200500104) [DOI] [PubMed] [Google Scholar]
- Mirza S. P., Olivier M. 2008. Methods and approaches for the comprehensive characterization and quantification of cellular proteomes using mass spectrometry. Physiol. Genomics 33, 3–11 (doi:org/10.1152/physiolgenomics.00292.2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouledous L., Hunt S., Harcourt R., Harry J., Williams K., Gutstein H. 2002. Lack of compatibility of histological staining methods with proteomic analysis of laser-capture microdissected brain samples. J. Biomol. Tech. 13, 258–264 [PMC free article] [PubMed] [Google Scholar]
- Oehlers L. P., Perez A. N., Walter R. B. 2007. Detection of hypoxia-related proteins in medaka (Oryzias latipes) brain tissue by difference gel electrophoresis and de novo sequencing of 4-sulfophenyl isothiocyanate-derivatized peptides by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 145, 120–133 (doi:10.1016/j.cbpc.2006.06.005) [DOI] [PubMed] [Google Scholar]
- Ong S.-E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 (doi:org/10.1074/mcp.M200025-MCP200) [DOI] [PubMed] [Google Scholar]
- Pozner-Moulis S., Cregger M., Camp R. L., Rimm D. L. 2007. Antibody validation by quantitative analysis of protein expression using expression of Met in breast cancer as a model. Lab. Invest. 87, 251–260 (doi:10.1038/labinvest.3700515) [DOI] [PubMed] [Google Scholar]
- Shaw J., Rowlinson R., Nickson J., Stone T., Sweet A., Williams K., Tonge R. 2003. Evaluation of saturation labelling two-dimensional difference gel electrophoresis fluorescent dyes. Proteomics 3, 1181–1195 (doi:10.1002/pmic.200300439) [DOI] [PubMed] [Google Scholar]
- Sitek B., Luttges J., Marcus K., Klöppel G. N., Schmiegel W., Meyer H. E., Hahn S. A., Stuhler K. 2005. Application of fluorescence difference gel electrophoresis saturation labelling for the analysis of microdissected precursor lesions of pancreatic ductal adenocarcinoma. Proteomics 5, 2665–2679 (doi:10.1002/pmic.200401298) [DOI] [PubMed] [Google Scholar]
- Sitek B., Potthoff S., Schulenborg T., Stegbauer J., Vinke T., Rump L. C., Meyer H. E., Vonend O., Stuhler K. 2006. Novel approaches to analyse glomerular proteins from smallest scale murine and human samples using DIGE saturation labelling. Proteomics 6, 4337–4345 (doi:10.1002/pmic.200500739) [DOI] [PubMed] [Google Scholar]
- Steen H., Mann M. 2004. The abc's (and xyz's) of peptide sequencing. Nat. Rev. Mol. Cell Biol. 5, 699–711 (doi:10.1038/nrm1468) [DOI] [PubMed] [Google Scholar]
- Suehara Y., et al. 2009. Anatomic site-specific proteomic signatures of gastrointestinal stromal tumors. Proteomics Clin. Appl. 3, 584–596 (doi:10.1002/prca.200800168) [Google Scholar]
- Tannu N. S., Hemby S. E. 2006. Two-dimensional fluorescence difference gel electrophoresis for comparative proteomics profiling. Nat. Protoc. 1, 1732–1742 (doi:10.1038/nprot.2006.256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannu N. S., Hemby S. T. 2007. De novo protein sequence analysis of Macaca mulatta. BMC Genomics 8, 270 (doi:10.1186/1471-2164-8-270) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templin M. F., Stoll D., Schrenk M., Traub P. C., Vohringer C. F., Joos T. O. 2002. Protein microarray technology. Trends Biotechnol. 20, 160–166 (doi:10.1016/S0167-7799(01)01910-2) [DOI] [PubMed] [Google Scholar]
- Unlu M., Morgan M. E., Minden J. S. 1997. Difference gel electrophoresis. A single gel method for detecting changes in protein extracts. Electrophoresis 18, 2071–2077 (doi:10.1002/elps.1150181133) [DOI] [PubMed] [Google Scholar]
- Viswanathan S., Unlu M., Minden J. S. 2006. Two-dimensional difference gel electrophoresis. Nat. Protoc. 1, 1351–1358 (doi:10.1038/nprot.2006.234) [DOI] [PubMed] [Google Scholar]
- Wiese S., Reidegeld K. A., Meyer H. E., Warscheid B. 2007. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics 7, 340–350 (doi:10.1002/pmic.200600422) [DOI] [PubMed] [Google Scholar]
- Wu L., Fan J., Belasco J. G. 2006. From the Cover: microRNAs direct rapid deadenylation of mRNA. Proc. Natl Acad. Sci. USA 103, 4034–4039 (doi:10.1073/pnas.0510928103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Wang G., Baek S. J., Shen R. 2005. Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. J Proteome Res. 5, 651–658 (doi:10.1021/pr050405o) [DOI] [PubMed] [Google Scholar]
- Xu J., Khor K. A., Sui J., Zhang J., Tan T. L., Chen W. N. 2008. Comparative proteomics profile of osteoblasts cultured on dissimilar hydroxyapatite biomaterials: an iTRAQ-coupled 2-D LC-MS/MS analysis. Proteomics 8, 4249–4258 (doi:10.1002/pmic.200800103) [DOI] [PubMed] [Google Scholar]
- Zellner M., Babeluk R., Diestinger M., Pirchegger P., Skeledzic S., Oehler R. 2008. Fluorescence-based Western blotting for quantitation of protein biomarkers in clinical samples. Electrophoresis 29, 3621–3627 (doi:10.1002/elps.200700935) [DOI] [PubMed] [Google Scholar]
- Zhang W., Carriquiry A., Nettleton D., Dekkers J. C. M. 2007. Pooling mRNA in microarray experiments and its effect on power. Bioinformatics 23, 1217–1224 (doi:10.1093/bioinformatics/btm081) [DOI] [PubMed] [Google Scholar]
- Zhou G., et al. 2002. 2D Differential in-gel electrophoresis for the identification of esophageal scans cell cancer-specific protein markers. Mol. Cell. Proteomics 1, 117–123 (doi:org/10.1074/mcp.M100015-MCP200) [DOI] [PubMed] [Google Scholar]
- Zolotarjova N., Martosella J., Nicol G., Bailey J., Boyes B. E., Barrett W. C. 2005. Differences among techniques for high-abundant protein depletion. Proteomics 5, 3304–3313 (doi:10.1002/pmic.200402021) [DOI] [PubMed] [Google Scholar]