Abstract
A proposed mechanism for magnetic compasses in animals is that systems of radical pairs transduce magnetic field information to the nervous system. One can show that perfectly ordered arrays of radical pairs are sensitive to the direction of the external magnetic field and can thus operate, in principle, as a magnetic compass. Here, we investigate how disorder, inherent in biological cells, affects the ability of radical pair systems to provide directional information. We consider biologically inspired geometrical arrangements of ensembles of radical pairs with increasing amounts of disorder and calculate the effect of changing the direction of the external magnetic field on the rate of chemical signal production by radical pair systems. Using a previously established signal transduction model, we estimate the minimum number of receptors necessary to allow for detection of the change in chemical signal owing to changes in magnetic field direction. We quantify the required increase in the number of receptors to compensate for the signal attenuation through increased disorder. We find radical-pair-based compass systems to be relatively robust against disorder, suggesting several scenarios as to how a compass structure can be realized in a biological cell.
Keywords: magnetic sensors, radical pair mechanism, signal-to-noise ratio
1. Introduction
Many animals use the geomagnetic field to obtain directional information (Wiltschko & Wiltschko 1995). The magnetite and radical pair models are currently discussed as possible biophysical mechanisms underlying magnetic sensing (Johnsen & Lohmann 2008). In the radical pair model (Schulten 1982; Ritz et al. 2000), which will be explored further here, the magnetic field affects a photochemical reaction step within a protein that involves a light-induced pair of radicals, i.e. molecules with unpaired electron spins. It has been shown that such reactions can be sensitive to earth-strength magnetic fields (Maeda et al. 2008). Birds were disoriented by artificial oscillating magnetic fields designed to disturb detection of the geomagnetic field by radical pair systems (Ritz et al. 2004; Wiltschko et al. 2005), suggesting that the magnetic compass of birds may be based on a radical pair mechanism. The light receptor cryptochrome has been suggested as a potential photo-magnetoreceptor candidate (Ritz et al. 2000) with some recent studies supporting a role of cryptochromes in magnetoreception (Ahmad et al. 2007; Gegear et al. 2008; Yoshii et al. 2009).
Here, we wish to abstract from any concrete candidate and investigate a key aspect of the radical pair mechanism. Radical pair reactions produce varying amounts of ligands, i.e. a chemical signal, depending on the intensity and the angle of the geomagnetic field with respect to the radical pairs. The identity of the ligands is unknown, but we can assume that they can be detected by receptors initiating neural transduction. To detect the small effects of the external magnetic field in a noisy biological environment, a multitude of receptors for this chemical signal is required (Weaver et al. 2000). In earlier studies (Ritz et al. 2000; Timmel et al. 2001; Wang et al. 2006), it was assumed that radical pairs are arranged in a crystalline geometry with all radical pairs perfectly aligned. A possible fundamental objection to the radical pair model in this simple formulation is that small amounts of disorder could drastically attenuate the signal, thus precluding directional sensing in realistic biological structures. Here, we study how robust the angular sensitivity of a system of radical pairs is to increased amounts of static disorder. We consider radical pair alignments drawn from Gaussian distributions with increasing variance and calculate radical pair recombination rates for different angles of the geomagnetic field. To relate the radical pair reaction rates to biologically meaningful numbers, we consider the number of ligand–receptor complexes required to detect a directional signal, i.e. to reach a signal-to-noise ratio of 1 for a change in geomagnetic field angle of 1°, using a previously established signal transduction model (Weaver et al. 2000). We find that a modest increase in receptor complexes can compensate for the effects of disorder up to variances of 30°, and that an increase of about a factor 100 in receptors is needed to compensate for variances of about 50°.
2. Theory
The theory of the radical pair mechanism, as for magnetic field detection, is by now well established (Schulten et al. 1978; Timmel et al. 1998, 2001; Ritz et al. 2000; Cintolesi et al. 2003; Rodgers & Hore 2009). After light-induced electron transfer, an intermediate radical pair state is created in which two separate electron spins evolve under the influence of the hyperfine interaction (HFI) with the nuclear spins, and the Zeeman interaction with the external magnetic field, according to
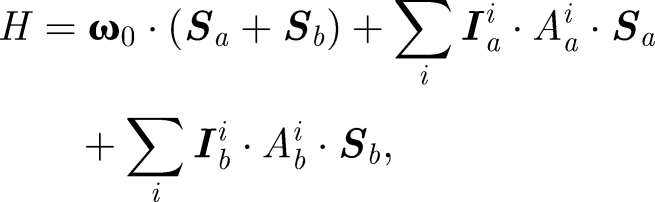 |
2.1 |
where Sa(b) is the electron spin for molecule a(b), Iia(b) is the ith nuclear spin in molecule a(b), Aa(b)i is the corresponding hyperfine coupling tensor, and ω0 is the Larmor frequency: ω0 = γeB. Axial anisotropic hyperfine couplings contribute stronger to the angular sensitivity of a radical pair than rhombic anisotropic or isotropic hyperfine couplings. It is reasonable to assume that Nature has optimized the hyperfine couplings in a magnetic sensor through evolution so as to provide maximal sensitivity. Therefore, we assume that the hyperfine couplings are all axially anisotropic, i.e. that the diagonal values of the hyperfine coupling tensor in its principal axis form obey the relationship
| 2.2 |
The singlet recombination rate constant is obtained by calculating the time dependence of the fraction of singlet radical pairs. In the simplest model, assuming that singlet and triplet states decay with equal rate constants, k = kS = kT, the singlet recombination rate constant can be calculated as
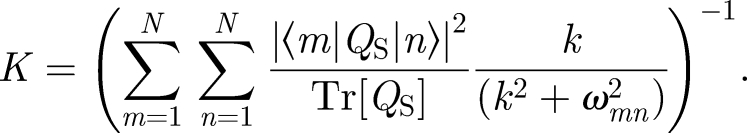 |
2.3 |
Here, QS denotes the singlet projection operator, |m〉 and |n〉 are eigenvectors of the Hamiltonian and ωmn = ωm − ωn, where ωm(n) is the eigenvalue of state m(n). The effect of the weak, external magnetic field is to alter the singlet (triplet) first-order recombination rate constant of the radical pair. The magnitude of the effect of the external magnetic field depends on its angle with respect to the radical pair since the HFI is anisotropic.
The signal transduction model of Weaver et al. (2000) assumes that radical pairs produce varying amounts of ligands that either bind to receptors on a neural substrate or are removed to a degenerative sink. In the original model, the goal was to determine how many ligand–receptor complexes are necessary to detect an intensity change in the magnetic field. Here, we are interested in the detection of the angle of the magnetic field. The above-defined singlet recombination rate constant K from equation (2.3) captures the dependence on the angle of the geomagnetic field and therefore replaces the intensity-dependent rate constant used originally (Weaver et al. 2000). The average number of ligand–receptor complexes in this signal transduction model is
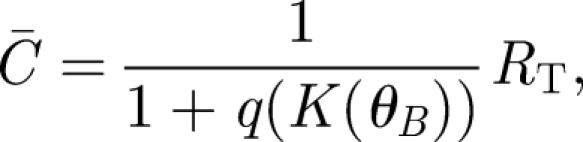 |
2.4 |
where RT is the total number of receptors and q is conveniently defined as the equilibrium dissociation constant divided by the concentration of signalling ligands. The ligand concentration is in turn dependent on the rate K; thus, q is a function of K(θB). The signal is defined as the shift in the number of complexes S = C̄1 − C̄op corresponding to the operating point angle θB,op and a second angle θB,1 chosen depending on the desired angular resolution. Defining for convenience the corresponding 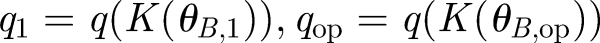 , we obtain the signal as a function of receptor number
, we obtain the signal as a function of receptor number
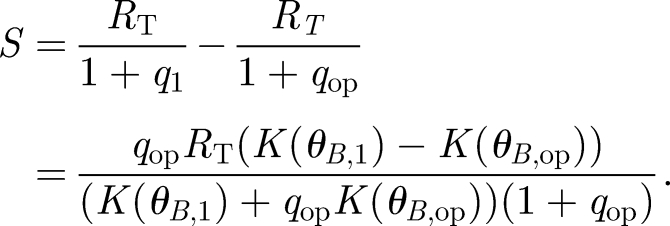 |
2.5 |
The noise, N, is considered, as in Weaver et al. (2000), to be the bound receptor fluctuation about the operating point and can be expressed as
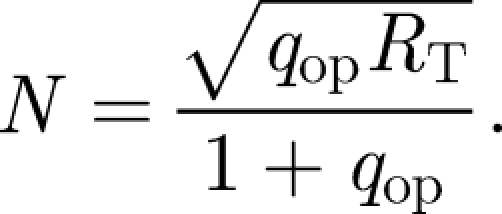 |
2.6 |
Setting the division of S by N equal to 1 to solve for RT, and finding that RT is minimized when qop = K(θB,1)/K(θB,op), the resultant dependence of the minimum total number of receptors on the angularly dependent rate constant is
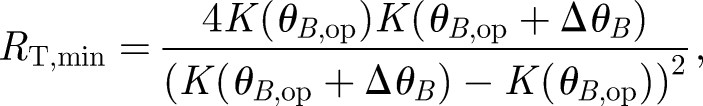 |
2.7 |
where θB,op is the initial, reference magnetic field angle or operating point of the receptor and ΔθB denotes the deviation from this reference angle, i.e. ΔθB = θB,1 − θB,op. The optimal operating point will correspond to the greatest change in the rate constant for small changes in angle. The largest slope in K depends on the type of radical pair and the surrounding environment. Therefore, the operating point is determined for each case by calculating the angle corresponding to the maximum of the derivative of K.
3. Modelled Geometries
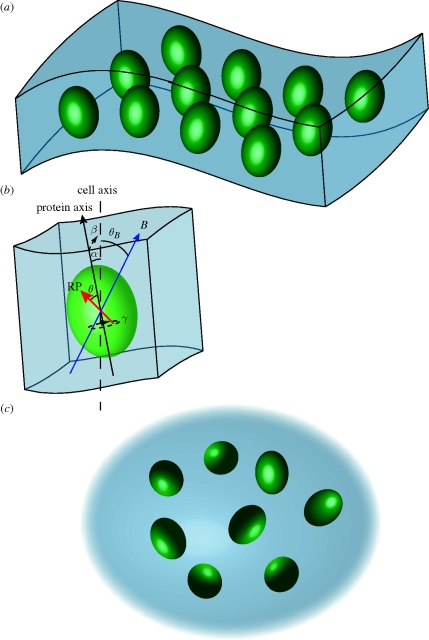
In order to computationally account for disorder in our model system, we consider two biologically inspired situations, a curved membrane containing magnetically sensitive radical pairs and an ensemble of magnetically sensitive radical pairs in cytosol, as depicted in figure 1. The magnetosensitive radical pair, denoted by the red arrow, is fixed within a protein (green ovaloid) with the same relative azimuth angle θ for all radical pairs. Since we assume the HFIs to be axial anisotropic, we can drop the polar angle ϕ from our considerations. The direction of the radical pair is defined as the z-axis of its dominant anisotropic HFI. For membrane proteins, the angles α and β would denote the direction of the protein axis with the cell axis, as indicated in the figure. The direction of the protein axis is taken to be identical to the membrane normal; hence α and β in the membrane case also denote the membrane normal at the location of each protein. For a perfectly flat membrane, α and β would be zero; only in curved membranes will α and β deviate from zero due to the changed membrane normals at different points in the membrane. On the other hand, membrane proteins are generally free to assume different polar angles γ with respect to the membrane normal and, hence, we assume that γ is completely randomized in our models. The magnetic field vector is denoted by B and makes an angle θB with respect to the cell axis.
Figure 1.
Visual representation of the five angles used in the theoretical computation to account for static disorder. A radical pair-containing protein is assumed to be associated with a structure inside a cell; however, the results hold for a general radical pair system. θ is the angle of the dominant axial HFI with respect to the protein axis. θB is the angle of the geomagnetic field with respect to the cell axis. Owing to disorder, all proteins will not be aligned with the same angles with respect to the cell axis. The changes due to disorder are quantified by the angles α, β and γ, as indicated in the figure. α and β are the x and y angles of the protein axis with respect to the cell axis. In membrane proteins, α and β would denote the membrane normal at the protein location. γ is the polar angle with respect to the protein axis (in membrane proteins) or the z-axis (in cytosolic proteins). The angle γ is irrelevant if the dominant axial HFI is in the direction of the protein axis in membrane proteins (θ = 0°), but it describes an additional level of randomization for other HFI orientations or in cytosolic proteins.
At first sight, it may seem surprising that we also consider a cytosol model, as one does not expect cytosolic proteins to be ordered. Besides being a valuable theoretical limiting case, we were motivated to include this model by the study of Maeda et al. (2008), in which they used polarized light to selectively excite a subset of molecules in solution that are aligned in the same plane. It may be possible that a similar strategy is employed in biological sensors as well, by filtering incident light on the magnetic sensory cells through material that polarizes light. After exciting an ordered subset of proteins, these proteins would then tumble due to thermal fluctuations and become increasingly disordered. The alignment of these selected cytosolic proteins within the cell after a certain time is then defined by the angles α, β and γ that would differ from protein to protein, as suggested in figure 1c.
In all of the following calculations, α and β, and in the cytosol models γ, were drawn from a Gaussian distribution with zero mean and standard deviation σ. The value of σ denotes the amount of disorder in the system. The recombination rate was calculated according to equation (2.3) at each magnetic field angle for ensembles of 2500 radical pairs with randomly drawn angles and then averaged.
We have performed calculations for membrane and cytosol models for several HFI angles θ. For the cytosol models, the HFI angle θ simply shifts the phase of the angular dependence, but it does not affect how the angular sensitivity is changed by different amounts of disorder. In contrast, for the membrane model, the HFI does affect how the angular sensitivity is changed by disorder. For the HFI angle θ = 0° the angle γ becomes irrelevant for the membrane protein model, as the HFI direction is independent of the value of the angle γ. For the HFI angle θ > 0°, and, most pronounced, for θ = 90°, the angle γ adds another level of randomization. However, we found that the membrane model with θ = 90° is an intermediate model in terms of its angular sensitivity between the membrane model with θ = 0° (highest angular sensitivity) and the cytosol model, regardless of the value of the HFI angle θ (lowest angular sensitivity). Therefore, we represent in the following only these two limiting cases, choosing the value θ = 0° for both membrane and cytosol models for easier comparison.
4. Results
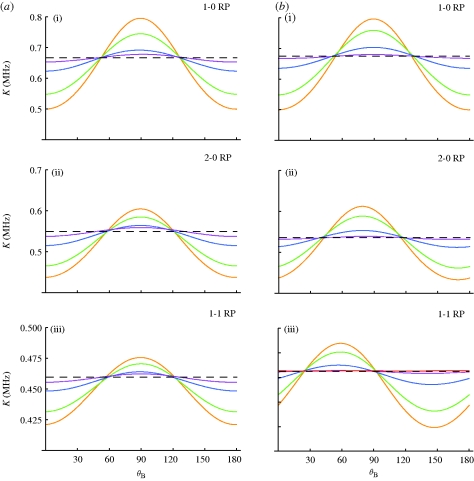
The effect of the static disorder on the radical pair system can be seen in figure 2, which shows the singlet recombination rate constant as it depends on the magnetic field angle for various amounts of disorder. The angular sensitivity of the yield, i.e. the difference between maximal and minimal yield values for different angles, provides an intuitive measure of the functionality of a radical-pair-based compass. As the angular sensitivity decreases, it becomes less likely that a compass can remain operational and, certainly, without any angular sensitivity (dotted lines), the compass would become dysfunctional. Figure 2a,b depicts the membrane and cytosol model geometries, respectively, whereas figure 2a(i),b(i), 2a(ii),b(ii) and 2a(iii),b(iii) distinguish between three radical pair models. Figure 2a(i),b(i) depicts angular sensitivities for radical pairs with only one anisotropic hyperfine coupling (1-0 radical pairs). In figure 2a(ii),b(ii), a second HFI was added on the same radical containing the first HFI, creating a (2-0) radical pair. In figure 2a(iii),b(iii), a second anisotropic HFI is added on the second radical, thereby creating a radical pair with one HFI on each radical (1-1 radical pair). In all calculations, the decay rate constant was 0.2 MHz. The HFI values were chosen based upon the strongest HFI within a flavin–tryptophan radical pair in DNA photolyase with parameters described as in a previous study (Cintolesi et al. 2003). DNA photolyase is similar to cryptochrome, a blue-light photoreceptor that has been discussed as a possible magnetoreceptor (Ritz et al. 2000, 2009; Cintolesi et al. 2003).
Figure 2.
Singlet recombination rate constant in different radical pair models versus the angle of the geomagnetic field for different disorder σ. The average recombination rate is calculated according to equation (2.3) for ensembles of radical pairs with randomized orientations. (a) In the membrane models, the radical pair orientations are randomized with respect to two angles drawn from a Gaussian distribution with standard deviation σ. (b) In the cytosol models, the radical pair orientations are randomized with respect to three angles drawn from a Gaussian distribution with standard deviation σ (see figure 1 for the definition of angles). The HFI angle θ is chosen to be 0°; in the membrane model, this represents the choice with the highest angular sensitivity. The cytosol model represents the opposite limiting case with lowest angular sensitivity, independent of HFI angle θ. Anisotropic HFIs are placed on one (1-0) and (2-0) models or both radicals (1-1) model. Purple line, 60°; blue line, 40°; green line, 20°; orange line, 0°.
All curves show the cos(2θB) dependence (Timmel et al. 2001; Maeda et al. 2008) typical for axially anisotropic HFIs with phases shifted by a few degrees between models with different HFI angles. As expected, the angular sensitivity decreases for increasing σ. The angular sensitivity decreases as the added HFIs become less aligned and the HFI angle with respect to the protein axis increases, such that randomization occurs in three instead of two dimensions. The angular dependence is not shifted for the membrane cases because γ is uniformly random, causing a larger range of averaging than the initially aligned, cytosolic proteins. After an initial fast decrease, resulting from the initial randomization in the angle γ, the angular sensitivity decreases more slowly for the membrane cases. Even at σ = 60°, the yield shows a small, remaining angular sensitivity for the membrane cases, whereas such angular sensitivity is barely noticeable for the cytosol cases. To generalize the results, we repeated the calculations varying the hyperfine coupling strengths and recombination rate constants. We found that the angular sensitivity remains essentially unchanged as long as hyperfine coupling strengths are larger than the external magnetic field and the decay rate is sufficiently slow (k < 2.0 MHz).
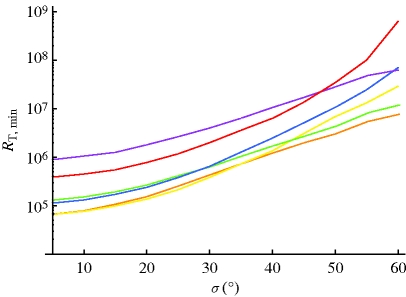
Using the recombination rate constants shown in figure 2, and the above-described signal transduction model, we estimate the minimum total number of receptors required to detect a change in geomagnetic field angle of 1° according to equation (2.7). Figure 3 shows the required number of receptors for all three radical pair cases in membrane and cytosol environments. The information quantified in figure 3 is based on and closely parallels the information from the angular sensitivity curves in figure 2. Initially, membrane models require a higher number of receptors, but, as disorder increases, the membrane models require lower numbers. It has been noted previously that radical pairs in which hyperfine couplings are located on one radical represent an optimal design for detecting weak magnetic fields (Rodgers et al. 2007; Ritz et al. 2009). This is borne out in the calculations, with the required number of receptors being smaller for the (1-0) and (2-0) radical pairs.
Figure 3.
Minimum number of receptors needed to detect a 1° change in angle as a function of disorder. The number is calculated according to equation (2.7) using the averaged recombination rates shown in figure 2 for the respective membrane (M) and cytosolic (C) radical pair models. Orange line, 1-0 RP-M; yellow line, 1-0 RP-C; green line, 2-0 RP-M; blue line, 2-0 RP-C; purple line, 1-1 RP-M; red line, 1-1 RP-C.
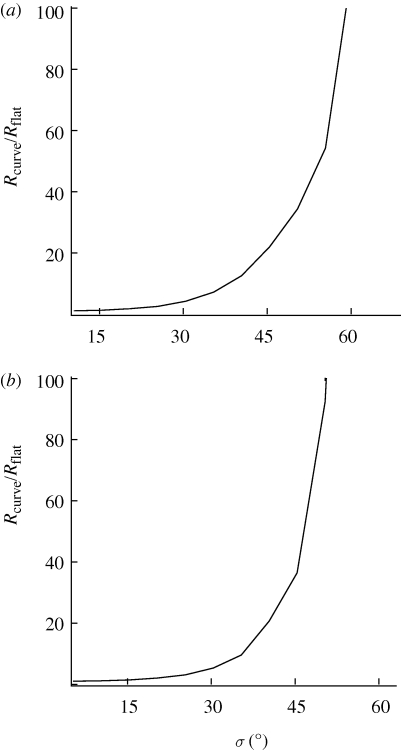
However, when normalized to the number of receptors for σ = 0°, the increase in the number of required receptors is essentially the same regardless of the case or model considered, as shown in figure 4 for the two extreme cases of (1-0) radical pairs in a membrane model and (1-1) radical pairs in a cytosol model, respectively. The dependence shown in figure 4 implies that the required number of receptors changes about a factor of 3.5 for disorder of about σ = 25°. Disorder of σ = 50° requires approximately a 100-fold increase in receptors. Larger disorder requires drastically increased numbers of receptors. The curves shown in figure 4 are independent of the desired accuracy.
Figure 4.
Minimum number of receptors needed to detect a signal-to-noise ratio of 1 normalized to the number of receptors for a radical pair system with no static disorder as a function of disorder. (a) 1-0 radical pair model in a membrane. (b) 1-1 radical pair model in cytosol.
To put these numbers in perspective, we first note that changing the radical pair composition from the least optimal (1-1) radical pair to the improved (2-0) radical pair, and the optimal (1-0) radical pair, reduces the required number of ligand–receptor complexes by a factor of 12, and 50, respectively. Thus, evolutionary selection for improved radical pair compositions can, at least in part, compensate for effects of disorder.
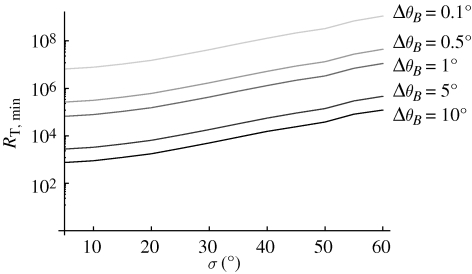
In our calculations so far, we have chosen 1° as the desired angular accuracy of the compass. It is illustrative to compare the effect of disorder and of changed angular accuracies on the required number of receptors. Figure 5 displays this effect for the 1-0 radical pair model in a membrane. Analogous curves can be found for the other radical pair compositions. Reduction of accuracy by a factor of 10 reduces the required number of receptors by about a factor of 100.
Figure 5.
Minimum number of receptors needed to detect changes in geomagnetic angle with different resolutions ΔθB evaluated for the 1-0 membrane radical pair model. The number is calculated according to equation (2.7) using the averaged recombination rate for the 1-0 membrane model shown in figure 2.
5. Discussion
We have extended the standard radical pair model by considering disorder in the geometric arrangement of radical pairs within biological systems. We found that the radical pair magnetic compass model is relatively robust against disorder. Effects of disorder of up to σ = 40° could be compensated for by a change in the chemical properties of the radical pair through the values of the HFIs, or by relaxing the angular accuracy of the compass from 1° to 5°. We have also shown that the effects of disorder on a physiological compass can be compensated for by a modest increase in the number of receptors. Within a factor of 100 increase, a compass can operate with the same accuracy even if the individual proteins in a receptor cell are distributed with a standard deviation of 50°.
We note that the required number of receptors to detect small changes in angles is significantly smaller than the required number of receptors to detect small intensity changes, using the same signal transduction model. Weaver calculated that 4 × 108 receptors are needed to detect an intensity change of 10−6 T (2% of the geomagnetic field) and that 4 × 1010 receptors are needed to detect an intensity change of 10−7 T (0.2% of the geomagnetic field) (Weaver et al. 2000). We find that about 4 × 106 receptors are sufficient to detect a 1° change in magnetic direction in the optimal (1-0) membrane model even for a disorder of σ = 60°, which is likely an overestimate of the disorder in a biological system. The required number of receptors increases to about 9 × 108 receptors in the worst case (1-1, cytosol) model with σ = 60°, which is of the same order as the number of receptors required to detect a 2 per cent change in magnetic intensity and much smaller than the number of receptors required to detect a 0.2 per cent change in magnetic intensity. This suggests that radical pair magnetic sensors are more sensitive to directional changes than to intensity changes.
It is not clear whether there is a maximal number of receptors beyond which a compass could not operate anymore. Weaver argued that 4 × 1010 receptors, required to detect a 0.2 per cent change in magnetic intensity, could be hosted in a small multicellular system. As pointed out above, a magnetic compass sensor can be realized with a significantly smaller number of receptors. It is perhaps a better question to ask whether a radical pair magnetic compass sensor could be realized within a single cell. Cells routinely express several million proteins. It is certainly not unreasonable to assume that about 50 000 proteins, or about 5 per cent of the total number of proteins, could serve as receptors in specialized receptor cells, with about half of them having ligands attached. Using 50 000 as an estimate for the number of receptors that could be realized in a single cell, we determine the best resolution that can be achieved for disordered systems with increasing σ. As shown in table 1, a resolution of nearly 1° is possible in the absence of disorder. The resolution decreases to about 5° for disorder of 40° and to about 15° for disorder of 60°. Certainly, birds can derive useful information about their direction from a compass with a resolution of 15° and this value compares reasonably well with standard deviations in bearings during orientation experiments.
Table 1.
Possible angular resolution ΔθB that can be achieved for different disorder by an ensemble of 50 000 receptors as could be hosted within a single cell.
| σ | 10° | 20° | 30° | 40° | 50° | 60° |
| ΔθB | 1.25° | 1.75° | 2.95° | 5.15° | 8.14° | 14.95° |
The absolute numbers should perhaps not be overinterpreted. Many parameters required for a solid estimate still remain unknown. However, once more information is available about the nature and concentration of receptors, the calculations in this manuscript provide a framework to evaluate what level of resolution can be achieved. While a relatively low-resolution compass sensor (of the order of 10°) may be realizable in a single cell, higher resolution, as would be necessary for a map sensor, would require multicellular systems. With regards to the fundamental question posed in the beginning, the relative numbers are illuminating: it would have been, in principle, possible to see signal attenuation by several orders of magnitude as one includes disorder in the radical pair model. Instead, we find a relatively modest effect of disorder on the functionality of a radical-pair-based compass. This effect can be compensated for by a variety of means, as discussed here, without severely affecting the functionality of a compass.
Acknowledgements
This work was supported by the Human Frontier Science Foundation (grant to T.R.). T.R. is a Cottrell Scholar of the Research Cooperation.
Footnotes
One contribution to the Theme Supplement ‘Magnetoreception’.
References
- Ahmad M., Galland P., Ritz T., Wiltschko R., Wiltschko W. 2007. Magnetic intensity affects cryptochrome-dependent responses in Arabidopsis thaliana. Planta 225, 615–624. ( 10.1007/s00425-006-0383-0) [DOI] [PubMed] [Google Scholar]
- Cintolesi F., Ritz T., Kay C., Timmel C., Hore P. 2003. Anisotropic recombination of an immobilized photoinduced radical pair in a 50-µT magnetic field: a model avian photomagnetoreceptor. Chem. Phys. 294, 385–399. ( 10.1016/S0301-0104(03)00320-3) [DOI] [Google Scholar]
- Gegear R. J., Casselman A., Waddell S., Reppert S. M. 2008. Cryptochrome mediates light-dependent magnetosensitivity in Drosophila. Nature 454, 1014–1018. ( 10.1038/nature07183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen S., Lohmann K. J. 2008. Magnetoreception in animals. Phys. Today 61, 29–35. ( 10.1063/1.2897947) [DOI] [Google Scholar]
- Maeda K., Henbest K. B., Cintolesi F., Kuprov I., Rodgers C. T., Liddell P. A., Gust D., Timmel C. R., Hore P. J. 2008. Chemical compass model of avian magnetoreception. Nature 453, 387–390. ( 10.1038/nature06834) [DOI] [PubMed] [Google Scholar]
- Ritz T., Adem S., Schulten K. 2000. A model for photoreceptor-based magnetoreception in birds. Biophys. J. 78, 707–718. ( 10.1016/S0006-3495(00)76629-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz T., Thalau P., Phillips J. B., Wiltschko R., Wiltschko W. 2004. Resonance effects indicate a radical-pair mechanism for avian magnetic compass. Nature 429, 177–180. ( 10.1038/nature02534) [DOI] [PubMed] [Google Scholar]
- Ritz T., Wiltschko R., Hore P. J., Rodgers C. T., Stapput K., Thalau P., Timmel C. R., Wiltschko W. 2009. Magnetic compass of birds is based on a molecule with optimal directional sensitivity. Biophys. J. 96, 3451–3457. ( 10.1016/j.bpj.2008.11.072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers C. T., Hore P. J. 2009. Chemical magnetoreception in birds: the radical pair mechanism. Proc. Natl Acad. Sci. USA 106, 353–360. ( 10.1073/pnas.0711968106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers C. T., Norman S. A., Henbest K. B., Timmel C. R., Hore P. J. 2007. Determination of radical re-encounter probability distributions from magnetic field effects on reaction yields. J. Am. Chem. Soc. 129, 6746–6755. ( 10.1021/ja068209l) [DOI] [PubMed] [Google Scholar]
- Schulten K. 1982. Magnetic field effects in chemistry and biology. Festkorperprobleme Adv. Solid State Phys. 22, 61–83. ( 10.1007/BFb0107935) [DOI] [Google Scholar]
- Schulten K., Swenberg C. E., Weller A. 1978. A biomagnetic sensory mechanism based on magnetic field modulated coherent electron spin motion. Z. Phys. Chem. NF111, 1–5. [Google Scholar]
- Timmel C., Till U., Brocklehurst B., McLauchlan K., Hore P. 1998. Effects of weak magnetic fields on free radical recombination reactions. Mol. Phys. 95, 71–89. ( 10.1080/00268979809483134) [DOI] [PubMed] [Google Scholar]
- Timmel C., Cintolesi F., Brocklehurst B., Hore P. 2001. Model calculations of magnetic field effects on the recombination reactions of radicals with anisotropic hyperfine reactions. Chem. Phys. Lett. 334, 387–395. ( 10.1016/S0009-2614(00)01436-6) [DOI] [Google Scholar]
- Wang K., Mattern E., Ritz T. 2006. On the use of magnets to disrupt the physiological compass of birds. Phys. Biol. 3, 220–231. ( 10.1088/1478-3975/3/3/007) [DOI] [PubMed] [Google Scholar]
- Weaver J., Vaughan T., Astumian R. 2000. Biological sensing of small field differences by magnetically sensitive chemical reactions. Nature 405, 707–709. ( 10.1038/35015128) [DOI] [PubMed] [Google Scholar]
- Wiltschko R., Wiltschko W. 1995. Magnetic orientation in animals, vol. 33 Berlin, Germany: Springer. [Google Scholar]
- Wiltschko R., Ritz T., Stapput K., Thalau P., Wiltschko W. 2005. Two different types of light-dependent responses to magnetic fields in birds. Curr. Biol. 15, 1518–1523. ( 10.1016/j.cub.2005.07.037) [DOI] [PubMed] [Google Scholar]
- Yoshii T., Ahmad M., Helfrich-Foerster C. 2009. Cryptochrome mediates light-dependent magnetosensitivity of Drosophila's circadian clock. PLoS Biol. 7, 813–819. ( 10.1371/journal.pbio.1000086) [DOI] [PMC free article] [PubMed] [Google Scholar]