Abstract
Natural killer (NK)–cell recognition of infected or neoplastic cells can induce cytotoxicity and cytokine secretion. So far, it has been difficult to assess the relative contribution of multiple NK-cell activation receptors to cytokine and chemokine production upon target cell recognition. Using Drosophila cells expressing ligands for the NK-cell receptors LFA-1, NKG2D, DNAM-1, 2B4, and CD16, we studied the minimal requirements for secretion by freshly isolated, human NK cells. Target cell stimulation induced secretion of predominately proinflammatory cytokines and chemokines. Release of chemokines MIP-1α, MIP-1β, and RANTES was induced within 1 hour of stimulation, whereas release of TNF-α and IFN-γ occurred later. Engagement of CD16, 2B4, or NKG2D sufficed for chemokine release, whereas induction of TNF-α and IFN-γ required engagement of additional receptors. Remarkably, our results revealed that, upon target cell recognition, CD56dim NK cells were more prominent cytokine and chemokine producers than CD56bright NK cells. The present data demonstrate how specific target cell ligands dictate qualitative and temporal aspects of NK-cell cytokine and chemokine responses. Conceptually, the results point to CD56dim NK cells as an important source of cytokines and chemokines upon recognition of aberrant cells, producing graded responses depending on the multiplicity of activating receptors engaged.
Introduction
Natural killer (NK) cells respond directly to infected or neoplastic cells through engagement of a multitude of germline-encoded receptors by ligands on target cells.1–3 Beside their ability to kill aberrant cells, NK cells are also critical components of the innate immune response by virtue of their capacity to produce a variety of cytokines and chemokines.4–6 Murine models have demonstrated a dependence on NK cell–derived cytokines in early responses to obligate intracellular parasites such as Listeria, Toxoplasma, and Leishmania and in resistance to cytomegalovirus infection.7–10 In many of these systems, NK cells respond to cues from sentinel immune cells, including dendritic cells, macrophages, and pathogen-infected tissue cells.11–13 These cues are communicated by release of cytokines, including interleukin-1 (IL-1), IL-10, IL-12, IL-15, and IL-18.14 Thus, secondary to triggering of innate immune cells by pattern recognition receptors, NK cells can relay and amplify cytokine signals. Among the most prominent cytokines produced by NK cells are tumor necrosis factor-α (TNF-α) and interferon γ (IFN-γ). Moreover, NK cells have been reported to secrete several other factors, including immunoregulatory cytokines such as IL-5, IL-10, IL-13, the growth factor GM-CSF, and the chemokines MIP-1α, MIP-1β, IL-8, and RANTES.15–22
In humans, NK cells are usually defined as CD3–CD56+ cells,23 and can be further subdivided based on CD56 expression. Typically, CD56dim NK cells constitute the majority (90%) of peripheral blood NK cells, whereas CD56bright NK cells are more abundant in secondary lymphoid tissues.14,24 CD56dim NK cells express high levels of the low-affinity Fc receptor CD16, display variegated expression of several types of inhibitory receptors for MHC class I, and express high levels of perforin. In contrast, CD56bright NK cells express no or low levels of CD16, exclusively express the inhibitory receptor CD94/NKG2A, and have 10-fold lower perforin expression than CD56dim NK cells.25–27 Because of these and other findings, CD56dim and CD56bright NK-cell subsets are considered to be developmentally distinct and to occupy different functional niches.12,28–30
Human NK-cell responses to exogenous cytokines have been extensively studied.21 In contrast, relatively less is known with respect to NK-cell cytokine and chemokine production upon target cell recognition. For example, the full spectrum of cytokines released by freshly isolated, resting NK cells upon target cell recognition has not been fully characterized. Furthermore, the minimal requirements for induction of cytokine secretion upon engagement of specific ligands on target cells are not known. To understand how NK cells may contribute to, and maybe even act as primary initiators of, immune responses upon target cell recognition, studies on how receptor-ligand interactions dictate qualitative and temporal aspects of cytokine and chemokine secretion are important. Here, we have set out to study in detail cytokine and chemokine production by human peripheral blood NK cells upon target cell recognition.
To overcome the complexity in receptor-ligand interactions between NK cells and target cells, we have developed a reconstitution system using Drosophila cells as targets.31 A notable advantage of such a system is that cytokine and chemokine secretion by primary, unmanipulated NK cells can be studied in the context of specific receptor-ligand interactions. This system has recently revealed cooperation among NK-cell receptors for discrete events in cytotoxicity, including NK-cell cytolytic granule polarization and exocytosis.32,33
Here, we addressed how specific engagement of the receptors NKG2D (CD314), DNAM-1 (CD226), 2B4 (CD244), LFA-1 (CD11a/CD18), and CD16, or combinations thereof, regulate cytokine and chemokine production by freshly isolated, resting human NK cells. The data provide insight into the regulation of cytokine and chemokine secretion by different NK-cell subsets upon target cell recognition.
Methods
Cells
Human NK cells were isolated from peripheral blood by negative selection (NK-cell isolation kit; Miltenyi Biotec). Approval was obtained from the Regional Ethics Review Board for the use of peripheral blood mononuclear cells from healthy donors. CD56dim and CD56bright NK-cell subsets were sorted based on CD56 expression by flow cytometry (FACSAria; BD Biosciences). Freshly isolated NK cells were maintained in complete medium (RPMI 1640 supplemented with 10% fetal bovine serum and 2mM l-glutamine; all from Invitrogen). NK-cell populations contained at least 99% CD3−CD56+ cells and were used within 2 days of isolation. The cell line K562 (ATCC) was maintained in complete medium. The transfection and maintenance of Drosophila S2 cells has been previously described and expression of the ligands for NK receptors was monitored prior to experiments.31,33 A rabbit serum raised against S2 cells and used to coat S2 cells with IgG has been described.32
Antibodies and fluorescent reagents
Fluorochrome-conjugated monoclonal antibodies (mAbs) used for flow cytometry were anti-CD3 (clone UCHT1; Dako), anti-CD56 (clone NCAM 16.2; BD Biosciences), anti-CD107a (clone H4A3; BD Biosciences), anti–MIP-1α (clone 93342; R&D Systems), anti–MIP-1β (clone D21-1351; BD Biosciences), anti–TNF-α (clone MP6-XT22; eBioscience), and anti–IFN-γ (clone 25723.11; BD Biosciences). Streptavidin-conjugated Qdot 605 was used to detect biotinylated anti-CD107a and a fixable cell viability dye (LIVE/DEAD 405 nm) was used to exclude dead cells from the analysis (both from Invitrogen).
Cytokine secretion measurements
Resting NK cells (2 × 105) were washed twice in and mixed with 4 × 105 S2 cells or K562 cells in 200 μL of complete medium. In experiments with sorted cells, 1 × 105 CD56dim or CD56bright NK cells were mixed with 1 × 105 S2 cells. Cells were incubated for the indicated time at 37°C in 5% CO2. Thereafter, supernatants were collected and stored at −20°C pending measurement. The concentrations of cytokines were quantified by a multiplex immunoassay (Luminex 100 IS; Invitrogen) using a kit that detected 25 different cytokines (Biosource; listed in Figure 1A). To address the kinetics of secretion of select cytokines, 5 simplexes (for IFN-γ, TNF-α, RANTES, MIP-1α, and MIP-1β) were mixed according to the manufacturer's instructions (Biosource).
Figure 1.
Profile and kinetics of NK-cell secretion upon interaction with K562 cells. (A) Resting NK cells were incubated alone or with K562 cells for 6 hours at 37°C. Supernatants were harvested, and the concentrations of indicated cytokines and chemokines were determined by a multiplex immunoassay. Values represent mean ± SD of 8 different donors. (B) NK cells were mixed with K562 cells and incubated at 37°C. Supernatants were harvested at different time points, as indicated, and the concentrations of IFN-γ, TNF-α, MIP-1β, MIP-1β, and RANTES were determined by a multiplex immunoassay. Values represent mean ± SD of 5 different donors.
Intracellular staining of cytokines
Resting NK cells (2 × 105) were added to 4 × 105 S2 cells in 200 μL of complete medium. IL-12, IL-15, and IL-18 (all from Peprotech) were added in some experiments. Cells were incubated for 1 hour at 37°C in 5% CO2. Thereafter, Brefeldin A (GolgiPlug; Becton Dickinson) was added to the cultures, which were incubated for 5 more hours. After 6 hours of incubation, the cells were spun down and resuspended in 50 μL of staining buffer (phosphate-buffered saline supplemented with 2% fetal bovine serum and 2mM ethylenediaminetetraacetic acid) added fluorochrome-conjugated mAbs for surface staining. Thereafter, cells were washed, fixed with 2% paraformaldehyde (Sigma-Aldrich) in phosphate-buffered saline, permeabilized, and stained intracellularly with fluorochrome-conjugated mAbs to cytokines and chemokines. Finally, cells were washed and analyzed on a CyAn ADP LX 9-color flow cytometer (Dako).34 Data were analyzed with FlowJo 8.6 software (TreeStar). Pie charts were generated with SPICE software, Version 4.1 (M. Roederer, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health).
Statistical analysis
All analyses were performed with GraphPad software (GraphPad Software). Statistical analyses of minimal requirements and comparisons between CD56dim and CD56bright responsiveness were performed using a 2-way analysis of variance (nonparametric) with Bonferroni posttest.
Results
Secretion profile of resting NK cells upon interaction with K562 cells
We first set out to determine the factors released by freshly isolated peripheral blood NK cells upon short incubations with susceptible K562 target cells. Resting NK cells were mixed with K562 cells for 6 hours. Supernatants were harvested and the concentration of 25 different soluble factors was determined by a multiplex immunoassay (Figure 1A). As anticipated, stimulation of NK cells by K562 cells induced IFN-γ and TNF-α secretion. In addition, stimulation induced ample secretion of several chemokines, including MCP-1 (CCL2), MIP-1α (CCL3), MIP-1β (CCL4), RANTES (CCL5), IL-8 (CXCL8), and IP-10 (CXCL10). Low levels of chemokines MIP-1α, MIP-1β, and RANTES were also detected in supernatants from NK cells incubated without target cells, suggesting a degree of constitutive secretion. However, incubation with K562 cells resulted in a 44-, 181-, and 6.5-fold increase in secretion of MIP-1α, MIP-1β, and RANTES, respectively, compared with supernatants from NK cells alone. Notably, stimulation of NK cells by K562 cells also induced release of soluble IL-2Rα (CD25). GM-CSF, IL-5, and IL-13 were weakly or not secreted after interaction of NK cells with K562 cells, contrasting previous studies using cytokine-cultured NK cells.15,17 Low levels of IL-1β, IL-6, IL-7, IL-10, IL-12p40, IFN-α, and MIG (CXCL9) secretion were measured after target cell stimulation. Secretion of IL-2, IL-4, IL-5, IL-13, IL-15, IL-17, and eotaxin (CCL26) was not detected.
Next, the temporal profile of cytokine and chemokine secretion was analyzed (Figure 1B). Chemokine secretion could be detected within 1 hour of mixing NK cells with K562 cells. In contrast, TNF-α and, in particular, IFN-γ secretion occurred later. After 6 hours of incubation, the highest levels of secreted cytokines were measured. Thereafter the concentrations of chemokines remained stable throughout the time course, whereas IFN-γ and TNF-α concentrations, if anything, decreased.
Thus, interactions with a susceptible target cell line can induce a profoundly proinflammatory profile of chemokine and cytokine secretion. Moreover, kinetic studies revealed a rapid release of chemokines, relative to cytokine secretion.
Minimal receptor engagement requirements for secretion of proinflammatory cytokines and chemokines by resting NK cells
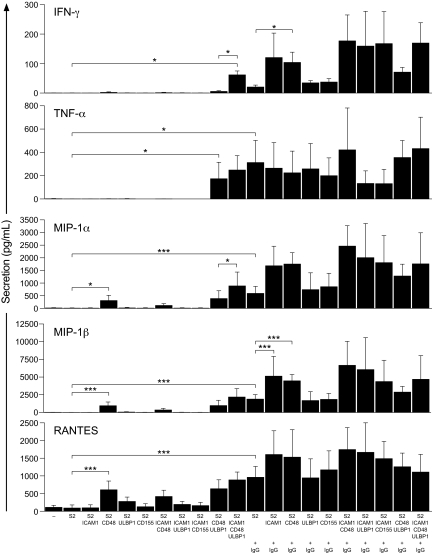
Having identified soluble factors that were secreted by resting NK cells upon interaction with a susceptible target cell, the contribution of individual receptor-ligand interactions to cytokine and chemokine secretion was assessed. Resting NK cells were mixed with Drosophila Schneider 2 (S2) cells expressing ICAM-1 (CD54), CD48, ULBP1, and CD155 (which bind receptors LFA-1, 2B4, NKG2D, and DNAM-1, respectively), or combinations thereof, and incubated for 6 hours. In these experiments, the concentrations of IFN-γ, TNF-α, MIP-1α, MIP-1β, and RANTES were determined using a multiplex immunoassay (Figure 2). Secretion of TNF-α required coexpression of CD48 and ULBP1 on S2 cells, whereas IFN-γ required combined expression of ICAM-1, CD48, and ULBP1. In contrast, expression of CD48 was sufficient to induce secretion of chemokines MIP-1α, MIP-1β, and RANTES. Coexpression of CD48 and ULBP1 on S2 cells did not enhance secretion of chemokines compared with S2 cells expressing CD48 alone. However, expression of ICAM-1, in addition to CD48 and ULBP1, on S2 cells could augment chemokine secretion, as observed for IFN-γ. To engage CD16 on NK cells, S2 cells were preincubated with an anti–S2 cell polyclonal serum.32 IgG coating of S2 cells was sufficient to induce significant TNF-α secretion, but much less IFN-γ secretion (Figure 2). In regard to IFN-γ secretion, expression of ICAM-1 or CD48 on S2 cells augmented IgG-mediated secretion. Chemokine secretion was readily induced by IgG-coated S2 cells. Expression of ICAM-1 or CD48 on S2 cells could augment IgG-induced chemokine secretion.
Figure 2.
Minimal requirements for receptor-ligand interactions for secretion of cytokines and chemokines by NK cells. Resting NK cells were mixed with S2 cells expressing ligands for NK-cell receptors, as indicated, and incubated for 6 hours at 37°C. For some stimulations, S2 cells were preincubated with diluted anti–S2 cell serum (+ IgG). Supernatants were harvested, and the concentrations of cytokines and chemokines were determined by a multiplex immunoassay. Values represent mean ± SD of 5 different donors. Data are representative of 3 independent experiments. For clarity, selected statistical analyses are indicated. *P < .05, ***P < .001.
Thus, in the absence of IgG, engagement of 2B4 was sufficient to induce chemokine secretion, whereas stronger signals evoked by 2B4 and NKG2D were required for secretion of TNF-α or IFN-γ. Coengagement of LFA-1 augmented cytokine and chemokine secretion induced by 2B4 and NKG2D. CD16 engagement could induce secretion of chemokines and cytokines. CD16-mediated secretion of IFN-γ and chemokine secretion, but not TNF-α, was augmented by LFA-1 coengagement.
Longer time points were also assessed and secretion patterns resembled that induced by K562 cell stimulation (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Notable features were a slight induction of chemokines by S2 cells expressing ULBP1, as previously described with cytokine-cultured NK cells,35 and that costimulation could accelerate chemokine secretion.
A range of S2 cell combinations engaging several activating receptors was assessed for the 25 factors listed in Figure 1A. Corroborating findings with NK-cell stimulation by K562 cells, profiles of secreted cytokines and chemokines were proinflammatory (data not shown). Thus, NK-cell activation by CD16, 2B4, and NKG2D, in addition to costimulation by DNAM-1 and LFA-1, induced secretion of proinflammatory, but not immunoregulatory, cytokines.
CD56dim NK cells are major producers of cytokines and chemokines upon target cell recognition
Whereas analysis of supernatants from cell-mixing experiments identified the predominant chemokines and cytokines secreted by freshly isolated NK cells in response to target cell recognition, a different approach was required to identify the phenotypic features of the cytokine- and chemokine-producing cells, to quantify the frequency of responding cells, and to examine to what extent individual cells could produce multiple soluble factors. To this end, multicolor flow cytometry was performed. Production of IFN-γ, TNF-α, MIP-1α, and MIP-1β was compared in CD56dim and CD56bright NK-cell subsets after 6 hours of incubation with different S2 cells (Figure 3A). Coexpression of ICAM-1, CD48, and ULBP1 on S2 cells was required to induce IFN-γ production by CD56dim NK cells. Expression of CD48 on S2 cells was sufficient to induce TNF-α production. Interestingly, despite LFA-1, 2B4, and NKG2D being expressed at similar levels on different NK-cell subsets (Bryceson et al36; data not shown), very few CD56bright NK cells expressed IFN-γ or TNF-α after incubation with target cells expressing ligands for these receptors. Expression of CD48 or ULBP1 on S2 cells was sufficient to induce MIP-1α and MIP-1β production by CD56dim NK cells (Figure 3A and supplemental Figure 2), corroborating results quantifying chemokine secretion. CD56bright NK cells did produce some MIP-1α and MIP-1β upon stimulation by S2 cells coexpressing CD48 and ULBP1. However, a consistently higher frequency of CD56dim NK cells produced chemokines upon such stimulations. For example, CD48 expression on S2 cells induced MIP-1α production in 9% (± 6%) of CD56bright NK cells, as opposed to 43% (± 7%) of CD56dim NK cells. Thus, strikingly, upon target cell recognition CD56dim NK cells were more prominent chemokine and cytokine producers than CD56bright NK cells. Strengthening this notion, greater proportions of CD56dim NK cells than CD56bright NK cells produced IFN-γ, TNF-α, MIP-1α, and MIP-1β upon K562 cell stimulation (Figure 3B). Moreover, experiments comparing cytokine and chemokine secretion from equal numbers of sorted CD56dim and CD56bright NK cells separately incubated with K562 cells or various S2 cell transfectants also showed greater secretion by CD56dim NK cells (Figure 3C and data not shown).
Figure 3.
CD56dim NK cells produce cytokines and chemokines upon target cell recognition. Resting NK cells were mixed with S2 cells expressing ligands for NK-cell receptors (A) or K562 cells (B), as indicated, and incubated for 6 hours at 37°C. After stimulation, the cells were surface stained with fluorochrome-conjugated anti-CD56 mAb, fixed, permeabilized, and stained intracellularly with fluorochrome-conjugated mAbs to cytokines and chemokines. The percentage of CD56dim or CD56bright NK cells producing IFN-γ, TNF-α, MIP-1α, and MIP-1β, as indicated, was determined by flow cytometry. Values represent mean ± SD of at least 6 different donors. (C) Sorted CD56dim or CD56bright NK cells were incubated alone or with K562 cells for 6 hours at 37°C. Supernatants were harvested, and the concentrations of cytokines and chemokines were determined by a multiplex immunoassay. Values represent mean ± SD of 5 different donors. (D) Resting NK cells were incubated alone or stimulated with 10 ng/mL IL-12, 100 ng/mL IL-15, or 100 ng/mL IL-18, or combinations thereof, for 24 hours at 37°C. After stimulation, the cells were surface stained with fluorochrome-conjugated anti-CD56 mAb, fixed, permeabilized, and stained intracellularly with fluorochrome-conjugated mAbs to cytokines and chemokines. The percentage of CD56dim or CD56bright NK cells producing IFN-γ, TNF-α, MIP-1α, and MIP-1β, as indicated, was determined by flow cytometry. Values represent mean ± SD of 5 different donors. For clarity, selected statistical analyses are indicated. *P < .05, **P < .01, ***P < .001.
Ig-coated S2 cells were sufficient to induce TNF-α and IFN-γ production by CD56dim, but not CD56bright, NK cells (supplemental Figures 2–3). However, Ig-coated S2 cells induced 29% (± 11%) MIP-1b+ CD56bright NK cells, demonstrating that engagement of CD16 can elicit responses by a subset of CD56bright NK cells (supplemental Figure 3).
Reports defining CD56bright NK cells as the major source of NK cell–derived cytokines have relied on exogenous cytokines for stimulation.26 To ensure that the low cytokine response of CD56bright NK cells upon target cell recognition was not an artifact of our experimental procedure, we evaluated the response of freshly isolated NK cells to 24 hours of stimulation with cytokines IL-12, IL-15, or IL-18, or combinations thereof (Figure 3D). These cytokines induce strong IFN-γ secretion by CD56bright NK cells in 72-hour assays.26 In agreement with other reports, a greater frequency of CD56bright NK cells, relative to CD56dim NK cells, produced IFN-γ and TNF-α in response to stimulation by exogenous cytokines. In comparison, production of MIP-1α and MIP-1β was equal or greater in CD56dim NK cells. Next, we tested whether the greater frequency of cytokine-producing CD56bright NK cells after 24 hours of stimulation could reflect temporal differences in responsiveness between the 2 subsets. After 6 hours of stimulation, somewhat higher frequencies of CD56bright NK cells, relative to CD56dim NK cells, produced IFN-γ and TNF-α in response to IL-12 plus IL-18 (Figure 4). Remarkably, as with 24 hours of stimulation, production of MIP-1α and MIP-1β was, if anything, more pronounced in CD56dim NK cells relative to CD56bright NK cells. Moreover, after 24 hours of stimulation with K562 cells, the frequency of cytokine- and chemokine-producing CD56bright NK cells was still lower than that of CD56dim NK cells (data not shown). Together, these results suggest that CD56bright NK cells are not inherently slower to respond to target cell recognition. Rather, CD56dim NK cells excel in cytokine and chemokine production upon recognition of target cells. Furthermore, a greater proportion of the CD56dim than the CD56bright subset of NK cells produced chemokines in response to exogenous cytokine stimulation.
Figure 4.
Costimulation of NK cells by exogenous cytokines enhances cytokine production by CD56dim NK cells upon interaction with K562 cells. Resting NK cells were incubated alone or stimulated with cytokines IL-12 and IL-18 (10 ng/mL and 100 ng/mL, respectively) for 6 hours at 37°C with or without K562 cells. After stimulation, the cells were surface stained with fluorochrome-conjugated anti-CD56 mAb, fixed, permeabilized, and stained intracellularly with fluorochrome-conjugated mAbs to cytokines and chemokines. The percentage of CD56dim or CD56bright NK cells producing IFN-γ, TNF-α, MIP-1α, and MIP-1β, as indicated, was determined by flow cytometry. Values represent mean ± SD of 7 different donors. For clarity, selected statistical analyses are indicated. *P < .05, **P < .01, ***P < .001.
IL-12 plus IL-18 potentiate cytokine secretion upon target cell recognition by CD56dim NK cells
Studies have shown that cytokines, such as IL-2, IL-12, IL-15, and IL-21, can enhance IFN-γ and chemokine secretion induced by target cell recognition.20,22,37–39 Therefore, we examined how exogenous cytokine stimulation by IL-12 plus IL-18 influenced NK-cell production of IFN-γ, TNF-α, MIP-1α, and MIP-1β upon interaction with K562 cells (Figure 4). Addition of IL-12 and IL-18 augmented the frequency of IFN-γ–producing CD56dim NK cells after 6 hours of incubation with K562 cells. Interestingly, IL-12 and IL-18 did not significantly increase MIP-1α and MIP-1β production by CD56dim NK cells in response to K562 cells. K562 cells alone did not induce cytokine and chemokine production by CD56bright NK cells, and addition of IL-12 and IL-18 did not increase the frequencies of cytokine- and chemokine-producing CD56bright NK cells relative to IL-12 plus IL-18 alone. In conclusion, exogenous cytokines potentiated cytokine production by CD56dim NK cells in response to K562 cells. In such settings, chemokine production by CD56dim NK cells could be ascribed mainly to target cell recognition, whereas cytokine and chemokine production by CD56bright NK cells could be ascribed to exogenous cytokines. These data suggest different requirements and signaling pathways for cytokine and chemokine production in CD56bright and CD56dim NK-cell subsets.
Costimulation signals lower the threshold for CD16-induced NK-cell cytokine and chemokine production
Next, we analyzed how the strength of stimulation through increased CD16 engagement affected the frequency of cytokine- and chemokine-producing CD56dim NK cells (Figure 5). By varying the concentration of anti-S2 serum with which S2 cells were coated, the density of ligands for CD16 could be modulated on target cells.32 Experiments assessing degranulation of CD56dim NK cells upon modulation of IgG density on target cells produced similar results as have been reported previously (supplemental Figure 4; Bryceson et al32). Increasing the concentration of IgG augmented cytokine and chemokine responses in a nonlinear fashion. Expression of CD48 or ICAM-1 on S2 cells lowered the concentrations of IgG required to augment the frequency of cytokine- and chemokine-producing CD56dim NK cells, with coexpression of CD48 and ICAM-1 on S2 cells providing the greatest response to low concentrations of IgG. NK-cell cytokine and chemokine responses reached a plateau at the highest concentrations of IgG. At a 10−3 dilution of anti-S2 serum, coexpression of ICAM-1 and CD48 increased the frequency of IFN-γ– and TNF-α–producing CD56dim NK cells 2.8- and 2.3-fold, respectively. Nonlinear regression analysis revealed that the half-maximal percentage of IFN-γ–producing CD56dim NK cells induced by CD16 engagement alone could be achieved with 10- and 63-fold lower IgG concentration when S2 cells expressed CD48 or ICAM-1 and CD48, respectively (data not shown). S2 cells expressing ICAM-1 achieved the half-maximal level of response of untransfected S2 cells with a 3-fold lower concentration of IgG. Similar observations were made for TNF-α secretion. In addition, the half-maximal MIP-1α or MIP-1β responses could be reached with 7- and 10-fold lower concentrations of IgG upon S2 cell expression of ICAM-1 and CD48, respectively. Thus, coengagement of different activating receptors can lower the activation threshold for CD56dim NK-cell cytokine and chemokine production, increasing the frequency of responding cells even in the context of high ligand densities for powerful NK cell–activating receptors such as CD16.
Figure 5.
Increasing activating ligand density on target cells augments the frequency of cytokine- and chemokine-producing CD56dim NK cells. Before mixing with NK cells, S2 cells expressing ligands for NK-cell receptors, as indicated, were preincubated with serial dilutions of anti–S2 cell serum. Resting NK cells were incubated with S2 cells for 6 hours at 37°C. After stimulation, the cells were surface stained with fluorochrome-conjugated anti-CD56 mAb, fixed, permeabilized, and stained intracellularly with fluorochrome-conjugated mAbs to cytokines and chemokines. The percentage of CD56dim NK cells producing IFN-γ, TNF-α, MIP-1α, and MIP-1β, as indicated, was determined by flow cytometry. Values represent mean ± SD of 3 different donors.
Interrelationships between different NK-cell responses induced by target cell recognition
In addition to determining the frequency and phenotype of responding cells, multicolor flow cytometric analysis can also provide information on how many different responses an individual cell can mediate. To gain insight into the relationships between different NK-cell responses and to what extent individual cells can perform multiple responses, intracellular expression of TNF-α, IFN-γ, MIP-1α, and MIP-1β and surface expression of CD107a were simultaneously assessed on CD56dim NK cells (Figure 6). Surface expression of CD107a (lysosome-associated membrane protein-1) is a marker of lytic granule exocytosis.32 In line with previous findings, untransfected S2 cells did not induce cytokine or chemokine expression, and CD107a was not expressed on the cell surface (Figure 6A). Stimulation of resting NK cells by S2 cells expressing ICAM-1, CD48, and ULBP1 induced intracellular expression of TNF-α, IFN-γ, MIP-1α, and MIP-1β and surface expression of CD107a on subsets of CD56dim NK cells (Figure 6A). The greatest frequency of responding cells was consistently observed upon staining with MIP-1β, closely followed by that of MIP-1α. Expression of MIP-1β correlated strongly with expression of MIP-1α. Expression of TNF-α, IFN-γ, and CD107a was confined to a MIP-1β+ NK-cell subset. CD107a expression did not necessarily correlate with expression of TNF-α or IFN-γ. However, IFN-γ expression was contained mostly within a TNF-α+ NK-cell subset. Thus, the data demonstrate specific interrelationships and a considerable degree of cellular overlap among different responses enacted by CD56dim NK cells. For example, IFN-γ production by individual CD56dim NK cells is likely to be accompanied by MIP-1β and TNF-α production by the same cell.
Figure 6.
Interrelationships between different NK-cell responses induced by target cell recognition. Resting NK cells were mixed with S2 cells expressing ligands for NK-cell receptors, as indicated, and incubated for 6 hours at 37°C. For some stimulations, S2 cells were preincubated with diluted anti–S2 cell serum (+ IgG). After stimulation, the cells were surface stained with fluorochrome-conjugated anti-CD56 and anti-CD107a mAbs, fixed, permeabilized, and stained intracellularly with fluorochrome-conjugated mAbs to cytokines and chemokines. (A) Lymphocytes were gated on forward scatter height (FSC-H) versus side scatter height plots (SSC-H). Single-cell events were gated on forward scatter height (FSC-H) versus forward scatter area plots (FSC-A). CD56dim NK cells were gated on CD56 versus dead cell marker (DCM) plots. The second and third rows show MIP-1α, TNF-α, IFN-γ, and CD107a staining in relation to MIP-1β staining after stimulation with S2 cells, as indicated. The bottom row shows MIP-1β, MIP-1α, TNF-α, and IFN-γ staining in relation to CD107a staining, and IFN-γ staining in relation to TNF-α staining (right panel). Gates were set using fluorochrome-conjugated isotype control mAbs. The plots are derived from one representative donor. (B) CD56dim NK cells were gated as described in panel A, and a Boolean gating strategy was used for analysis. Pie charts represent the frequency of cells positive for the given number of measured responses (MIP-1β, TNF-α, IFN-γ, and CD107a). Thus, cells can be categorized into the number of responses they display. Arcs depict the relative frequency of cells specifically positive for MIP-1β, TNF-α, IFN-γ, and/or CD107a staining, as indicated. Values represent the mean of 6 different donors.
Further analysis was performed to enumerate responses by individual CD56dim NK cells upon recognition of target cells expressing increasing numbers of ligands for activating receptors (Figure 6B). Given the high correlation between expression of MIP-1α and MIP-1β, MIP-1α was omitted from this analysis. Incubation of resting NK cells alone, with untransfected S2 cells, or S2 cells expressing ICAM-1 did not induce responses by CD56dim NK cells. Expression of CD48 or ULBP1 on S2 cells induced MIP-1β and weak degranulation by CD56dim NK cells. Coexpression of CD48 and ULBP1 on S2 cells specifically synergized for degranulation, as neither the frequency of cells producing MIP-1β nor TNF-α was increased relative to S2 cells only expressing CD48. A small fraction of cells coexpressed MIP-1β, TNF-α, and surface CD107a. Coexpression of ICAM-1, CD48, and ULBP1 further increased the frequency of cells producing MIP-1β and TNF-α. Moreover, IFN-γ was produced by a sizable fraction of CD56dim NK cells. If anything, surface expression of CD107a decreased, compared with stimulation with S2 cells coexpressing CD48 and ULBP1. Upon stimulation with S2 cells coexpressing ICAM-1, CD48, and ULBP1, a small fraction of cells responded with all 4 responses examined, whereas a greater fraction of cells responded with 3 responses. Last, when NK cells were stimulated with IgG-coated S2 cells expressing CD48 and ULBP1, more than 90% of the CD56dim NK cells responded in terms of at least one parameter and 16% of the cells responded in terms of all parameters examined. Analogously, increasing densities of IgG on target cells augmented the frequencies of CD56dim NK cells displaying multiple different responses, and costimulation reduced the density of IgG required for NK cells to display multiple responses (supplemental Figure 5).
Together, these data provide insights into the heterogeneous response of NK cells to target cell recognition, revealing specific interrelationships and a hierarchy among the NK-cell responses for degranulation of lytic granules and production of chemokines and cytokines. Moreover, the data illustrate how increasing signal strength can induce multiple different effector responses from individual NK cells.
Discussion
With a multiplicity of receptors evolved to sense cellular homeostasis and distress, NK cells are well equipped to act as primary initiators of immune responses upon recognition of infected or neoplastic cells.40 Such responses are not confined to cytotoxic effector mechanisms, but also involve the secretion of cytokines and chemokines. Here, we define the cytokines and chemokines that are secreted by normal, freshly isolated human peripheral blood NK cells upon interaction with target cells expressing complex or defined sets of ligands for specific activating receptors. We demonstrate that engagement of the activating receptors CD16, 2B4, or NKG2D sufficed for rapid secretion of chemokines, and that coengagement of additional activating receptors could accelerate and increase chemokine secretion. In contrast, secretion of IFN-γ required engagement of multiple different receptors and occurred later, revealing a tighter control and a higher activation threshold for induction of cytokine secretion (Figure 7). Unexpectedly, the results revealed CD56dim, rather than CD56bright, NK cells to be more prominent producers of cytokines upon target cell recognition. Furthermore, upon both target cell recognition and exogenous cytokine stimulation, CD56dim NK cells excelled in production of the chemokine MIP-1β. These experiments highlight CD56dim NK cells as an important proinflammatory cytokine source during early immune responses to aberrant cells.
Figure 7.
Schematic representation of activation thresholds and kinetics of resting CD56dim NK-cell responses. (A) Approximate times required for induction of different NK-cell responses such as degranulation (surface expression of CD107a), chemokine secretion (MIP-1α and MIP-1β), and cytokine secretion (IFN-γ and TNF-α) are indicated on the time scale. (B) The figure depicts the relative signal strength required for induction of different NK-cell responses such as degranulation (surface expression of CD107a), chemokine secretion (MIP-1α and MIP-1β), and cytokine secretion (IFN-γ and TNF-α).
A current view of NK-cell biology regards CD56bright and CD56dim NK cells as developmentally distinct subsets occupying different functional niches.41,42 In general, CD56bright NK cells are considered to be the major source of cytokines, whereas CD56dim NK cells are regarded as specialized for cytotoxic function.12,30,43 The anatomic distribution of the 2 subsets underlies this perceived dichotomy. Enriched in secondary lymphoid tissues, CD56bright NK cells can intimately interact with other immune cells, influencing ongoing adaptive immune responses through secretion of soluble factors. Conversely, in the periphery, CD56dim NK-cell cytotoxic activity can provide immunosurveillance of infected or neoplastic cells. Notably, CD56bright NK cells produce more cytokines in response to phorbol myristate acetate and ionomycin stimulation than CD56dim NK cells.26,27 This fact has been ascribed to the relatively low expression of the phosphatase SHIP-1 and the high expression of the phosphatase inhibitor SET in CD56bright NK cells, facilitating a lower activation threshold for cytokine secretion.44,45 Although we also found that exogenous cytokines induced a greater frequency of IFN-γ– and TNF-α–producing CD56bright NK cells in 6- and 24-hour assays, our data highlight a prominent role for CD56dim NK cells in producing cytokines and chemokines upon target cell recognition. In fact, CD56dim NK cells responded more vigorously in terms of cytokine and chemokine production relative to CD56bright NK cells. These data substantiate and extend previous findings demonstrating that CD56dim NK cells can produce cytokines in response to a tumor cell line.46 Here, cytokines and chemokines were induced by 2B4 and NKG2D synergy or K562 cells that express ligands for the natural cytotoxicity receptor NKp30, NKG2D, and DNAM-1.47,48 Comparatively, CD56bright and CD56dim NK-cell subsets express similar levels of activating receptors.36 Therefore, differential expression of molecules conveying signals from receptors such as 2B4, NKG2D, and NKp30 might underlie the relatively more potent response of CD56dim NK cells to target cell recognition. Division of NK-cell subsets on the basis of variegated inhibitory receptor expression or maturation markers such as CD27 may further distinguish CD56dim NK-cell subsets in terms of cytokine production and will be addressed in further experiments.46,49,50
NK cells have previously been reported to secrete a plethora of proinflammatory and immunoregulatory cytokines. In our experimental setting, using resting human NK cells freshly isolated from peripheral blood, K562 target cell recognition consistently induced secretion of a proinflammatory cytokine profile characterized by MIP-1α, MIP-1β, RANTES, TNF-α, and IFN-γ. This profile was observed also with S2 cells expressing ligands for NKG2D and 2B4, IgG-coated S2 cells triggering CD16, and K562 cells triggering NKp30. NKG2D and 2B4 synergy is immunoreceptor tyrosine-based activation motif independent, whereas CD16 and NKp30 are associated with immunoreceptor tyrosine-based activation motif–containing adaptor proteins.51–53 Thus, diverse signals induce proinflammatory cytokine and chemokine secretion by NK cells. Strikingly, upon NK-cell recognition of target cells, we did not detect secretion of immunoregulatory cytokines, such as IL-5, IL-10, IL-13, or GM-CSF. Such immunoregulatory cytokines are more often reported to be secreted by NK cells upon stimulation with exogenous cytokines. For example, IL-10 is secreted predominately by CD56bright NK cells upon stimulation by IL-12 with IL-2 or IL-15 (Fehniger et al21; Wolk et al54; C.F. and Y.T.B. unpublished observations, March 2009). In mice, such NK-cell secretion of IL-10 requires STAT4,55 which signals downstream of the IL-12 and IL-23 receptors.56 Our data suggest that NK cells are initially wired to promote immune responses by secreting proinflammatory cytokines upon target cell recognition.
With Drosophila S2 cells expressing ligands for NK-cell receptors, the minimal requirements for cytokine and chemokine production were dissected. Interestingly, IFN-γ secretion required the coengagement by LFA-1, 2B4, and NKG2D, reflecting a minimal requirement previously identified for natural cytotoxicity.33 Likewise TNF-α secretion also required receptor coengagement, although here 2B4 and NKG2D synergy sufficed. Data corroborate an earlier study using antibody-coated beads, where mAbs for different NK cell activating receptors could not induce secretion of TNF-α and IFN-γ on their own by freshly isolated NK cells.36 However, S2 cells revealed that secretion of IFN-γ is particularly dependent on LFA-1 engagement, as opposed to secretion of TNF-α. This might reflect not only LFA-1–mediated adhesion, but also signaling.57 Remarkably, chemokines such as MIP-1α, MIP-1β, and RANTES could be induced by engagement of 2B4 or NKG2D. In contrast to other activating receptors, engagement of CD16 was sufficient to induce some IFN-γ and TNF-α secretion, in addition to chemokine secretion. The strong propensity of CD16 to induce NK-cell activation reflects the role of antibody-dependent cell-mediated cytotoxicity as an effector arm of the adaptive immune system. Thus, in vivo, selection of B cells can safeguard robust activation mediated by CD16 engagement. Notably, upon stimulation of receptors such as 2B4 and NKG2D, intracellular cytokine and chemokine expression was apparent, sometimes without much secretion detected. The divergence in intracellular expression versus secretion after stimulation might reflect regulation of cytokine and chemokine exocytosis. This facet of regulation will be interesting to explore in more detail.
Temporally, the secretion of chemokines always preceded that of TNF-α and IFN-γ. Comparing the frequency of cytokine- and chemokine-producing cells revealed that a higher proportion of NK cells produced chemokines relative to cytokines after 6 hours of stimulation. Costimulation of receptors by engagement of LFA-1 or other coactivation receptors could, in many instances, accelerate and increase cytokine and chemokine secretion, and increase the frequency of cells producing these factors. As we could simultaneously monitor production of MIP-1α, MIP-1β, TNF-α, and IFN-γ, as well as CD107a, in individual cells, the relationships between production of specific chemokines, cytokines, and degranulation could be assessed. These experiments revealed that production of TNF-α and IFN-γ and degranulation were contained within the chemokine-producing NK-cell subset. Moreover, most IFN-γ–producing cells also expressed TNF-α, but the inverse relationship did not hold. Production of TNF-α and IFN-γ did not necessarily correlate with degranulation, but this could in part reflect differences in kinetics of these responses (Figure 7A). Taken together, these results reveal different activation thresholds for distinct responses. Chemokine production is an early feature of NK-cell responses and is triggered with relatively weak activating signals, requiring less stimulation than that necessary for degranulation or cytokine production. Induction of TNF-α and IFN-γ is more stringently controlled, with a greater requirement for receptor cooperation and release at later time points after stimulation. Thus, the data reveal a hierarchy among factors released upon NK-cell interactions with target cells, with graded responses according to the degree of ligand expression for activating receptors (Figure 7B).
Altogether, this study provides detailed insight into regulation of NK-cell cytokine secretion upon target cell recognition. Engagement of individual activating receptors on resting NK cells suffices for chemokine secretion, to alert and recruit other immune cells. More complex interactions, upon which multiple activating NK-cell receptors are engaged, can induce production of TNF-α and IFN-γ, to promote cellular resistance to infections and shape adaptive immune responses. These chemokines and cytokines are readily produced by the CD56dim NK-cell subset. Chemokine and cytokine production by CD56dim NK cells may thus be an important component of immunosurveillance.
Acknowledgments
We thank S. Wood for critical reading of the paper and J. Michaëlsson for helpful discussions.
This work was supported by grants from the Swedish Foundation for Strategic Research, Research Council, and Cancer Society (C.F., H.-G.L., and Y.T.B.), by the Intramural Research Program at National Institutes of Health, National Institute of Allergy and Infectious Diseases (E.O.L.), and Mary Beve's Foundation, David & Astrid Hagelen's Foundation, the Karolinska Institute Research Foundation, and Jonas Söderquist's Stipend (Y.T.B.).
Footnotes
A Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: C.F. designed research, performed experimental work, analyzed and interpreted data, and drafted the paper; E.O.L and H.-G.L. designed research and contributed to drafting the paper; and Y.T.B. designed research, analyzed and interpreted data, and drafted the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Cyril Fauriat or Yenan T. Bryceson, Center for Infectious Medicine, Department of Medicine, Karolinska Institute, Karolinska University Hospital Huddinge, S-14186 Stockholm, Sweden; e-mail: cfauriat@wanadoo.fr or yenan.bryceson@ki.se.
References
- 1.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 2.Bottino C, Castriconi R, Moretta L, Moretta A. Cellular ligands of activating NK receptors. Trends Immunol. 2005;26(4):221–226. doi: 10.1016/j.it.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Bryceson YT, March ME, Ljunggren HG, Long EO. Activation, coactivation, and costimulation of resting human natural killer cells. Immunol Rev. 2006;214(1):73–91. doi: 10.1111/j.1600-065X.2006.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lodoen MB, Lanier LL. Natural killer cells as an initial defense against pathogens. Curr Opin Immunol. 2006;18(4):391–398. doi: 10.1016/j.coi.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SH, Miyagi T, Biron CA. Keeping NK cells in highly regulated antiviral warfare. Trends Immunol. 2007;28(6):252–259. doi: 10.1016/j.it.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9(5):503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 7.Wherry JC, Schreiber RD, Unanue ER. Regulation of gamma interferon production by natural killer cells in scid mice: roles of tumor necrosis factor and bacterial stimuli. Infect Immun. 1991;59(5):1709–1715. doi: 10.1128/iai.59.5.1709-1715.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gazzinelli RT, Hieny S, Wynn TA, Wolf S, Sher A. Interleukin 12 is required for the T-lymphocyte-independent induction of interferon gamma by an intracellular parasite and induces resistance in T-cell-deficient hosts. Proc Natl Acad Sci U S A. 1993;90(13):6115–6119. doi: 10.1073/pnas.90.13.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scharton TM, Scott P. Natural killer cells are a source of interferon gamma that drives differentiation of CD4+ T cell subsets and induces early resistance to Leishmania major in mice. J Exp Med. 1993;178(2):567–577. doi: 10.1084/jem.178.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orange JS, Wang B, Terhorst C, Biron CA. Requirement for natural killer cell-produced interferon gamma in defense against murine cytomegalovirus infection and enhancement of this defense pathway by interleukin 12 administration. J Exp Med. 1995;182(4):1045–1056. doi: 10.1084/jem.182.4.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 12.Caligiuri MA. Human natural killer cells. Blood. 2008;112(3):461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zucchini N, Crozat K, Baranek T, Robbins SH, Altfeld M, Dalod M. Natural killer cells in immunodefense against infective agents. Expert Rev Anti Infect Ther. 2008;6(6):867–885. doi: 10.1586/14787210.6.6.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22(11):633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 15.Cuturi MC, Anegon I, Sherman F, et al. Production of hematopoietic colony-stimulating factors by human natural killer cells. J Exp Med. 1989;169(2):569–583. doi: 10.1084/jem.169.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smyth MJ, Zachariae CO, Norihisa Y, Ortaldo JR, Hishinuma A, Matsushima K. IL-8 gene expression and production in human peripheral blood lymphocyte subsets. J Immunol. 1991;146(11):3815–3823. [PubMed] [Google Scholar]
- 17.Warren HS, Kinnear BF, Phillips JH, Lanier LL. Production of IL-5 by human NK cells and regulation of IL-5 secretion by IL-4, IL-10, and IL-12. J Immunol. 1995;154(10):5144–5152. [PubMed] [Google Scholar]
- 18.Bluman EM, Bartynski KJ, Avalos BR, Caligiuri MA. Human natural killer cells produce abundant macrophage inflammatory protein-1 alpha in response to monocyte-derived cytokines. J Clin Invest. 1996;97(12):2722–2727. doi: 10.1172/JCI118726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peritt D, Robertson S, Gri G, Showe L, Aste-Amezaga M, Trinchieri G. Differentiation of human NK cells into NK1 and NK2 subsets. J Immunol. 1998;161(11):5821–5824. [PubMed] [Google Scholar]
- 20.Oliva A, Kinter AL, Vaccarezza M, et al. Natural killer cells from human immunodeficiency virus (HIV)-infected individuals are an important source of CC-chemokines and suppress HIV-1 entry and replication in vitro. J Clin Invest. 1998;102(1):223–231. doi: 10.1172/JCI2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fehniger TA, Shah MH, Turner MJ, et al. Differential cytokine and chemokine gene expression by human NK cells following activation with IL-18 or IL-15 in combination with IL-12: implications for the innate immune response. J Immunol. 1999;162(8):4511–4520. [PubMed] [Google Scholar]
- 22.Roda JM, Parihar R, Magro C, Nuovo GJ, Tridandapani S, Carson WE., III Natural killer cells produce T cell-recruiting chemokines in response to antibody-coated tumor cells. Cancer Res. 2006;66(1):517–526. doi: 10.1158/0008-5472.CAN-05-2429. [DOI] [PubMed] [Google Scholar]
- 23.Lanier LL, Phillips JH, Hackett J, Jr, Tutt M, Kumar V. Natural killer cells: definition of a cell type rather than a function. J Immunol. 1986;137(9):2735–2739. [PubMed] [Google Scholar]
- 24.Ferlazzo G, Munz C. NK cell compartments and their activation by dendritic cells. J Immunol. 2004;172(3):1333–1339. doi: 10.4049/jimmunol.172.3.1333. [DOI] [PubMed] [Google Scholar]
- 25.Lanier LL, Le AM, Civin CI, Loken MR, Phillips JH. The relationship of CD16 (Leu-11) and Leu-19 (NKH-1) antigen expression on human peripheral blood NK cells and cytotoxic T lymphocytes. J Immunol. 1986;136(12):4480–4486. [PubMed] [Google Scholar]
- 26.Cooper MA, Fehniger TA, Turner SC, et al. Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood. 2001;97(10):3146–3151. doi: 10.1182/blood.v97.10.3146. [DOI] [PubMed] [Google Scholar]
- 27.Jacobs R, Hintzen G, Kemper A, et al. CD56bright cells differ in their KIR repertoire and cytotoxic features from CD56dim NK cells. Eur J Immunol. 2001;31(10):3121–3127. doi: 10.1002/1521-4141(2001010)31:10<3121::aid-immu3121>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 28.Münz C. Non-cytotoxic protection by human NK cells in mucosal secondary lymphoid tissues. Eur J Immunol. 2008;38(11):2946–2948. doi: 10.1002/eji.200838849. [DOI] [PubMed] [Google Scholar]
- 29.Di Santo JP. Natural killer cells: diversity in search of a niche. Nat Immunol. 2008;9(5):473–475. doi: 10.1038/ni.f.201. [DOI] [PubMed] [Google Scholar]
- 30.Colonna M. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity. 2009;31(1):15–23. doi: 10.1016/j.immuni.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 31.Barber DF, Long EO. Coexpression of CD58 or CD48 with intercellular adhesion molecule 1 on target cells enhances adhesion of resting NK cells. J Immunol. 2003;170(1):294–299. doi: 10.4049/jimmunol.170.1.294. [DOI] [PubMed] [Google Scholar]
- 32.Bryceson YT, March ME, Barber DF, Ljunggren HG, Long EO. Cytolytic granule polarization and degranulation controlled by different receptors in resting NK cells. J Exp Med. 2005;202(7):1001–1012. doi: 10.1084/jem.20051143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bryceson YT, Ljunggren HG, Long EO. Minimal requirement for induction of natural cytotoxicity and intersection of activation signals by inhibitory receptors. Blood. 2009;114(13):2657–2666. doi: 10.1182/blood-2009-01-201632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez VD, Bjorkstrom NK, Malmberg KJ, et al. Application of nine-color flow cytometry for detailed studies of the phenotypic complexity and functional heterogeneity of human lymphocyte subsets. J Immunol Methods. 2008;330(1–2):64–74. doi: 10.1016/j.jim.2007.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutherland CL, Chalupny NJ, Schooley K, VandenBos T, Kubin M, Cosman D. UL16-binding proteins, novel MHC class I-related proteins, bind to NKG2D and activate multiple signaling pathways in primary NK cells. J Immunol. 2002;168(2):671–679. doi: 10.4049/jimmunol.168.2.671. [DOI] [PubMed] [Google Scholar]
- 36.Bryceson YT, March ME, Ljunggren HG, Long EO. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood. 2006;107(1):159–166. doi: 10.1182/blood-2005-04-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anegón I, Cuturi MC, Trinchieri G, Perussia B. Interaction of Fc receptor (CD16) ligands induces transcription of interleukin 2 receptor (CD25) and lymphokine genes and expression of their products in human natural killer cells. J Exp Med. 1988;167(2):452–472. doi: 10.1084/jem.167.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carson WE, Giri JG, Lindemann MJ, et al. Interleukin (IL) 15 is a novel cytokine that activates human natural killer cells via components of the IL-2 receptor. J Exp Med. 1994;180(4):1395–1403. doi: 10.1084/jem.180.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roda JM, Parihar R, Lehman A, Mani A, Tridandapani S, Carson WE., III Interleukin-21 enhances NK cell activation in response to antibody-coated targets. J Immunol. 2006;177(1):120–129. doi: 10.4049/jimmunol.177.1.120. [DOI] [PubMed] [Google Scholar]
- 40.Bryceson YT, Long EO. Line of attack: NK cell specificity and integration of signals. Curr Opin Immunol. 2008;20(3):344–352. doi: 10.1016/j.coi.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freud AG, Caligiuri MA. Human natural killer cell development. Immunol Rev. 2006;214:56–72. doi: 10.1111/j.1600-065X.2006.00451.x. [DOI] [PubMed] [Google Scholar]
- 42.Romagnani C, Juelke K, Falco M, et al. CD56brightCD16- killer Ig-like receptor- NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J Immunol. 2007;178(8):4947–4955. doi: 10.4049/jimmunol.178.8.4947. [DOI] [PubMed] [Google Scholar]
- 43.Di Santo JP. Functionally distinct NK-cell subsets: developmental origins and biological implications. Eur J Immunol. 2008;38(11):2948–2951. doi: 10.1002/eji.200838830. [DOI] [PubMed] [Google Scholar]
- 44.Trotta R, Parihar R, Yu J, et al. Differential expression of SHIP1 in CD56bright and CD56dim NK cells provides a molecular basis for distinct functional responses to monokine costimulation. Blood. 2005;105(8):3011–3018. doi: 10.1182/blood-2004-10-4072. [DOI] [PubMed] [Google Scholar]
- 45.Trotta R, Ciarlariello D, Dal Col J, et al. The PP2A inhibitor SET regulates natural killer cell IFN-gamma production. J Exp Med. 2007;204(10):2397–2405. doi: 10.1084/jem.20070419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anfossi N, Andre P, Guia S, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25(2):331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 47.Brandt CS, Baratin M, Yi EC, et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med. 2009;206(7):1495–1503. doi: 10.1084/jem.20090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carlsten M, Bjorkstrom NK, Norell H, et al. DNAX accessory molecule-1 mediated recognition of freshly isolated ovarian carcinoma by resting natural killer cells. Cancer Res. 2007;67(3):1317–1325. doi: 10.1158/0008-5472.CAN-06-2264. [DOI] [PubMed] [Google Scholar]
- 49.Kim S, Sunwoo JB, Yang L, et al. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc Natl Acad Sci U S A. 2008;105(8):3053–3058. doi: 10.1073/pnas.0712229105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silva A, Andrews DM, Brooks AG, Smyth MJ, Hayakawa Y. Application of CD27 as a marker for distinguishing human NK cell subsets. Int Immunol. 2008;20(4):625–630. doi: 10.1093/intimm/dxn022. [DOI] [PubMed] [Google Scholar]
- 51.Lanier LL, Yu G, Phillips JH. Co-association of CD3 zeta with a receptor (CD16) for IgG Fc on human natural killer cells. Nature. 1989;342(6251):803–805. doi: 10.1038/342803a0. [DOI] [PubMed] [Google Scholar]
- 52.Kurosaki T, Ravetch JV. A single amino acid in the glycosyl phosphatidylinositol attachment domain determines the membrane topology of Fc gamma RIII. Nature. 1989;342(6251):805–807. doi: 10.1038/342805a0. [DOI] [PubMed] [Google Scholar]
- 53.Pende D, Parolini S, Pessino A, et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med. 1999;190(10):1505–1516. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolk K, Kunz S, Asadullah K, Sabat R. Cutting edge: immune cells as sources and targets of the IL-10 family members? J Immunol. 2002;168(11):5397–5402. doi: 10.4049/jimmunol.168.11.5397. [DOI] [PubMed] [Google Scholar]
- 55.Grant LR, Yao ZJ, Hedrich CM, et al. Stat4-dependent, T-bet-independent regulation of IL-10 in NK cells. Genes Immun. 2008;9(4):316–327. doi: 10.1038/gene.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watford WT, Hissong BD, Bream JH, Kanno Y, Muul L, O'Shea JJ. Signaling by IL-12 and IL-23 and the immunoregulatory roles of STAT4. Immunol Rev. 2004;202:139–156. doi: 10.1111/j.0105-2896.2004.00211.x. [DOI] [PubMed] [Google Scholar]
- 57.Perez OD, Mitchell D, Jager GC, et al. Leukocyte functional antigen 1 lowers T cell activation thresholds and signaling through cytohesin-1 and Jun-activating binding protein 1. Nat Immunol. 2003;4(11):1083–1092. doi: 10.1038/ni984. [DOI] [PubMed] [Google Scholar]