Abstract
Background:
The incidence of human papillomavirus-associated vulval neoplasia is increasing worldwide; yet the associated genetic changes remain poorly understood.
Methods:
We have used single-nucleotide polymorphism microarray analysis to perform the first high-resolution investigation of genome-wide allelic imbalance in vulval neoplasia. Our sample series comprised 21 high-grade vulval intraepithelial neoplasia and 6 vulval squamous cell carcinomas, with paired non-lesional samples used to adjust for normal copy number variation.
Results:
Overall the most common recurrent aberrations were gains at 1p and 20, with the most frequent deletions observed at 2q, 3p and 10. Copy-neutral loss of heterozygosity at 6p was a recurrent event in vulval intraepithelial neoplasia. The pattern of genetic alterations differed from the characteristic changes we previously identified in cutaneous squamous cell carcinomas. Vulval neoplasia samples did not exhibit gain at 5p, a frequent recurrent aberration in a series of cervical tumours analysed elsewhere using an identical protocol.
Conclusion:
This series of 27 vulval samples comprises the largest systematic genome-wide analysis of vulval neoplasia performed to date. Despite shared papillomavirus status and regional proximity, our data suggest that the frequency of certain genetic alterations may differ in vulval and cervical tumours.
Keywords: SNP array, vulva, VIN, VSCC, human papillomavirus, chromosome aberration
Vulval squamous cell carcinoma (VSCC) is a relatively uncommon malignancy in women, accounting for less than 5% of gynaecological cancer (Landis et al, 1999). It is now established that there are two distinct types of VSCCs, those associated with high-risk α human papillomavirus (hrHPV) and non-hrHPV-associated variants (McCluggage, 2009). HPV-positive tumours are frequently contiguous with classic vulval intraepithelial neoplasia (VIN) and usually arise in younger women. The incidence of HPV-positive VIN and VSCC is steadily increasing worldwide (Joura et al, 2000). An association has been reported (Hording et al, 1996; de Bie et al, 2009) between the development of classic VIN/VSCC and cervical intraepithelial neoplasia (CIN)/cervical SCC (CxSCC) but little is known of whether, in addition to their positive HPV status and regional proximity, these lesions share genetic similarities.
Limited data are available on the genetic alterations associated with VSCC and VIN precursor lesions; previous studies have used microsatellite marker analysis and metaphase comparative genomic hybridisation (CGH) rather than higher resolution array-based techniques (Pinto et al, 1999; Jee et al, 2001; Rosenthal et al, 2001; Allen et al, 2002; Micci et al, 2003; Bryndorf et al, 2004; Huang et al, 2005). In contrast, the genetic changes associated with CIN and CxSCC have been extensively studied, most recently using array-based methods (Hidalgo et al, 2005; Wilting et al, 2006, 2009; Choi et al, 2007; Kloth et al, 2007; Scotto et al, 2008a, 2008b).
Microarray-based single-nucleotide polymorphism (SNP) analysis facilitates the accurate and rapid identification of genome-wide allelic changes in tumour DNA samples. It has been used to investigate the molecular pathogenesis of various human cancers, including cutaneous SCC (Purdie et al, 2007, 2009) and cervical cancer (Kloth et al, 2007; Scotto et al, 2008a, 2008b). Here, for the first time, we have applied high-resolution microarray analysis to vulval squamous neoplasia to compare the pattern of genetic changes occurring during the development of hrHPV-associated VSCC with those alterations previously identified during cervical carcinogenesis. This reported investigation of 27 VIN and VSCC samples is the largest systematic genome-wide analysis of vulval cancer performed to date.
Materials and methods
Study patients
Patients attending a dedicated vulval clinic provided written, informed consent to undergo a diagnostic or therapeutic biopsy for either VIN or VSCC. Additional written consent was obtained for two 4-mm punch biopsies to be taken, one from the lesion and one from upper inner arm skin. Full demographic information was collected through a detailed questionnaire. Ethical approval for this investigation was obtained from the East London and City Health Authority local ethics committee and the study was conducted according to the Declaration of Helsinki Principles. All patients biopsied have remained under consultant dermatologist care (KG). Average duration of follow-up is 5 years (range 2–7 years).
Samples
Routine histopathological evaluation was performed on formalin-fixed vulval lesional sections stained with hematoxylin and eosin, with only samples identified as classic VIN or classic VIN-associated SCC selected for inclusion in this study. Staging of SCC was performed according to Federation of Gynaecology and Obstetrics and American Joint Committee on Cancer TNM criteria, in which Stage I/II is defined as carcinoma confined to the vulva and perineum, whereas Stage III/IV tumours are those that have spread to another anogenital region and/or regional lymph node(s). All diagnoses were confirmed by a single experienced gynaecological pathologist (NS). All punch biopsies collected were immediately snap-frozen in liquid nitrogen.
Microdissection and DNA extraction
Lesional tissue was microdissected to overcome the problem of stromal/lymphocyte contamination that can lead to a reduction in tumour DNA purity. Laser capture microdissection was performed using the PALM Microbeam system (Carl Zeiss, Welwyn Garden City, Hertfordshire, UK) on 8 μm fresh snap-frozen tissue sections cut onto Zeiss 1.0 PEN membrane slides (Carl Zeiss). DNA was extracted from lesional and non-lesional control skin biopsies using the Qiagen DNA Micro Kit (Qiagen, Crawley, West Sussex, UK) according to the manufacturer's instructions.
HPV detection and genotyping
The presence of hrHPV DNA was investigated in vulval samples using the digene Reverse Hybridisation Assay (Qiagen) that detects 18 high-risk mucosotropic HPV types. This study focused on HPV-associated vulval neoplasia, hence only hrHPV-positive samples were selected for inclusion in this study.
SNP array analysis
Samples were subjected to 250 K Nsp SNP array analysis (Affymetrix, Santa Clara, CA, USA) and the proprietary Genome Oriented Laboratory File (GOLF) system was used for the analysis and display of SNP call and signal data as previously described (Purdie et al, 2007).
Comparison with cervical cancer data
250 K Nsp SNP array data from a previously published study on cervical carcinomas (Scotto et al, 2008b) were downloaded from the NCBI GEO database (Edgar et al, 2002; GEO accession GSE10092) and analysed by GOLF software. This study was chosen for comparative purposes because the experimental protocol mirrored the current investigation, both in terms of the use of fresh frozen samples and the array-based technique. Furthermore, detailed clinical and histological information, including tumour stage, were available for the samples. Data from paired non-lesional samples were not available for analysis, hence raw data from seven normal cervical epithelial samples analysed by Scotto et al (2008b) were combined to make a pooled normal control. Vulval squamous cell carcinoma samples were reanalysed similarly and the results compared with those from paired sample analysis to confirm that this approach permitted the accurate identification of gross chromosomal aberrations.
Statistical analysis
The two-sided Fisher's exact test was used to calculate P-values, which were adjusted for multiple testing using Hochberg's method (Hochberg, 1988).
Results
Majority of HPV-positive vulval lesions contained HPV-16
The presence of hrHPV DNA was detected in 27 vulval lesional samples, comprising 21 classic high-grade VIN (VIN2/3) and 6 VIN-associated VSCC (all Stage IA/B). HPV-16 was the most frequent type, identified in 90% (19 out of 21) VIN samples as well as in 33% (2 out of 6) VSCC samples (Supplementary Table 1). HPV-33 was detected in three samples (two VIN, one VSCC) and HPV-18 was identified in one VSCC lesion. Lesions were associated with a single HPV type with the exception of 2 multiply-infected VSCC that contained HPV types 45 and 56 and types 39, 58, 59 and 66, respectively. Two high-grade VIN samples tested negative for HPV DNA and were excluded from further analysis.
Gain events were the most frequent recurrent chromosomal aberrations
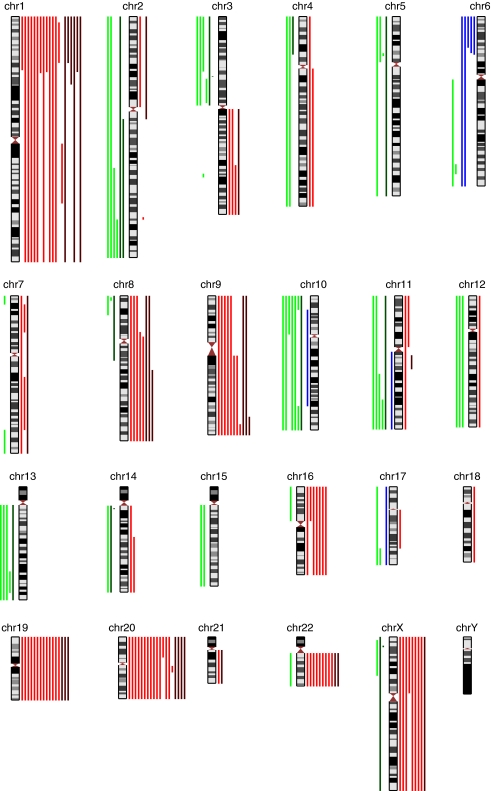
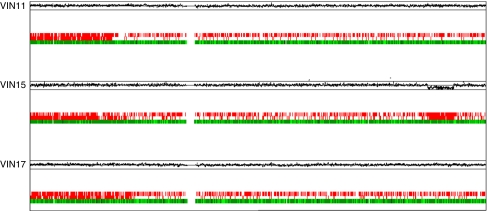
SNP microarray analysis was performed on lesional and paired non-lesional DNA samples to adjust for normal copy number variation (Redon et al, 2006). In high-grade VIN, gains at chromosomes 1p, 19 and 20 were the most frequent recurrent events, occurring in a majority of samples (13 out of 21, 62% for 1p and 19 and 14 out of 21, 67% for 20; Table 1, Figure 1). Some recurrent losses were observed, with the most frequent occurring at 10p (6 out of 21, 29%), 10q (5 out of 21, 24%) and 2q, 3p and 11q (all 4 out of 21, 19% samples). Comparison of SNP genotypes in lesional and paired normal samples revealed that 5 of 21 samples (24%) displayed extensive loss of heterozygosity (LOH) at 6p in the absence of copy number change. In three cases this copy-neutral LOH, also known as uniparental disomy (UPD), involved the distal region from 6pter-6p21.3 (Figure 2). The remaining two samples showed UPD across the whole of chromosome 6. In VSCC, gains remained the most frequent events, with all six samples showing 1p gain (Table 1, Figure 1). Gain of whole chromosome 20 was observed in four of six samples (67%) and gains at 1q, 8q, 9q and whole chromosome 19 all occurred in three samples (50%). Recurrent losses occurred at 2q, 3p, 14q and Xp, all in two of six (33%) samples.
Table 1. Recurrent aberrations in VIN and VSCC.
|
Frequency
a
|
|||
|---|---|---|---|
| Chromosome | Copy number change | VIN (n=21) | VSCC (n=6) |
| 1p | Gain | 13 (62%) | 6 (100%) |
| 1q | Gain | 10 (48%) | 3 (50%) |
| 2q | Deletion | 4 (19%) | 2 (33%) |
| 3p | Deletion | 4 (19%) | 2 (33%) |
| 6p | UPD | 5 (24%) | 0 |
| 8p | Gain | 4 (19%) | 2 (33%) |
| 8q | Gain | 5 (24%) | 3 (50%) |
| 9p | Gain | 5 (24%) | 2 (33%) |
| 9q | Gain | 8 (38%) | 3 (50%) |
| 10p | Deletion | 6 (29%) | 1 (17%) |
| 10q | Deletion | 5 (24%) | 1 (17%) |
| 14q | Deletion | 1 (5%) | 2 (33%) |
| 16p | Gain | 7 (33%) | 0 |
| 16q | Gain | 6 (29%) | 0 |
| 19 | Gain | 13 (62%) | 3 (50%) |
| 20p | Gain | 14 (66%) | 4 (67%) |
| 20q | Gain | 14 (66%) | 4 (67%) |
| 22q | Gain | 9 (43%) | 2 (33%) |
| Xp | Deletion | 1 (5%) | 2 (33%) |
| Xp | Gain | 8 (38%) | 1 (17%) |
| Xq | Gain | 7 (33%) | 1 (17%) |
Abbreviations: UPD=uniparental disomy or copy-neutral loss of heterozygosity; VIN=vulval intraepithelial neoplasia; VSCC=vulval squamous cell carcinoma.
Events are only listed if they occurred in 20% or more of one sample group.
Figure 1.
Ideogram summarising allelic imbalance in high-grade VIN (light coloured lines) and VSCC (dark coloured lines). Loss of heterozygosity events are indicated to the left of chromosomes with deletion shown in green and uniparental disomy in blue and gains are indicated to the right in red.
Figure 2.
Copy-neutral LOH at chromosome 6pter-6p21.3 in three high-grade VIN samples. Upper panels indicate lesional/non-lesional copy number ratios plotted on a log2 scale according to chromosomal position. Upper line represents log2(2) and lower line represents log2(0.5). Lower panels depict a comparison of SNP genotypes in VIN and paired normal samples. Single-nucleotide polymorphism loci shown in green indicate calls conserved between the two samples, loci shown in red immediately above indicate LOH in the VIN3 sample and loci shown in red on the top row indicate SNPs not called in the VIN3 sample. Localised areas of low call rates may imply LOH in a subpopulation of cells. Note that sample VIN 15 additionally exhibits LOH accompanied by loss of copy number (deletion) at 6q24.3-6q25.3.
Most frequent recurrent aberration in CxSCC was not detected in vulval neoplasia
To examine whether the genetic alterations observed here were shared by other HPV-associated genital neoplasia, we used the same software platform to analyse previously published data from a series of 70 cervical carcinomas that had been subjected to an identical microarray analysis protocol (Scotto et al, 2008b). Final analysis was restricted to 53 HPV-positive CxSCC with high call rates, including 10 tumours classified as Stage IA/IB according to Federation of Gynaecology and Obstetrics cervical carcinoma staging criteria (defined as carcinoma confined to the cervix). Gain at 5p, which had been identified as the most frequent recurrent change in the series of 70 CxSCC (Scotto et al, 2008b), was observed in 58% (31 out of 53) of the re-analysed cervical tumours compared with none of the 27 vulval neoplasia samples. Statistical analysis revealed that this aberration was significantly more frequent in CxSCC than VSCC, P-value=0.0083. The difference between the two groups did not reflect the earlier tumour stage of the VSCC series because this event occurred at comparable frequency in Stage I cervical tumours (50% Table 2). Other differences between the two tumour groups were not significant at the 5% level after adjustment for multiple testing.
Table 2. Comparison of aberrations in VSCC and CxSCC.
| Event | VSCC | All CxSCC a | Stage Ib CxSCC |
|---|---|---|---|
| Gain at 1p | 6/6 (100%) | 24/53 (45%) | 4/10 (40%) |
| Gain at 1q | 3/6 (50%) | 29/53 (55%) | 5/10 (50%) |
| Loss at 2q | 2/6 (33%) | 22/53 (43%) | 3/10 (30%) |
| Loss at 3p | 2/6 (33%) | 34/53 (64%) | 4/10 (40%) |
| Gain at 3q | 1/6 (17%) | 37/53 (70%) | 6/10 (60%) |
| Loss at 4p | 1/6 (17%) | 26/53 (49%) | 6/10 (60%) |
| Loss at 4q | 0/6 | 24/53 (45%) | 7/10 (70%) |
| Gain at 5p | 0/6 | 31/53 (58%) | 5/10 (50%) |
| Loss at 5q | 1/6 (17%) | 17/53 (32%) | 2/10 (20%) |
| Gain at 6p | 0/6 | 13/53 (25%) | 3/10 (30%) |
| Gain at 7q | 3/6 (50%) | 10/53 (19%) | 2/10 (20%) |
| Gain at 8p | 2/6 (33%) | 16/53 (30%) | 2/10 (20%) |
| Gain at 8q | 3/6 (50%) | 23/53 (43%) | 2/10 (20%) |
| Loss at 9p | 0/6 | 10/53 (19%) | 2/10 (20%) |
| Gain at 9q | 3/6 (50%) | 14/53 (26%) | 1/10 (10%) |
| Loss at 11q | 1/6 (17%) | 24/53 (45%) | 3/10 (30%) |
| Loss at 13q | 1/6 (17%) | 23/53 (43%) | 2/10 (20%) |
| Loss at 14q | 2/6 (33%) | 2/53 (4%) | 0/10 (0%) |
| Gain at 15q | 0/6 | 16/53 (30%) | 1/10 (10%) |
| Gain at 19q | 3/6 (50%) | 27/53 (50%) | 6/10 (60%) |
| Gain at 20q | 4/6 (67%) | 22/53 (42%) | 3/10 (30%) |
| Gain at 22q | 2/6 (33%) | 13/53 (25%) | 2/10 (20%) |
Reanalysed data from a previous study on cervical carcinomas (Scotto et al, 2008b).
According to FIGO criteria, Stage I CxSCC is carcinoma restricted to the cervix.
Discussion
This study is the first to investigate the genome-wide genetic changes in HPV-associated VIN and VSCC using high-resolution SNP array analysis. In a series of 27 VIN/VSCC samples with matched normal tissue, we have shown gains to be the most frequent chromosomal aberrations in both VIN and VSCC. The most common recurrent aberrations were gains, at 1p and 20 in particular. Some recurrent deletions were also observed, most frequently at 2q, 3p and 10. Copy-neutral LOH at 6p was a recurrent event in VIN. Despite our use of high-resolution SNP analysis, gross chromosomal aberrations comprised the majority of the recurrent events, with a few exceptions such as 3p and14q microdeletions, each observed in single VSCC samples (Figure 1). None of the 27 samples displayed gain at 5p, the most frequent genetic alteration previously reported in a series of CxSCC analysed using an identical protocol.
Comparison with previous studies on vulval neoplasia
This study is the first to use a high-resolution array-based technique to analyse the genetic alterations associated with hrHPV-positive VIN and VSCC. Indeed, few studies have previously examined the genetic changes associated with high-grade VIN using lower resolution techniques. One investigation (Rosenthal et al, 2001) reported that, of 6 loci (3p, 4q, 5p, 9p, 11p and 17p) examined for LOH using microsatellite marker analysis, 3p was the most frequently affected with LOH in 8 of 28 (29%) informative HPV-positive VIN3 samples, comparable to this study. Elsewhere, metaphase CGH was used to analyse the genetic events in nine HPV-positive VIN3 samples (Bryndorf et al, 2004). In line with this study, gains of whole chromosomes 1 and 20 were the most frequent aberrations observed in six (67%) and five (56%) of nine VIN3 samples, respectively. In contrast, gains at chromosome 19 were not detected; however, metaphase CGH has been shown to be unreliable for analysis of chromosome 19 aberrations (Kallioniemi et al, 1994).
The genetic alterations associated with hrHPV-positive VSCC have been analysed previously in several studies using various lower resolution techniques (summarised in Supplementary Table 2). Differences exist between these earlier investigations both in the nature of the samples (formalin-fixed paraffin embedded vs fresh frozen) and the methodologies used (microsatellite marker analysis that solely detects LOH vs metaphase CGH that has greater sensitivity for gain events). Nevertheless, all seven previous studies identified loss at 3p as an important event, occurring in 14% (1 out of 7) to 67% (4 out of 7) of samples, and recurrent gain of 8q was observed in all five CGH studies in 20% (2 out of 10) to 67% (4 out of 6) samples. Direct comparisons with our data are limited by methodological differences; however, both of these findings were consistent with our data. Some dissimilarities were apparent, in particular the lower frequency of gains reported elsewhere at chromosomes 1, 19 and 20. One explanation for these discrepancies may be the comparatively low overall level of the gains; in our samples the copy number ratios of these gains were often considerably lower than 1.5, suggesting that the frequency of these events may have been underestimated elsewhere had a higher cutoff value been used. Alternatively, because the low copy number ratio implied that the gains were present in only a subset of tumour cells and the samples examined here were early stage (Stage IA/B) cancers, it is possible that this subpopulation might decrease during VSCC progression. This would be consistent with data from one previous study showing a high frequency of chromosome 1 and 20 gains in VIN3 but not VSCC samples (Bryndorf et al, 2004). The fact that another investigation (Huang et al, 2005) observed gains at chromosomes 1 and 20 in only 17% of metastatic tumours provides further support for this theory. Although the remaining studies provided no information on tumour stage, the authors reported frequent recurrent gains at 3q and 5p, which have been shown to correlate with pathological staging of SCC (Oga et al, 2001; Yen et al, 2001; Noguchi et al, 2003; Klussmann et al, 2009); hence, it seems likely that many of these samples were also late stage tumours. Finally, it should be noted that two of the CGH studies included in the comparison (Jee et al, 2001; Micci et al, 2003) did not investigate the HPV status of the VSCC analysed; hence, it remains unclear whether their results are relevant to HPV-associated vulval neoplasia.
Vulval and CxSCC may be associated with different genetic alterations
Our data suggest that vulval and cervical tumours may exhibit different patterns of chromosomal aberrations. Specifically, reanalysis of previously published CxSCC data revealed frequent recurrent gain at 5p. Other studies have reported a similar finding in early stage cervical tumours. For example, Wilting et al (2006) identified recurrent 5p gain in 33% (3 out of 9) non-metastatic CxSCC and Scotto et al (2008b) reported that 26% (5 out of 19) of high-grade CIN samples showed gain at 5p. In contrast, none of the series of 27 high-grade VIN and VSCC in this study displayed this aberration. Statistical analysis revealed a significant difference between CxSCC and VSCC samples with respect to 5p gain, although any definite conclusions must be limited by the small number of VSCC examined. After controlling for multiple testing, the other differences observed between the two tumour groups were not significant. However, it would be of interest to investigate some of these differences further in a larger series of vulval samples. For example, 3q gain was detected in 17% (1 out of 6) VSCC compared with 70% (37 out of 53) of the CxSCC series analysed concomitantly (including 60%, 6 out of 10 of the Stage I tumours) and 100% (9 out of 9) of the Stage I and II cervical tumours investigated by Wilting et al (2006). Furthermore, frequent recurrent gain of 3q has been reported in cervical neoplasia before the acquisition of the invasive phenotype: Kirchhoff et al (1999) and Umayahara et al (2002) identified 3q gain in 35% (6 out of 17) and 61% (11 out of 18), respectively, of CIN3 samples compared with 14% (3 out of 21) high-grade VIN in this study. These combined data suggest that chromosomal aberrations characteristic of advanced SCC at several sites, including hrHPV-associated cancers such as tonsillar SCC (Oga et al, 2001; Yen et al, 2001; Dahlgren et al, 2003; Noguchi et al, 2003; Klussmann et al, 2009), commonly appear at an early stage of cervical transformation.
It is well established that the incidence of cervical carcinoma is 5- to 10-fold greater than that of other lower genital tract cancers and peaks at least 10 years earlier (Watson et al, 2008). HPV infections are commonly detected throughout the region (Hording et al, 1996), implying that cervical epithelium may be more susceptible to transformation than other anogenital epithelia. The cervix bears a unique anatomical feature, the transformation zone, where squamous metaplasia occurs as a normal physiological phenomenon. It is this area that is particularly susceptible to HPV infection and where both infection and CIN begin. The reasons for this remain unknown. This may be because the cellular microenvironment of replicating non-maturing reserve cells or unique cell-surface receptors on this cell population favour HPV infection (Crum, 2000). The prevalence of HPV-18 infection is higher in cervical than in other anogenital SCC (De Vuyst et al, 2009), and it has been speculated that HPV-18 infection may be more aggressive clinically (Bulk et al, 2007). However, differences in type-specific prevalence are not sufficiently great to account for the disparity in cancer incidence. Another possible explanation is that the susceptibility to hrHPV-mediated transformation of the various anogenital epithelia may in part reflect their immune or hormonal microenvironment. It has recently been found that the clinical outcome of preinvasive cervical lesions can be accurately predicted by the physical status of the hrHPV genome (integrated vs episomal) in conjunction with viral load (Wanram et al, 2009). The transition from low-grade dysplastic lesion harbouring episomal genomes to late-stage invasive carcinoma containing only integrated copies has been shown to occur by an intermediate step in which episomal and integrated genomes co-exist (Pett et al, 2004). Cells harbouring both genome types were characterised by copy number imbalances that predominantly comprised whole chromosome gains or losses, similar to the genetic events observed in vulval neoplasia in this study. In contrast, cells containing HPV integrant alone displayed frequent structural as well as numerical chromosomal instability, with recurrent aberrations including gain at 3q and 5p, mirroring those identified in the cervical tumours analysed here as well as in high-grade CIN (Scotto et al, 2008b). During the intermediate stage, integrants are subject to transcriptional repression by the episomal genomes (Dall et al, 2008). Selective advantage has been found to be insufficient for the progression of the integrant-bearing cell (Pett et al, 2006); rather there appears to be a requirement for episomal clearance, which can be hastened by treatment with various immunomodulators including type I interferon (Herdman et al, 2006). The physical status of the hrHPV genome in vulval neoplasia remains unclear. Data suggest that HPV integrants are present in a lower proportion of high-grade VIN than CIN (Hudelist et al, 2004; Hillemanns and Wang, 2006). The presence in VSCC of both integrated and episomal forms has been shown (Ueda et al, 2004), although analysis was restricted to a single sample. A larger study identified HPV integrants in the majority of VSCC examined (van de Nieuwenhof et al, 2009); however, the possibility of co-existent viral episomes was not excluded. Further work is clearly required to examine this hypothesis.
HPV-associated VSCC display HPV-specific aberrations
Previous studies have reported significant genetic differences between hrHPV-associated and hrHPV-independent SCC at a range of anatomical sites, including the oral cavity, oropharynx and cervix (Dahlgren et al, 2003; Braakhuis et al, 2004; Smeets et al, 2006; Klussmann et al, 2009; Wilting et al, 2009). The alterations identified as specific for hrHPV-independent tumours, namely recurrent losses at 5q and 9p and gains at 7q and 11q, were not observed in our VSCC series. Conversely, recurrent gain at chromosome 20, observed in 67% (4 out of 6) VSCC, was found to be a specific marker of HPV-associated cancer. All five earlier studies also reported that hrHPV-positive SCC were characterised by a lower level of chromosomal aberrations overall. Our laboratory has performed preliminary analysis (data not shown) on three HPV-independent VSCC, categorised thus because tumours developed in a background of lichen sclerosus and/or differentiated VIN and tested negative for hrHPV DNA. These data suggest that the significant genetic differences between hrHPV-positive and hrHPV-negative tumours observed at other sites may be mirrored in the vulva. The most marked difference was the finding of 9p loss in all three HPV-independent tumours. In addition, allelic imbalance was observed at a mean number of 10 chromosomes, compared with 6.5 in the HPV-associated VSCC. In contrast to these data, an earlier study from our laboratory (Purdie et al, 2009) examining the genetics of cutaneous SCC found no significant differences between HPV-positive and HPV-negative tumours. However, it must be noted that skin cancers are overwhelmingly associated with HPV from the β rather than α genus. The most common recurrent chromosomal aberration identified in cutaneous SCC was LOH at 9p, with similar frequencies in HPV-positive and HPV-negative SCC (32 out of 43, 74% and 13 out of 17, 76% respectively), whereas gain at chromosome 20 was observed at much lower frequency in 16% (7 out of 43) and 18% (3 out of 17) of HPV-positive and HPV-negative tumours, respectively. These combined data suggest that HPV from the β and α genera are associated with distinct patterns of chromosomal aberrations.
Possible candidate genes targeted by genetic alterations
Despite considerable methodological differences, combined data from the current and previous investigations suggest a characteristic pattern of genetic changes within VSCC. In particular, deletion at 3p was reported as a frequent recurrent event in all studies. This aberration is likely to target fragile histidine triad (FHIT), a recognised tumour suppressor gene known to be inactivated in other SCC (Yoshino et al, 2000; Lee et al, 2006; Purdie et al, 2009). Indeed, one of the VSCC samples exhibited a microdeletion restricted to the FHIT locus at 3p14.2. FHIT is located at the fragile site FRA3B, which has been identified as an integration hotspot for HPV (Huang et al, 2005), although FHIT also appears to be targeted during HPV-independent carcinogenesis. Gain at 8q, resulting in amplification of the oncogene c-Myc (Issing et al, 1993), was also consistently identified, as was gain at chromosome 20, a specific marker of HPV-associated cancer that has been reported to result in elevated mRNA levels of the de novo DNA methyltransferase DNMT3B at 20q (Wilting et al, 2006). In vitro studies (Savelieva et al, 1997; Klingelhutz et al, 2005) have shown that gain at 20q is an early event in hrHPV-mediated immortalisation of epithelial cells, suggesting that increased DNMT3B expression may contribute to the sequential tumour suppressor gene promoter hypermethylation that has been identified during HPV-induced transformation (Henken et al, 2007). Overall, the most frequent event across our series was gain at 1p, shown elsewhere to correlate with increased expression of the JUN protooncogene (Taniguchi et al, 2007). Gain at 3q, a marker of late stage HPV-associated cancer, was less frequently observed in the VSCC series. This aberration has been shown to result in the amplification of hTERC, the RNA component of human telomerase (Heselmeyer-Haddad et al, 2003), thereby complementing the activity of the hrHPV E6 oncoprotein that transcriptionally activates hTERT, the catalytic subunit of telomerase (Veldman et al, 2001). Uniparental disomy at 6p was the only recurrent copy-neutral LOH event identified in the series. Although this aberration was restricted to VIN samples, VSCC numbers were too small to determine whether this represented a genuine difference between preinvasive and invasive lesions. In cervical neoplasia, LOH at 6p is frequent in both high-grade CIN and CxSCC (Chatterjee et al, 2001; Kloth et al, 2007). Evidence suggests that HLA class I genes are the target of this aberration (Koopman et al, 2000), thereby permitting tumours to escape immune surveillance.
In summary, we have used high-resolution SNP microarray analysis to investigate genome-wide allelic imbalance in hrHPV-associated vulval neoplasia. Across the spectrum, gains of chromosomal material were more frequent than losses. Despite shared HPV status and regional proximity, our data suggest that vulval and cervical tumours may differ in their pattern of associated genetic alterations. A better understanding of any differences is likely to provide insight into mechanisms of HPV-associated epithelial carcinogenesis and may prove important in future studies of diagnostic, predictive and therapeutic biomarkers relevant to VSCC treatment and prevention.
Acknowledgments
This work was supported by Cancer Research UK, British Skin Foundation Small Grant Award reference S535. We thank Dr David Mesher and Professor Peter Sasieni, Centre for Epidemiology, Mathematics and Statistics, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, for their statistical advice.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Allen DG, Hutchins AM, Hammet F, White DJ, Scurry JP, Tabrizi SN, Garland SM, Armes JE (2002) Genetic aberrations detected by comparative genomic hybridisation in vulvar cancers. Br J Cancer 86: 924–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakhuis BJ, Snijders PJ, Keune WJ, Meijer CJ, Ruijter-Schippers HJ, Leemans CR, Brakenhoff RH (2004) Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J Natl Cancer Inst 96: 998–1006 [DOI] [PubMed] [Google Scholar]
- Bryndorf T, Kirchhoff M, Larsen J, Andreasson B, Bjerregaard B, Westh H, Rose H, Lundsteen C (2004) The most common chromosome aberration detected by high-resolution comparative genomic hybridization in vulvar intraepithelial neoplasia is not seen in vulvar squamous cell carcinoma. Cytogenet Genome Res 106: 43–48 [DOI] [PubMed] [Google Scholar]
- Bulk S, Berkhof J, Rozendaal L, Fransen Daalmeijer NC, Gok M, de Schipper FA, van Kemenade FJ, Snijders PJ, Meijer CJ (2007) The contribution of HPV18 to cervical cancer is underestimated using high-grade CIN as a measure of screening efficiency. Br J Cancer 96: 1234–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Pulido HA, Koul S, Beleno N, Perilla A, Posso H, Manusukhani M, Murty VV (2001) Mapping the sites of putative tumor suppressor genes at 6p25 and 6p21.3 in cervical carcinoma: occurrence of allelic deletions in precancerous lesions. Cancer Res 61: 2119–2123 [PubMed] [Google Scholar]
- Choi YW, Bae SM, Kim YW, Lee HN, Park TC, Ro DY, Shin JC, Shin SJ, Seo JS, Ahn WS (2007) Gene expression profiles in squamous cell cervical carcinoma using array-based comparative genomic hybridization analysis. Int J Gynecol Cancer 17: 687–696 [DOI] [PubMed] [Google Scholar]
- Crum CP (2000) Contemporary theories of cervical carcinogenesis: the virus, the host, and the stem cell. Mod Pathol 13: 243–251 [DOI] [PubMed] [Google Scholar]
- Dahlgren L, Mellin H, Wangsa D, Heselmeyer-Haddad K, Bjornestal L, Lindholm J, Munck-Wikland E, Auer G, Ried T, Dalianis T (2003) Comparative genomic hybridization analysis of tonsillar cancer reveals a different pattern of genomic imbalances in human papillomavirus-positive and -negative tumors. Int J Cancer 107: 244–249 [DOI] [PubMed] [Google Scholar]
- Dall KL, Scarpini CG, Roberts I, Winder DM, Stanley MA, Muralidhar B, Herdman MT, Pett MR, Coleman N (2008) Characterization of naturally occurring HPV16 integration sites isolated from cervical keratinocytes under noncompetitive conditions. Cancer Res 68: 8249–8259 [DOI] [PubMed] [Google Scholar]
- de Bie RP, van de Nieuwenhof HP, Bekkers RL, Melchers WJ, Siebers AG, Bulten J, Massuger LF, de Hullu JA (2009) Patients with usual vulvar intraepithelial neoplasia-related vulvar cancer have an increased risk of cervical abnormalities. Br J Cancer 101: 27–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vuyst H, Clifford GM, Nascimento MC, Madeleine MM, Franceschi S (2009) Prevalence and type distribution of human papillomavirus in carcinoma and intraepithelial neoplasia of the vulva, vagina and anus: a meta-analysis. Int J Cancer 124: 1626–1636 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henken FE, Wilting SM, Overmeer RM, van Rietschoten JG, Nygren AO, Errami A, Schouten JP, Meijer CJ, Snijders PJ, Steenbergen RD (2007) Sequential gene promoter methylation during HPV-induced cervical carcinogenesis. Br J Cancer 97: 1457–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdman MT, Pett MR, Roberts I, Alazawi WO, Teschendorff AE, Zhang XY, Stanley MA, Coleman N (2006) Interferon-beta treatment of cervical keratinocytes naturally infected with human papillomavirus 16 episomes promotes rapid reduction in episome numbers and emergence of latent integrants. Carcinogenesis 27: 2341–2353 [DOI] [PubMed] [Google Scholar]
- Heselmeyer-Haddad K, Janz V, Castle PE, Chaudhri N, White N, Wilber K, Morrison LE, Auer G, Burroughs FH, Sherman ME, Ried T (2003) Detection of genomic amplification of the human telomerase gene (TERC) in cytologic specimens as a genetic test for the diagnosis of cervical dysplasia. Am J Pathol 163: 1405–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo A, Baudis M, Petersen I, Arreola H, Pina P, Vazquez-Ortiz G, Hernandez D, Gonzalez J, Lazos M, Lopez R, Perez C, Garcia J, Vazquez K, Alatorre B, Salcedo M (2005) Microarray comparative genomic hybridization detection of chromosomal imbalances in uterine cervix carcinoma. BMC Cancer 5: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillemanns P, Wang X (2006) Integration of HPV-16 and HPV-18 DNA in vulvar intraepithelial neoplasia. Gynecol Oncol 100: 276–282 [DOI] [PubMed] [Google Scholar]
- Hochberg Y (1988) A sharper Bonferroni procedure for multiple tests of significance. Biometrika 75: 800–802 [Google Scholar]
- Hording U, Daugaard S, Junge J, Lundvall F (1996) Human papillomaviruses and multifocal genital neoplasia. Int J Gynecol Pathol 15: 230–234 [DOI] [PubMed] [Google Scholar]
- Huang FY, Kwok YK, Lau ET, Tang MH, Ng TY, Ngan HY (2005) Genetic abnormalities and HPV status in cervical and vulvar squamous cell carcinomas. Cancer Genet Cytogenet 157: 42–48 [DOI] [PubMed] [Google Scholar]
- Hudelist G, Manavi M, Pischinger KI, Watkins-Riedel T, Singer CF, Kubista E, Czerwenka KF (2004) Physical state and expression of HPV DNA in benign and dysplastic cervical tissue: different levels of viral integration are correlated with lesion grade. Gynecol Oncol 92: 873–880 [DOI] [PubMed] [Google Scholar]
- Issing WJ, Wustrow TP, Heppt WJ (1993) Oncogenes related to head and neck cancer. Anticancer Res 13: 2541–2551 [PubMed] [Google Scholar]
- Jee KJ, Kim YT, Kim KR, Kim HS, Yan A, Knuutila S (2001) Loss in 3p and 4p and gain of 3q are concomitant aberrations in squamous cell carcinoma of the vulva. Mod Pathol 14: 377–381 [DOI] [PubMed] [Google Scholar]
- Joura EA, Losch A, Haider-Angeler MG, Breitenecker G, Leodolter S (2000) Trends in vulvar neoplasia. Increasing incidence of vulvar intraepithelial neoplasia and squamous cell carcinoma of the vulva in young women. J Reprod Med 45: 613–615 [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Piper J, Tanner M, Stokke T, Chen L, Smith HS, Pinkel D, Gray JW, Waldman FM (1994) Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci USA 91: 2156–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff M, Rose H, Petersen BL, Maahr J, Gerdes T, Lundsteen C, Bryndorf T, Kryger-Baggesen N, Christensen L, Engelholm SA, Philip J (1999) Comparative genomic hybridization reveals a recurrent pattern of chromosomal aberrations in severe dysplasia/carcinoma in situ of the cervix and in advanced-stage cervical carcinoma. Genes Chromosomes Cancer 24: 144–150 [DOI] [PubMed] [Google Scholar]
- Klingelhutz AJ, Qian Q, Phillips SL, Gourronc FA, Darbro BW, Patil SR (2005) Amplification of the chromosome 20q region is associated with expression of HPV-16 E7 in human airway and anogenital epithelial cells. Virology 340: 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloth JN, Oosting J, van Wezel T, Szuhai K, Knijnenburg J, Gorter A, Kenter GG, Fleuren GJ, Jordanova ES (2007) Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex genetic alterations in cervical cancer. BMC Genomics 8: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klussmann JP, Mooren JJ, Lehnen M, Claessen SM, Stenner M, Huebbers CU, Weissenborn SJ, Wedemeyer I, Preuss SF, Straetmans JM, Manni JJ, Hopman AH, Speel EJ (2009) Genetic signatures of HPV-related and unrelated oropharyngeal carcinoma and their prognostic implications. Clin Cancer Res 15: 1779–1786 [DOI] [PubMed] [Google Scholar]
- Koopman LA, Corver WE, van der Slik AR, Giphart MJ, Fleuren GJ (2000) Multiple genetic alterations cause frequent and heterogeneous human histocompatibility leukocyte antigen class I loss in cervical cancer. J Exp Med 191: 961–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis SH, Murray T, Bolden S, Wingo PA (1999) Cancer statistics, 1999. CA Cancer J Clin 49: 8–31, 1 [DOI] [PubMed] [Google Scholar]
- Lee EJ, Lee BB, Kim JW, Shim YM, Hoseok I, Han J, Cho EY, Park J, Kim DH (2006) Aberrant methylation of Fragile Histidine Triad gene is associated with poor prognosis in early stage esophageal squamous cell carcinoma. Eur J Cancer 42: 972–980 [DOI] [PubMed] [Google Scholar]
- McCluggage WG (2009) Recent developments in vulvovaginal pathology. Histopathology 54: 156–173 [DOI] [PubMed] [Google Scholar]
- Micci F, Teixeira MR, Scheistroen M, Abeler VM, Heim S (2003) Cytogenetic characterization of tumors of the vulva and vagina. Genes Chromosomes Cancer 38: 137–148 [DOI] [PubMed] [Google Scholar]
- Noguchi T, Kimura Y, Takeno S, Chujo M, Uchida Y, Mueller W, Gabbert HE (2003) Chromosomal imbalance in esophageal squamous cell carcinoma: 3q gain correlates with tumor progression but not prognostic significance. Oncol Rep 10: 1393–1400 [PubMed] [Google Scholar]
- Oga A, Kong G, Tae K, Lee Y, Sasaki K (2001) Comparative genomic hybridization analysis reveals 3q gain resulting in genetic alteration in 3q in advanced oral squamous cell carcinoma. Cancer Genet Cytogenet 127: 24–29 [DOI] [PubMed] [Google Scholar]
- Pett MR, Alazawi WO, Roberts I, Dowen S, Smith DI, Stanley MA, Coleman N (2004) Acquisition of high-level chromosomal instability is associated with integration of human papillomavirus type 16 in cervical keratinocytes. Cancer Res 64: 1359–1368 [DOI] [PubMed] [Google Scholar]
- Pett MR, Herdman MT, Palmer RD, Yeo GS, Shivji MK, Stanley MA, Coleman N (2006) Selection of cervical keratinocytes containing integrated HPV16 associates with episome loss and an endogenous antiviral response. Proc Natl Acad Sci USA 103: 3822–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto AP, Lin MC, Mutter GL, Sun D, Villa LL, Crum CP (1999) Allelic loss in human papillomavirus-positive and -negative vulvar squamous cell carcinomas. Am J Pathol 154: 1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdie KJ, Lambert SR, Teh MT, Chaplin T, Molloy G, Raghavan M, Kelsell DP, Leigh IM, Harwood CA, Proby CM, Young BD (2007) Allelic imbalances and microdeletions affecting the PTPRD gene in cutaneous squamous cell carcinomas detected using single nucleotide polymorphism microarray analysis. Genes Chromosomes Cancer 46: 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdie KJ, Harwood CA, Gulati A, Chaplin T, Lambert SR, Cerio R, Kelly GP, Cazier JB, Young BD, Leigh IM, Proby CM (2009) Single nucleotide polymorphism array analysis defines a specific genetic fingerprint for well-differentiated cutaneous SCCs. J Invest Dermatol 129: 1562–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME (2006) Global variation in copy number in the human genome. Nature 444: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal AN, Ryan A, Hopster D, Surentheran T, Jacobs IJ (2001) High frequency of loss of heterozygosity in vulval intraepithelial neoplasia (VIN) is associated with invasive vulval squamous cell carcinoma (VSCC). Int J Cancer 94: 896–900 [DOI] [PubMed] [Google Scholar]
- Savelieva E, Belair CD, Newton MA, DeVries S, Gray JW, Waldman F, Reznikoff CA (1997) 20q gain associates with immortalization: 20q13.2 amplification correlates with genome instability in human papillomavirus 16 E7 transformed human uroepithelial cells. Oncogene 14: 551–560 [DOI] [PubMed] [Google Scholar]
- Scotto L, Narayan G, Nandula SV, Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright JD, Pothuri B, Mansukhani M, Murty VV (2008a) Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: potential role in progression. Genes Chromosomes Cancer 47: 755–765 [DOI] [PubMed] [Google Scholar]
- Scotto L, Narayan G, Nandula SV, Subramaniyam S, Kaufmann AM, Wright JD, Pothuri B, Mansukhani M, Schneider A, Arias-Pulido H, Murty VV (2008b) Integrative genomics analysis of chromosome 5p gain in cervical cancer reveals target over-expressed genes, including Drosha. Mol Cancer 7: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets SJ, Braakhuis BJ, Abbas S, Snijders PJ, Ylstra B, van de Wiel MA, Meijer GA, Leemans CR, Brakenhoff RH (2006) Genome-wide DNA copy number alterations in head and neck squamous cell carcinomas with or without oncogene-expressing human papillomavirus. Oncogene 25: 2558–2564 [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Karnan S, Fukui T, Yokoyama T, Tagawa H, Yokoi K, Ueda Y, Mitsudomi T, Horio Y, Hida T, Yatabe Y, Seto M, Sekido Y (2007) Genomic profiling of malignant pleural mesothelioma with array-based comparative genomic hybridization shows frequent non-random chromosomal alteration regions including JUN amplification on 1p32. Cancer Sci 98: 438–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y, Enomoto T, Miyatake T, Shroyer KR, Yoshizaki T, Kanao H, Ueno Y, Sun H, Nakashima R, Yoshino K, Kimura T, Haba T, Wakasa K, Murata Y (2004) Analysis of clonality and HPV infection in benign, hyperplastic, premalignant, and malignant lesions of the vulvar mucosa. Am J Clin Pathol 122: 266–274 [DOI] [PubMed] [Google Scholar]
- Umayahara K, Numa F, Suehiro Y, Sakata A, Nawata S, Ogata H, Suminami Y, Sakamoto M, Sasaki K, Kato H (2002) Comparative genomic hybridization detects genetic alterations during early stages of cervical cancer progression. Genes Chromosomes Cancer 33: 98–102 [DOI] [PubMed] [Google Scholar]
- van de Nieuwenhof HP, van Kempen LC, de Hullu JA, Bekkers RL, Bulten J, Melchers WJ, Massuger LF (2009) The etiologic role of HPV in vulvar squamous cell carcinoma fine tuned. Cancer Epidemiol Biomarkers Prev 18: 2061–2067 [DOI] [PubMed] [Google Scholar]
- Veldman T, Horikawa I, Barrett JC, Schlegel R (2001) Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J Virol 75: 4467–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanram S, Limpaiboon T, Leelayuwat C, Yuenyao P, Guiney DG, Lulitanond V, Jearanaikoon P (2009) The use of viral load as a surrogate marker in predicting disease progression for patients with early invasive cervical cancer with integrated human papillomavirus type 16. Am J Obstet Gynecol 201: 79, e1–7 [DOI] [PubMed] [Google Scholar]
- Watson M, Saraiya M, Ahmed F, Cardinez CJ, Reichman ME, Weir HK, Richards TB (2008) Using population-based cancer registry data to assess the burden of human papillomavirus-associated cancers in the United States: overview of methods. Cancer 113: 2841–2854 [DOI] [PubMed] [Google Scholar]
- Wilting SM, Snijders PJ, Meijer GA, Ylstra B, van den Ijssel PR, Snijders AM, Albertson DG, Coffa J, Schouten JP, van de Wiel MA, Meijer CJ, Steenbergen RD (2006) Increased gene copy numbers at chromosome 20q are frequent in both squamous cell carcinomas and adenocarcinomas of the cervix. J Pathol 209: 220–230 [DOI] [PubMed] [Google Scholar]
- Wilting SM, Smeets SJ, Snijders PJ, van Wieringen WN, van de Wiel MA, Meijer GA, Ylstra B, Leemans CR, Meijer CJ, Brakenhoff RH, Braakhuis BJ, Steenbergen RD (2009) Genomic profiling identifies common HPV-associated chromosomal alterations in squamous cell carcinomas of cervix and head and neck. BMC Med Genomics 2: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CC, Chen YJ, Chen JT, Hsia JY, Chen PM, Liu JH, Fan FS, Chiou TJ, Wang WS, Lin CH (2001) Comparative genomic hybridization of esophageal squamous cell carcinoma: correlations between chromosomal aberrations and disease progression/prognosis. Cancer 92: 2769–2777 [DOI] [PubMed] [Google Scholar]
- Yoshino K, Enomoto T, Nakamura T, Sun H, Ozaki K, Nakashima R, Wada H, Saitoh J, Watanabe Y, Noda K, Murata Y (2000) FHIT alterations in cancerous and non-cancerous cervical epithelium. Int J Cancer 85: 6–13 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.