Abstract
Human p53R2 (hp53R2) is a 351 residue p53-inducible ribonucleotide reductase (RNR) small subunit. It shares >80% sequence identity with hRRM2, the small RNR subunit responsible for normal maintenance of the deoxyribonucleotide (dNTP) pool used for DNA replication, which is active during the S-phase in a cell-cycle dependent fashion. But rather than cyclic dNTP synthesis, hp53R2 has been shown to supply dNTPs for DNA repair to cells in G0-G1 in a p53-dependent fashion. The first x-ray crystal structure of hp53R2 is solved to 2.6 Å, in which monomers A and B exhibit mono- and bi-nuclear iron occupancy, respectively. The pronounced structural differences at three regions between hp53R2 and hRRM2 highlight the possible regulatory role in iron assimilation, and help explain previously observed physical and biochemical differences in the mobility and accessibility of the radical-iron center, as well as radical transfer pathways between the two enzymes. The sequence-structure-function correlations that differentiate hp53R2 and hRRM2 are revealed for the first time. Insight gained from this structural work will be used toward the identification of biological function, regulation mechanism and inhibitors selection in RNR small subunits.
RNR catalyzes the reduction of all four ribonucleotides to their corresponding deoxyribonucleotides, the building blocks for DNA biosynthesis (1). There are currently three identified classes of RNRs. Class I RNRs are biologically active as α2β2 tetramers. Three class I RNR subunits have been identified in mammals. The large (α) subunit, M1, contains the enzyme active site and allosteric effector sites where substrate reduction is mediated by a cysteine thiyl radical, and a pair of redox active cysteines (1). The small (β) subunit, M2, contains a dinuclear iron site that instigates formation of a stable tyrosyl radical via the four electron reduction of molecular oxygen to water (2, 3). The hRRM2/M1 holo complex provides dNTPs to proliferating cells in an S-phase dependent fashion (4), where hRRM2 is under the transcriptional regulation of cell-cycle associated factors (5, 6, 7). p53R2, identified in 2000 by Tanaka et al., is a small subunit exhibiting many conserved features of M2 (>80% identical). Like M2, p53R2 contains the di-iron/dityrosyl cofactor, but p53R2, and not M2, is transactivated by p53 in response to DNA damage to cells in G0-G1 in a p53-dependent fashion (8, 9). Additionally, p53R2-null mice demonstrated enhanced frequency of spontaneous mutations and activation of p53-dependent apoptotic pathways (10). The hp53R2 gene contains a 20 nucleotide p53 binding site in intron 1, and two putative stretches of nuclear localization sequences on the gene product (8). Both hRRM2 and hp53R2 were shown to interact with hRRM1 through the C-terminal binding domain, and converted CDP to dCDP (11). Further, the highly conserved diiron/dityrosyl pockets afford both hp53R2 and hRRM2 the ability to form the tyrosyl radical necessary for NDP reduction at hRRM1 (12, 13). Despite the high sequence identity, hRRM2 and hp53R2 exhibit many differences that are reflected in their different biological roles in assisting DNA biosynthesis in two distinct pathways described above. The hRRM2 crystal structure is available (PDB ID: 2UW2). However, past attempts to identify compounds that selectively inhibit either hRRM2 or hp53R2 have been severely hampered by the lack of the hp53R2 crystal structure. In this work, we present the 2.6 Å x-ray crystal structure of hp53R2, the first x-ray crystal structure of human p53R2 enzyme. Extensive structural comparisons between the hp53R2 and other mammalian R2s offer a high resolution rationale for differences in susceptibilities to iron extruding and radical scavenging agents.
MATERIALS & METHODS
Materials
All chemicals were purchased from Sigma-Aldrich Chemical Co. and were the highest grade available. pET28a (+), Escherichia coli strain BL21 (DE3) was purchased from Novagen.
Protein Expression and Purification
His6-tagged hp53R2 was expressed as previously reported (13); the purification was modified as follows. All steps were performed at 4 °C. Harvested cells were suspended in lysis buffer (Tris pH 7.5, 150 mM NaCl, 50 mM imidazole, 10% glycerol), sonicated, and clarified. This was followed by TALON® metal affinity resin purification. The partially purified protein was concentrated to 10 mg/mL, and further purified by gel filtration on a Superdex® 200 HR 10/300 GL column with 20 mM Tris pH 7.5 and 150 mM NaCl to afford >99% pure protein.
Crystallization and Data Collection
hp53R2 was crystallized via the sitting drop vapor diffusion method at 25 °C. 2 μL of 4.5 mg/mL protein in 20 mM Tris, pH 7.5, with 150 mM NaCl, were added to 2 μL precipitant (0.1 M sodium citrate pH 6.45, 1.3 M Li2SO4, and 0.5 M (NH4)2SO4). Reservoir volume was 250 μL. Crystals were visible after 7 days, with full size reached between 10 and 14 days. Ferrous ammonium sulfate was added to crystal drops for a final concentration of 5 mM one hour prior to harvesting. Prior to liquid N2 flash cooling, crystals were cryoprotected in a solution of 70%: 30% (v/v) of the crystallization precipitant : glycerol. A 2.6 Å resolution data set was collected at the Advanced Light Source (ALS, beamline 8.2.1) at −160 °C. A 3.4 Å SAD dataset was collected at the Fe peak (1.74 Å) after observing a strong iron fluorescence signal to confirm iron presence. All data were processed with HKL2000 (14). Statistics for both datasets are listed in Table S1.
Structure Determination
hp53R2 was solved via MR using mouse R2 structure (PDB ID: 1XSM) (15) as a template with the program EPMR (16). Refinement began with CNS (17) using torsion angle SA followed by energy minimization, with positional and individual B-factor refinement. Subsequent iterative rounds of model rebuilding with (Win) Coot (18) and refinement in CNS with the maximum likelihood approach were used to lower the free and crystalline R-values. Finally, waters and crystallization agents were added to non-protein density.
Iron Sites
Upon synchrotron irradiation of the crystal (back-soaked in iron-free cryoprotectant prior to freezing) at the iron-absorption edge, a strong fluorescence signal was detected. Two large anomalous peaks were observed from the anomalous difference map (FT (fom (|f_a+|−|f_a−|) exp(i[phase-90]) calculated using CNS (17), with the SAD experimental amplitudes (1.74 Å) (Table S1). The model phase calculated from the refined hp53R2 model was used. A simulated annealing composite omit map was subsequently generated by CNS that corroborated the peak from the anomalous FT map with a defined patch of electron density outside the current protein model, and overlaying right on the highest anomalous difference peaks (8.92 and 6.93 I/sigma, respectively) that is positioned at the expected iron-coordinating residues, where iron is expected to bind hp53R2 by homology to the mRRM2 coordinates. The monomer B Fe1 and Fe2 sites were treated as two separate iron atoms, and refined, with energy minimization followed by positional and individual B-factor refinement settings. Iron occupancies were set to 1 for both Fe2 sites, and the individual B-factors refined to 60.77 for Fe2 in monomer A, and 82.11 for Fe2 in monomer B. The iron occupancy of Fe1 in monomer B was refined to 0.45 and the individual B-factor refined to 79.07. Upon removal of the iron coordinates from the refined structure, as well as calculating a composite omit map and an Fo-Fc map, a patch of robust electron density is observed at the Fe1 site (composite omit map contoured at 1.0 sigma, and Fo-Fc map contoured at 3.0 sigma). These data are further corroborated with the anomalous difference map, which displays clear density overlapping the same vicinities – strong density contoured at 3.0 sigma.
RESULTS AND DISCUSSION
Overall Fold
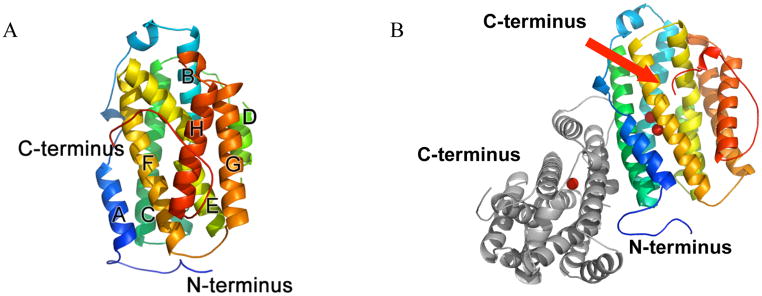
The hp53R2 structure was resolved to 2.6 Å, and refined to an Rcrys of 22.2% (Rfree, 27.3%). The natural dimer is observed per asymmetric unit (Figs. 1B, S5). The coordinates exhibit a highly similar topology to that of the hRRM2 (PDB ID: 2UW2 – pending publication with coordinates released) and mRRM2 structures (PDB ID: 1XSM; 1W68; 1W69) (15, 19). The structure also maintains a similar pattern of disorder at the N- and C-termini. The structure consists of helices and loops, and 8 of the 11 helices form a central bundle, for which a secondary structure identification scheme has been described with the mRRM2 structure (Fig. 1A), (19). Both monomers of hp53R2 can be superimposed onto mRRM2 and hRRM2 with 0.80–0.93 Å RMSD. The two monomers in hp53R2 have a 0.69 Å RMSD. The electron density map for monomer A has gaps (missing both backbone and sidechain density) from 1–28, 101–104, and 312–351, while in monomer B there are gaps from 1–29, 160–161, and 318–351. Additionally, though the backbone density is clear, D100 of monomer B is missing sidechain density.
Figure 1.
(A) Overall topology of hp53R2 monomer B. The structure is entirely helical (helices A-H as described (15)), beginning at residue S29 and ending at residue L317. The helices form a central helical bundle around the dityrosyl/diiron active site, for which structural details are described in the text. (B) Overall dimer showing monomer B in similar orientation to panel (A), along with iron sites, and monomer A (gray). The lower iron in the image is Fe1.
Iron Site: Mono- versus Di-iron in Monomers A and B
The hp53R2 iron-binding site is created by α-helices B, C, E, and F, as in the mRRM2 and hRRM2 structures (Fig. 1A–B) (15, 19). However, the active site iron coordination environment (E131, H134, D100, E194, E228 and H231) is different between monomers A and B (Fig. 2A–D). Monomer A exhibits one iron in the active site (Fe2 by convention), whereas monomer B exhibits occupancy of both iron sites. The Fe sites were confirmed to be iron based on a SAD dataset collected at the iron absorption peak (1.74 Å). The two highest peaks in the anomalous Fourier map correspond to one of the two previously reported iron locations, Fe2, in both monomers A and B (Fig. 2C). In monomer B, the second iron site (Fe1 by convention) is also observed in a Fo-Fc omit map (Fig. 2B). In the Fourier transform electron density map, the monomer B Fe1–Fe2 density appears as two overlapping distorted spheres over a much larger region than would be expected for a single iron (Fig. 2C). The 3.5 Å resolution of the SAD dataset may be too low to resolve the distance between the two iron sites. As found in the Fo-Fc omit map (Fig. 2B), these atoms are approximately 5 Å apart. The active site iron coordination environment (E131, H134, D100, E194, E228 and H231) is different between monomers A and B (Fig. 2A–D). This reflects the mono- and di-iron occupancies in monomers A and B, respectively. This crystal structure captures the mono-/di-iron occupied hp53R2 for the first time.
Figure 2.
(A) Overlay of monomer A (green) and monomer B (gray) iron coordination sites. Subtle shifts in the positions of all residues, and dramatic shift in the position of D100 backbone. (BI) Electron density of iron coordination residues in monomer B. Blue density is 2Fo-Fc contoured at 1 sigma. Green spheres around iron sites: Fo-Fc omit map contoured at 3 sigma. (BII) Composite Omit map, contoured at 1 sigma, calculated after removal of the iron atoms from the model to show the Fe1 density. (C) Anomalous difference peaks of iron atoms, calculated with refined model phase and the 3.5 Å SAD (1.74 Å) amplitudes. Electron density is contoured at 3.0 sigma. (DI) Irons and coordination residues/water in monomer B. (DII) Schematic displaying the same environment/similar orientation as in DI, with distances (Å). The D100 side chain is hypothetical (logical stereochemical and conformational constraints) as it is not seen in the electron density. (EI) Overlay of reduced mRRM2 (1W69) in orange and hp53R2 monomer B in gray to show significant positional differences among many of the active site residues (similar orientation to images in Fig. D). (EII) Image displaying iron coordination distances (Å) for 1W69 to highlight where they are substantially different between the active sites of hp53R2 and 1W69 (similar orientation to images in Fig. D; coordination distances can be compared to those of hp53R2 in Fig. DII). Water: hp53R2 structure; acetate: mRRM2 structure. hp53R2 irons, green; mRRM2 irons, purple.
The Shifted Fe1 Position
The superposition of Fe1–Fe2 sites in hp53R2 monomer B and mRRM2 (PDB 1W69, also with two Fe sites) suggests that the irons are under a reducing condition due to the lack of oxide density in the putative μ-oxo iron bridge position (Fig. 2E) (15). This could be a result of the excess FeII in the crystallization drop. The reduced active site is not the enzymatically active form of the enzyme. The active subunit is the di-ferric form in which the active tyrosyl radical is created via the following reaction:
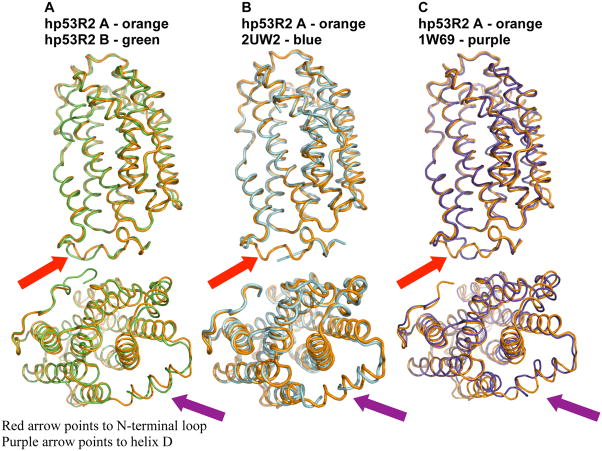
where the first three reducing electrons are iron-derived, and the fourth is from oxidation of the active tyrosine. Just as in the two monomers of hp53R2, the major observable difference between the hp53R2 and hRRM2 active sites is the position of hp53R2 D100 relative to D138 of hRRM2, as well as the position of Fe1 (Fig. 2E). hp53R2 monomer B exhibits electron density for the main chain atoms of D100, though no sidechain density could be observed for monomer B at residue 100 (thus a glycine is included at this position for monomer B in Fig. 2A–C, E). Figure 2D displays a hypothetical positioning of the D100 sidechain in monomer B based on stereo and conformational constraints, and logical orientation with regard to the Fe1 site. High D100 flexibility in this environment could be the reason for the shift of the Fe1 site (Fig. 2D&E). For example, the mRRM2 E170 is 2.03 Å from the Fe1 iron site, while the corresponding hp53R2 E131 is 2.80 Å from the Fe1 iron position in hp53R2 monomer B (Fig. 2D&E). Further, hp53R2 H134 is 2.74 Å away from the same site, while the corresponding mRRM2 H173 is only 2.19 Å from the mouse Fe1 iron position. From in vitro biochemical studies, hp53R2’s diferric iron center was 158-fold more susceptible to the iron chelator deferoxamine mesylate than was hMMR2, while 2.5 times less sensitive than hRRM2 to the radical scavenger, hydroxyurea (10). In addition, the bivalency of E228 in hp53R2 is not observed in mRRM2 or hRRM2 (15, 19). These observations, taken with the structural details lead to a proposal that there are differences in gating capabilities/mechanisms regarding iron assimilation and susceptibilities to radical scavenging agents between the hp53R2 and other mammalian RRM2s. For clarity, Figure 3 (overlays of C-α traces) is provided to highlight the regions of the most substantial differences between hp53R2 and different RRM2 structures.
Figure 3.
(A) C-α overlay of hp53R2 monomer A (orange), and hp53R2 monomer B (green) with similar orientation to Figure 1. The N-terminus and helix G display clear conformational differences between the monomers, and are the regions of greatest structural difference between the homologs as shown in panels B and C. (B) C-α overlay of hp53R2 monomer A and hRRM2 (2UW2). (C) C-α overlay of hp53R2 monomer A and mRRM2 (1W69).
Structural basis of the B-helix disorder: the N-terminal Swivel Region
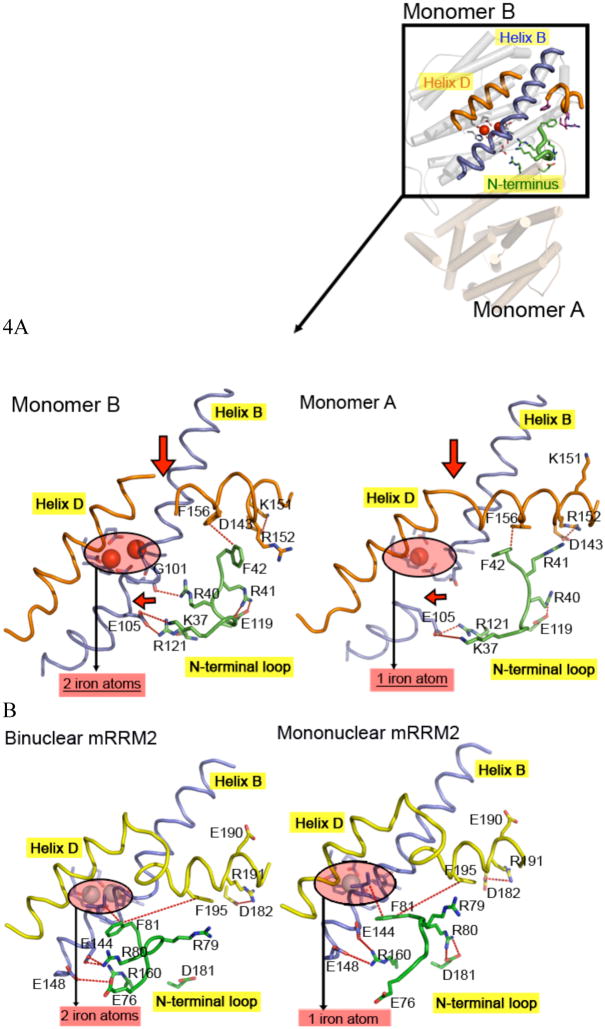
The crystal exhibits the dimer per asymmetric unit. To verify that this structure contains the true biological unit, we have provided an FPLC trace from a gel filtration result (Fig. S5), and we extend a comparison of hp53R2 to the E.coli x-ray crystal structure, both in the Supporting Information section. Similarly, use of the homologous structure dimers is described in the Supporting Information. The hp53R2 structure offers two snapshots of protein conformation trapped in mono- and di-iron occupancies for monomers A and B, respectively (Fig. 4A). N-terminal residues 37–42 from one monomer can swivel between two conformations and impose significant influences on helix D (Fig. 4F–G) and helix B of the opposite monomer (Fig. 4D–E). This change ultimately affects the orientation of D100, and thus the integrity of the binuclear iron environment (Fig. 4A). Specifically, D100 in monomer A is swung away from the active site, compared to monomer B (D100 sidechain in monomer B artificially modeled in image 2D; it is not actually in the density map. In the deposited structure, only the backbone density is modeled, as is displayed in all other panels of Figure 2). Note that though the sidechain for D100 in monomer B is missing, we do clearly see the backbone density. Because there is clear electron density of both backbone and sidechain density from D101 to D104 in monomer B, the difference in D100 main-chain position between monomers A and B would create a different environment in this region. We will refer to the N-terminal residues (residues 37–42) as the N-terminal swivel region throughout this manuscript.
Figure 4.
(A) Bird’s eye view of N-terminal swivel region and 2° structural regions it contacts within hp53R2 dimer, and also zoom views of monomers A and B. Major conformational differences highlighted with red arrows, and highlighted iron atoms. (B) The homologous structural regions as in (A), but for mouse structures 1W69 (left), and 1XSM (right). (C) Residues of the N-terminal swivel region, overlaying hp53R2 monomers A and B (left), and mRRM2 mononuclear (1XSM) and binuclear occupied iron (1W69) structures (right). The major conformational changes for the sequence-identical Arg residues are denoted with red arrows. (D) Interactions between monomer B N-terminal swivel region and monomer A (helix B) that lead to destabilization of helix B, and ultimately mononuclear iron occupancy. Red arrows indicate directionality of helix. (E) Interactions between monomer A N-terminal swivel region and monomer B (helix B) that lead to stabilization of helix B, and ultimately binuclear iron occupancy. Red arrows indicate directionality of helix. Note: for clarity a water molecule was left out of this panel. There is a water spanning the gap between the lys 37 ammonium of monomer A, and E105 of helix B on monomer B (2.34 Å from the lys 37 to the water and 3.20 Å from the water to the carboxylate of E105). (F) Interactions between monomer B N-terminal swivel region and helix D of monomer A that stabilize helix D. Additionally highlighted is the salt bond between R152 and D143 in the D-loop region that exists in the stabilized D-helix conformation. Red arrows indicate directionality of helix. (G) Interactions between monomer A N-terminal swivel region and helix D of monomer B that destabilize helix D. Additionally highlighted is the salt bond between K151 and D143 in the D-loop region that exists in the destabilized D-helix conformation. Red arrows indicate directionality of helix.
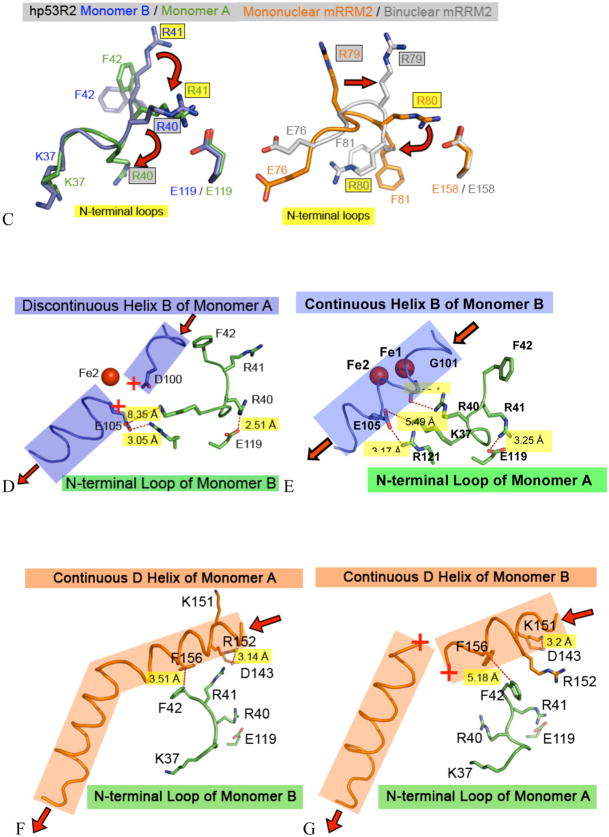
Looking to the N-terminal swivel region in Fig. 4C, in hp53R2 monomer A, R41 is swung inward by about 90° from its position in monomer B (i.e. it interacts with the same monomer) and forms a salt bridge with the E119 side chain of monomer A. In this conformation, F42 is flipped inward, and R40 of monomer A is swung outward to H-bond with G101 in monomer B (4.05 Å from G101) (Fig. 4E). Furthermore, K37 in monomer A spans the dimer interface to form a salt bridge with E105 to stabilize helix B of monomer B. The interactions of the monomer A N-terminal swivel region stabilize the B-helix of monomer B, and allow D100 of monomer B to be oriented in the proper vicinity to bind iron (Fe1), thereby stabilizing the full binuclear iron environment. Conversely, in the N-terminal swivel region of monomer B, R40 flips inward and H-bonds with the E119 side chain, and F42 flips outward and interrupts Helix B, and D100 no longer interacts favorably with the iron center (Fig. 4D). Therefore, interactions of the N-terminal residues in monomer B with the B-helix in monomer A are correlated with the helix B disorder, which presumably prevents D100 of monomer A from binding the second Fe.
hp53R2 Helix D and D-loop
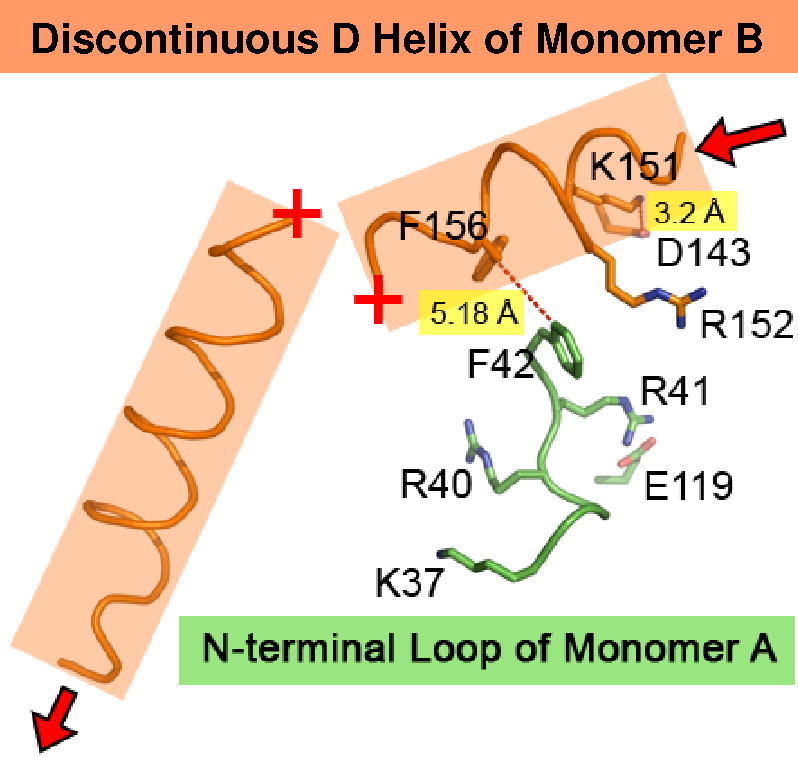
The N-terminal swivel region also affects the order at helix D. As seen in Fig. 4F, when R40 is bridging to E119 of monomer B, F42 is positioned 3.51 Å from F156 on helix D of monomer A, forming a hydrophobic interaction. With this interaction intact, the remainder of helix D is ordered. As seen in Fig. 4G, when R41 bridges to E119 of monomer A, F42 is now flipped 5.18 Å away from F156 of helix D on monomer B, and no longer creates a favorable hydrophobic interaction. The loss of this Phe:Phe interaction is concurrent with the loss of order in the middle of helix D (missing E160 and T161).
The third structural region affected by the two N-terminal swivel conformations is the D-loop (residues D143 – R152). As seen in Fig. 4F, when F42 stacks with F156 of monomer A, R152 is bridging to D143 at a 3.14 Å distance, and K151 is facing solvent. The R152→D143 salt bridge exists when there is order in helix D. The alternative conformation is seen in Fig. 4G, where no favorable hydrophobic interaction is maintained between F42 of the monomer A and F156 of monomer B. This translates to the K151 of monomer B bridging to D143 at a 3.2 Å distance, and R152 is facing solvent. The K151→D143 salt bridge coincides with the disorder of helix D.
mRRM2 N-terminal Swivel on Helices B, D, and D loop
Unlike the hp53R2 N-terminal swivel region, in the mRRM2 swivel, only R80 (equivalent to hp53R2 R41) adopts two different conformations (Fig. 4C). R79 merely shifts a small distance and does not create any new interactions. As in hp53R2, F81 of mRRM2 also flips between two conformations. The N-terminal swivel region on the hRRM2 structure is likely similar to the same region on the mRRM2 structure, where the coordinates (2UW2) for this region (residues 78–81) are missing. For clarity, this region is highlighted in Fig. 3, identifying it as one of the two regions where the most differences are seen between hp53R2 and m/hRRM2.
Helix B in each of the mononuclear and binuclear iron occupied mRRM2 structures is ordered (Fig. 5B). When R80 bridges to E144, it aids in pulling N-terminal and helix B closer together (Fig. 5B, 1W69) where, in addition to this salt bridge, R160 is brought closer to E144 and E148 by 2.35 (5.86–3.51=2.35) Å and 0.11 (5.18–5.07=0.11) Å, respectively. E76 in mRRM2 cannot create any favorable interactions with E144 or E148 on helix B. In contrast, hp53R2 has K37 (the equivalent residue of E76 in mRRM2), which forms a salt bridge with E105 on helix B, resulting in the disordered helix B in monomer A (Fig. 4D).
Figure 5.
(A) Overlay of D-helix regions of both mono and binuclear occupied mRRM2 structures (1XSM and 1W69) highlighting the lack of conformational change between the two, owed to lack of crosstalk between the F81 residue at the N-terminal swivel region of the structures. (B) Comparison of the interactions between the N-terminal swivel region and the B-helix of both structures. Structural integrity of helix B is preserved in both mono and binuclear occupied structures, because the N-terminal swivel region does not impose strong enough differences from one swivel conformation to the other in its interactions with helix B.
As seen in Fig. 4B, helix D in each of the mononuclear and binuclear iron occupied mRRM2 structures is ordered. Fig. 5A displays the overlaid ‘would-be’ interacting residues between the N-terminal swivel region and helix D of both mRRM2 structures. In both mRRM2 structures, F81 and F195 (equivalent to F42 and F156 of hp53R2) are about 9 Å apart, and form no hydrophobic interactions. Therefore, the N-terminal swivel region of mRRM2 does not dictate conformational change/order at helix D. As for the D-loop conformation, K151 and R152 in hp53R2 are equivalent to E190 and R191 in mRRM2, respectively. In comparison, both mono- and di-iron mRRM2 structures (Fig. 5A) show that the D loop (residues 182–190) has only one conformation, where R191 bridges to D182.
hp53R2/mRRM2 N-terminal swivel region comparative summary
The N-terminal swivel region and its two conformations in the hp53R2 (monomers A versus B) and mRRM2 (mono- versus di-Fe) structures showed that the N-terminal region communicates directly to three different regions (helix B, D-loop, and helix D) on the opposite monomer (Fig. 4A-B). Through F42/F156 hydrophobic interaction, the N-terminal swivel region can stabilize or destabilize helix D in hp53R2, but not in mRRM2 (Fig. 4F–G and Fig. 5A). In addition to the lack of Phe-Phe communication between the N-terminal swivel region of mRRM2 and the D-helix, the lack of K151 in mRRM2 (which has an E190 at this position) also precludes mRRM2 from forming the swivel at the D-loop. Only in hp53R2 do the two swivel conformations lead to a significant effect at helix B. This difference is essentially owed to the sequence variation between K37 in hp53R2, and its sequence equivalent E76 on mRRM2.
When synthesizing all these components, it is clear that hp53R2 has greater crosstalk between its secondary structural regions than its eukaryotic counterparts, owed to unique swivel points and favorable charge interactions. These regions work to either open or close helix B, a motion that acts as a gate keeper to the iron coordination environment. The fact that helix B can exist in an open conformation in hp53R2 implies, not only a different mechanism of iron incorporation from homologous RRM2 structures, but is consistent with the observation that hp53R2 binds iron less well, and that it is more susceptible to the iron chelating deferoxamine mesylate (13). We propose this to be a unique gating mechanism to allow iron in and out of the hp53R2 active site. This is also corroborated by the observation that hp53R2 is 158-fold more susceptible to iron extrusion by deferoxamine mesylate, than hRRM2 (13). A main point to be clarified is that, though both mRRM2 structures maintain order at helix B, only one exhibits binuclear iron occupancy. The mononuclear iron occupied mRRM2 was crystallized at pH 4.7; even though the structural integrity might allow for residence of both iron sites, iron is acid labile, and the condition is too acidic.
Additional Attributes of Helix D, Cys Versus Tyr
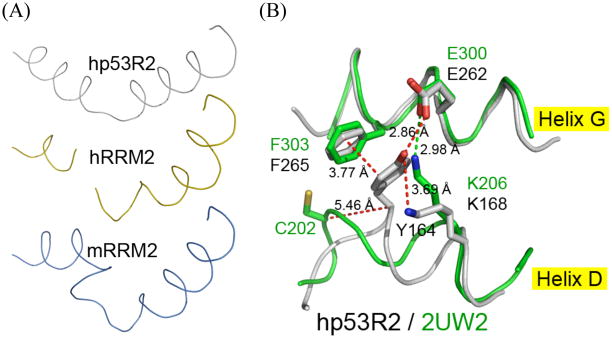
As an integral part of the gating mechanism, helix D can adopt two conformations in hp53R2, but not in mRRM2. Looking from a bird’s eye view of helix D (Fig. 6A), in hp53R2 it maintains a regular helical order but is bent in the middle, in hRRM2 this region has more loop-like characters than α-helices, and in mRRM2 it has an additional kink. The variation of this topology goes beyond the previous discussion of helix D, and looks farther down the helix toward the C-terminal direction, where a critical sequence variation of C203/202 in m/hRRM2 is equivalent to Y164 in hp53R2 (Fig. 6B). When overlapping the structures, the cysteine C-α in hRRM2 is 5 Å away from the C-α of hp53R2 Y164, which forms a 2.86 Å hydrogen bond with E262 (Fig. 6B). Because m/hRRM2 has a Cys instead of a Tyr at this position, this H-bond does not occur in the RRM2 structures. Rather, K206 of the RRM2 structures forms a salt bridge with the sequence equivalent glutamate (E300). Y164 in hp53R2 also forms pi-stacking interactions with F265 that do not occur in the RRM2 structures (Fig. 6B). The interactions mediated by Y164 of hp53R2 bring helix D and helix G closer together in hp53R2 than they do in RRM2 structures. As K206 of the RRM2 structures reaches to bridge E300, thereby occupying the space where a tyrosine would sit, it kinks the helix, and leads to the helix D conformation seen in Fig. 6A. The sequence equivalent K168 in hp53R2 does not form this salt bridge, because Y164 bonds with E262, though K168 does still H-Bond with Y164 (Fig. 6B).
Figure 6.
(A) Overlaid stretch of helix D residues from identical vantage point of homologous structures. From top to bottom: hp53R2 monomer A, hRRM2 (2UW2), mRRM2 (1XSM). (B) hp53R2 structure in gray; hRRM2 in green. The switch from hRRM2 C202 to hp53R2 Y164 at this sequence equivalent position confers dramatic conformational changes among the compared structures, because of different non-covalent interactions that can or cannot happen in hp53R2 versus hRRM2. Y164 H-bonds to E262, and thus K168 does not form a salt bond there. In contrast, in the hRRM2 structure, K206 (sequence equivalent of hp53R2 K168) forms a salt bridge with the equivalent glutamate (E300). The conformation of all mRRM2 structures is nearly identical at the displayed positions in the 2UW2 coordinates, therefore only the hRRM2 is depicted for comparison to hp53R2.
The switch from a Cys in the RRM2 structures to a Tyr in hp53R2 leads to a major structural difference: hp53R2 has an open channel (Fig. 7A–B) never reported in RRM2 before. This open channel helps explain why hp53R2 has drastically different iron chelator susceptibilities from those of hRRM2. Both hp53R2 monomers have an opening that runs through the Fe2 site; the Fe1 site in monomer B also resides in this open channel (Fig. S1 B–D). At the surface, h/mRRM2 has F237/236 that corresponds to hp53R2 F198 (Fig. 7A). However, in hp53R2, Y164 spans helices D and G and alters the conformations of a series of Phe residues, ultimately moving them out of the pore vicinity. Because the other RRM2 structures have a Cys in this position that does not alter the equivalent Phe positions, those Phe conformations are such that the pore is occluded in the RRM2 structures (Figs. 8A, B; S3A–C, S4A–C). Therefore, the open channel is linked to the Cys versus Tyr difference observed in the D helix. That this pore is in the immediate vicinity of the iron binding site of hp53R2, and is not seen in m/hRRM2, is a second explanation for why hp53R2 is 158-fold more susceptible to the iron chelator, deferoxamine mesylate, than is hRRM2 (13). Aside from possibly conferring increased susceptibility to iron chelators, the channel may play a role in the regulation of the enzyme, though much has yet to be determined about the hp53R2 regulatory pathway. Site directed mutagenesis specific to residues in the tunnel region could offer insight into the physiological importance of this region.
Figure 7.
(A) Surface representations displaying identical vantage points, and 180° rotations of hp53R2 monomers A & B. hp53R2 exhibits a hole proximal to the iron coordination site that traverses the entire structure, that is not seen in the compared structures (see Fig. S2 for hRRM2, mRRM2 surface representations).
Figure 8.
(A) View of hp53r2 monomer A pore with 2UW2 phenylalanine 236 overlaid to show how this residue occludes the pore in both hRRM2 and mRRM2 structures. (B) View of hp53R2 monomer B overlaid with 1W68 displaying the conformational changes at specific phenylalanines that lead to the pore occlusion in the hRRM2 and mRRM2 structures, but not the hp53R2 structure. The conformational changes can be traced back to the sequence switch from C202/203 in hRRM2/mRRM2 → Y164 in hp53R2.
Structural Insights and Potential Anti-cancer Relevance
RNR is paramount for cancer survival, and is a validated target for anti-cancer agents. There are currently many RNR-targeting anti-cancer molecules in the clinic or in various clinical trials. However, the broad specificity of many such RNR inhibitors remains a major shortcoming. The idea to inhibit the RNR small subunits selectively is one with high appeal and momentum,, because different cancers use the two dNTP producing pathways differently. For example, inhibiting p53R2 but not R2 can specifically target tumors that over-express p53R2. Further, the inactivation of hp53R2-dependent DNA synthesis will activate p53-dependent apoptosis (8, 9).
Several insights have been gleaned from the hp53R2 crystal structure. Three major structural regions have been highlighted to impart major differences between the iron-protective environments, in addition to an enzyme spanning pore that passes directly to one side of the hp53R2 iron core. With greater than 80% sequence identity between the two subunits, the differences observed in the x-ray crystal structures are differences that could not have been clearly or readily identified by other methods.
The most likely region to be exploited from a rational drug design perspective is the channel spanning each monomer. Figure S1 shows that this region is exposed in both the monomeric and dimeric hp53R2 structures. In silico inhibitor screening efforts that limit the search parameters to this pocket could first filter molecules based on size and geometric constraints, then optimize the identified subset of compounds on the basis of charge properties. These criteria could exclude a large number of inhibitor-like candidates and identify a manageable subset for subsequent in vitro inhibition assays. Therefore, the hp53R2 crystal structure provides a template for in silico inhibitor screening effort that will impact the development of RNR small-subunit-specific anti-cancer therapeutics.
Biological Significance
hp53R2, the first p53R2 x-ray crystal structure, exhibits many distinct structural features not reported before in the m/hRRM2 structures. The sequence switch from m/hRRM2 C203/C202 to Y164 in hp53R2 results in a dramatic change in the surrounding phenylalanine conformations creating a central channel through hp53R2, not seen in reported homologous structures. This feature helps to explain the increased susceptibility of hp53R2 to iron sequestering deferoxamine mesylate (13). Additionally, in hp53R2, the communication between the N-terminal swivel residues, and the opposite B- and D-helices, allows two different conformations that alternatively swivel at residues R40 and R41, as well as K151 & R152. These conformations are structural evidence for a possible gating mechanism in iron assimilation. Though the swivel can occur at the equivalent N-terminal positions in the mRRM2 structures, it does not lead to the far sweeping differences seen in hp53R2 due to sequence variations. That helix B can exist in an open conformation in hp53R2 implies a different mechanism of iron incorporation from homologous RRM2 structures, and offers further evidence to bolster the observation that hp53R2 binds iron less well, and that it is more susceptible to the iron chelating deferoxamine mesylate (13). This also lends insight to a possible role for iron availability and occupancy relative to hp53R2 pathway regulation. Though the full biological cascade and control mechanisms relating to hp53R2 have yet to be determined, it is possible that the enzyme is under some form of iron-dependent activation/repression. For the first time, the structural differences are presented between the two mammalian small subunits. These insights pave the foundation for understanding the biological function, regulation mechanism, and the development of selective inhibitors of these RNR small subunits.
Supplementary Material
Acknowledgments
This work was supported partially by NCI grant CA 127541 and Sino-American Cancer Foundation
Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Abbreviations
- RNR
ribonucleotide reductase
- M1
human ribonucleotide reductase large subunit
- M2
human ribonucleotide reductase small subunit
- NDP
nucleoside diphosphate
- NTP
nucleoside triphosphate
- ATP
adenosine-5′-triphosphate
- CDP
cytidine-5′-diphosphate
- dNDP
deoxynucleoside diphosphate
- dNTP
deoxynucleoside triphosphate
- MR
molecular replacement
- SA
simulated annealing
Footnotes
The atomic coordinates have been deposited in the Protein Data Bank (accession code 3HF1)
Supporting Information Available. Included in the Supporting Information are data statistics, and graphics that further detail important structural features of hp53R2 in comparison with the homologous mammalian structures described in the main text. Additionally, there is discussion of assumptions used in the homologous structure comparisons. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jordan A, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 1998;67:71–98. doi: 10.1146/annurev.biochem.67.1.71. [DOI] [PubMed] [Google Scholar]
- 2.Stubbe J, Riggs-Gelasco P. Harnessing free radicals: formation and function of the tyrosyl radical in ribonucleotide reductase. Trends Biochem Sci. 1998;23:438–443. doi: 10.1016/s0968-0004(98)01296-1. [DOI] [PubMed] [Google Scholar]
- 3.Stubbe J. Di-iron-tyrosyl radical ribonucleotide reductases. Curr Opin Chem Biol. 2003;7:183–188. doi: 10.1016/s1367-5931(03)00025-5. [DOI] [PubMed] [Google Scholar]
- 4.Chabes A, Thelander L. Controlled protein degradation regulates ribonucleotide reductase activity in proliferating mammalian cells during the normal cell cycle and in response to DNA damage and replication blocks. J Biol Chem. 2000;275:17747–17753. doi: 10.1074/jbc.M000799200. [DOI] [PubMed] [Google Scholar]
- 5.Filatov D, Thelander L. Role of a proximal NF-Y binding promoter element in S phase-specific expression of mouse ribonucleotide reductase R2 gene. J Biol Chem. 1995;270:25239–25243. doi: 10.1074/jbc.270.42.25239. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, et al. Nuclear factor Y regulation and promoter transactivation of human ribonucleotide reductase subunit M2 gene in gemcitabine resistant KB clone. Biochem Pharmacol. 2004;67:1499–1511. doi: 10.1016/j.bcp.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 7.Chabes AL, Bjorklund S, Thelander L. S phase-specific transcription of the mouse ribonucleotide reductase R2 gene requires both a proximal repressive E2F-binding site and an upstream promoter activating region. J Biol Chem. 2004;279:10796–10807. doi: 10.1074/jbc.M312482200. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka H, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi T, et al. p53R2-dependent pathway for DNA synthesis in a p53-regulated cell cycle checkpoint. Cancer Res. 2001;61:8256–8262. [PubMed] [Google Scholar]
- 10.Kimura T, et al. Impaired function of p53R2 in Rrm2b-null mice causes severe renal failure through attenuation of dNTP pools. Nat Genet. 2003;34:440–445. doi: 10.1038/ng1212. [DOI] [PubMed] [Google Scholar]
- 11.Guittet O, et al. Mammalian p53R2 protein forms an active ribonucleotide reductase in vitro with the R1 protein, which is expressed both in resting cells in response to DNA damage and in proliferating cells. J Biol Chem. 2001;276:40647–40651. doi: 10.1074/jbc.M106088200. [DOI] [PubMed] [Google Scholar]
- 12.Zhou B, et al. A dityrosyl-diiron radical cofactor center is essential for human ribonucleotide reductases. Mol Cancer Ther. 2005;4:1830–1836. doi: 10.1158/1535-7163.MCT-05-0273. [DOI] [PubMed] [Google Scholar]
- 13.Shao J, et al. In vitro characterization of enzymatic properties and inhibition of the p53R2 subunit of human ribonucleotide reductase. Cancer Res. 2004;64:1–6. doi: 10.1158/0008-5472.can-03-3048. [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 15.Strand KR, et al. Crystal structural studies of changes in the native dinuclear iron center of rionucleotide reductase protein R2 from mouse. J Biol Chem. 2004;279:46794–46801. doi: 10.1074/jbc.M407346200. [DOI] [PubMed] [Google Scholar]
- 16.Kissinger CR, Gehlhaar DK, Fogel DB. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr D. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 17.Brunger AT, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 18.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 19.Kauppi B, et al. The three-dimensional structure of mammalian ribonucleotide reductase protein R2 reveals a more-accessible iron-radical site than Escherichia coli R2. J Mol Biol. 1996;262:706–720. doi: 10.1006/jmbi.1996.0546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.