Abstract
A homozygous H493R mutation in the active site of tyrosyl-DNA phosphodiesterase (TDP1) has been implicated in hereditary spinocerebellar ataxia with axonal neuropathy (SCAN1), an autosomal recessive neurodegenerative disease. However, it is uncertain how the H493R mutation elicits the specific pathologies of SCAN1. To address this question, and to further elucidate the role of TDP1 in repair of DNA end modifications and general physiology, we generated a Tdp1 knockout mouse and carried out detailed behavioral analyses as well as characterization of repair deficiencies in extracts of embryo fibroblasts from these animals. While Tdp1−/− mice appear phenotypically normal, extracts from Tdp1−/− fibroblasts exhibited deficiencies in processing 3′-phosphotyrosyl single-strand breaks and 3′-phosphoglycolate double-strand breaks, but not 3′-phosphoglycolate single-strand breaks. Supplementing Tdp1−/− extracts with H493R TDP1 partially restored processing of 3′-phosphotyrosyl single-strand breaks, but with evidence of persistent covalent adducts between TDP1 and DNA, consistent with a proposed intermediate-stabilization effect of the SCAN1 mutation. However, H493R TDP1 supplementation had no effect on phosphoglycolate termini on 3′ overhangs of double-strand breaks; these remained completely unprocessed. Altogether, these results suggest that for 3′-phosphoglycolate overhang lesions, the SCAN1 mutation confers loss of function, while for 3′-phosphotyrosyl lesions, the mutation uniquely stabilizes a reaction intermediate.
Keywords: TDP1, DNA repair, mouse models, NHEJ, TopoisomeraseI, end modifications
1. INTRODUCTION
A homozygous mutation in the active site of tyrosyl-DNA phosphodiesterase (TDP1) has been identified as the causative mutation in hereditary spinocerebellar ataxia with axonal neuropathy (SCAN1). SCAN1 is inherited as an autosomal recessive disorder that becomes apparent at adolescence; clinical features in these individuals include distal muscle weakness, absence of deep tendon reflexes, gait disturbances, and mild brain atrophy [1].
TDP1 has primarily been characterized as resolving trapped topoisomerase I cleavable complexes (TopIcc) by hydrolyzing the normally transient tyrosyl linkage that forms between the active site of topoisomerase I (Top1) and the 3′ DNA terminus; this occurs during DNA relaxation that facilitates replication and transcription [2–4]. TDP1 can also process protruding 3′-phosphoglycolate (PG) termini on DNA double-strand breaks (DSBs) [5,6] that are formed in response to oxidative stress [7], ionizing radiation [8], and specific chemotherapeutic agents such as bleomycin [9].
Because the H493R TDP1 mutation associated with SCAN1 (hereafter referred to as SCAN1 TDP1) retains partial activity, SCAN1 cells do not provide a true null model of TDP1 deficiency. Moreover, it is uncertain how the mutation elicits the specific pathologies of SCAN1, and in particular whether unresolved TOPI-linked breaks, unrepaired PG-terminated breaks, or covalent TDP1-DNA intermediates are the critical toxic lesions. As reported below, we generated a Tdp1 knockout mouse, carried out detailed behavioral analyses, and derived embryo fibroblast cell lines from these mice. These cells provided an in vitro system that permitted investigation of end-processing of these candidate DNA lesions.
2. MATERIALS AND METHODS
2.1 Generation of Tdp1 knockout mice
To generate both constitutive and conditional Tdp1 knockout (Tdp1−/−) mice, a Tdp1 targeting vector was constructed using pKO NTKV-1901 (Stratagene). This vector was modified by the insertion of loxP sites on either side of the PGK/neo/BGH cassette. A 4.8-kb region of the Tdp1 gene containing exons 5–7 (with 3.2 kb of intron 6 deleted) was generated by PCR from a 129/Sv BAC clone, and a third loxP site (with an associated NheI site) was inserted into intron 5 (Figure 1, panel ii). This arm was then inserted between the BglII and XhoI sites of NTKV-1901. A 4.7-kb XmaI/SmaI BAC clone fragment containing Tdp1 exons 8–12 was inserted into the NTKV-1901 SmaI site. The Tdp1 targeting vector was linearized with SmaI and electroporated into 129/SV embryonic stem (ES) cells. To identify the desired homologous recombinants, genomic DNA from ES cell clones resistant to both G418 and gangcyclovir was screened by long-range PCR and Southern blotting. To delete the neo gene in the targeted Tdp1 allele and generate knockout and conditional knockout alleles (as shown in Figure 1 panels iv and v), ES cells from a correctly targeted clone were electroporated with an MC1-cre expression vector and G418-sensitive clones selected. Knockout recombinants were identified by PCR and injected into blastocysts that were implanted into pseudopregnant females. Tdp1+/− mice were then generated from the resulting chimeras and were interbred to generate homozygous knockouts, as verified by PCR and Southern blotting (see Supplemental Methods and Supplemental Figure 1 for additional details). These mice were subjected to a variety of behavioral tests, also described in detail in Supplemental Methods. In addition to a functional observational battery assessment [10], assays were employed to assess specific differences in motor performance and possible ataxia, including rotarod performance and general locomotor activity measured in standard activity chambers.
Figure 1. Generation of Tdp1−/− mice.
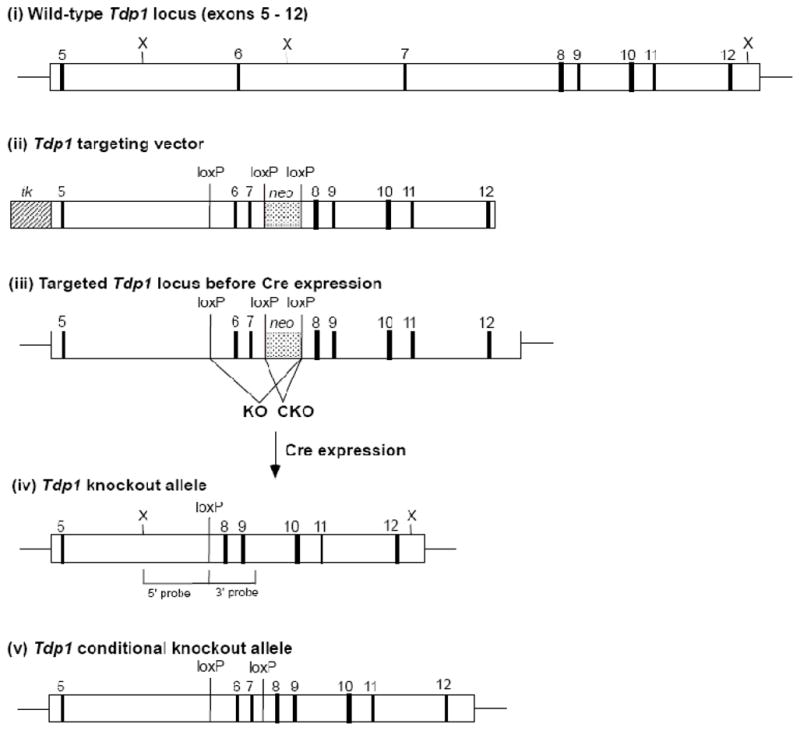
Tdp1 targeting strategy. (i) Structure of the mouse Tdp1 gene, exons 5–12. (ii) Targeting vector for generation of both conditional and total knockout of the Tdp1 gene. (iii) Structure of the initial targeted Tdp1 allele prior to Cre expression. Structure of the knockout (iv) or conditional knockout (v) allele of Tdp1 following transient expression of Cre in the targeted ES cells.
All animal studies were approved by the Institutional Animal Care and Use Committee at Virginia Commonwealth University, and were conducted in accordance with the Animal Welfare Act, the PHS Policy on Humane Care and Use of Laboratory Animals, and the U.S. Government Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training.
2.2 Generation and culture of embryonic fibroblasts
For generation of Tdp1+/+, Tdp1+/−, and Tdp1−/− mouse embryonic fibroblasts, males and females of the same or opposite genotype were mated, and 14-day embryos were dissociated, trypsinized and plated on 0.1% gelatin. Cells were immortalized by continuous culture for 6 months in DMEM with 10% fetal bovine serum. A single Tdp−/− clone was expanded and infected with a lentivirus expressing FLAG-tagged TDP1, and stable integration was verified by PCR. This infected clone was subsequently passaged as a separate cell clone, Tdp1comp, and screened for expression of hTDP1 by anti-TDP1 western blotting and an enzyme activity assay, described below. (See Supplemental Methods for further details.)
2.3 Preparation of Tyrosyl and 3′ PG DNA Substrates
The 3′-PG-terminated 17-mer 5′-CGAGGAACGCGAAAACG-3′ was prepared by bleomycin-mediated cleavage of 5′-CGAGGAACGCGAAAACGCCC-3′, and purified by polyacrylamide gel electrophoresis and HPLC [11,12]. The 3′-PG oligomers and a 3′-pTyr 18-mer (obtained from Midland Certified Reagents) and unmodified marker oligomers were 5′-32P labeled with [γ-32P]ATP and T4 polynucleotide kinase, then annealed to complementary unlabeled oligos to create single strand-break substrates (SSBs) [13]. A plasmid substrate containing a DNA double-strand break (DSB) with PG-terminated ACG 3′ overhang was constructed as described [14]. The substrate containing a DNA DSB with one 3′-PG-terminated blunt end and one recessed 3′-PG end [15] was constructed similarly.
2.4 Site-Directed Mutagenesis and Enzyme purification
A pET plasmid expressing His-tagged human TDP1 was kindly provided by Howard Nash, NIMH. Mutants were generated using the QuickChange site directed-mutagenesis kit from Stratagene and primers selected by the QuickChange Primer Design Program. Wild-type or mutant plasmids were freshly transformed into E. coli BL21(DE3) strain and cells were grown in 1 L LB medium supplemented with 100 μg/ml ampicillin. When the OD600 reached 0.4–0.5, 1 mM IPTG was added for induction. After 4 h, cells were harvested and dry pellets were stored at −80°C.
Cell pellets were re-suspended in 12 ml lysis buffer (50 mM NaH2PO4, pH 8.0, 0.3 M NaCl, 10 mM imidazole, 1 mg/ml lysozyme, 1 mM phenylmethylsulphonyl fluoride). After 30 min on ice, cells were sonicated 6×15s using a sonicator on 30% power from Ultrasonic Inc. Crude extracts were centrifuged at 14000xg at 4°C for 30 min. Soluble cell proteins were mixed with 1.5 ml Ni-NTA Superflow (Qiagen) and gently stirred 1h at 4°C. The suspension was then added to a chromatography column and washed with 25 ml of wash buffer (50 mM NaH2PO4, pH 8.0, 0.3 M NaCl, 20 mM imidazole 1 mM phenylmethylsulphonyl fluoride). Bound proteins were then eluted four times with the same buffer containing 0.5 M imidazole. All four fractions were analyzed by SDS-PAGE electrophoresis and peak fractions were dialyzed against 2 L 20 mM Tris-HCl pH 8.0 overnight, passed through a 0.22 μm filter, loaded on a Pharmacia MonoQ FPLC column at 22°C and eluted with a 15-min gradient of 0–0.8 M NaCl in 20 mM Tris-HCl pH 8.0. Fractions were collected and analyzed by SDS-PAGE. Protein concentration was determined using a Pierce BCA assay (Thermo Fisher Scientific Inc.). Proteins were stored in 50% glycerol at −20°C.
2.5 Analysis of DNA End Processing and Repair
Organ homogenates were prepared by sacrificing mice of each genotype and dissecting the liver and brain, which were then flash frozen. Tissues were thawed and minced, then resuspended in lysis buffer (10 mM Hepes pH 7.8, 60 mM KCl, 1 mM EDTA, 0.5% NP 40, 0.5 mM DTT, 100 mM PMSF, and Sigma protease and phosphatase inhibitor cocktails (catalogue numbers P8340 and P5726)) and homogenized with a Polytron tissue homogenizer (Kinematica). Extracts of MEFs were prepared according to procedures described previously [16]. Protein concentrations of both tissue lysates and cell extracts were assessed with a BCA Assay (Pierce). Organ homogenates were diluted in buffer containing 50 mM Tris pH 8.0, 5 mM DTT, 100 mM NaCl, 5 mM EDTA, 10% glycerol, and 500 μg/mL BSA. A 0.5 μL aliquot of each dilution was allowed to react with 50 fmol of a radioactive 3′-pTyr oligomer substrate (see Figure 2B), synthesized by Midland Certified Reagent Co. by a procedure described previously [17], in 5 μL of reaction buffer containing 50 mM triethanolamine pH 7.5, 1 mM DTT, 5 mM EDTA, 100 μM dNTPs, and 1 mM ATP. Following incubation at 37°C for 1 h, samples were diluted in an equal volume of formamide containing 20 mM EDTA and denatured at 90°C for 5 min. For determining activity toward a pTyr nick substrate (see Figure 3A), whole-cell extracts were serially diluted and incubated with 50 fmol of the substrate in the same reaction buffer described above, with the exception of 10 mM Mg(OAc)2 substituting for 5 mM EDTA. Typically, 3 – 5 μL of extract was added to a total reaction volume of 5 – 10 μL, for 1 h unless otherwise noted. In some cases, serial dilutions of FLAG-tagged TDP1 purified from 293T cells was added to Tdp1−/− extracts [6]. To test the temporal requirements for pTyr processing, 50 ng of human TDP1 produced in E. coli was added to Tdp1−/− extracts in 5 μL reaction buffer and incubated with the radiolabeled substrate from 1 – 60 min, as indicated (Figure 3D).
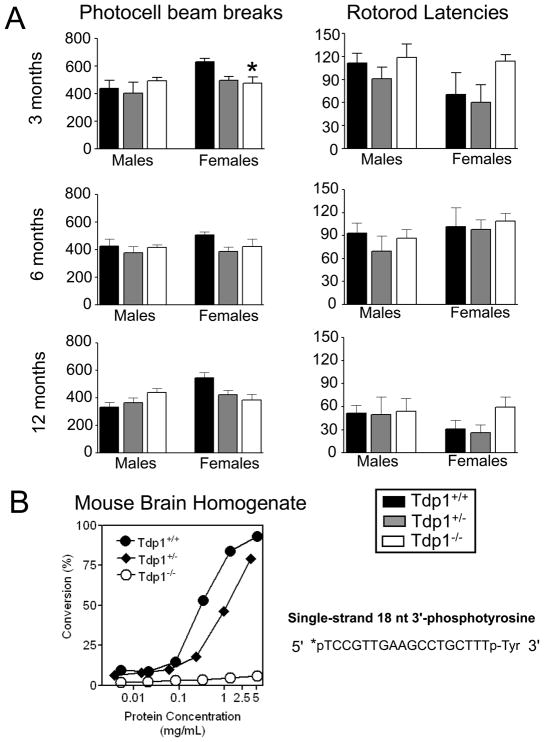
Figure 2. Behavior of Tdp1−/− mice and deficiency in 3′ tyrosyl processing.
(A) Motor activity and latency to fall from the rotarod was determined for male and female mice of each genotype and at each age: 3, 6, and 12 months in top, middle and bottom panels, respectively. Asterisk indicates that at the 3 month assessment, Tdp1−/− female mice were significantly less active than Tdp1+/+ mice, p < 0.05. (B) A radiolabeled (*) 3′-pTyr oligomeric substrate was treated with four-fold serial dilutions of brain tissue homogenates from a Tdp1+/+, Tdp1+/−, or Tdp1−/− mouse for 1 h, subjected to denaturing gel electrophoresis, and phosphorimaged. The percent conversion from the tyrosyl substrate to its phosphate product was calculated by densitometry.
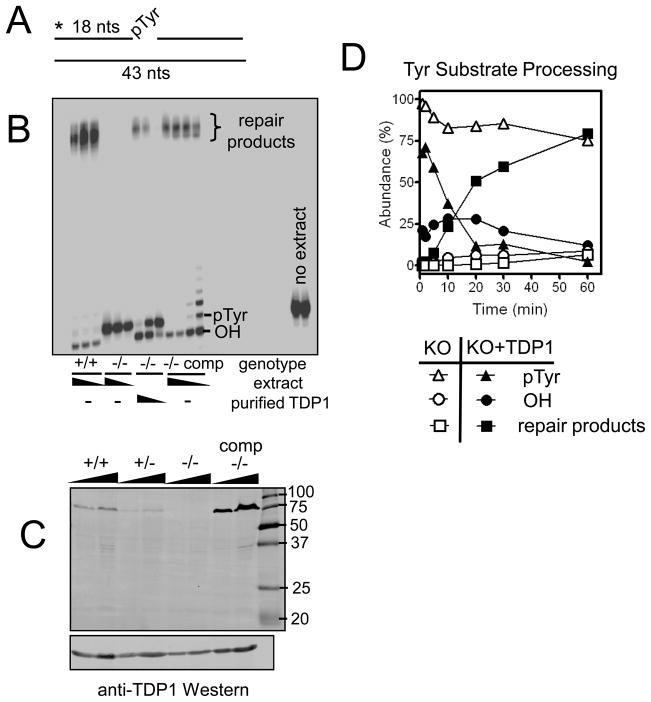
Figure 3. Fibroblast cell lines derived from embryos of each genotype possess corresponding capacities for 3′ tyrosyl processing and can be biochemically or genetically complemented.
Whole cell extracts from mouse embryonic fibroblast cell lines derived from Tdp1+/+ or Tdp1−/− mouse embryos were incubated with a substrate mimicking a SSB with a 3′-pTyr end modification. Processed and unprocessed forms of the substrate are indicated. (A) Diagram of 3′-pTyr SSB substrate; asterisk represents the radioactive 5′ tag on the 3′-pTyr 18-mer. There is no gap between the tyrosyl modification and the adjoining 25-mer. The substrate was treated with (B) 40, 20, and 10 μg of whole cell extracts from Tdp1+/+ and Tdp1−/− cell lines, and 40, 20, 10, and 5 μg of whole cell extract from the complemented Tdp1−/− cell line for 1 h. Lanes 7–9 contain reactions with 40 μg of Tdp1−/− extract, and 5, 1, and 0.2 ng of affinity-purified FLAG-TDP1, respectively. Presence of Tdp1 was also determined in MEF lysates from all three Tdp1 genotypes, as well as MEFs stably complemented by LV-FLAGhTDP1-Red transduction (“−/− comp”), by western blotting with anti-hTDP1 antibody (C). The gel contains 15 and 30 μL of each MEF lysate, and an identical membrane was loaded and probed with anti-β-actin as a loading control. The SSB substrate was treated from 1 to 60 min with 35 μg Tdp1−/− MEF extract, with (filled symbols) and without (open symbols) supplementation with 50 ng of purified wild-type His-TDP1. The percent conversion from the tyrosyl substrate to its hydroxyl intermediate and repair products was calculated by densitometry (D).
To assess activity toward PG nick and gap substrates, 20 μg of Tdp1+/+ and Tdp1−/− whole cell extract was incubated with 50 fmol of either substrate in 5 μL of reaction buffer containing 50 mM triethanolamine pH 7.5, 1 mM DTT, 100 μM dNTPs, and 1 mM ATP, and either 5 mM EDTA, 2, or 10 mM Mg(OAc)2. For the DSB substrates, extracts were incubated in 32 μL reaction buffer containing 50 mM triethanolamine pH 7.5, 1 mM DTT, 100 μM dNTPs, and 2 mM ATP, and 1.3 mM Mg(OAc)2 with substrate for 6 h, unless otherwise noted. Some Tdp1−/− extracts were supplemented with 25 or 125 ng of wildtype, SCAN1, or H493N TDP1 protein purified from E. coli as noted. Samples were deproteinized with proteinase K, extracted with phenol and chloroform, nucleic acids were precipitated, and then treated with BstXI and TaqI [18]. Samples were analyzed on a denaturing 20% polyacrylamide gel, wet gels were frozen and exposed on PhosphorImager screens for 1–2 days at −20°C. Phosphor images were developed with the Typhoon 9410 (General Electric) and analyzed with ImageQuant 3.3 software (Molecular Dynamics).
For fibroblast extract repair assays, unpaired two-tailed t tests were performed on triplicate data points using GraphPad Prism 3.0 (GraphPad Software, Inc). P values are indicated as; * < 0.05, ** < 0.01, ***<0.001.
2.6 Western blotting
Extracts used in the DNA repair assays were separated by SDS-PAGE and transferred to PVDF membranes. Membranes were exposed to anti-FLAG antibody from Sigma at a dilution of 1:1000, AbNova anti-human TDP1 (cat no. H00055775-A01) at a dilution of 1:2500, or Santa Cruz B-actin (cat no. sc-1615) antibody. Specific protein bands were detected using infrared emitting conjugated secondary antibodies; anti-mouse 680 Alexa (Invitrogen), or anti-goat IRDYE 800 (Rockland Immunochemicals), using the Odyssey infrared imaging system (LI-COR Biosciences).
3. RESULTS
3.1 Generation of Tdp1-deficient mice
To assess the function of Tdp1 in vivo and to provide a source of Tdp1-deficient cell lines, we disrupted the endogenous mouse Tdp1 gene in ES cells by deleting exons 6 and 7, which also causes a frame-shift in translation of sequences encoded in the downstream exons (Figure 1). The Tdp1 protein contains two HKD sequence motifs, encoded in exons 6–7 and 13–14, which together comprise the single active site of the enzyme [19]. Thus, the predicted truncated protein product of the targeted allele, which should contain only the first 253 amino acids of Tdp1 plus 10 out-of-frame amino acids, completely lacks the catalytic site. Our initial targeting vector also allowed for the generation of conditional Tdp1 knockout mice in which a loxP site has been inserted into both introns 5 and 7 (Figure 1, panel v), allowing for cell type-specific deletion of exons 6 and 7 upon interbreeding to lines of mice expressing Cre recombinase in specific tissues. However, only the conventional knockout mice (hereafter referred to as Tdp1−/− mice) were used in the studies described below. Tdp1−/− mice were born in the expected Mendelian ratio upon interbreeding of Tdp1+/− mice, indicating that Tdp1 deficiency does not result in embryonic or neonatal lethality. In addition, Tdp1−/− mice were fertile and had a normal lifespan. Reverse transcriptase-real-time PCR confirmed the presence of Tdp1 mRNA encoding exons 3–5 and exons 13–14, but not exons 6–7, in brain and liver tissue from Tdp1−/− mice (Supplemental Figure 1C).
3.2 Tdp1−/− mice are phenotypically and behaviorally indistinguishable from Tdp+/+ littermates
Visual inspection of the mice did not reveal any notable differences between genotypes. At each age, average weights were similar across genotype for each sex, as were average body temperature and reflex sensitivity to a thermal stimulus (Supplemental Table 1). Similarly, phenotypic differences were not seen in average stride length or in any measure within the five domains of the functional observation battery, including CNS excitability, CNS activity, muscle tone/equilibrium, autonomic effects or sensorimotor effects (data not shown). Figure 2A shows motor activity and latency to fall from the rotarod in male and female mice of each genotype and at each age. Statistical analyses of these data revealed that Tdp1−/− female mice (but not male mice) were significantly less active than Tdp1+/+ mice at the 3-month assessment [F(2,12)=4.1, p<0.05]. However, this difference was transient, as a difference in motor activity across genotype was not observed at later assessment times. In addition, although rotarod performance declined markedly with age, rotarod latencies were not significantly different across genotype at any age in either sex. Microscopic examination of brains sections did not reveal any overt differences between mice of any genotype. In particular, both granule cells and Purkinje cells in the cerebellum were present in equal numbers and appeared morphologically similar in brains of wild-type and Tdp1 knockout mice, both at 12 and 22 months of age (not shown). Altogether, under normal conditions, Tdp1−/− mice had no detectable behavioral phenotype.
3.3 Tdp1−/− cell extracts cannot process 3′-pTyr linkages
To confirm that Tdp1 activity was abolished by disruption of the Tdp1 allele, Tdp1 enzymatic activity was measured in tissue homogenates from Tdp1−/− mice (Figure 2). Brain (Figure 2B, Supplemental Figure 2B) and liver (Supplemental Figure 2C and D) tissue homogenates from a Tdp1+/+ and Tdp1+/− mouse efficiently converted a 3′-pTyr terminus on an 18 nucleotide oligomer (Figure 2B) to a 3′-phosphate, while liver and brain homogenates from a Tdp1−/− mouse failed to process the oligomers at any detectable level. This 18-base 5′-labeled 3′-pTyr substrate mimics the covalent DNA-protein linkage that would be formed between a tyrosine in TOP1 and DNA at sites of torsional stress; these linkages must be resolved and effectively rejoined by DNA ligase to prevent these sites from converting to DSBs. All the above reactions were performed in the presence of 5 mM EDTA, which prevents 3′-phosphate removal by PNKP, a magnesium-requiring enzyme.
To assess 3′-pTyr processing in the context of SSB repair, the nicked duplex shown in Figure 3A was constructed and incubated in whole-cell extracts of MEFs derived from mice of each genotype. Accurate repair of this break would require cleaving the tyrosine-DNA linkage, removing a phosphate group, and ligating the reformed 3′ hydroxyl group to the 5′ phosphate. Ligated repair products can be seen in samples treated with +/+ and +/− extracts, and these products display several distinct mobilities, possibly due to slight 3′ resection at the end of the duplex (Figure 3B; +/− extract shown in Supplemental Figure 3A). No such repair products were generated in −/− extracts; instead these samples showed only a single band corresponding to the unprocessed 3′-pTyr 18-mer. Reactions with the SSB substrate were performed in buffer containing 10 mM Mg++, sufficient for Ape1 and PNKP activity. Thus, the band migrating slightly faster than the untreated substrate in the +/+ and +/− reactions is likely a 3′-hydroxyl 18-mer, generated by the combined action of Tdp1 and PNKP.
To verify that the lack of processing in −/− MEFs was due to Tdp1 deficiency, the MEFs were stably transduced with a lentivirus expressing FLAG-tagged hTDP1 (Supplemental Figure 3B). As expected, only cells infected with the FLAG-hTDP1 vector expressed FLAG at the expected molecular weight of TDP1, uninfected cells or cells infected with control viruses (LVTHM-DsRed, LV-IRES-DSRed) gave no signal. These same FLAG-TDP1 infected cells were subsequently shown to overexpress TDP1 in comparison to MEFs of other genotypes (lanes ‘−/− comp’ in Figure 3C). Additionally, there was no evidence of any expression of a full-length or truncated Tdp1 protein being expressed from the targeted knockout allele in −/− MEFs. The ectopic TDP1 expression rescued both 3′ processing and formation of repair products from the tyrosyl-modified SSB substrate (Figure 3B). Lower concentrations of the complemented cell extract (5 and 10 μg of extract in a 10 μL reaction volume) produced bands that migrated just above the pTyr; these are likely the result of displacement synthesis after removal of the pTyr from the SSB substrate. Supplementing −/− MEF extracts with purified hTDP1 protein also effectively reinstated the ability to process and repair a 3′-pTyr SSB (Figure 3B). Quantitative analysis of a reaction timecourse for the supplemented extract showed that the hydroxyl intermediate appeared almost immediately, substantial repair product was visible within 10 min, and repair appeared complete at 60 min (Figure 3D). It is notable that under these conditions, a low level of slow processing and conversion to the repair product occurred in the unsupplemented −/− MEF extracts. This small amount of processing was seen in two independent experiments and may indicate an alternate, though less efficient, pathway of processing tyrosyl modifications in the context of SSBs. In summary, rescuing the TDP1 deficiency either by lentiviral infection or simply adding exogenous protein to cell lysates effectively reinstates TDP1-dependent 3′-pTyr processing.
Similar experiments were performed with duplexes that contained model free-radical-mediated SSBs, i.e., bearing 3′-PG and 5′-phosphate termini, either with or without a 1-base gap (Supplemental Figure 4). There was no deficiency in repair of PG-terminated SSBs in Tdp1−/− cells, presumably reflecting processing of these lesions by Ape1 [20]. However, the results also indicated that Tdp1 could process 3′ termini at these lesions when Ape1 activity was suppressed by Mg++ chelation (Supplemental Figure 4B and C).
3.4 Tdp1 is required for processing PG modifications on a subset of DNA DSBs
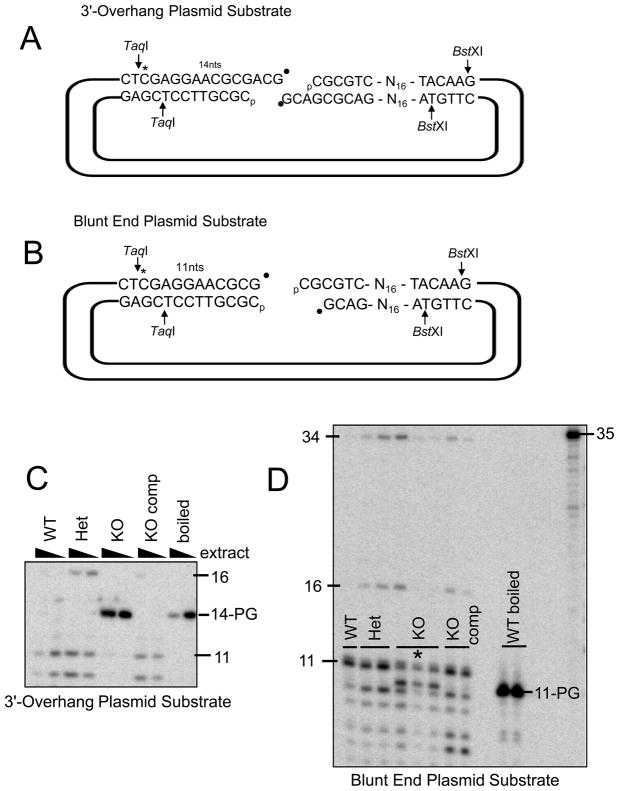
To assess whether Tdp1 status affects processing of PG-modified ends on DSBs, model DSBs were constructed (Figure 4, A and B). These substrates were designed to mimic breaks that would commonly result from DNA sugar damage induced by either bleomycin [9], neocarzinostatin, or radiation [8], in which the deoxyribose fractures, a base is lost, and a 3′-PG group remains. Accurate repair of a break with a 3′ overhang (Figure 4A) would require annealing the complementary CG sequences in each overhang, filling in the 1-base gap using the opposite overhang as a template, removal of the PG, and ligation. Substrates were constructed such that they were labeled with 32P either 14 (Figure 4A) or 11 (Figure 4B) bases from the 3′ termini. These substrates were incubated in whole-cell extracts and then digested with restriction enzymes, which released short fragments that were analyzed on sequencing gels. The proposed accurate repair would therefore generate fragments of 42 nucleotides in the case of the 3′ overhang DSB substrate, and 38 nucleotides in the case of the blunt-end DSB substrate.
Figure 4. Tdp1−/− fibroblasts are deficient in processing PG on DSBs.
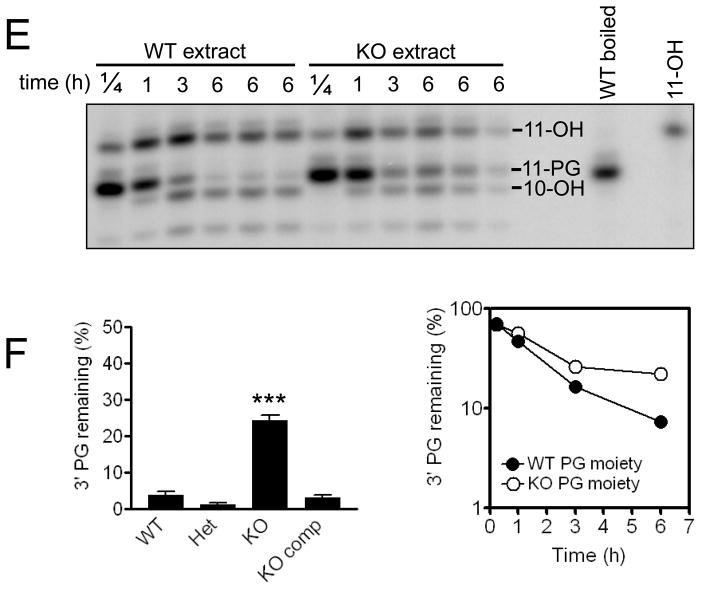
Plasmid substrates mimicking DSBs with either partially complementary 3′ overhang (A) or blunt (B) DNA ends bearing PG modifications were incubated with whole-cell extract (or boiled extract) from Tdp1+/+, Tdp1+/−, and Tdp1−/− and Tdp1comp MEF cell lines for 6 h. Reactions were deproteinized, nucleic acids were precipitated, digested with BstXI and TaqI, and subjected to denaturing gel electrophoresis (C and D). In (C), the overhang substrate was incubated with 2.5 or 5 μg extract per μL reaction volume. In (D), the blunt ended substrate was incubated with 5 μg (or 10 μg *) extract per uL reaction volume. In (D), rightmost lane contains radiolabeled marker of 35 bases. The blunt-end DSB substrate was also incubated for 15 min, 1 h, 3 h, or 6 h with 7 μg of Tdp1+/+ or Tdp1−/− cell extract per μL reaction volume, then deproteinized, precipitated, and digested (E). Rightmost lane contains radiolabeled marker of 11 bases. After a 6 h incubation of the 3′-PG blunt end DSB substrate with extract, the percent of PG remaining in the lane was calculated by densitometry (F). Graph on left includes data from 4 independent experiments, performed with multiple preparations of cell extract per genotype; error bars indicate standard error and *** indicates p<0.001. Rightmost graph shows quantification of (E), time course of PG processing with Tdp1+/+ or Tdp1−/− cell extract. The 6-h time point, conducted in triplicate, includes error bars that are contained within the symbols.
When extracts from +/+, +/−, −/− and −/− comp MEFs were incubated for 6 h with the 3′ overhang substrate, the +/+, +/−, and −/− comp extract all produced shorter products that represented processing and resection of the 3′-PG 14-mer (Figure 4C), and longer products of 16 and 34 nucleotides that represent repair of the DSB substrate (Figure 4C and 5B), whereas no processing or repair could be observed in the −/− extract. In contrast, when extracts of all aforementioned genotypes were incubated with a radiolabeled plasmid substrate constructed to mimic a blunt-ended DSB, reactions of all genotypes formed processed intermediates and repair products (Figure 4D). However, the −/− extract reactions uniquely exhibited a significant fraction of persistent unprocessed PG after 6 h of incubation, (Figure 4D – 4F), indicating a deficiency of DNA blunt-end processing in the −/− MEFs.
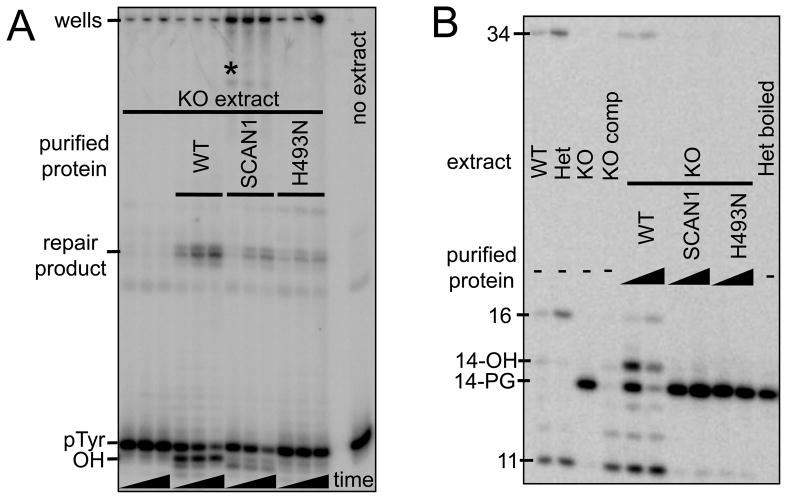
Figure 5. Complementation with either SCAN1 or H493N mutant Tdp1 fails to restore the wild-type phenotype for processing either pTyr or PG end modifications.
The pTyr substrate mimicking a SSB depicted in Figure 3A was treated with 30 μg of whole-cell extract from Tdp1+/+, Tdp1−/− MEFs for 5, 10, and 20 min (A). The substrate was also treated with 30 μg of Tdp1−/− MEF extract that had been supplemented with 50 ng of either wild type purified TDP1, the SCAN1 mutant form of TDP1, or the mutant H493N TDP1. Processed (OH), unprocessed (pTyr), and repaired forms of the substrate are indicated. The radioactivity remaining in the SCAN1 wells presumably represents an unresolved DNA-TDP1 adduct; bands running beneath the wells and above the repair bands may represent a proteolyzed form of the adduct (*). The substrate mimicking a DSB with partially complementary 3′ overhangs as depicted in Figure 4A was treated with 60 μg whole-cell extract from Tdp1+/+, Tdp1+/−, and Tdp1−/− and Tdp1comp MEFs for 6 h (B). The substrate was also treated with Tdp1−/− MEF extract that had been supplemented with either 25 ng (1X) or 125 ng (5X) of either wild type purified TDP1, the SCAN1 mutant form of TDP1, or the mutant H493N TDP1.
The longer repair product shown in Figure 5B was seen in three independent experiments with the 3′ overhang substrate and always migrated at approximately 34 bases, 8 bases shorter than the expected accurate repair product that dominates repair reactions in human cell extracts [14]. This result suggests that in MEF extracts a resection-based end joining pathway is utilized instead. Both the 3′ overhang and blunt-ended substrate can be converted by 3′ resection to an intermediate with self-complementary CGCG 5′-overhangs, the alignment of which would result in a 34-base repair product [14] containing an MluI cleavage site (ACGCGT). This scenario was confirmed by treating the DNA from these repair reactions with MluI, which completely eliminated the 34-base product, thus confirming its identity (data not shown).
Together, these results indicate that TDP1 is required for processing 3′-PG on overhangs, and also processes 3′-PG on blunt ends. In the absence of TDP1, other enzymes can process 3′-PG on blunt ends, but less efficiently than TDP1.
3.5 H493R (SCAN1) or H493N TDP1 cannot effectively rescue the deficits in processing either 3′-pTyr SSBs or 3′-PG DSBs
The mutant H493R TDP1 enzyme associated with SCAN1 has at least 25-fold lower activity than wild type, but it can under certain conditions cleave 3′-pTyr linkages and form a persistent (t1/2~13 min) TDP1-DNA adduct intermediate [21]. In yeast, it has been shown that under conditions of elevated TOP1, H432N TDP1 (the yeast equivalent of H493N human TDP1 mutation) confers greater cytotoxicity than SCAN1 TDP1 [22]. To examine the action of mutant TDP1 in the presence of other DSB/SSB repair proteins, wild-type, SCAN1, and H493N forms of human TDP1 were generated in bacteria and purified. Since deficits had been identified in both 3′-pTyr and 3′ overhang PG processing in Tdp1−/− extracts, these assays were repeated in order to compare the effects of SCAN1 TDP1 and H493N TDP1 to that of complete Tdp1 deficiency.
As shown previously in Figure 3B, supplementing −/− extract with wild-type TDP1 reinstates the processing and repair of the pTyr nick substrate depicted in Figure 3A. However, supplementing −/− extract with SCAN1 TDP1 leads to an accumulation of radioactive signal in the wells of the gel, consistent with the presence of a protein-DNA adduct too large to migrate into the gel (Figure 5A). Extract supplemented with SCAN1 TDP1 also shows more pTyr processing and more repair product than the unsupplemented −/− extract, and there is an additional novel band running beneath the wells and above the repair bands, which may represent a proteolyzed form of the protein(TDP1)-DNA adduct (denoted by the asterisk in Figure 5A). A smaller increase in the repair product is seen when −/− extract is supplemented with H493N TDP1, consistent with this less conservative amino acid change. However, there is little or no increase in the band representing 18-hydroxyl, the product of the removal of the pTyr group. Thus, the H493N mutant retains a low level of pTyr-processing activity but does not form a persistent DNA adduct like SCAN1 TDP1.
Supplementing −/− extract with wild type TDP1 also reinstates the processing and repair of the 3′ overhang PG DSB substrate, with the addition of more TDP1 resulting in greater conversion of the 14-PG to its processed intermediates, 14-hydroxyl, as well as repair products (Figure 5B). Supplementing −/− extract with either SCAN1 TDP1 or H493N TDP1 does not lead to the appearance of either processed intermediates or repair bands; as in the −/− extract by itself, the unprocessed 3′-PG persists for at least 6 h of incubation in the extract.
DISCUSSION
While there is little doubt that an H493R TDP1 mutation confers SCAN1 [1], it is still unclear how the mutation elicits symptoms of the disease, and why symptoms are largely neurological. In an attempt to address these questions, we generated a Tdp1 knockout mouse. However, extensive behavioral analysis of Tdp1−/− mice up to 12 months of age did not reveal any detectable deficits in experimental correlates of human SCAN1, such as grip strength (muscle weakness), stride length or rotarod latency (ataxia), and indeed did not show any behavioral differences from wild-type mice (Figure 2). A similar lack of overt behavioral symptoms was noted in two other independently derived Tdp1−/− mice, although in neither case were quantitative data reported [23,24]. In one of these studies [23], there was a progressive age-dependent reduction of cerebellar size in Tdp1−/− mice when compared to their wildtype littermates. When comparing brain mass and gross brain histology, no differences were observed between genotypes in our studies, corroborating the results of Hirano et al.. However, no attempt was made to quantify the cerebellar to whole brain ratio as was performed by Katyal et al., who noted small differences in brain mass, but also that cerebellar morphology and foliation was indistinguishable between genotypes. Katyal et al. also observed in vivo hypersensitivity of the Tdp1−/− mice to the effects of the TopI inhibitor topotecan, displaying significant weight loss due to intestinal progenitor cell hypersensitivity after six daily doses [23]. Hirano and colleagues found similar in vivo Tdp1−/− hypersensitivity to both camptothecin and the camptothecin derivative topotecan, and in both cases the sensitivity required repeated doses in a relatively short amount of time to manifest [24]. In either study, even lethal treatments with TopI inhibitors still failed to elicit any SCAN1-like symptoms. Indeed, rapidly proliferating cells in the gut, liver, and spleen, rather than postmitotic neurons, appeared to be particularly sensitive. Thus, overall, the Tdp1−/− mouse phenotype bears little resemblance to human SCAN1.
While fundamental differences between human and mouse phenotypes of a given gene are not unusual, it is possible that rather than the simple loss of TDP1 function, the symptoms of SCAN1 are dependent on the unique features of the H493R mutant enzyme. In particular, it has been proposed that the residual activity of this mutant enzyme may be sufficient to process many if not most Top1cc that fail to religate, converting them to persistent TDP1-DNA adducts that may be more toxic and/or less amenable to alternative repair pathways than the initial TopI-DNA adducts [21]. On the other hand, there is evidence that the initial TopI-DNA adducts are also more persistent in SCAN1 than in normal cells, and that the difference is comparable to the overall deficit in SSB repair [4]. In either scenario, the neuronal-specific phenotype can be explained by the paucity of alternative repair pathways in terminally-differentiated, non-replicating neuronal cells, the high levels of oxidative stress in neuronal tissue [25], and elevated levels of transcription in neuronal cells. Furthermore, it has been shown that in yeast the toxicity of SCAN1 TDP1 is dependent upon elevated levels of Top1 [22] and that Top1 is particularly highly expressed in Purkinje neurons of adolescence human brain [26,27]. Although there have been to date no autopsy studies on SCAN1 patients, other neurodegenerative disorders that feature ataxia among their symptoms have specific Purkinje neuron pathology [28,29]. Thus, the ataxia seen in SCAN1 patients may be directly caused by Purkinje neuron pathology that results from the interaction of SCAN1 TDP1 in the presence of high levels of Top1.
In vitro complementation studies showed that extracts from Tdp1−/− mouse fibroblasts largely could not process a tyrosyl-DNA substrate; these results correlated with previous observations made in extracts from SCAN1 lymphoblasts [30], astrocyte and cerebellar extracts [23], and mouse neurospheres [24]. When supplemented with wild-type TDP1, processing of the tyrosyl-DNA substrate was reinstated, as also shown by Katyal et al. [23]. The generation of products of extremely low mobility in Tdp1−/− extracts supplemented with TDP1-H493R (Figure 5A), similar to the results seen by Interthal, et al. [21], under conditions where the wild-type enzyme promotes SSB repair, suggests that substantial amounts of persistent TDP1-DNA adducts would be formed during attempted repair of TopI lesions in SCAN1 cells. Cosedimentation of TDP1 protein with DNA in CsCl-fractionated extracts of SCAN1 but not normal fibroblasts [24] suggests that such complexes form in vivo.
Extracts of Tdp1−/− cells are also completely deficient in processing of protruding 3′-PG DSB termini, and recombinant TDP1 can rescue that deficiency (Figure 5). Previously, we showed similar lack of processing of PG-terminated 3′ overhangs in extracts of TDP1-mutant human SCAN1 cells [6], but due to low ligation efficiency in those extracts, generation of ligated products was not demonstrated. However, in contrast to the tyrosyl lesion, there is no evidence that the mutant enzyme can form covalent adducts at 3′-PG termini (Figure 5). Thus, presumably because the 3′-PG lesion is a much less favorable substrate than a 3′-pTyr [5], SCAN1 TDP1 displays a hypomorphic or perhaps even a null phenotype in PG processing. Tdp1−/− extracts also show partial deficiency in processing blunt 3′-PG DSBs, but no deficiency in processing 3′-PG SSBs. Inasmuch as bleomycin induces almost exclusively 3′-PG SSBs and (predominantly blunt-ended) 3′-PG DSBs [31], it appears likely that unrepaired DSBs are responsible for the previously reported bleomycin sensitivity of Tdp1−/− mice and Tdp1−/− MEFs [24]. While neither Tdp1−/− astrocytes, Tdp1−/− cerebellar granule cells [23], nor SCAN1 lymphoblasts [32] show any detectable deficit in repair of radiation-induced DSBs, a recent mass spectral analysis suggests that the fraction of radiation-induced breaks with 3′-PG termini may be as small as 10% [33], much lower than the previous estimate of ~50% [34]. Although SCAN1 cells have an apparent defect in repair of radiation-induced SSB [32], this deficit may reflect persistence of TopI-mediated lesions formed at sites of base damage [35,36], rather than failure to process direct free radical-mediated, PG-terminated SSBs.
At present, the most likely etiology of SCAN1 is that oxidative lesions in DNA promote transcription-mediated formation of trapped TopIcc that are then converted to persistent SSBs with a unique covalent 3′ linkage between DNA and the TDP1-H493R enzyme. These lesions could lead to cytotoxicity either by promoting apoptosis or simply by inhibiting transcription. If this is the case, however, the SCAN1 phenotype may not be very informative with respect to the normal function of human TDP1. The finding that a mouse Tdp1−/− phenotype is only seen under conditions of severe genotoxic stress suggests that there are redundant repair pathways for the major functions of Tdp1, as is likely the case for the human enzyme as well.
Supplementary Material
Acknowledgments
This work was supported by NIH R01 AG023783, a pilot project grant from the Massey Cancer Center, NIH P01 CA72955, NIH R01 CA40615, and NIH P30 CA16059, which provides support to both the Massey Cancer Center Molecular Biology Core and the Transgenic/Knockout Mouse Core. We thank Kimberly Stratton of the Molecular Biology Core for construction of the Tdp1 targeting vector, Heju Zhang of the Transgenic/Knockout Mouse Core for knock-out mouse generation, and Mary Tokarz for behavioral testing.
Abbreviations
- DSB
double-strand break
- HRR
homologous recombination repair
- IR
ionizing radiation
- MEFs
mouse embryonic fibroblasts
- NHEJ
Non-Homologous End-Joining
- PG
phosphoglycolate
- pTyr
phosphotyrosyl
- SSB
single-strand break
- TDP1
tyrosyl-DNA phosphodiesterase
- TopI
Topoisomerase I
- TopIcc
Topoisomerase I cleavage complex
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, Stockton DW, Lupski JR. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 2.Yang SW, Burgin AB, Jr, Huizenga BN, Robertson CA, Yao KC, Nash HA. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc Natl Acad Sci U S A. 1996;93:11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 4.Miao ZH, Agama K, Sordet O, Povirk L, Kohn KW, Pommier Y. DNA Repair. Vol. 5. Amst: 2006. Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes; pp. 1489–1494. [DOI] [PubMed] [Google Scholar]
- 5.Inamdar KV, Pouliot JJ, Zhou T, Lees-Miller SP, Rasouli-Nia A, Povirk LF. Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J Biol Chem. 2002;277:27162–27168. doi: 10.1074/jbc.M204688200. [DOI] [PubMed] [Google Scholar]
- 6.Zhou T, Lee JW, Tatavarthi H, Lupski JR, Valerie K, Povirk LF. Deficiency in 3′-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1) Nucleic Acids Res. 2005;33:289–297. doi: 10.1093/nar/gki170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertoncini CR, Meneghini R. DNA strand breaks produced by oxidative stress in mammalian cells exhibit 3′-phosphoglycolate termini. Nucleic Acids Res. 1995;23:2995–3002. doi: 10.1093/nar/23.15.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henner WD, Rodriguez LO, Hecht SM, Haseltine WA. Gamma ray induced deoxyribonucleic acid strand breaks. 3′ glycolate termini. J Biol Chem. 1983;258:711–713. [PubMed] [Google Scholar]
- 9.Povirk LF. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: Bleomycin, neocarzinostatin and other enediynes. Mutat Res. 1996;355:71–89. doi: 10.1016/0027-5107(96)00023-1. [DOI] [PubMed] [Google Scholar]
- 10.U.S. Environmental Protection Agency. Developmental neurotoxicity study, series 83–6, addendum 10 (neurotoxicity), subdivision F: Hazard evaluation: Human and domestic animals. 1991. EPA Publication No. 540/09-91-123; NTIS Publication No. PB 91–154617. [Google Scholar]
- 11.Chen S, Hannis JC, Flora JW, Muddiman DC, Charles K, Yu Y, Povirk LF. Homogeneous preparations of 3′-phosphoglycolate-terminated oligodeoxynucleotides from bleomycin-treated DNA as verified by electrospray ionization fourier transform ion cyclotron resonance mass spectrometry. Anal Biochem. 2001;289:274–280. doi: 10.1006/abio.2000.4936. [DOI] [PubMed] [Google Scholar]
- 12.Povirk LF, Zhou T, Zhou R, Cowan MJ, Yannone SM. Processing of 3′-phosphoglycolate-terminated DNA double strand breaks by artemis nuclease. J Biol Chem. 2007;282:3547–3558. doi: 10.1074/jbc.M607745200. [DOI] [PubMed] [Google Scholar]
- 13.Suh D, Wilson DM, 3rd, Povirk LF. 3′-phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res. 1997;25:2495–2500. doi: 10.1093/nar/25.12.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S, Inamdar KV, Pfeiffer P, Feldmann E, Hannah MF, Yu Y, Lee JW, Zhou T, Lees-Miller SP, Povirk LF. Accurate in vitro end joining of a DNA double strand break with partially cohesive 3′-overhangs and 3′-phosphoglycolate termini: Effect of ku on repair fidelity. J Biol Chem. 2001;276:24323–24330. doi: 10.1074/jbc.M010544200. [DOI] [PubMed] [Google Scholar]
- 15.Gu XY, Bennett RA, Povirk LF. End-joining of free radical-mediated DNA double-strand breaks in vitro is blocked by the kinase inhibitor wortmannin at a step preceding removal of damaged 3′ termini. J Biol Chem. 1996;271:19660–19663. doi: 10.1074/jbc.271.33.19660. [DOI] [PubMed] [Google Scholar]
- 16.Baumann P, West SC. DNA end-joining catalyzed by human cell-free extracts. Proc Natl Acad Sci U S A. 1998;95:14066–14070. doi: 10.1073/pnas.95.24.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Houten B, Chen Y, Nicklas JA, Rainville IR, O’Neill JP. Development of long PCR techniques to analyze deletion mutations of the human hprt gene. Mutat Res. 1998;403:171–175. doi: 10.1016/s0027-5107(98)00076-1. [DOI] [PubMed] [Google Scholar]
- 18.Povirk LF, Zhou RZ, Ramsden DA, Lees-Miller SP, Valerie K. Phosphorylation in the serine/threonine 2609–2647 cluster promotes but is not essential for DNA-dependent protein kinase-mediated nonhomologous end joining in human whole-cell extracts. Nucleic Acids Res. 2007;35:3869–3878. doi: 10.1093/nar/gkm339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies DR, Interthal H, Champoux JJ, Hol WG. The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1. Structure. 2002;10:237–248. doi: 10.1016/s0969-2126(02)00707-4. [DOI] [PubMed] [Google Scholar]
- 20.Parsons JL, Dianova II, Dianov GL. APE1 is the major 3′-phosphoglycolate activity in human cell extracts. Nucleic Acids Res. 2004;32:3531–3536. doi: 10.1093/nar/gkh676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, Champoux JJ. SCAN1 mutant Tdp1 accumulates the enzyme--DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 2005;24:2224–2233. doi: 10.1038/sj.emboj.7600694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He X, van Waardenburg RC, Babaoglu K, Price AC, Nitiss KC, Nitiss JL, Bjornsti MA, White SW. Mutation of a conserved active site residue converts tyrosyl-DNA phosphodiesterase I into a DNA topoisomerase I-dependent poison. J Mol Biol. 2007;372:1070–1081. doi: 10.1016/j.jmb.2007.07.055. [DOI] [PubMed] [Google Scholar]
- 23.Katyal S, el-Khamisy SF, Russell HR, Li Y, Ju L, Caldecott KW, McKinnon PJ. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007;26:4720–4731. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirano R, Interthal H, Huang C, Nakamura T, Deguchi K, Choi K, Bhattacharjee MB, Arimura K, Umehara F, Izumo S, Northrop JL, Salih MA, Inoue K, Armstrong DL, Champoux JJ, Takashima H, Boerkoel CF. Spinocerebellar ataxia with axonal neuropathy: Consequence of a Tdp1 recessive neomorphic mutation? EMBO J. 2007;26:4732–4743. doi: 10.1038/sj.emboj.7601885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caldecott KW. DNA single-strand break repair and spinocerebellar ataxia. Cell. 2003;112:7–10. doi: 10.1016/s0092-8674(02)01247-3. [DOI] [PubMed] [Google Scholar]
- 26.Holden JA, Rahn MP, Jolles CJ, Vorobyev SV, Bronstein IB. Immunohistochemical detection of DNA topoisomerase I in formalin fixed, paraffin wax embedded normal tissues and in ovarian carcinomas. Mol Pathol. 1997;50:247–253. doi: 10.1136/mp.50.5.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorodetsky E, Calkins S, Ahn J, Brooks PJ. DNA Repair. Vol. 6. Amst: 2007. ATM, the Mre11/Rad50/Nbs1 complex, and topoisomerase I are concentrated in the nucleus of purkinje neurons in the juvenile human brain; pp. 1698–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Q, Hashizume Y, Yoshida M, Wang Y, Goto Y, Mitsuma N, Ishikawa K, Mizusawa H. Morphological purkinje cell changes in spinocerebellar ataxia type 6. Acta Neuropathol. 2000;100:371–376. doi: 10.1007/s004010000201. [DOI] [PubMed] [Google Scholar]
- 29.Sugawara M, Wada C, Okawa S, Kobayashi M, Sageshima M, Imota T, Toyoshima I. Purkinje cell loss in the cerebellar flocculus in patients with ataxia with ocular motor apraxia type 1/early-onset ataxia with ocular motor apraxia and hypoalbuminemia. Eur Neurol. 2008;59:18–23. doi: 10.1159/000109256. [DOI] [PubMed] [Google Scholar]
- 30.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 31.Povirk LF, Han YH, Steighner RJ. Structure of bleomycin-induced DNA double-strand breaks: Predominance of blunt ends and single-base 5′ extensions. Biochemistry. 1989;28:5808–5814. doi: 10.1021/bi00440a016. [DOI] [PubMed] [Google Scholar]
- 32.El-Khamisy SF, Hartsuiker E, Caldecott KW. DNA Repair. Vol. 6. Amst: 2007. TDP1 facilitates repair of ionizing radiation-induced DNA single-strand breaks; pp. 1485–1495. [DOI] [PubMed] [Google Scholar]
- 33.Chen B, Zhou X, Taghizadeh K, Chen J, Stubbe J, Dedon PC. GC/MS methods to quantify the 2-deoxypentos-4-ulose and 3′-phosphoglycolate pathways of 4′ oxidation of 2-deoxyribose in DNA: Application to DNA damage produced by gamma radiation and bleomycin. Chem Res Toxicol. 2007;20:1701–1708. doi: 10.1021/tx700164y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henner WD, Grunberg SM, Haseltine WA. Sites and structure of gamma radiation-induced DNA strand breaks. J Biol Chem. 1982;257:11750–11754. [PubMed] [Google Scholar]
- 35.Daroui P, Desai SD, Li TK, Liu AA, Liu LF. Hydrogen peroxide induces topoisomerase I-mediated DNA damage and cell death. J Biol Chem. 2004;279:14587–14594. doi: 10.1074/jbc.M311370200. [DOI] [PubMed] [Google Scholar]
- 36.Pourquier P, Ueng LM, Fertala J, Wang D, Park HJ, Essigmann JM, Bjornsti MA, Pommier Y. Induction of reversible complexes between eukaryotic DNA topoisomerase I and DNA-containing oxidative base damages. 7, 8-dihydro-8-oxoguanine and 5-hydroxycytosine. J Biol Chem. 1999;274:8516–8523. doi: 10.1074/jbc.274.13.8516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.