Abstract
The role played by long chain fatty acids (LCFA) in promoting energy expenditure is confounded by their dual function as substrates for oxidation and as putative classic uncouplers of mitochondrial oxidative phosphorylation. LCFA analogs of the MEDICA (MEthyl-substituted DICarboxylic Acids) series are neither esterified into lipids nor β-oxidized and may thus simulate the uncoupling activity of natural LCFA in vivo, independently of their substrate role. Treatment of rats or cell lines with MEDICA analogs results in low conductance gating of the mitochondrial permeability transition pore (PTP), with 10–40% decrease in the inner mitochondrial membrane potential. PTP gating by MEDICA analogs is accounted for by inhibition of Raf1 expression and kinase activity, resulting in suppression of the MAPK/RSK1 and the adenylate cyclase/PKA transduction pathways. Suppression of RSK1 and PKA results in a decrease in phosphorylation of their respective downstream targets, Bad(Ser-112) and Bad(Ser-155). Decrease in Bad(Ser-112, Ser-155) phosphorylation results in increased binding of Bad to mitochondrial Bcl2 with concomitant displacement of Bax, followed by PTP gating induced by free mitochondrial Bax. Low conductance PTP gating by LCFA/MEDICA may account for their thyromimetic calorigenic activity in vivo.
Keywords: Bioenergetics, Diseases/Obesity, Lipid/Fatty Acid, Membrane/Energetics, Metabolism/Fatty Acid, Metabolism/Metabolic Syndrome, Phosphorylation/Kinases/Serine-Threonine, Signal Transduction
Introduction
Uncoupling of mitochondrial oxidative phosphorylation by long chain fatty acids (LCFA)3 has been extensively verified in isolated mitochondria and reported to result in increased mitochondrial state 4 respiration and decreased P/O ratio of state 3. Mitochondrial uncoupling by LCFA has been ascribed to their protonophoric activity, mediated by fatty acids crossing the mitochondrial inner membrane in their protonated form followed by efflux of the fatty acid anion through anion channels (Ref. 1; reviewed in Ref. 2). Mitochondrial uncoupling by LCFA in isolated mitochondria has been further reported to be partly abrogated by cyclosporin A (CsA), indicating gating of the mitochondrial permeability transition pore (PTP) (Refs. 3, 4 and references therein). Low conductance PTP gating (LC-PTP) may allow for passage of ions <300 Da across the inner mitochondrial membrane, resulting in restrained uncoupling and thermogenesis (4–8), while high conductance PTP gating (HC-PTP) may result in passage of solutes up to 1500 Da, mitochondrial swelling, cytochrome c efflux, formation of a functional apoptosome, and apoptosis (9–11).
In contrast to the well reported uncoupling effect of LCFA in isolated mitochondria, their uncoupling activity in vivo is still unresolved. Fatty acids are well known to stimulate respiration of isolated hepatocytes and of perfused liver and heart. However, it remains disputed whether stimulation of respiration by fatty acids in vivo is accounted for by their intrinsic mitochondrial uncoupling activity, or due to their availability as substrates for oxidation, combined with stimulation of extramitochondrial ATP-consuming reactions. Indeed, increased oxygen consumption induced by fatty acids in the perfused liver or in isolated liver cells has been reported to be essentially or partly eliminated upon blocking oxidative phosphorylation by added oligomycin or atractyloside, thus refuting classic mitochondrial uncoupling (12–15). Furthermore, inner mitochondrial membrane potential measured in isolated hepatocytes, or phosphorylation potential measured in situ in the perfused heart were found to be unaffected or rather increased by added fatty acids. Hence, mitochondrial uncoupling by LCFA in vivo is still unresolved, being confounded by their dual role as substrates for oxidation and as putative genuine uncouplers of mitochondrial oxidative phosphorylation.
Hexadecanedioic acids, tetramethyl-substituted in the αα′ or ββ′ carbons (MEDICA (M) analogs Mαα or Mββ (16): HOOC-C(α′)-C(β′)-(CH2)10-C(β)-C(α)-COOH) may dissociate between the substrate role and the putative uncoupling activity of LCFA. MEDICA analogs may be thioesterified endogenously into their respective mono-acyl-CoA thioesters, as verified both in vivo and in cultured cells (17). However, MEDICA analogs are not esterified into lipids, whereas the methyl substitutions at the αα′ or ββ′ positions block β-oxidation. ATP-dependent CoA thioesterification of MEDICA analogs to yield MEDICA-CoA does not result in sequestration of cellular CoA and does not limit CoA thioesterification of endogenous LCFA, as previously verified by profiling the content of liver acyl-CoAs in animals and cell cultures (17). Analogs of the MEDICA series and their respective CoA thioesters may thus simulate the in vivo mode of action of natural LCFA under conditions where respiration of the putative uncoupler does not confound its inherent uncoupling activity. Indeed, treatment of lean rats with MEDICA analogs results in pronounced increase in oxygen consumption accompanied by decrease in liver mitochondrial phosphate potential and cytosolic redox potential, thus reflecting mitochondrial uncoupling in vivo (18). Furthermore, treatment of obese leptin receptor-deficient rats (e.g. Zucker, cp/cp) with MEDICA analogs results in increase in oxygen consumption and food consumption with concomitant decrease in weight gain, implying increased total body energy expenditure (19, 20).
The uncoupling mode of action of MEDICA analogs has previously been studied in isolated liver and heart mitochondria as well as in freshly isolated hepatocytes and adipocytes. Under Ca+2-free conditions, MEDICA analogs induced a saturable, oligomycin-insensitive, atractylate-sensitive, 30% decrease in basal (state 4), succinate-driven (state 3), or ATP-driven mitochondrial membrane potential, proton gradient and proton motive force, with concomitant increase in oxygen consumption (21, 22). Similarly, MEDICA analogs were found to induce uncoupling protein 1 (UCP1)-mediated increase in oxygen consumption of cultured brown fat adipocytes (23). Furthermore, in line with PTP gating by LCFA (reviewed in Refs. 3, 4 and references therein), MEDICA analogs were found to induce Ca+2-dependent, CsA-sensitive decrease in mitochondrial membrane potential in isolated mitochondria, implying PTP gating by MEDICA analogs in vitro (22). In this report, we have dissected the mode of action of MEDICA analogs in inducing PTP gating in vivo. PTP gating by MEDICA analogs in vivo was found to be apparently similar to that of thyroid hormone (T3) (24, 25).
EXPERIMENTAL PROCEDURES
Animals
Male albino rats, weighing 150–180 × g, were fed a standard laboratory chow diet (diet 19520, Koffolk, Israel). Animals were treated by gavage once daily with MEDICA analogs suspended in 1% carboxyl methyl cellulose, as indicated. Control animals were treated with the vehicle only. Fed animals from all groups were killed between 900 a.m. and 1100 a.m. by injection of 0.1 ml/100 × g of body weight of a mixture containing 85 mg/ml ketamine and 3 mg/ml xylazine. Animal care and experimental procedures were in accordance with guidelines of the accredited animal ethics committee of the Hebrew University.
Cultured Cells
Jurkat cells were cultured in RPMI 1640, 10 mm HEPES, 2.2 mm glutamine, 100 units of penicillin-streptomycin/ml, supplemented with 10% fetal calf serum (Biological Industries, Beit Haemek, Israel). Unless otherwise specified, MEDICA effects in Jurkat cells were verified by culturing the cells with 150 μm Mαα or Mββ for 24 h. One-half volume of the Jurkat culture medium was replaced every other day. HeLa and COS-1 cells were cultured in Dulbecco's modified Eagle's medium, 1.1 mm glutamine, 100 units of penicillin/ml, and 0.1 mg of streptomycin/ml medium, supplemented with 10% fetal calf serum (Biological Industries). Unless otherwise specified, MEDICA effects in HeLa or COS-1 cells were verified by culturing the cells with 250 μm Mαα or Mββ for 24 h.
Mitochondria Isolation
Rat liver mitochondria were isolated as previously described (22). Livers were homogenized in buffer A, containing 300 mm sucrose, 20 mm Tris-HCl (pH 7.4), 2 mm EGTA, 0.1 μg/ml phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 0.7 μg/ml pepstatin, 200 μm sodium orthovanadate, and 1 mm β-glycerol phosphate, followed by centrifugation at 1,100 × g for 10 min at 4 °C. The supernatant was filtered through gauze and centrifuged at 1,100 × g for 10 min at 4 °C, followed by precipitating the crude mitochondrial fraction at 10,800 × g for 10 min at 4 °C. The mitochondrial fraction was resuspended in buffer A and re-precipitated.
For isolating mitochondria of Jurkat cells, 60 × 106 cells were harvested by centrifugation at 1,000 × g for 2 min at 4 °C. Cell pellets were washed twice with cold phosphate-buffered saline and resuspended in 1 ml of buffer B, containing 250 mm sucrose, 20 mm HEPES-KOH (pH 7.4), 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 200 μm sodium orthovanadate, 40 nm bis-peroxo(1,10-phenanthroline)oxovanadate(V) (Calbiochem #203695), 0.1 μg/ml okadaic acid, and 1 μg/ml protease inhibitor mix (Sigma-Aldrich). The cells were incubated on ice for 5 min and then homogenized on ice using Teflon pestle. The homogenate was centrifuged at 750 × g for 10 min at 4 °C to remove nuclei and cell fragments, followed by 10,000 × g for 20 min at 4 °C to yield the crude mitochondrial fraction.
Mitochondria and Cell Extracts
Mitochondrial extracts were prepared by dissolving respective mitochondrial fractions in lysis buffer containing 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1% Triton X-100, 60 mm octyl-β-d-glucopyranoside (O-8001, Sigma-Aldrich), 200 μm sodium orthovanadate, 50 mm NaF, 1 mm phenylmethylsulfonyl fluoride, 0.1 μg/ml okadaic acid, 40 nm bis-peroxo(1,10-phenanthroline)oxovanadate(V), 1 mm β-glycerol phosphate, and 1.0 μg/ml protease inhibitor mix (Sigma-Aldrich). Total cellular extracts were prepared by homogenizing Jurkat (1–3 × 107 cells/sample) or liver samples in 3 vol of lysis buffer by glass homogenizer or Polytron (Brinkmann Instruments, Westbury, NY), respectively. Following incubation in lysis buffer for 30 min at 4 °C, respective lysates were cleared by centrifugation at 15,000 × g for 10 min, and the supernatants were kept at (−70) °C. Protein content of cellular and mitochondrial extracts was determined by the BCA protein assay (23225, Pierce).
Mitochondrial Bcl2-Bad Heterodimer Analysis
Mitochondrial Bcl2-Bad heterodimers were analyzed as described by Gross (26) and Mikhailov (27) with slight modifications. Briefly, a 1-mg sample of liver or Jurkat mitochondria was dissolved in 100 μl of modified buffer A (20 mm HEPES (pH 7.4) replacing Tris-HCl) or in buffer B, respectively, supplemented with 2% CHAPS and 20 mm disuccinimidyl suberate (21655, Pierce). Samples were incubated with gentle shaking at room temperature for 30 min followed by blocking the cross-linking reaction by adding Tris-HCl (pH 7.5) buffer to a final concentration of 40 mm. Samples were further incubated for additional 15 min, cleared by centrifugation at 12,000 × g for 10 min at 4 °C, and subjected to SDS-PAGE under reducing conditions followed by Western blotting.
Western Blot Analysis
Equal amounts of protein were used in each lane for Western blot analysis. Proteins were resolved by using 7–12.5% SDS-PAGE under reducing conditions (28) and transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, MA) or cellulose nitrate membranes (Schleicher & Schuell, Dassel, Germany). Blots were probed with the indicated first antibody, followed by horseradish peroxidase-labeled second antibody. Bands were analyzed by ImageQuant software (Molecular Devices, Sunnyvale, CA). Cross-linked Bcl2-Bad heterodimer (49 kDa) of mitochondrial lysates was analyzed by reacting respective blots with anti-Bad antibody, followed by stripping with 4 m guaninethiocyanate and reacting with anti-Bcl2 antibody.
Mitochondrial Membrane Potential (ΔΨ)
Jurkat cells (1 × 106 cells per sample) were incubated in the dark at 37 °C for 60 min in RPMI 1640 medium containing 25 nm tetramethylrhodamine methylester perchlorate (TMRM, Molecular Probes Inc., Junction City, OR). Following incubation with TMRM the cells were centrifuged at 1,000 × g for 2 min, the pellet was resuspended in 400 μl of phosphate-buffered saline and then immediately analyzed by flow cytometric scanning (FACS, FL-2 setting, 20,000 events/analysis in triplicates) using CellQuest software (BD Biosciences, Mountain View, CA). Uncoupling by 40 μm carbonyl cyanide m-chlorophenylhydrazone was used as positive control. Unless otherwise indicated, effects of CsA or Sanglifherin A (SfA) on mitochondrial membrane potential were evaluated by adding them with the TMRM dye 1 h prior to measuring mitochondrial TMRM.
Cellular Apoptosis
Jurkat cells (3 × 105 cells) were incubated at 37 °C for 60 min in RPMI 1640 medium containing 10 μm CaspACETM FITC-VAD-FMK (G746, Promega) with additions as indicated. Following incubation with FITC-VAD-FMK the cells were pelleted, rinsed with phosphate-buffered saline (1000 × g for 2 min), resuspended in 300 μl of phosphate-buffered saline containing 1 μg of propidium iodide (Sigma), and subjected to fluorescence measurement within the following 10 min. Cellular green (FL1; FITC-VAD-FMK) and red (FL3; propidium iodide) fluorescence was measured by FACScan (BD Biosciences) and analyzed using CellQuest software (BD Biosciences).
cAMP
COS-1 cells (2 × 105 cells/well in 24-well plates) or Jurkat cells (5–7 × 105 cells/ml in T-25 flasks) were incubated with additions as indicated. cAMP was determined using an enzyme immunoassay kit (RPN225, Amersham Biosciences) according to manufacturer instructions.
Transient Transfection
COS-1 cells, cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, were transfected with pEGFP-Raf-1 or pEGFP-GFP expression vectors (29) (T. Balla, Bethesda, MD), using TransIT-LT1 transfection reagent (#MIR23090, Mirus Bio LLC). Following 6 h of transfection, the medium was replaced with a fresh one, and the cells were incubated for an additional 18 h to allow for the expression of transfected proteins. Transfected cells were further incubated for 24 h in the presence of additions as indicated.
Real-time PCR
Total RNA was prepared using the TRI reagent (Sigma) according to manufacturer instructions. First strand cDNA used as template was synthesized by reverse transcription using oligo(dT) or random hexamers mix as primer and the Reverse-iTMAX First Strand Kit (ABgene, Epson, UK). Raf1, CHOP, BiP, and GADD34 expression normalized by S18 or tubulin expression was quantified by real-time PCR (Rotor Gene RG-3000A, Australia) using SYBER green MasterMix (Absolute Syber Green ROX Mix, ABgene, Epson, UK) and the following primers: Human Raf1, forward (5′-TTTCCTGGATCATGTTCCCCT-3′) and reverse (5′-ACTTTGGTGCTACAGTGCTCA-3′); human CHOP, forward (5′-CAGAACCAGCAGAGGTCACA-3′) and reverse (5′-AGCTGTGCCACTTTCCTTTC-3′); human BiP, forward (5′-CATCACGCCGTCCTATGTCG-3′) and reverse (5′-CGTCAAAGACCGTGTTCTCG-3′); human GADD34, forward (5′-TGAAAACCAGCAGTTCCCTTC-3′) and reverse (5′-CCATCTGCAAATTGACTTCCCTG-3′); 18 S rRNA, forward (5′-GATGGGCGGCGGAAAATAG-3′) and reverse (5′-GCGTGGATTCTGCATAATGGT-3′); and tubulin, forward (5′-TAGCAGAGATCACCAATGCC-3′) and reverse (5′-GGCAGCAAGCCATGTATTTA-3′).
Materials and Methods
Jurkat cells stably transfected with pEF-puro-Bcl2 expression plasmid or with the respective empty vector (30) were from G. Hacker (Technische Universitat, Munich, Germany). Rabbit anti-rat and anti-human Bcl2 (sc-492), rabbit anti-human Raf1 (sc-227), rabbit anti-human ERK (sc-93), and mouse anti-human P-ERK1/2(Y204) (sc-7383) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-human P-Bcl2(S70) (#2871), rabbit anti-human Bad (#9292), rabbit anti-human P-Bad(Ser-155) (#9297), rabbit anti-human P-Bad(Ser-112) (#9291), rabbit anti-human PARP (#9542), and rabbit anti-human P-Raf1(S338) (#9427) antibodies were from Cell Signaling Technology (Danvers, MA). Mouse monoclonal anti-α-tubulin antibody (T6074) was from Sigma-Aldrich. Rabbit anti-rat Bax antibody was from Dr. Atan Gross (Weizmann Institute of Science, Rehovot, Israel). Mouse monoclonal anti-rat Bak antibody (AM04) was from Oncogene Research Products (San Diego, CA). Horseradish peroxidase-labeled anti-rabbit and anti-mouse secondary antibodies were from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). CsA and SfA were from Novartis Pharmaceuticals (East Hanover, NJ). Forskolin, 3-isobutyl-1-methylxanthine (IBMX), N6,2′-O-dibutyryladenosine 3′,5′-cyclic monophosphate sodium salt (Bt2cAMP), 4-phenylbutyric acid, propidium iodide solution, tunicamycin, thapsigargin, and cholera toxin were from Sigma-Aldrich. Pertussis toxin was from Calbiochem. MEDICA analogs were synthesized as previously described (16).
Data Analysis
Statistical analysis was performed by one-way repeated-measure analysis of variance with a Student-Newman-Keuls test. When only two groups were compared, significance was analyzed by paired t test.
RESULTS
PTP Gating by MEDICA Analogs
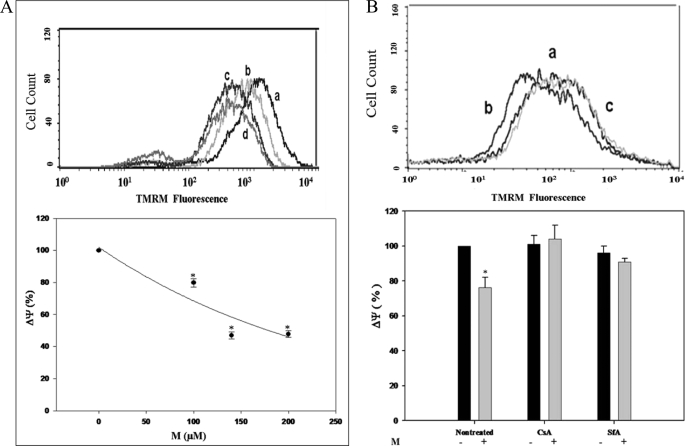
PTP gating by MEDICA (M) analogs has been verified interchangeably for Mαα and Mββ, yielding essentially similar findings for both. Hence, unless otherwise specified, results presented here for M analogs refer to Mαα (M). Treatment of Jurkat cells with M analogs for 24 h resulted in concentration-dependent 10–40% decrease in mitochondrial membrane potential (Fig. 1A). Jurkat mitochondrial uncoupling by M analogs, used at concentrations of up to 150 μm for 24 h, was sensitive to CsA or SfA added together with the TMRM dye 1 h prior to measuring mitochondrial TMRM (Fig. 1B), implying LC-PTP gating. Full abrogation by CsA or SfA may indicate that Jurkat mitochondrial uncoupling by M analogs was essentially accounted for by PTP gating, rather than by their protonophoric activity. PTP gating by M analogs has been further verified in a variety of cell lines (e.g. primary rat hepatocytes and COS-1 and HeLa cells), implying a broad spectrum of PTP gating. The micromolar concentrations required for M-induced PTP gating reflect the high binding affinity of M analogs to medium albumin (estimated to be higher than 99%, independently of Mαα or Mββ concentrations in the range of 0–0.9 mm),4 (unpublished results)), resulting in nanomolar concentrations of the free MEDICA acid in the culture medium at the concentrations of M analogs used herewith.
FIGURE 1.
Mitochondrial PTP gating induced by M analogs in Jurkat cells. A, percent TMRM fluorescence intensity of Jurkat cells incubated with increasing M concentrations as indicated. The mean TMRM fluorescence intensity of non-treated Jurkat cells is defined as 100%. Values are mean ± S.E. of three independent experiments. *, significant as compared with non-treated cells (p < 0.05). Inset: representative histogram of cells treated with 0 (a), 100 μm (b), 140 μm (c), or 200 μm of added M (d). B, percent TMRM fluorescence intensity of M-treated Jurkat cells, in the absence or presence of 6.0 μg/ml CsA or 1.0 μm SfA added 1 h prior to TMRM measurement, as indicated. The mean TMRM fluorescence intensity of non-treated Jurkat cells is defined as 100%. Values are mean ± S.E. of three independent experiments. *, significant as compared with non-treated controls (p < 0.05). Inset: representative histogram of non-treated cells (a), M-treated cells (b), and M-treated cells in the presence of added CsA (c).
Mitochondrial Bcl2 Family Protein Profile Induced by M Analogs
LC-PTP gating is strongly affected by the mitochondrial availability of Bcl2-family proteins (25, 31; reviewed in Ref. 32), rather than the expression level of intrinsic PTP components (e.g. adenine nucleotide translocase, voltage-dependent anion channel, and cyclophilin D) (reviewed in Ref. 33). Hence, PTP gating by M analogs was further evaluated by the content and phosphorylation profile of mitochondrial Bcl2-family proteins. Treatment of rats with Mββ resulted in pronounced decrease in liver mitochondrial Bcl2 with concomitant increase in mitochondrial Bax, Bak, and Bcl2-Bad heterodimer (Fig. 2A). Similar changes in mitochondrial Bcl2-family proteins were observed in Jurkat cells (Fig. 2B). The observed changes in mitochondrial Bcl2-family proteins were not accounted for by their respective cellular contents (not shown), indicating that these were not due to changes in their expression levels. Furthermore, overexpression of Bcl2 resulted in abrogating the decrease in mitochondrial membrane potential induced by M analogs (Fig. 2C), implying an essential role of mitochondrial Bax and Bak sequestration by Bcl2 in PTP gating by M analogs.
FIGURE 2.
Profile of liver and Jurkat mitochondrial Bcl2-family proteins induced by M analogs. A, liver mitochondrial Bcl2 (26 kDa), Bax (21 kDa), Bak (25 kDa), and the Bad constituent of disuccinimidyl suberate cross-linked Bcl2/Bad heterodimer (49 kDa) of rats treated with 480 mg/kg of body weight for 21 days. The densitometric intensity of respective Bcl2-family proteins of non-treated animals is defined as 1.0. Values are mean ± S.E. of 4 rats. *, significant as compared with non-treated rats (p < 0.05). B, mitochondrial Bcl2 (26 kDa) of M-treated Jurkat cells, and the Bad constituent of Bcl2-Bad heterodimer cross-linked by disuccinimidyl suberate (49 kDa), determined by SDS-PAGE/Western blot analysis. Cross-linked Bcl2-Bad heterodimer (49 kDa) of mitochondrial lysates was analyzed by reacting respective blots with anti-Bad antibody, followed by stripping with 4 m guanine thiocyanate and reacting with anti-Bcl2 antibody. The densitometric intensity of respective Bcl2-family proteins of non-treated Jurkat cells is defined as 1.0. Values are mean ± S.E. of three independent experiments. *, significant as compared with non-treated cells (p < 0.05). Inset: representative blots. C, percent TMRM fluorescence intensity of Mββ-treated Jurkat cells overexpressing Bcl2. The mean TMRM fluorescence intensity of non-treated Jurkat cells is defined as 100%. Values are mean ± S.E. of three independent experiments. *, significant as compared with respective non-treated cells (p < 0.05). Histograms are representative of non-treated (a) and Mββ-treated (b) wild-type (left upper panel) and Bcl2-overexpressing (right upper panel) Jurkat cells. Inset: representative blot of overexpressed Bcl2.
Increase in mitochondrial free Bax and Bak induced by M analogs, with concomitant increase in Bcl2-Bad heterodimer (Fig. 2), may indicate displacement of Bax (and Bak) from the Bcl2-Bax heterodimer, due to binding of non-phosphorylated Bad(Ser-112, Ser-155) to Bcl2. In its phosphorylated form, Bad associates with cytoplasmic 14-3-3 proteins allowing for the survival activity of free Bcl2 (or Bcl-XL), while in its non-phosphorylated form, Bad is targeted to the mitochondria where it binds Bcl2 (or Bcl-XL) with displacement of Bax or Bak (34, 35). Hence, PTP gating by M was further pursued in terms of P-Bad(Ser-112, Ser-155).
Bad(Ser-112) and Bad(Ser-155) phosphorylation level is determined by their respective kinases (31–35) as well as by dephosphorylation of P-Bad(Ser-112, Ser-155) by the phosphatases PP2A and/or PP2B (36, 37). Jurkat mitochondrial PTP gating by M analogs still prevailed in the presence of added okadaic acid or FK506 (not shown), implying suppression of the respective protein kinase activities, rather than activation of respective protein phosphatases. Lack of effect of FK506 on PTP gating by M analogs is of particular interest in light of PP2B obligatory role in LC-PTP gating by thyroid hormone (T3), mediated by dephosphorylation of mitochondrial P-Bcl2(S70) by T3-activated PP2B (25).
Suppression of P-Bad(Ser-112, Ser-155) by M Analogs
Phosphorylation of Bad(Ser-155) is preferentially determined in vitro and exclusively determined in vivo by PKA (38–41), whereas phosphorylation of Bad(Ser-112) is due to activated RSK1 (42, 43). RSK1 may be activated in Jurkat cells by the PMA/PKC/MAPK/RSK1 transduction pathway (44). Hence, suppression of P-Bad(Ser-112, Ser-155) by M analogs was studied in terms of both Bad(Ser-155) phosphorylation by PKA as well as of Bad(Ser-112) phosphorylation by activated RSK1.
Mitochondrial PTP gating by M analogs was indeed accompanied by a pronounced decrease in P-Bad(Ser-155) of Jurkat or COS-1 cells (Fig. 3A). M-induced decrease in P-Bad(Ser-155) was evident under basal PKA conditions as well as in the presence of added forskolin/IBMX, implying suppression of PKA activity. Furthermore, decrease in mitochondrial membrane potential by M analogs was accompanied by a pronounced decrease in cAMP levels (Fig. 3B) and was partly abrogated by added Bt2cAMP (Fig. 3C), indicating that suppression of PKA activity by M was due to limiting cellular cAMP. M-induced decrease in cAMP and P-Bad(Ser-155) prevailed under conditions of inhibiting the phosphodiesterase by added IBMX (Fig. 3, A and B), implying that the decrease in cAMP levels induced by M analogs was accounted for by inhibition of the adenylate cyclase activity, rather than activation of the phosphodiesterase. Inhibition of the adenylate cyclase was maintained in the presence of added cholera or pertussis toxins (Fig. 3D), indicating that inhibition of the cyclase did not result from targeting the Gαs or Gαi transducers of the cyclase by M analogs (Scheme 1). Concomitantly with suppression of P-Bad(Ser-155), PMA-induced P-Bad(Ser-112) and P-ERK(Y204) were pronouncedly suppressed by M analogs (Fig. 3E), implying suppression of the MAPK/RSK1 transduction pathway.
FIGURE 3.
Suppression of P-Bad(Ser-155, Ser-112) by M analogs. A, P-Bad(Ser-155) of M-treated Jurkat and COS-1 cells, determined by SDS-PAGE/Western blot analysis, in the presence or absence of 10 μm Forskolin (Fors) and 30 μm IBMX added 1 h prior to sampling, as indicated. The P-Bad(Ser-155)/Bad ratio of extracts of non-treated controls is defined as 1.0. Values are mean ± S.E. of three independent experiments. *, significant as compared with non-treated controls (p < 0.05). #, significant as compared with Forskolin/IBMX-treated controls (p < 0.05). Inset: representative blots. B, cAMP content of Jurkat and COS-1 cells treated as described in A. The cAMP content of extracts of non-treated controls is defined as 1.0. Values are mean ± S.E. of four independent experiments. *, significant as compared with non-treated controls (p < 0.05). #, significant as compared with Forskolin/IBMX-treated controls (p < 0.05). C, percent TMRM fluorescence intensity of M-treated Jurkat cells in the presence or absence of 500 μm Bt2cAMP added 4 h prior to sampling, as indicated. The mean TMRM fluorescence intensity of non-treated controls is defined as 100%. Values are mean ±S.E. of five independent experiments. *, significant as compared with non-treated controls (p < 0.05). #, significant as compared with Mαα-treated controls (p < 0.05). Inset: representative histogram of non-treated controls (a) and cells treated with Bt2cAMP (b), M (c), or M + Bt2cAMP (d). D, cAMP content of M-treated Jurkat cells in the presence and absence of 5 μg/ml cholera toxin (CTX) (left ordinate, mean of two independent experiments differing by no more than 10%) or 50 ng/ml pertussis toxin (PTX) (right ordinate, mean ± S.E. of three independent experiments) added 3 h prior to sampling, as indicated. The cAMP content of extracts of non-treated controls is defined as 1.0. *, significant as compared with non-treated controls (p < 0.05); #, significant as compared with PTX-treated controls (p < 0.05). E, Bad, P-Bad(Ser-112), ERK, and P-ERK(Y204) of M-treated Jurkat cells, determined by SDS-PAGE/Western blot analysis, in the presence or absence of 50 nm PMA added 1 h prior to sampling. The -Bad(Ser-112)/Bad and P-ERK(Y204)/ERK ratio of extracts of non-treated controls is defined as 1.0. Values are mean ± S.E. of three independent experiments. *, significant as compared with non-treated controls (p < 0.05); #, significant as compared with PMA-treated controls (p < 0.05). Inset: representative blots.
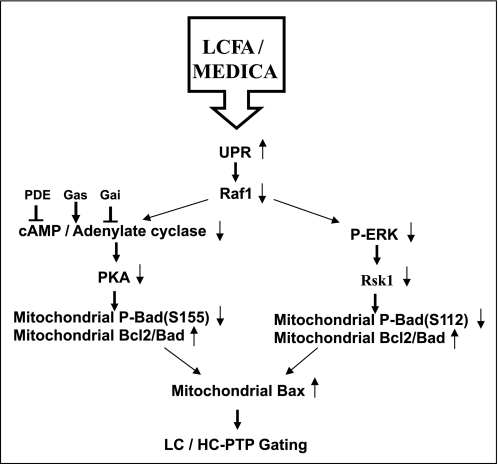
SCHEME 1.
Gating of mitochondrial PTP by LCFA/MEDICA. Suppression of Raf1 expression by LCFA/MEDICA-induced UPR is proposed to result in inhibition of both the Raf1/MEK/ERK/RSK1 and the Raf1/adenylate cyclase/PKA transduction pathways. Suppression of RSK1 and PKA may result in decreased mitochondrial P-Bad(Ser-112) and P-Bad(Ser-155), respectively. Mitochondrial nonphosphorylated Bad(Ser-112, Ser-155) may bind to Bcl2 resulting in displacement of Bax or Bak. Increased mitochondrial Bax or Bak may induce LC-PTP gating and calorigenesis that may drift to HC-PTP gating and apoptosis.
Suppression of Raf1 by M Analogs
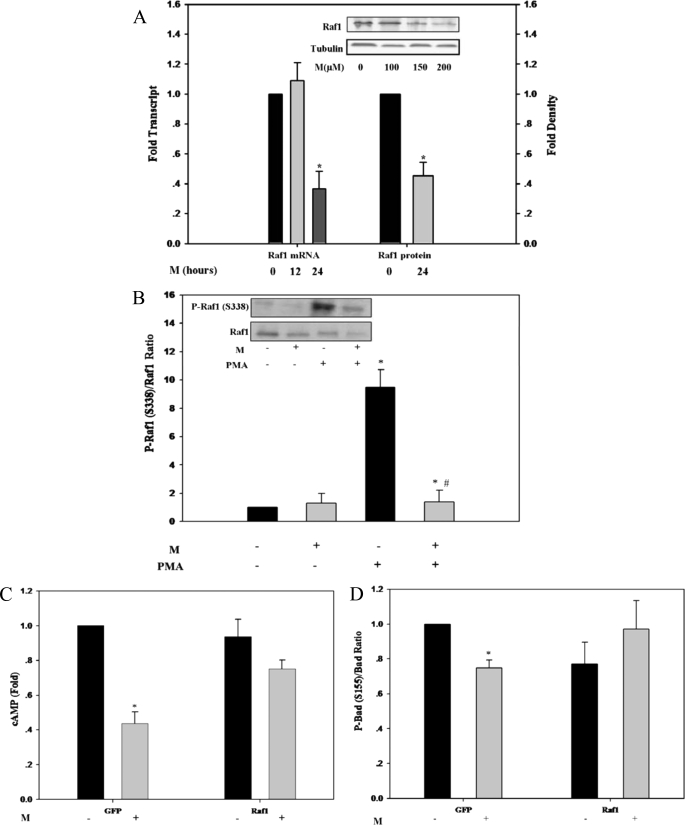
Suppression of the adenylate cyclase/cAMP/PKA/P-Bad(Ser-155) pathway, together with suppression of the MAPK/RSK1/Bad(Ser-112) pathway, implied a putative MEDICA target shared by both transduction pathways. Raf1 could serve as upstream candidate target, in light of its dual role in phosphorylating and activating both, MEK1,2 of the MAPK pathway (44, 45) and the adenylate cyclase (46–52) (Scheme 1). Raf1 transcript and protein were indeed suppressed dose- and time-dependently by M analogs in Jurkat (Fig. 4A) and in a variety of other cell lines (e.g. COS-1 and 3T3-L1) (not shown). Furthermore, M analogs suppressed PMA-induced as well as basal P-Raf1(Ser-338) (Fig. 4B), indicating inhibition of Raf1 kinase activity (53). Hence, inhibition of Raf1 by M analogs may be ascribed to both suppression of Raf1 expression as well as its kinase activity. The role played by Raf1 as upstream target for M analogs was further verified by evaluating the cAMP levels and P-Bad(Ser-155) in COS-1 cells that overexpress Raf1. Overexpression of Raf1 resulted in abrogating the decrease in cAMP (Fig. 4C) and in P-Bad(Ser-155) (Fig. 4D), indicating that Raf1 suppression may indeed account for LC-PTP gating by M analogs.
FIGURE 4.
Suppression of Raf1 by M analogs. A, Raf1 transcript and Raf1 protein levels of M-treated Jurkat cells, determined by quantitative RT-PCR relative to tubulin expression (left ordinate) and by SDS-PAGE/Western blot analysis relative to tubulin levels (right ordinate), respectively. Raf1 transcript and protein levels of extracts of non-treated cells are defined as 1.0. Values are mean ± S.E. of three independent experiments. *, significant as compared with non-treated controls (p < 0.05). Inset: representative blots. B, Raf1 and P-Raf1(Ser-338) of M-treated Jurkat cells, determined by SDS-PAGE/Western blot analysis, in the presence or absence of 50 nm PMA added 1 h prior to sampling. The P-Raf1(Ser-338)/Raf1 ratio of extracts of non-treated controls is defined as 1.0. Values are mean ± S.E. of three independent experiments. *, significant as compared with non-treated controls (p < 0.05); #, significant as compared with PMA-treated controls (p < 0.05). Inset: representative blots. C, cAMP content of M-treated COS-1 cells, transfected with GFP-Raf1 expression vector or with the respective GFP plasmid as indicated. Values are corrected for transfection efficiency, estimated by the percentage of GFP-expressing cells. The cAMP content of extracts of non-treated controls is defined as 1.0. Values are mean ± S.E. of four independent experiments. *, significant as compared with non-treated controls (p < 0.05). D, P-Bad(Ser-155)/Bad ratio of M-treated COS-1 cells, transfected with GFP-Raf1 expression vector or with the respective GFP plasmid as indicated. The P-Bad(Ser-155)/Bad ratio of extracts of non-treated controls is defined as 1.0. Values are mean ± S.E. of four independent experiments. *, significant as compared with non-treated controls (p < 0.05).
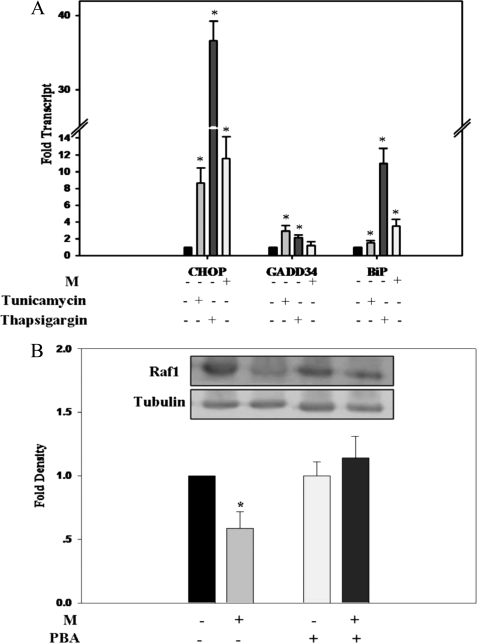
In addition to mitochondrial PTP gating, LCFAs have repeatedly been reported to induce the unfolded protein response (UPR) in a variety of cell lines (54–56). Furthermore, the HSP90 chaperone of the UPR has previously been reported to associate with Raf1 (57), thus prompting us to evaluate a possible link between UPR and suppression of Raf1 expression by M analogs. Treatment of Jurkat cells with M analogs resulted in induction of the UPR markers CHOP, GADD34, and BiP transcripts to an extent similar to that induced by tunicamycin, but less as compared with thapsigargin (Fig. 5A). Furthermore, suppression of Raf1 expression by M was abrogated by the chemical chaperone 4-phenylbutyric acid, previously reported to alleviate UPR (58, 59) (Fig. 5B), pointing to a causal linkage between M-induced UPR and suppression of Raf1 expression.
FIGURE 5.
M-induced UPR. A, CHOP, GADD34, and BiP transcripts of Jurkat cells, determined by quantitative RT-PCR relative to tubulin expression, in the absence or presence of M, 5 μg/ml tunicamycin, or 0.4 μm thapsigargin, as indicated. Transcripts of respective non-treated controls are defined as 1.0. Values are mean ± S.E. of three independent experiments. *, significant as compared with respective non-treated controls (p < 0.05). B, Raf1 protein levels of M-treated Jurkat cells, determined by SDS-PAGE/Western blot analysis relative to tubulin, in the presence or absence of 0.5 mm 4-phenylbutyric acid (PBA) added 6 h prior to M. Raf1 protein levels of extracts of respective non-treated controls are defined as 1.0. Values are mean ± S.E. of four independent experiments. *, significant as compared with non-treated controls (p < 0.05).
MEDICA-induced Apoptosis
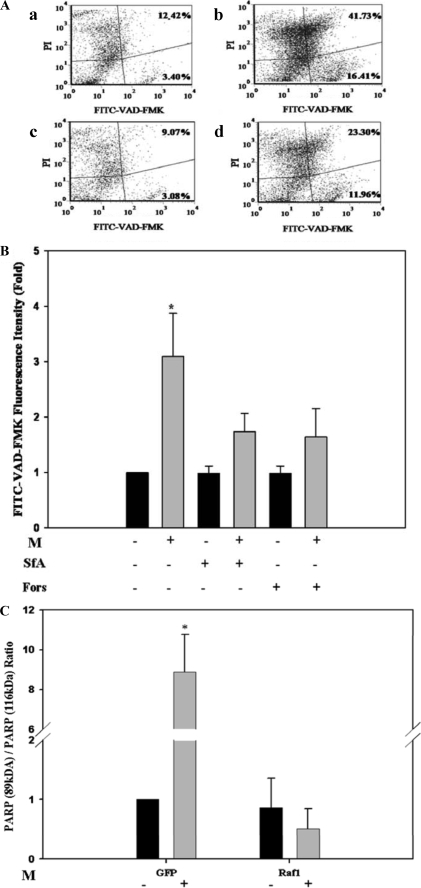
Saturated LCFAs have been reported to induce high conductance (HC)-PTP gating, characterized by increase in effector caspases and apoptosis (3). Indeed, treatment of Jurkat or COS-1 cells with concentrations of M analogs higher than 150 and 250 μm, respectively, resulted in HC-PTP gating and apoptosis. This was verified by FITC-VAD-FMK staining for activated caspase (Fig. 6, A and B) or by truncated PARP (Fig. 6C). In contrast to LC-PTP induced by M analogs, which was rescued by CsA or SfA added with the TMRM dye prior to measuring mitochondrial TMRM (Fig. 1B), apoptosis induced by high concentrations of M analogs was partly abrogated by SfA or by forskolin/IBMX (Fig. 6, A and B), if present throughout the incubation period with M. Similarly to M-induced LC-PTP gating (Fig. 4D), M-induced PARP cleavage was essentially rescued by overexpressed Raf1 (Fig. 6C), indicating that both, LC- and HC-PTP gating, induced by M analogs may share the same transduction pathway leading from Raf1 inhibition to dephosphorylation of Bad(Ser-112, Ser-155) and increase in mitochondrial free Bax, followed by Bax-induced PTP gating.
FIGURE 6.
M-induced apoptosis. A, representative scattergrams of FITC-VAD-FMK and propidium iodide (PI)-bound Jurkat cells. Non-treated controls (a), cells treated for 24 h with 200 μm of M (b), 1 μm SfA (c), or M plus SfA (d). B, relative FITC-VAD-FMK fluorescence intensity of the upper right (late apoptotic cells) plus lower right (early apoptotic cells) quadrants of respective scattergrams. Jurkat cells were treated for 24 h with 200 μm of M, 1 μm SfA, 10 μm Forskolin (Fors), M plus SfA, or M plus Forskolin. The mean FITC-VAD-FMK fluorescence intensity of respective non-treated controls is defined as 1. Values are mean ± S.E. of three independent experiments. *, significant as compared with respective non-treated controls (p < 0.05). C, PARP (89 kDa)/PARP (116 kDa) ratio of COS-1 cells transfected with GFP-Raf1 plasmid or with the respective GFP plasmid and incubated for 24 h with 300 μm of M, as indicated. PARP (89 kDa)/PARP (116 kDa) ratio of extracts of non-treated controls is defined as 1.0. Values are mean ± S.E. of three independent experiments. *, significant as compared with non-treated controls (p < 0.05).
DISCUSSION
The protonophoric and PTP gating activity of LCFA has previously been extensively verified in isolated mitochondria. However, the uncoupling activity of LCFA in vivo remained questionable, being confounded by their dual role as substrates for oxidation and as putative classic uncouplers of oxidative phosphorylation. Analogs of the MEDICA series may simulate the in vivo mode of action of natural LCFA under conditions where respiration of the putative uncoupler does not compromise its inherent uncoupling activity. The present report dissects the transduction pathway of M analogs in gating the mitochondrial PTP in vivo.
PTP gating by LCFA/M analogs is proposed to be transduced as described in Scheme 1. Suppression of Raf1 by UPR induced by M analogs is proposed to result in suppression of the Raf1/MEK/ERK/RSK1 and the Raf1/adenylate cyclase/cAMP/PKA transduction pathways, resulting in decreased P-Bad(Ser-112) and P-Bad(Ser-155), respectively. Decrease in P-Bad(Ser-112, Ser-155) may result in decreased binding of Bad to 14-3-3 with a concomitant increase in its binding to mitochondrial Bcl2 (34, 35). Bad binding to Bcl2 may result in Bax (or Bak) displacement followed by Bax- or Bak-induced PTP gating (30). Mitochondrial PTP gating by LCFA/MEDICA may offer a unified paradigm for LCFA/MEDICA in inducing calorigenesis or apoptosis by mitochondrial LC-PTP or HC-PTP gating, respectively. The respective outcome may depend on LCFA/MEDICA concentrations as well as on additional factors that may drift LC- to HC-PTP gating. In contrast to most previous studies that analyzed mitochondrial PTP in its apoptotic context, this study underscores the physiological aspects of mitochondrial PTP in modulating metabolic rate. Furthermore, the proposed role played by Bad phosphorylation in regulating metabolic rates may add to its recently reported role in modulating pancreatic insulin secretion beyond its apoptotic function (60).
The proposed transduction pathway conforms to the following observations: (a) mitochondrial PTP gating by M analogs was accompanied by an increase in CHOP, GADD34, and BiP transcripts with a concomitant suppression of Raf1 expression. Raf1 suppression by M was abrogated by abrogating UPR by chemical chaperone; (b) Raf1 suppression by M analogs was accompanied by a decrease in P-ERK1,2(Tyr-204) and in its downstream P-Bad(Ser-112) target; (c) Raf1 suppression by M analogs was accompanied by a decrease in cAMP/PKA and in its downstream P-Bad(Ser-155) target. A decrease in cAMP and P-Bad(Ser-155) by M analogs was abrogated by overexpression of Raf1, indicating causal linkage between suppression of Raf1 and inhibition of the adenylate cyclase; (e) PTP gating by M analogs was partly rescued by added forskolin/IBMX or Bt2cAMP; (f) PTP gating by M analogs was accompanied by increase in mitochondrial Bcl2-Bad heterodimer, Bax, and Bak and was abrogated by overexpressing Bcl2; and (g) the proposed transduction pathway is in line with the reported activities of Raf1/RSK1 (reviewed in Ref. 61) and P-Bad(Ser-112, Ser-155) (reviewed in Ref. 62) in promoting cell survival. The mode of action of M analogs in inducing UPR as well as the mode of suppression of Raf1 expression and kinase activity still remain to be investigated. Preliminary profiling of UPR markers induced by MEDICA analogs in cell lines and in vivo has indicated a specific profile of UPR markers rather than an exhaustive response.
Both, thyroid hormone (T3) and M analogs are calorigenic in vivo, and the non-protonophoric mitochondrial activity of M analogs is apparently similar to that of T3 (25). Thus, T3 induces CsA-sensitive decrease in phosphate and redox potentials with concomitant increase in oxygen consumption in cultured cells as well as in vivo (22, 24, 63–65), indicating that both M analogs and T3 do converge onto LC/HC-PTP gating. Furthermore, PTP gating by M analogs or T3 is mediated by modulating the profile of mitochondrial Bcl2-family proteins, resulting in increase in mitochondrial free Bax (25, 31). The concerned transduction pathways differ, however, in their mode of promoting the dissociation of the Bcl2-Bax heterodimer. Thus, dissociation of the Bcl2-Bax heterodimer by T3 is driven by dephosphorylation of Bcl2(Ser-70) by T3-activated PP2B (31), whereas dissociation of the Bcl2/Bax heterodimer by M analogs is driven by Bax displacement due to binding of non-phosphorylated Bad(Ser-112, Ser-155). Indeed, in contrast to T3 (31), PTP gating induced by M analogs remained unaffected by added FK506. Also, in contrast to M analogs, PTP gating by T3 was accompanied by an increase in P-Bad(Ser-155).5 Hence, the two transduction pathways converge at their downstream Bax target but diverge upstream of the Bcl2/Bax heterodimer.
LC-PTP gating by M analogs may account for their calorigenic activity in vivo (17, 18), and may imply a similar calorigenic activity of natural non-esterified LCFA upon reaching high enough intracellular concentrations in vivo. Hence, the surprising efficacy of low carbohydrate high fat diets in counteracting obesity (66) may partly be accounted for by the high concentration of intracellular non-esterified LCFA reached under conditions of limited insulin levels. However, in contrast to low carbohydrate high fat diets where weight loss may still be compromised by the esterification of the LCFA into lipids, M analogs are not esterified into lipids nor β-oxidized to yield energy, thus dissociating between the substrate role of LCFA and their mitochondrial PTP gating activity. MEDICA analogs may thus serve the pharmacological counterpart of low carbohydrate high fat diets in treating obesity.
Acknowledgments
The kind generosity of Dr. G Hacker (Bcl2 Jurkat cells), Dr. A. Gross (rabbit anti-rat Bax antibody), and Dr. T. Balla (pEGFP-Raf-1) is deeply acknowledged.
This work was supported by the Israeli Science Foundation.
J. Bar-Tana, unpublished results.
B. Kalderon, unpublished observation.
- LCFA
- long chain fatty acid
- CsA
- cyclosporine A
- Bt2cAMP
- N6,2′-O-dibutyryladenosine 3′,5′-cyclic monophosphate sodium salt
- HC-PTP
- high conductance PTP
- IBMX
- 3-isobutyl-1-methylxanthine
- LC-PTP
- low conductance PTP
- MAPK
- mitogen-activated protein kinase
- MEDICA
- methyl-substituted hexadecanedioic acids
- Mαα
- 2,2,15,15-tetramethylhexadecanedioic acid
- Mββ
- 3,3,14,14-tetramethylhexadecanedioic acid
- PTP
- permeability transition pore
- PMA
- phorbol 12-myristate 13-acetate
- PTX
- pertussis toxin
- SfA
- sanglifherin A
- TMRM
- tetramethylrhodamine methylester
- UCP
- uncoupling protein
- UPR
- unfolded protein response
- PKA
- protein kinase A
- T3
- thyroid hormone
- PARP
- poly(ADP-ribose) polymerase
- ERK
- extracellular signal-regulated kinase
- MEK
- MAPK/ERK kinase
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- FITC
- fluorescein isothiocyanate
- FMK
- fluoromethyl ketone
- GFP
- green fluorescence protein
- EGFP
- enhanced GFP
- M
- MEDICA analog
- RSK
- ribosomal s6 Kinase.
REFERENCES
- 1.Skulachev V. P. (1991) FEBS Lett. 294, 158–162 [DOI] [PubMed] [Google Scholar]
- 2.Wojtczak L., Wieckowski M. R., Schönfeld P. (1998) Arch. Biochem. Biophys. 357, 76–84 [DOI] [PubMed] [Google Scholar]
- 3.Bernardi P., Penzo D., Wojtczak L. (2002) Vitam. Horm. 65, 97–126 [DOI] [PubMed] [Google Scholar]
- 4.Koshkin V., Dai F. F., Robson-Doucette C. A., Chan C. B., Wheeler M. B. (2008) J. Biol. Chem. 283, 7936–7948 [DOI] [PubMed] [Google Scholar]
- 5.Novgorodov S. A., Gudz T. I. (1996) J. Bioenerg. Biomembr. 28, 139–146 [DOI] [PubMed] [Google Scholar]
- 6.Ichas F., Mazat J. P. (1998) Biochim. Biophys. Acta 1366, 33–50 [DOI] [PubMed] [Google Scholar]
- 7.Brustovetsky N., Dubinsky J. M. (2000) J. Neurosci. 20, 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hausenloy D., Wynne A., Duchen M., Yellon D. (2004) Circulation 109, 1714–1717 [DOI] [PubMed] [Google Scholar]
- 9.Zoratti M., Szabò I. (1995) Biochim. Biophys. Acta 1241, 139–176 [DOI] [PubMed] [Google Scholar]
- 10.Crompton M. (1999) Biochem. J. 341, 233–249 [PMC free article] [PubMed] [Google Scholar]
- 11.Halestrap A. P., McStay G. P., Clarke S. J. (2002) Biochimie 84, 153–166 [DOI] [PubMed] [Google Scholar]
- 12.Scholz R., Schwabe U., Soboll S. (1984) Eur. J. Biochem. 141, 223–230 [DOI] [PubMed] [Google Scholar]
- 13.Soboll S., Gründel S., Schwabe U., Scholz R. (1984) Eur. J. Biochem. 141, 231–236 [DOI] [PubMed] [Google Scholar]
- 14.Kingsley-Hickman P. B., Sako E. Y., Uðurbil K., From A. H., Foker J. E. (1990) J. Biol. Chem. 265, 1545–1550 [PubMed] [Google Scholar]
- 15.Schönfeld P., Wojtczak A. B., Geelen M. J., Kunz W., Wojtczak L. (1988) Biochim. Biophys. Acta 936, 280–288 [DOI] [PubMed] [Google Scholar]
- 16.Bar-Tana J., Ben-Shoshan S., Blum J., Migron Y., Hertz R., Pill J., Rose-Khan G., Witte E. C. (1989) J. Med. Chem. 32, 2072–2084 [DOI] [PubMed] [Google Scholar]
- 17.Kalderon B., Sheena V., Shachrur S., Hertz R., Bar-Tana J. (2002) J. Lipid Res. 43, 1125–1132 [DOI] [PubMed] [Google Scholar]
- 18.Kalderon B., Hertz R., Bar-Tana J. (1992) Endocrinology 131, 1629–1635 [DOI] [PubMed] [Google Scholar]
- 19.Russell J. C., Shillabeer G., Bar-Tana J., Lau D. C., Richardson M., Wenzel L. M., Graham S. E., Dolphin P. J. (1998) Diabetes 47, 770–778 [DOI] [PubMed] [Google Scholar]
- 20.Mayorek N., Kalderon B., Itach E., Bar-Tana J. (1997) Diabetes 46, 1958–1964 [DOI] [PubMed] [Google Scholar]
- 21.Hermesh O., Kalderon B., Bar-Tana J. (1998) J. Biol. Chem. 273, 3937–3942 [DOI] [PubMed] [Google Scholar]
- 22.Hermesh O., Kalderon B., Berman B., Bar-Tana J. (2000) Biochim. Biophys. Acta 1457, 166–174 [DOI] [PubMed] [Google Scholar]
- 23.Shabalina I. G., Backlund E. C., Bar-Tana J., Cannon B., Nedergaard J. (2008) Biochim. Biophys. Acta 1777, 642–650 [DOI] [PubMed] [Google Scholar]
- 24.Kalderon B., Hertz R., Bar-Tana J. (1992) Endocrinology 131, 400–407 [DOI] [PubMed] [Google Scholar]
- 25.Yehuda-Shnaidman E., Kalderon B., Bar-Tana J. (2005) Endocrinology 146, 2462–2472 [DOI] [PubMed] [Google Scholar]
- 26.Gross A., Jockel J., Wei M. C., Korsmeyer S. J. (1998) EMBO J. 17, 3878–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mikhailov V., Mikhailova M., Degenhardt K., Venkatachalam M. A., White E., Saikumar P. (2003) J. Biol. Chem. 278, 5367–5376 [DOI] [PubMed] [Google Scholar]
- 28.Garfin D. E. (1990) Methods Enzymol. 182, 425–441 [DOI] [PubMed] [Google Scholar]
- 29.Balla A., Tuymetova G., Barshishat M., Geiszt M., Balla T. (2002) J. Biol. Chem. 277, 20041–20050 [DOI] [PubMed] [Google Scholar]
- 30.Huang D. C., Cory S., Strasser A. (1997) Oncogene 14, 405–414 [DOI] [PubMed] [Google Scholar]
- 31.Yehuda-Shnaidman E., Kalderon B., Azazmeh N., Bar-Tana J. (2010) FASEB J. 24, 93–104 [DOI] [PubMed] [Google Scholar]
- 32.Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., Strasser A., Kluck R. M., Adams J. M., Huang D. C. (2007) Science 315, 856–859 [DOI] [PubMed] [Google Scholar]
- 33.Halestrap A. P. (2009) J. Mol. Cell Cardiol. 46, 821–831 [DOI] [PubMed] [Google Scholar]
- 34.Yang E., Zha J., Jockel J., Boise L. H., Thompson C. B., Korsmeyer S. J. (1995) Cell 80, 285–291 [DOI] [PubMed] [Google Scholar]
- 35.Zha J., Harada H., Yang E., Jockel J., Korsmeyer S. J. (1996) Cell 87, 619–628 [DOI] [PubMed] [Google Scholar]
- 36.Chiang C. W., Kanies C., Kim K. W., Fang W. B., Parkhurst C., Xie M., Henry T., Yang E. (2003) Mol. Cell. Biol. 23, 6350–6362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H. G., Pathan N., Ethell I. M., Krajewski S., Yamaguchi Y., Shibasaki F., McKeon F., Bobo T., Franke T. F., Reed J. C. (1999) Science 284, 339–343 [DOI] [PubMed] [Google Scholar]
- 38.Tan Y., Demeter M. R., Ruan H., Comb M. J. (2000) J. Biol. Chem. 275, 25865–25869 [DOI] [PubMed] [Google Scholar]
- 39.Lizcano J. M., Morrice N., Cohen P. (2000) Biochem. J. 349, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Datta S. R., Katsov A., Hu L., Petros A., Fesik S. W., Yaffe M. B., Greenberg M. E. (2000) Mol. Cell 6, 41–51 [PubMed] [Google Scholar]
- 41.Zhou X. M., Liu Y., Payne G., Lutz R. J., Chittenden T. (2000) J. Biol. Chem. 275, 25046–25051 [DOI] [PubMed] [Google Scholar]
- 42.Bonni A., Brunet A., West A. E., Datta S. R., Takasu M. A., Greenberg M. E. (1999) Science 286, 1358–1362 [DOI] [PubMed] [Google Scholar]
- 43.Tan Y., Ruan H., Demeter M. R., Comb M. J. (1999) J. Biol. Chem. 274, 34859–34867 [DOI] [PubMed] [Google Scholar]
- 44.Mor A., Philips M. R. (2006) Annu. Rev. Immunol. 24, 771–800 [DOI] [PubMed] [Google Scholar]
- 45.Alejandro E. U., Johnson J. D. (2008) J. Biol. Chem. 283, 2407–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beazely M. A., Watts V. J. (2006) Eur. J. Pharmacol. 535, 1–12 [DOI] [PubMed] [Google Scholar]
- 47.Yue X., Varga E. V., Stropova D., Vanderah T. W., Yamamura H. I., Roeske W. R. (2006) Eur. J. Pharmacol. 540, 57–59 [DOI] [PubMed] [Google Scholar]
- 48.Osawa Y., Yim P. D., Xu D., Panettieri R. A., Emala C. W. (2007) Am. J. Physiol. Lung Cell Mol. Physiol. 292, L1414–L1421 [DOI] [PubMed] [Google Scholar]
- 49.Feldman R. D., Gros R. (2007) Life Sci. 81, 267–271 [DOI] [PubMed] [Google Scholar]
- 50.Tan C. M., Kelvin D. J., Litchfield D. W., Ferguson S. S., Feldman R. D. (2001) Biochemistry 40, 1702–1709 [DOI] [PubMed] [Google Scholar]
- 51.Beazely M. A., Alan J. K., Watts V. J. (2005) Mol. Pharmacol. 67, 250–259 [DOI] [PubMed] [Google Scholar]
- 52.Ding Q., Gros R., Gray I. D., Taussig R., Ferguson S. S., Feldman R. D. (2004) Mol. Pharmacol. 66, 921–928 [PubMed] [Google Scholar]
- 53.Wellbrock C., Karasarides M., Marais R. (2004) Nat. Rev. Mol. Cell Biol. 5, 875–885 [DOI] [PubMed] [Google Scholar]
- 54.Wei Y., Wang D., Topczewski F., Pagliassotti M. J. (2006) Am. J. Physiol. Endocrinol. Metab. 291, E275–E281 [DOI] [PubMed] [Google Scholar]
- 55.Karaskov E., Scott C., Zhang L., Teodoro T., Ravazzola M., Volchuk A. (2006) Endocrinology 147, 3398–3407 [DOI] [PubMed] [Google Scholar]
- 56.Cnop M., Igoillo-Esteve M., Cunha D. A., Ladrière L., Eizirik D. L. (2008) Biochem. Soc. Trans. 36, 909–915 [DOI] [PubMed] [Google Scholar]
- 57.Schulte T. W., Blagosklonny M. V., Ingui C., Neckers L. (1995) J. Biol. Chem. 270, 24585–24588 [DOI] [PubMed] [Google Scholar]
- 58.Ozcan U., Yilmaz E., Ozcan L., Furuhashi M., Vaillancourt E., Smith R. O., Görgün C. Z., Hotamisligil G. S. (2006) Science 313, 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choi S. E., Lee Y. J., Jang H. J., Lee K. W., Kim Y. S., Jun H. S., Kang S. S., Chun J., Kang Y. (2008) Arch. Biochem. Biophys. 475, 109–114 [DOI] [PubMed] [Google Scholar]
- 60.Danial N. N., Walensky L. D., Zhang C. Y., Choi C. S., Fisher J. K., Molina A. J., Datta S. R., Pitter K. L., Bird G. H., Wikstrom J. D., Deeney J. T., Robertson K., Morash J., Kulkarni A., Neschen S., Kim S., Greenberg M. E., Corkey B. E., Shirihai O. S., Shulman G. I., Lowell B. B., Korsmeyer S. J. (2008) Nat. Med. 14, 144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anjum R., Blenis J. (2008) Nat. Rev. Mol. Cell Biol. 9, 747–758 [DOI] [PubMed] [Google Scholar]
- 62.Danial N. N. (2008) Oncogene 27, Suppl. 1, S53–S70 [DOI] [PubMed] [Google Scholar]
- 63.Soboll S. (1993) Biochim. Biophys. Acta. 1144, 1–16 [DOI] [PubMed] [Google Scholar]
- 64.Harper M. E., Brand M. D. (1993) J. Biol. Chem. 268, 14850–14860 [PubMed] [Google Scholar]
- 65.Bobyleva V., Pazienza T. L., Maseroli R., Tomasi A., Salvioli S., Cossarizza A., Franceschi C., Skulachev V. P. (1998) FEBS Lett. 430, 409–413 [DOI] [PubMed] [Google Scholar]
- 66.Foster G. D., Wyatt H. R., Hill J. O., McGuckin B. G., Brill C., Mohammed B. S., Szapary P. O., Rader D. J., Edman J. S., Klein S. (2003) N. Engl. J. Med. 348, 2082–2090 [DOI] [PubMed] [Google Scholar]