Abstract
Many human epithelial cancers are characterized by abnormal activation of the epidermal growth factor receptor (EGFR), which is often caused by its excessive expression in tumor cells. The abundance of EGFR is modulated, in part, by its ubiquitination, which targets it for degradation. The components responsible for adding ubiquitin to EGFR are well characterized, but this is a reversible process, and the mechanisms that modulate the removal of ubiquitin from the EGFR are not well known. We found that de-ubiquitination of EGFR was regulated by diacylglycerol kinase δ (DGKδ), a lipid kinase that terminates diacylglycerol signaling. In DGKδ-deficient cells, ubiquitination of EGFR was enhanced, which attenuated the steady-state levels of EGFR and promoted its ligand-induced degradation. These effects were not caused by changes in the ubiquitinating apparatus, but instead were due to reduced expression of the de-ubiquitinase, ubiquitin-specific protease 8 (USP8). Depletion of protein kinase Cα (PKCα), a target of diacylglycerol, rescued the levels of USP8 and normalized EGFR degradation in DGKδ-deficient cells. Moreover, the effects of PKCα were caused by its inhibition of Akt, which stabilizes USP8. Our data indicate a novel mechanism where DGKδ and PKCα modulate the levels of ubiquitinated EGFR through Akt and USP8.
Keywords: Enzymes/Lipid, Lipid, Lipid/Diacylglycerol, Lipid/Phospholipid, Receptors/Tyrosine Kinase, Signal Transduction
Introduction
The EGFR2 is an important cancer target that is often abnormally active in human tumors. Its role in carcinogenesis has been firmly established in numerous animal models of cancer. For example, mice harboring mutations in the tumor suppressor adenomatous polyposis coli gene develop 50–90% fewer polyps when EGFR signaling is disrupted either genetically (1) or through the use of EGFR inhibitors (1–3). Its importance in tumorigenesis makes it critical that we understand in more detail the mechanisms by which EGFR can be modulated so that we can identify additional avenues to inhibit this important signaling pathway.
Upon binding to its ligands the EGFR activates phospholipase C enzymes that generate diacylglycerol (DAG), an important lipid second messenger that recruits and can activate numerous signaling proteins, including PKC enzymes. DAG is metabolized by a family of proteins called the diacylglycerol kinases (DGKs), which consume DAG in a spatially discrete manner that allows some DGK isoforms to regulate specific DAG target proteins. DGKδ is a type II DGK with two alternatively spliced products, DGKδ1 and DGKδ2, that differ at their amino termini (4). This alternative splicing does not alter their DGK activity, but appears to affect their sub-cellular localization (4). We disrupted the DGKδ gene in mice and found that DGKδ null mice displayed a phenotype almost identical to that of EGFR null mice: the pups were born with open eyelids, developed respiratory difficulty, and died within 24 h after birth (5). This phenotype was specific to DGKδ: other DGK knock-out mice (α, ϵ, ζ, ι, and θ) did not display these defects (Refs. 5–9 and data not shown). In DGKδ null mice, we found that EGFR expression was reduced and that the kinase activity of EGFR was attenuated (5). In DGKδ null mice we also observed excessive phosphorylation of threonine 654 (Thr-654) in EGFR, which led us to hypothesize that this change altered both the expression and the activity of EGFR (5). In this report, we show that Thr-654 is not responsible for the changes in EGFR abundance. Instead, we found that DGKδ deficiency reduces steady-state levels of EGFR and accelerates its ligand-induced degradation by modulating the expression of USP8, a de-ubiquitinase. We show that the effects of DGKδ on USP8 and EGFR are mediated through PKCα, which we found was excessively active in DGKδ-deficient cells. PKCα, in turn, inhibits Akt activity that is necessary for proper USP8 expression. Collectively, these observations contribute novel mechanistic insights linking DGKδ, PKCα, Akt, USP8, and EGFR that could identify additional mechanisms to disrupt EGFR signaling in tumors.
EXPERIMENTAL PROCEDURES
Expression Plasmids, Cell Culture, and Transfection
Full-length human wt, T654A, or Y1045F EGFR were cloned into pcDNA3.1/Myc-His (Invitrogen), and human DGKδ1 and DGKδ2 were cloned into p3XFLAG (Sigma). V5-c-Cbl was a kind gift from Dr. S. Kuwada (University of Utah). V5-USP8 (murine) was from Dr. K. L. Carraway 3rd (University of California Davis). HeLa, SCC-9, H441, H460, SW1271, H1650, and H1975 cell lines were from ATCC and were grown according to their instructions. Primary keratinocytes were isolated and grown as described (5). Transfection of expression vectors was performed using Lipofectamine (Invitrogen) according to the instructions. Prior to treating cells with recombinant EGF (#236-EG from R&D Systems), all cells were starved for 4–24 h. In some cases, 250 nm Gö6976 (Calbiochem) was added prior to the agonist. DGKδ RNAi was performed using Oligofectamine (Invitrogen) and the siRNA duplex: 5′-GGCCAUGGUUCACACAUCGTT-3′ and 5′-CGAUGUGUGAACCAUGGCCTT-3′. The PKCα siRNA duplexes were: 5′-GGCUUCCAGUGCCAAGUUUTT-3′ and 5′-AAACUUGGCACUGGAAGCCTT-3′. Scrambled siRNA duplexes were used as controls.
Antibodies, Western Blots, and Immunoprecipitation
Western blotting was performed according to instructions provided by the suppliers. Anti-EGFR (#2232), anti-phospho-EGFR (#2234 or #2237), anti-pThr (#9381), and anti-phospho-MARCKS (#2741) antibodies were from Cellular Signaling. Anti-PKCα (sc-208, for Western blotting), anti-Myc (sc-40), anti-MARCKS (sc-6454), anti-c-Cbl (sc-170), anti-Ub (sc-8017), and anti-ERK2 (sc-154) were from Santa Cruz Biotechnology. Anti-FLAG M2 was from Sigma, anti-V5 for immunoprecipitation (ab9116) was from Abcam, anti-PKCα (P16520, for immunoprecipitation) and anti-transferrin receptor (612124) were from BD Transduction Laboratories, anti-V5 for Western blotting (#46-0705) was from Invitrogen, and anti-DGKδ has been described (10). Anti-pY774-c-Cbl (#1160) was from Epitomics. Anti-USP8 was from Dr. K. Carraway (University of California Davis). Horseradish peroxidase-labeled secondary antibodies (Cellular Signaling) were used to detect the primary antibodies, and multiple exposures were obtained to ensure that the signal was in the linear range. To quantify bands, scanned Western blot images were analyzed using National Institutes of Health ImageJ. Mean ± S.D. were calculated, and statistical significance was established by paired, one-tailed t tests.
To immunoprecipitate endogenous EGFR or transfected EGFR-Myc, cells were collected in lysis buffer (Cellular Signaling #9803), incubated on ice for 10 min, and then centrifuged to remove debris. The lysates (200–1500 μg of protein) were incubated with anti-Myc or anti-EGFR overnight (4 °C) and then protein A/G plus agarose (25 μl, Santa Cruz Biotechnology) was added for 1 h. After three washes with lysis buffer, the pellets and lysates (10–50 μg) were separated by SDS-PAGE and then immunoblotted to detect Ub or EGFR. To immunoprecipitate V5-Cbl or V5-USP8, anti-V5 antibodies were used in the above protocol.
EGFR Endocytosis and Recycling Assays
To measure the rate of EGFR endocytosis, HeLa cells were transfected with scrambled control or DGKδ siRNA duplexes, and then internalization of [125I]EGF (0.5 ng/ml, PerkinElmer Life Sciences) was detected in duplicate each minute for 6 min as described before (11). The internal:surface ratio of radioactivity was plotted versus time, and the endocytosis rate constant (ke) was calculated from the plots (11). To measure recycling of EGFR, HeLa cells were transfected with scrambled control or DGKδ siRNA duplexes, and then recycling of EGFR was measured in duplicate by loading the cells with [125I]EGF (0.5 ng/ml) for 10 min, stripping the surface-bound EGF, adding excess cold EGF (100 ng/ml), and then measuring the amount of intact [125I]EGF released into the medium over 40 min (12).
Cell Viability Assay
Lung cancer cells were plated in 96-well plates at a density of 4000 cells/well. On the following day, the medium was changed to RPMI medium with 2% fetal bovine serum (80 μl/well). A mixture of 1 μl of siRNA duplex in 16 μl of Opti-Mem (Invitrogen) plus 0.4 μl of Oligofectamine in 2.6 μl of Opti-Mem was added to each well. Cell viability was measured 72 h later using the Cell Proliferation Kit II (XTT) from Roche Applied Science according to their instructions.
RESULTS
DGKδ Deficiency Accelerates EGFR Degradation
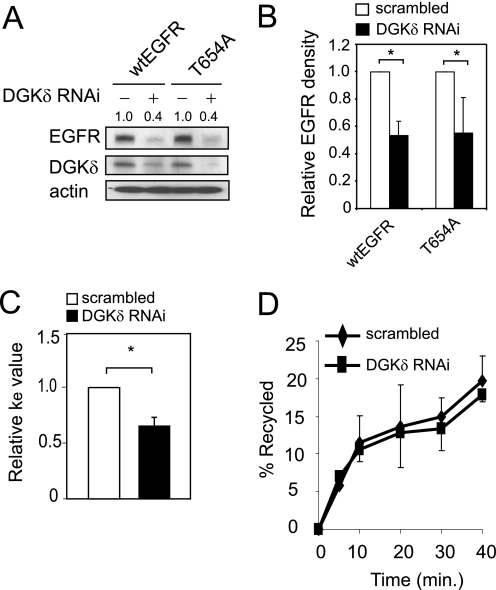
In our prior work we found reduced expression of EGFR in DGKδ-deficient cells and tissues that was not due to changes in the abundance of EGFR mRNA (5). We also found higher levels of phosphorylated Thr-654 in DGKδ-deficient tissues (5). Because phosphorylation of Thr-654 in EGFR modulates EGFR trafficking (12–15), we hypothesized that this excessive phosphorylation of Thr-654 was responsible for reducing the levels of EGFR. To test this possibility, we transfected wt EGFR or a T654A EGFR mutant into HeLa cells (or SCC-9 cells, not shown) and then knocked down the expression of DGKδ using RNAi. We found that DGKδ deficiency still reduced expression of the T654A EGFR mutant (Fig. 1, A and B) and concluded that phosphorylation of Thr-654 in EGFR does not substantially contribute to the changes in EGFR expression caused by DGKδ.
FIGURE 1.
EGFR endocytosis and recycling in DGKδ-deficient cells. A, HeLa cells were transfected with Myc-tagged wt or T654A EGFR, and then DGKδ was knocked down using siRNA. After 24 h EGFR (anti-Myc), DGKδ, and actin were detected by immunoblotting. EGFR band densities (arbitrary units) are shown above the blot. B, the band densities of wtEGFR or T654A EGFR from three experiments similar to Fig. 1A were determined and plotted. Shown are mean values with S.D. (*, p < 0.05). B, HeLa cells were transfected with scrambled control or DGKδ siRNA duplexes, and the endocytosis rate constant (ke) was determined using [125I]EGF (11). Shown are mean, relative ke values (±S.D.) from four experiments. The asterisk indicates a statistically significant difference compared with scrambled control cells (p < 0.01). C, HeLa cells were transfected with scrambled control or DGKδ siRNA duplexes, and then recycling of EGFR was measured (12). Shown are mean values (±S.D.) from three experiments.
In our prior work, we also found that the reduced steady-state expression levels of EGFR that were caused by DGKδ deficiency were rescued by concurrent knockdown of dynamin II (5), a protein that is required for the initial steps of endocytosis. Because the abundance of EGFR is highly regulated by endosome trafficking, and endocytosis is essential for this process (16), we next examined the effects of DGKδ deficiency on EGFR trafficking. First, we measured the rate of ligand-induced endocytosis using radiolabeled EGF. Based on the reduced levels of EGFR in DGKδ-deficient cells, we predicted that the internalization rate (ke) of EGFR might be increased by DGKδ deficiency. But, contrary to this expected outcome, we found that DGKδ deficiency reduced the rate of EGFR endocytosis (Fig. 1C). This surprising result is consistent with published data showing reduced clathrin-mediated endocytosis in DGKδ-deficient cells (17, 18) and was likely caused in this context either by PKC phosphorylation of Thr-654 in EGFR (12) or by effects on the adaptor protein AP2 (17). The slower rates of internalization that we measured in DGKδ-deficient cells did not account for the defects in steady-state EGFR expression levels, so we next examined additional steps in the EGFR trafficking pathway.
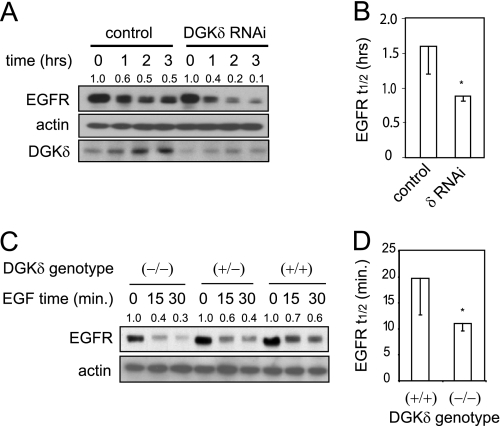
Once internalized, EGFR is either recycled back to the plasma membrane or is directed toward lysosomes for degradation. When we measured EGFR recycling using radiolabeled EGF (12), we found slightly reduced recycling of EGFR in DGKδ-deficient cells (Fig. 1D). But this change was not statistically significant. Finally, we measured the rate of ligand-induced EGFR degradation by Western blotting and found more rapid EGFR decay in DGKδ-deficient cells (Fig. 2A). The half-life of EGFR in cells depleted of DGKδ was about one-half that of control cells (Fig. 2B). We found similar changes in the presence of cycloheximide, a protein synthesis inhibitor (data not shown and see Fig. 6A). In primary keratinocytes, which degrade EGFR more rapidly, we also observed that DGKδ deficiency accelerated EGFR decay (Fig. 2, C and D). Collectively, these data demonstrated that DGKδ deficiency promotes ligand-induced EGFR decay. It is important to note that, because DGKδ deficiency reduced the rate of EGFR internalization (Fig. 1C), the rate at which EGFR is degraded once it is internalized in DGKδ-deficient cells is probably in excess of what we measured by Western blotting.
FIGURE 2.
Accelerated decay of EGFR in DGKδ-deficient cells. A, HeLa cells were transfected with scrambled or DGKδ siRNA duplexes, starved for 16 h, and then treated with EGF (5 ng/ml) for 0–3 h. EGFR, actin, and DGKδ were then detected in cell lysates by Western blotting. EGFR band densities (arbitrary units) are shown above the blot. Similar results were obtained when cyclohexamide (5 μm) was included to minimize new protein synthesis. B, the half-life (t½) of EGFR was measured in three experiments like that shown in A. Shown are mean values with S.D. (*, p < 0.05). C, EGFR decay in primary keratinocytes from wild-type mice or mice with heterozygous or homozygous deletion of DGKδ was measured after treatment with EGF (10 ng/ml) for the indicated times. EGFR band densities (arbitrary units) are shown above the blot. D, the half-life (t½) of EGFR was measured in three experiments like that shown in C. Shown are mean values with S.D. (*, p < 0.05).
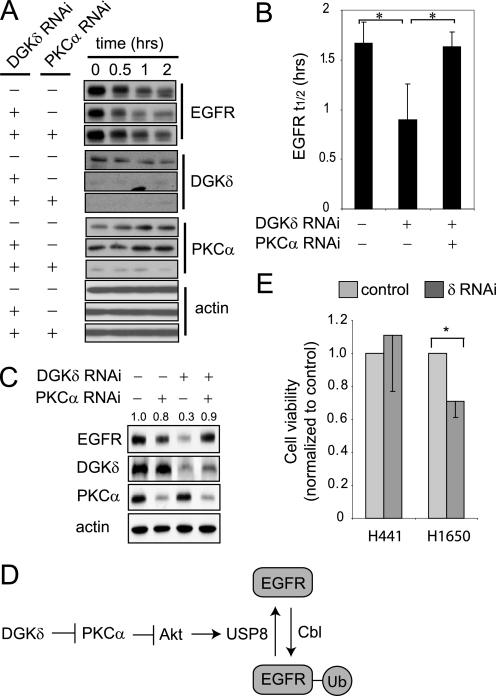
FIGURE 6.
Depleting PKCα rescues the EGFR defects caused by DGKδ deficiency. A, HeLa cells were transfected with scrambled or the indicated siRNA duplexes, starved, treated with cycloheximide (5 μm, 20 min), and then exposed to EGF (5 ng/ml for 0–2 h). EGFR, DGKδ, PKCα, and actin were detected in cell lysates by Western blotting. Samples are from the same blot, but are presented to allow comparison. B, the half-life (t½) of EGFR was determined from Western blots in three independent experiments. The asterisk indicates statistical significance (p < 0.05, n = 3). C, HeLa cells were transfected with scrambled, DGKδ, and/or PKCα siRNA duplexes, grown in medium with 10% serum, and then steady-state levels of EGFR, actin, DGKδ, and PKCα were detected by Western blotting. EGFR band densities (arbitrary units) are indicated above the blot. D, our data support a model where DGKδ and PKCα regulate the activity of Akt, which in turn modulates the levels of USP8 and consequently the amount of ubiquitin attached to EGFR. E, DGKδ was knocked down using RNAi in H441 lung cancer cells (which are resistant to EGFR inhibitors) or H1650 lung cancer cells (which are sensitive) grown in medium with 2% serum. At 72 h after knockdown, cell viability was examined. Viability (mean ± S.D.) is shown normalized to cells exposed to scrambled siRNA. The asterisk indicates statistical significance (p < 0.01, n = 4). Western blotting demonstrated similar knockdown of DGKδ and EGFR in both cell lines (not shown).
DGKδ Deficiency Enhances Both Steady-state and Ligand-induced Ubiquitination of EGFR
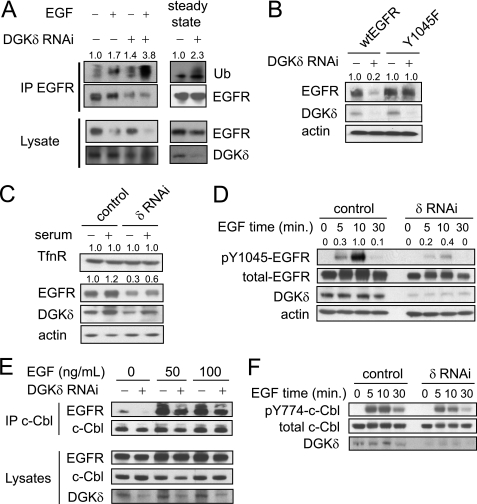
EGFR is marked for degradation by ubiquitin, a modification that directs EGFR toward lysosomes (19). Because attaching ubiquitin to EGFR is critical for its subsequent degradation, our data indicated that DGKδ might alter EGFR degradation by affecting the levels of ubiquitinated EGFR. To test this possibility, we used RNAi to knock down DGKδ in HeLa cells, treated the cells with EGF, and then measured the levels of ubiquitinated EGFR (EGFR-Ub). We found that DGKδ deficiency increased the amount of EGFR-Ub (Fig. 3A). We also found elevated levels of EGFR-Ub in DGKδ-deficient cells under steady-state conditions (Fig. 3A). Because ubiquitination of EGFR promotes its degradation, these results indicated that DGKδ deficiency modulates EGFR degradation in both ligand-activated and steady-state conditions.
FIGURE 3.
Enhanced ubiquitination of EGFR in DGKδ-deficient cells is not due to an effect on c-Cbl. A, HeLa cells were transfected with scrambled or DGKδ siRNA duplexes. In the left panel, cells were starved for 16 h and then treated with EGF (10 ng/ml) for 5 min. In the right panel (steady-state), cells were grown in medium with 10% serum for 48 h. EGFR was immunoprecipitated from cell lysates (1 mg of protein for EGF-treated cells and 1.5 mg of protein for steady-state cells) and then ubiquitin (Ub), EGFR, and DGKδ were detected by Western blotting the immunoprecipitation or cell lysates. Ub band densities (arbitrary units) are shown above the blot. B, HeLa cells were transfected with Myc-tagged wild-type or Y1045F EGFR and then DGKδ was knocked down using siRNA. After 48 h EGFR (anti-Myc), DGKδ, and actin were detected by immunoblotting. EGFR band densities (arbitrary units) are shown above the blot. C, HeLa cells were transfected with scrambled or DGKδ siRNA duplexes, and then grown in complete medium or starved for 24 h. DGKδ, EGFR, transferrin receptor (TfnR), or actin were then detected in cell lysates by Western blotting. TfnR and EGFR band densities (arbitrary units) are shown above the blots. D, HeLa cells were transfected with scrambled or DGKδ siRNA duplexes, starved for 16 h, and then treated with EGF (5 ng/ml) for 0–30 min. Total EGFR, pY1045EGFR, actin, and DGKδ were detected in cell lysates by Western blotting. Band densities (arbitrary units) of pY1045EGFR normalized to total EGFR are shown above the blot. E, HeLa cells were transfected with V5-tagged c-Cbl and scrambled or DGKδ siRNA duplexes. After starvation for 16 h, the cells were exposed to 50 or 100 ng/ml EGF for 10 min, and then c-Cbl was immunoprecipitated from 1 mg of cell lysates. EGFR, c-Cbl (anti-V5), and DGKδ were detected in the precipitates and cell lysates by Western blotting. F, HeLa cells were transfected with scrambled or DGKδ siRNA duplexes, starved for 16 h, and then treated with EGF (5 ng/ml) for 0–30 min. Total c-Cbl, pY774-c-Cbl, and DGKδ were detected in cell lysates by Western blotting. All blots are representative of at least three independent experiments.
The process of attaching ubiquitin to EGFR is mediated by the E3 ubiquitin ligase, c-Cbl, which docks on phosphotyrosine 1045 in EGFR. Mutating tyrosine 1045 to phenylalanine (Y1045F) impairs the ability of Cbl to bind to EGFR and consequently reduces the levels of EGFR-Ub (19, 20). Our data showing elevated levels of EGFR-Ub in steady-state conditions suggested that excessive ubiquitination led to reduced steady-state levels of EGFR. To demonstrate that modifying EGFR with ubiquitin was required for DGKδ deficiency to alter the steady-state expression levels of EGFR, we expressed wild-type EGFR or Y1045F EGFR in HeLa cells, reduced expression of DGKδ using RNAi, and then measured the levels of EGFR. We found that, although the expression of wild-type EGFR was attenuated in DGKδ-deficient cells, the Y1045F mutant was resistant to the effects of DGKδ deficiency (Fig. 3B). As another approach to show that the effects of DGKδ require ubiquitination, we examined the transferrin receptor that, like EGFR, undergoes clathrin-mediated endocytosis, but unlike EGFR it is recycled back to the plasma membrane because it is not ubiquitinated. We found that DGKδ deficiency did not affect the levels of the transferrin receptor (Fig. 3C). Collectively, these data indicated that ubiquitination of EGFR is required for DGKδ deficiency to reduce its steady-state levels and were consistent with a model where DGKδ deficiency enhances the levels of ubiquitin on EGFR, which accelerates its degradation.
DGKδ Modulates the Levels of an Enzyme That Removes Ubiquitin from EGFR
The excessive levels of EGFR-Ub in DGKδ-deficient cells suggested that c-Cbl might be abundantly active in DGKδ-deficient cells. To assess this possibility, we first examined expression of c-Cbl in cell lysates and found similar levels of c-Cbl in DGKδ-deficient and control cells (Fig. 3F). Next, we assayed the extent of phosphorylated Y1045 in EGFR, the major Cbl binding site, and found reduced phosphorylation of EGFR in DGKδ-deficient cells (Fig. 3D). This result is consistent with our previous data showing reduced levels of pY1068 and pY992 EGFR in conditions of DGKδ deficiency (see Fig. 5B and Ref. 5). Because Cbl can bind other tyrosines in EGFR and can also bind indirectly to EGFR, we additionally examined association of c-Cbl with EGFR and found slightly reduced coimmunoprecipitation of c-Cbl and EGFR in DGKδ-deficient cells (Fig. 3E). This change, however, could not account for the elevated levels of ubiquitin on EGFR. Finally, to measure changes in c-Cbl activation, we assayed the levels of tyrosine-phosphorylated c-Cbl and discovered that c-Cbl phosphorylation was reduced in DGKδ-deficient cells (Fig. 3F), indicating that c-Cbl was less active in conditions of DGKδ deficiency. Less phosphorylated c-Cbl was consistent with the reduced levels of phosphorylated EGFR in DGKδ-deficient cells. None of these changes could account for the excessive levels of EGFR-Ub in DGKδ-deficient cells, but there are other Cbl isoforms that we did not test (21). However, the significant attenuation of pY1045 and other EGFR phosphorylations in DGKδ-deficient cells argues against the possibility that other Cbl isoforms might contribute to the excessive ubiquitination of EGFR. Thus, we concluded that DGKδ deficiency did not enhance ubiquitination of EGFR through an effect on Cbl proteins.
FIGURE 5.
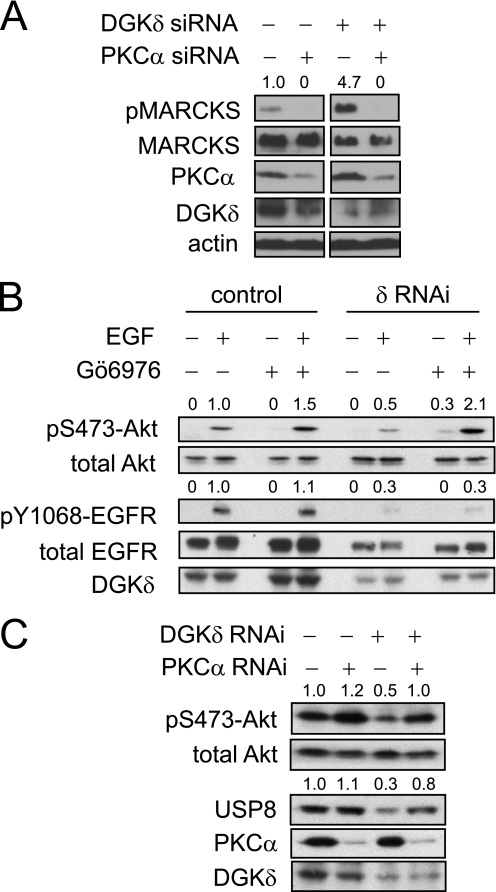
DGKδ and PKCα modulate Akt phosphorylation and USP8 levels. A, DGKδ and/or PKCα were depleted in HeLa cells using RNAi and then the levels of phosphorylated MARCKS (pMARCKS), MARCKS, PKCα, DGKδ, and actin were determined by Western blotting. Lanes were removed from the blot for clarity. The densities of the pMARCKS bands (arbitrary units) are indicated above the blot. B, HeLa cells were transfected with scrambled or DGKδ siRNA duplexes, starved for 16 h, exposed to 250 nm Gö6976 and then to EGF (10 ng/ml) for 10 min. The levels of the indicated proteins in cell lysates were detected by Western blotting and the densities of phosphorylated Akt and EGFR (arbitrary units) are indicated above the blots. C, HeLa cells were transfected with scrambled, PKCα, and/or DGKδ siRNA duplexes, grown in medium with serum, and then the levels of the indicated proteins in cell lysates were detected by Western blotting and the densities of phosphorylated Akt and USP8 (arbitrary units) are indicated above the blots.
Ubiquitination of EGFR is also modulated by de-ubiquitinases that remove ubiquitin from EGFR. Current data suggest that USP8 (which is also called UBPY) is primarily responsible for removing ubiquitin from EGFR (22, 23). To determine if USP8 was affected by DGKδ deficiency, we used RNAi to knock down DGKδ in HeLa cells and then assayed expression levels of USP8 by Western blotting. We consistently found reduced expression of USP8 in DGKδ-deficient cells (Fig. 4A). Thus, instead of affecting the process of attaching ubiquitin to EGFR, DGKδ appears to modulate its de-ubiquitination by helping maintain the levels of USP8.
FIGURE 4.
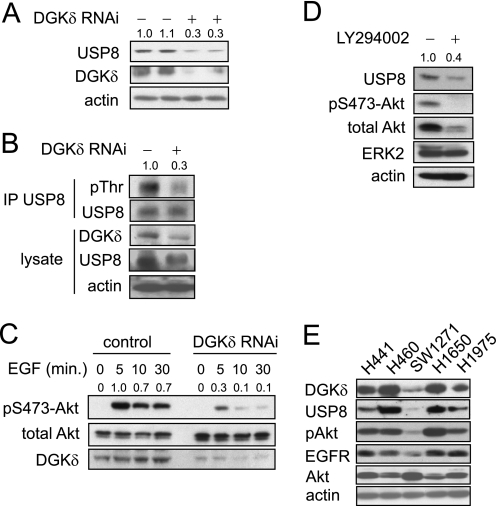
DGKδ deficiency reduces the levels of USP8 and attenuates Akt phosphorylation. A, in two separate experiments, HeLa cells were transfected with scrambled or DGKδ siRNA duplexes and then USP8, DGKδ, and actin were detected in cell lysates by Western blotting. USP8 band densities (arbitrary units) are shown above the blot. B, HeLa cells were transfected with V5-USP8 and then with DGKδ or control siRNA duplexes. Cells grown in medium with serum were treated with calyculin A (100 nm) for 5 min, and then lysates were harvested and V5-USP8 was immunoprecipitated. The indicated proteins were detected by Western blotting, and pThr band densities (arbitrary units) are shown above the blot. C, HeLa cells were transfected with scrambled or DGKδ siRNA duplexes, starved for 16 h, and then treated with EGF (5 ng/ml) for the indicated times. Akt, p473-Akt, and DGKδ were detected in cell lysates by Western blotting. Band densities (arbitrary units) of phosphorylated Akt are shown above the blot. D, HeLa cells growing in medium with serum were treated with 20 μm LY294002 for 24 h, and then the levels of the indicated proteins were detected in cell lysates by Western blotting. USP8 band densities (arbitrary units) are shown above the blot. E, lung cancer cell lines (noted above each lane) were grown in complete medium and harvested, and then the indicated proteins were detected by Western blotting.
DGKδ Affects USP8 by Modulating the Activation of Akt
We next performed a series of experiments aimed at determining the cause of altered USP8 expression. First, to determine if the effects of DGKδ on the levels of USP8 were due to transcriptional regulation, we used reverse transcription-PCR to assay the abundance of USP8 mRNA in control or DGKδ knockdown cells. We observed that there was no difference in the level of USP8 mRNA (not shown), indicating that the changes were due to post-transcriptional alterations. The abundance of USP8 can be modulated by Akt, which phosphorylates a threonine in USP8 that is thought to stabilize the protein by an unknown mechanism (24). We examined the levels of threonine-phosphorylated USP8 and found reduced levels in DGKδ-deficient cells (Fig. 4B), indicating that this might cause the attenuated expression of USP8. To test if this reduced phosphorylation of USP8 resulted from altered Akt activity, we measured Akt phosphorylation in response to EGF and found reduced levels of phosphorylated Akt in DGKδ-deficient cells (Fig. 4C). The levels of phosphorylated Akt were also reduced in DGKδ-deficient cells under steady-state conditions (Fig. 5C).
These data indicated that the reduced Akt activity caused by DGKδ deficiency might contribute to the attenuated levels of USP8. To test this possibility, we exposed cells to LY294002, a phosphatidylinositol 3-kinase inhibitor, for 24 h and then measured USP8 expression by Western blotting. We found that this inhibitor attenuated Akt phosphorylation and reduced the levels of USP8 (Fig. 4D). Total Akt was also reduced by the inhibitor, but ERK2 and actin levels were not altered. These changes in phosphorylated Akt and USP8 were similar to the effects of DGKδ deficiency. Finally, to determine if the levels of EGFR, DGKδ, USP8, and phosphorylated Akt in cancer cells paralleled our experimental observations, we examined five lung cancer cell lines and found that the levels of EGFR, USP8, and phosphorylated Akt correlated closely with those of DGKδ in most of the lines (Fig. 4E). Collectively, these data indicate that, by reducing Akt activation, DGKδ modulates the expression of USP8.
PKCα Is Necessary for DGKδ to Modulate USP8 Levels
Work in our laboratory has demonstrated that, by removing DAG, DGKs inhibit the function of DAG-activated proteins (7, 25, 26). We found evidence of augmented PKCα activity in DGKδ null cells and tissues (5), and others have observed that phorbol esters, which activate PKCs, reduced phosphorylation of Akt by inhibiting phosphatidylinositol 3-kinases (27). Together, these observations suggested that the excessive activation of PKCα in DGKδ-deficient cells might account for the reduced Akt phosphorylation that we observed.
To test this possibility, we first assayed the activity of PKCα under these conditions by measuring phosphorylated myristoylated alanine-rich protein kinase C substrate (MARCKS), a major PKC substrate. Knockdown of PKCα almost completely reduced MARCKS phosphorylation (Fig. 5A), indicating that PKCα is responsible for the majority of phosphorylated MARCKS in the cells. In DGKδ knockdown cells, we measured higher levels of phosphorylated MARCKS compared with control cells (Fig. 5A), demonstrating that knockdown of DGKδ enhanced PKCα activity. Next, we attempted to rescue Akt phosphorylation in DGKδ-deficient cells by inhibiting PKCα with Gö6976. We found that this inhibitor normalized Akt phosphorylation in DGKδ-deficient cells (Fig. 5B). In parallel, we also measured phosphorylation of EGFR and found that it was not rescued by Gö6976 (Fig. 5B). Together, these data indicated that the effects on Akt in DGKδ-deficient cells were due to PKCα and occurred downstream of EGFR.
As another approach to modulate the activity of PKCα and to measure Akt phosphorylation under steady-state conditions, we used RNAi to reduce the levels of PKCα and/or DGKδ, and then we measured phosphorylated Akt. Similar to ligand-activated conditions (Fig. 5B), the steady-state levels of phosphorylated Akt in DGKδ-deficient cells were reduced, but were rescued by PKCα knockdown (Fig. 5C). Collectively, these data suggested that, by modulating the function of PKCα, DGKδ regulates Akt signaling and consequently the levels of USP8. Finally, to determine if PKCα cooperated with DGKδ to modulate USP8 levels, we tested whether we could rescue USP8 expression in DGKδ-deficient cells by reducing the expression of PKCα. We found that knockdown of PKCα increased the expression of USP8 in DGKδ-deficient cells (Fig. 5C). Together, these data indicated that, through PKCα, DGKδ modulates the levels of USP8.
Depleting PKCα Rescues the EGFR Defects in DGKδ-deficient Cells
Our experiments suggested a model in which DGKδ regulates EGFR ubiquitination and consequent degradation by modulating the activity of PKCα. To test if PKCα was necessary for DGKδ deficiency to accelerate EGFR degradation, we simultaneously knocked down both PKCα and DGKδ using RNAi and then followed EGFR degradation by Western blotting. We found that PKCα knockdown normalized EGFR decay in DGKδ-deficient cells (Fig. 6, A and B), indicating that it was necessary for DGKδ to accelerate EGFR degradation. We found similar results with Gö6976 (not shown). In addition to testing ligand-induced degradation of EGFR, we also determined the effects of PKCα knockdown under steady-state conditions. We found that, like its effects on ligand-induced EGFR decay, RNAi knockdown of PKCα normalized steady-state levels of EGFR in DGKδ-deficient cells (Fig. 6C). Taken together, our data indicate that PKCα and DGKδ collectively regulate expression levels of EGFR during both steady-state and ligand-activated conditions by modulating its ubiquitination through Akt and USP8 (Fig. 6D).
Depleting DGKδ Reduces Growth of Lung Cancer Cells That Are Sensitive to EGFR Inhibitors
Finally, we tested whether reducing the expression of DGKδ affected the viability of lung cancer cell lines that were either resistant (H441) or sensitive (H1650) to EGFR inhibitors (28, 29). To examine this issue, we knocked down DGKδ using RNAi and then measured cell viability 72 h later. We found that depleting DGKδ reduced the viability of the sensitive H1650 cells but did not affect the resistant H441 cell line (Fig. 6D).
DISCUSSION
The EGFR has a central role in normal cell survival, differentiation, and proliferation. As such, its activity must be precisely regulated. This is accomplished, in part, through its removal via endocytic pathways that terminate in lysosomes where EGFR is degraded. A critical step in this process is the conjugation of ubiquitin to EGFR. The mechanisms responsible for attaching ubiquitin to EGFR have been studied in great detail. However, this is a reversible process, and evidence indicates that de-ubiquitinases also have important roles in maintaining the levels of EGFR and modulating its rate of degradation. Although there is some controversy about whether USP8 promotes or inhibits EGFR degradation (23, 30), evidence strongly supports that USP8 most likely stabilizes EGFR, and it appears to be a critical modulator of the levels of ubiquitin on EGFR. For example, USP8 de-ubiquitinates EGFR in vitro and depletion of USP8 by RNAi increases EGFR ubiquitination and enhances its degradation (23). Additionally, conditional deletion of USP8 in mice causes reduced steady-state levels of EGFR (22).
We found that DGKδ modulated the expression of USP8, altered the levels of ubiquitin on EGFR, modified the rate of EGFR degradation, and controlled the steady-state levels of EGFR. That DGKδ modulates USP8 is consistent with data showing that several features are shared by both USP8-deficient cells and DGKδ-deficient cells: both have reduced steady-state levels of EGFR (5, 22), increased ubiquitination and degradation of EGFR (Figs. 2 and 3) (23), and enlarged early endosomes (18, 23). Collectively, these data provide strong evidence that DGKδ is an important regulator of USP8 and of its effects on EGFR.
Little is known about how USP8 is regulated, but several phosphorylation sites on USP8 have been identified, and some of them alter its activity or stability. For example, phosphorylation of serine 680 in murine USP8 inhibits its activity by promoting association of USP8 with 14-3-3 proteins (31). We assayed the levels of phosphorylated serine 680 in overexpressed murine USP8 using a 14-3-3 binding site antibody and did not detect any differences between wild-type and DGKδ-deficient cells (not shown). Nor did we find evidence of changes in the affinity of USP8 for 14-3-3 proteins (not shown). Together, these data indicated that DGKδ does not affect USP8 activity by modifying serine 680 phosphorylation. Threonine 907 in murine USP8 is an Akt substrate, and phosphorylation of this residue stabilizes USP8 by an unknown mechanism (24). Several of the changes that we found in DGKδ-deficient cells are consistent with the possibility that DGKδ modifies USP8 levels by affecting phosphorylation of threonine 907. For example, both USP8 threonine phosphorylation and Akt phosphorylation were significantly reduced in DGKδ-deficient cells, and there were lower levels of USP8 protein. Moreover, inhibition of phosphatidylinositol 3-kinase attenuated the expression of USP8. Together, these data strongly argue that DGKδ modifies the levels of USP8 by modulating Akt activation.
The link between DGKδ, Akt, and USP8 appears to be PKCα, which was excessively active in DGKδ-deficient cells. The abnormal activity of PKCα that we discovered was most likely due to the elevated levels of DAG that we observed in DGKδ-deficient cells (5). How PKCα modulates the activation of Akt is an area that we are currently investigating. But prior data showing that activation of PKCs with phorbol ester reduced both Akt phosphorylation and phosphatidylinositol 3-kinase activity (27) suggest that this might be an effect on phosphatidylinositol 3-kinases. PKCs have long been known to modulate the activity and trafficking of many receptors, including EGFR, but the mechanisms by which they do so have been somewhat nebulous. Our data indicate that some of the effects on EGFR trafficking were caused by changes in its ubiquitination that, in turn, were mediated through PKCα and DGKδ.
Because we have also shown that DGKζ, a type IV DGK, regulates PKCα (26), we tested whether reducing the expression of DGKζ using RNAi altered EGFR degradation. We found that DGKζ had no effect on EGFR degradation (not shown), indicating that these two DGKs likely modulate the activity of PKCα under different circumstances. Indeed, their association with PKCα occurs in different contexts: while DGKζ binds PKCα only in non-stimulated conditions (26, 32), DGKδ binds PKCα predominantly in cells activated by EGFR ligands (not shown).
There are two DGKδ splice variants that are both associated with clathrin-coated vesicles (33). Using reverse transcription-PCR, we were able to detect only the DGKδ2 splice variant in HeLa cells, suggesting that it is responsible for modulating EGFR decay. Unfortunately, we have not been able to selectively knock down each splice variant in a suitable cell line to understand their specific effects on EGFR, but we are currently preparing the necessary reagents for rescue experiments.
In summary, our experiments show that DGKδ participates in regulating the expression levels of EGFR. However, the effect of depleting DGKδ on the viability of lung cancer cells was modest (Fig. 6D), calling into question the physiological significance of our findings. But one must consider several technical issues that might have diminished the effects of depleting DGKδ in those experiments. For example, the levels of DGKδ protein were not significantly reduced until about 24 h after the experiment began, providing only 48 h of depletion. Additionally, we only achieved a ∼75% reduction in DGKδ protein. Our DGKδ knock-out mice have a phenotype very similar to EGFR knock-out mice (5), suggesting that more robust and prolonged depletion of DGKδ might have more pronounced effects on EGFR signaling. Thus, it is reasonable to proceed with experiments testing the effects of prolonged depletion of DGKδ in appropriate cancer cell lines. It is interesting to note that DGKη, which is structurally similar to DGKδ, was recently shown to affect EGFR signaling, and depletion of DGKη reduced cell viability by a similar amount compared with DGKδ. However, these effects occurred by a different mechanism that affected Raf signaling (34). DGKα has also been shown to modulate receptor tyrosine kinase signaling (35–37), but DGKα null mice have no evidence of altered EGFR signaling (5–9). We and others have additionally generated mouse knockouts of DGKs ϵ, ζ, ι, and θ, all of which show no evidence of EGFR signaling defects (Refs. 5–9 and data not shown). Thus, it appears that regulation of EGFR signaling might be a unique property of the type II DGKs δ and η. Further studies to determine the suitability of these DGKs as therapeutic targets appear to be warranted.
Acknowledgment
We are indebted to Fumio Sakane for providing the DGKδ antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-CA95463 (to M. K. T.), P01-CA73992 (to D. M. S.), and R01-CA123541 (to K. L. C.). This work was also supported by the Huntsman Cancer Foundation, the R. Harold Burton Foundation, and by the Cancer Center Support Grant P30-CA042014 for support of core facilities.
- EGFR
- epidermal growth factor receptor
- DGK
- diacylglycerol kinase
- USP8
- ubiquitin-specific protease 8
- DAG
- diacylglycerol
- PKC
- protein kinase C
- pY1068-EGFR
- phosphotyrosine 1068 EGFR
- EGFR-Ub
- ubiquitinated EGFR
- ke
- endocytosis rate constant
- MARCKS
- myristoylated alanine-rich C-kinase substrate
- RNAi
- RNA interference
- siRNA
- small interference RNA
- wt
- wild type
- E3
- ubiquitin-protein isopeptide ligase.
REFERENCES
- 1.Roberts R. B., Min L., Washington M. K., Olsen S. J., Settle S. H., Coffey R. J., Threadgill D. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1521–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torrance C. J., Jackson P. E., Montgomery E., Kinzler K. W., Vogelstein B., Wissner A., Nunes M., Frost P., Discafani C. M. (2000) Nat. Med. 6, 1024–1028 [DOI] [PubMed] [Google Scholar]
- 3.Buchanan F. G., Holla V., Katkuri S., Matta P., DuBois R. N. (2007) Cancer Res. 67, 9380–9388 [DOI] [PubMed] [Google Scholar]
- 4.Sakane F., Imai S., Yamada K., Murakami T., Tsushima S., Kanoh H. (2002) J. Biol. Chem. 277, 43519–43526 [DOI] [PubMed] [Google Scholar]
- 5.Crotty T., Cai J., Sakane F., Taketomi A., Prescott S. M., Topham M. K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15485–15490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong X. P., Hainey E. A., Olenchock B. A., Jordan M. S., Maltzman J. S., Nichols K. E., Shen H., Koretzky G. A. (2003) Nat. Immunol. 4, 882–890 [DOI] [PubMed] [Google Scholar]
- 7.Regier D. S., Higbee J., Lund K. M., Sakane F., Prescott S. M., Topham M. K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 7595–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olenchock B. A., Guo R., Carpenter J. H., Jordan M., Topham M. K., Koretzky G. A., Zhong X. P. (2006) Nat. Immunol. 7, 1174–1181 [DOI] [PubMed] [Google Scholar]
- 9.Rodriquez de Turco E. B., Tang W., Topham M. K., Sakane F., Marcheselli V. L., Chen C., Taketomi A., Prescott S. M., Bazan N. G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4740–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakane F., Imai S., Kai M., Wada I., Kanoh H. (1996) J. Biol. Chem. 271, 8394–8401 [DOI] [PubMed] [Google Scholar]
- 11.Wiley H. S., Cunningham D. D. (1982) J. Biol. Chem. 257, 4222–4229 [PubMed] [Google Scholar]
- 12.Bao J., Alroy I., Waterman H., Schejter E. D., Brodie C., Gruenberg J., Yarden Y. (2000) J. Biol. Chem. 275, 26178–26186 [DOI] [PubMed] [Google Scholar]
- 13.Beguinot L., Hanover J. A., Ito S., Richert N. D., Willingham M. C., Pastan I. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 2774–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin C. R., Chen W. S., Lazar C. S., Carpenter C. D., Gill G. N., Evans R. M., Rosenfeld M. G. (1986) Cell 44, 839–848 [DOI] [PubMed] [Google Scholar]
- 15.Lund K. A., Lazar C. S., Chen W. S., Walsh B. J., Welsh J. B., Herbst J. J., Walton G. M., Rosenfeld M. G., Gill G. N., Wiley H. S. (1990) J. Biol. Chem. 265, 20517–20523 [PubMed] [Google Scholar]
- 16.Wiley H. S. (2003) Exp. Cell Res. 284, 78–88 [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki T., Kobayashi T., Ueyama T., Shirai Y., Saito N. (2008) Biochem. J. 409, 471–479 [DOI] [PubMed] [Google Scholar]
- 18.Pelkmans L., Fava E., Grabner H., Hannus M., Habermann B., Krausz E., Zerial M. (2005) Nature 436, 78–86 [DOI] [PubMed] [Google Scholar]
- 19.Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Mol. Cell 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 20.Levkowitz G., Waterman H., Ettenberg S. A., Katz M., Tsygankov A. Y., Alroy I., Lavi S., Iwai K., Reiss Y., Ciechanover A., Lipkowitz S., Yarden Y. (1999) Mol. Cell 4, 1029–1040 [DOI] [PubMed] [Google Scholar]
- 21.Schmidt M. H., Dikic I. (2005) Nat. Rev. Mol. Cell Biol. 6, 907–918 [DOI] [PubMed] [Google Scholar]
- 22.Niendorf S., Oksche A., Kisser A., Löhler J., Prinz M., Schorle H., Feller S., Lewitzky M., Horak I., Knobeloch K. P. (2007) Mol. Cell. Biol. 27, 5029–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizuno E., Iura T., Mukai A., Yoshimori T., Kitamura N., Komada M. (2005) Mol. Biol. Cell 16, 5163–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao Z., Wu X., Yen L., Sweeney C., Carraway K. L., 3rd (2007) Mol. Cell. Biol. 27, 2180–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Topham M. K., Prescott S. M. (2001) J. Cell Biol. 152, 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo B., Prescott S. M., Topham M. K. (2003) J. Cell Biol. 160, 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan L., Song K., Pysz M. A., Curry K. J., Hizli A. A., Danielpour D., Black A. R., Black J. D. (2007) J. Biol. Chem. 282, 14213–14225 [DOI] [PubMed] [Google Scholar]
- 28.Paez J. G., Jänne P. A., Lee J. C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F. J., Lindeman N., Boggon T. J., Naoki K., Sasaki H., Fujii Y., Eck M. J., Sellers W. R., Johnson B. E., Meyerson M. (2004) Science 304, 1497–1500 [DOI] [PubMed] [Google Scholar]
- 29.Sordella R., Bell D. W., Haber D. A., Settleman J. (2004) Science 305, 1163–1167 [DOI] [PubMed] [Google Scholar]
- 30.Row P. E., Prior I. A., McCullough J., Clague M. J., Urbé S. (2006) J. Biol. Chem. 281, 12618–12624 [DOI] [PubMed] [Google Scholar]
- 31.Mizuno E., Kitamura N., Komada M. (2007) Exp. Cell Res. 313, 3624–3634 [DOI] [PubMed] [Google Scholar]
- 32.Luo B., Prescott S. M., Topham M. K. (2003) J. Biol. Chem. 278, 39542–39547 [DOI] [PubMed] [Google Scholar]
- 33.Imai S., Yasuda S., Kai M., Kanoh H., Sakane F. (2009) Biochim. Biophys. Acta 1791, 246–253 [DOI] [PubMed] [Google Scholar]
- 34.Yasuda S., Kai M., Imai S., Takeishi K., Taketomi A., Toyota M., Kanoh H., Sakane F. (2009) J. Biol. Chem. 284, 29559–29570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chianale F., Cutrupi S., Rainero E., Baldanzi G., Porporato P. E., Traini S., Filigheddu N., Gnocchi V. F., Santoro M. M., Parolini O., van Blitterswijk W. J., Sinigaglia F., Graziani A. (2007) Mol. Biol. Cell 18, 4859–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bacchiocchi R., Baldanzi G., Carbonari D., Capomagi C., Colombo E., van Blitterswijk W. J., Graziani A., Fazioli F. (2005) Blood 106, 2175–2182 [DOI] [PubMed] [Google Scholar]
- 37.Baldanzi G., Mitola S., Cutrupi S., Filigheddu N., van Blitterswijk W. J., Sinigaglia F., Bussolino F., Graziani A. (2004) Oncogene 23, 4828–4838 [DOI] [PubMed] [Google Scholar]