Abstract
Interleukin-12 (IL-12), p80, and IL-23 are structurally related cytokines sharing a p40 subunit. We have recently demonstrated that celecoxib and its COX-2-independent analogue 4-trifluoromethyl-celecoxib (TFM-C) inhibit secretion but not transcription of IL-12 (p35/p40) and p80 (p40/p40). This is associated with a mechanism involving altered cytokine-chaperone interaction in the endoplasmic reticulum (ER). In the present study, we found that celecoxib and TFM-C also block secretion of IL-23 (p40/p19 heterodimers). Given the putative ER-centric mode of these compounds, we performed a comprehensive RT-PCR analysis of 23 ER-resident chaperones/foldases and associated co-factors. This revealed that TFM-C induced 1.5–3-fold transcriptional up-regulation of calreticulin, GRP78, GRP94, GRP170, ERp72, ERp57, ERdj4, and ERp29. However, more significantly, a 7-fold up-regulation of homocysteine-inducible ER protein (HERP) was observed. HERP is part of a high molecular mass protein complex involved in ER-associated protein degradation (ERAD). Using co-immunoprecipitation assays, we show that TFM-C induces protein interaction of p80 and IL-23 with HERP. Both HERP siRNA knockdown and HERP overexpression coupled to cycloheximide chase assays revealed that HERP is necessary for degradation of intracellularly retained p80 by TFM-C. Thus, our data suggest that targeting cytokine folding in the ER by small molecule drugs could be therapeutically exploited to alleviate inappropriate inflammation in autoimmune conditions.
Keywords: Cytokines/Interleukins, Protein/Degradation, Protein/Secretion, Chaperone Chaperonin, Endoplasmic Reticulum (ER), Celecoxib, HERP, Interleukin-23
Introduction
The interleukin-12 (IL-12)2 subfamily of cytokines is structurally based around a shared p40 subunit, covalently linked to p35 in IL-12 and to p19 in IL-23. In addition, p40 can be secreted both as a monomer and as a homodimer, named p80. Further members of this family include IL-27, which is a heterodimer consisting of the p40-related protein “Epstein-Barr virus-induced gene 3” (EBI3) and the p35-related subunit p28 and IL-35 (EBI3/p35) (1). IL-12 and IL-23 have been identified at elevated levels in a number of autoimmune diseases, including multiple sclerosis (MS) and its animal model EAE (2, 3), psoriasis (4), and inflammatory bowel disease (5). p80 has recently been shown to control macrophage chemotaxis and dendritic cell migration in lung inflammatory disease pathogenesis (1, 6–8). There is widespread interest in the assessment of IL-12-type cytokines as therapeutic targets for preventing or attenuation of disease. Monoclonal p40 antibodies (9, 10) have shown promise in limiting inflammatory disease severity. Various small molecule drugs have been reported that are capable of blocking IL-12, IL-23, and p80 at distinct stages of their biological pathways (transcription, assembly, receptor binding, and signal transduction) (for review see Ref. 11). One such compound is celecoxib (12).
Celecoxib is a non-steroidal anti-inflammatory drug originally designed to specifically inhibit cyclooxygenase-2 (COX-2), up-regulated in cancer and sites of inflammation (13). However, COX-2 inhibitors have been subject to critical analysis because of cardiovascular problems, and a number of functions related to celecoxib are now known to be attributable to COX-2-independent mechanisms (12–15). One such effect is the ability of celecoxib to increase cytoplasmic calcium concentrations through inhibition of ER Ca2+-ATPases (SERCA; Refs. 12, 16–18). Experiments with other COX-2 inhibitors such as rofecoxib, ibuprofen, aspirin, naproxen, DuP697, and NS298 have all shown that this calcium-mobilizing property is unique to celecoxib (12, 16).
We have shown that both celecoxib and a celecoxib analogue from which COX-2 inhibitory properties have been knocked out, i.e. 4-[5-(4-trifluoromethylphenyl)-3-(trifluoromethyl)-1H-pyrazol-1-yl]benzenesulfonamide, referred to hereafter as 4-trifluoromethyl-celecoxib (TFM-C), block IL-12 and p80 secretion through a post-transcriptional mechanism involving retention of the cytokines in the ER (12). TFM-C shares the effect of celecoxib in depleting ER Ca2+ stores in noncancerous HEK293 cells, i.e. the recombinant cytokine producer cells used in our study (12). For p80, this secretion block is accompanied with altered interaction with the ER chaperones calreticulin (CRT) and ERp44 (12). In the present study, we show that celecoxib and TFM-C block secretion of a third dimeric member of the IL-12 subfamily, namely IL-23, concomitant with intracellular retention of the IL-23 subunit p19. In an attempt to identify the ER factors crucially involved in this process we analyzed the effect of TFM-C on expression of 23 ER chaperones and cofactors. We identified homocysteine-inducible endoplasmic reticulum protein (HERP) as the gene transcript most dramatically up-regulated by TFM-C. HERP is a 54 kDa ubiquitously expressed ER membrane protein that is up-regulated as part of the ER-unfolded protein response (UPR) (19–23). HERP is required for destruction of a number of proteins via the ER-associated degradation (ERAD) pathway, such as CD3-delta, connexin 43, non-secreted Igκ LC, mutant Igγ LC, and truncated Igγ HC (24–26). HERP is a component of a larger ERAD retrotranslocation complex comprising the E3 ligase HRD1, Derlin-1, VIMP, and the ATPase p97 (26, 27). HERP knock-out also results in increased susceptibility to ER stress-related cell death by thapsigargin, tunicamycin, or A23187 (24). The proteasome inhibitor lactacystin accelerates cell death in HERP knock-out cells and blocks the degradation of connexin 43, Igκ LC, Igγ LC, and Igγ HC, indicating that HERP is involved in delivering ERAD-targeted proteins to the proteasome (24, 25). In this study, we show that TFM-C induces protein interaction of HERP with intracellularly retained p80 and IL-23, and that HERP is functionally required for the degradation of intracellularly retained dimeric p80.
EXPERIMENTAL PROCEDURES
Cell Culture Conditions
All tissue culture reagents were purchased from Invitrogen (Paisley, UK) unless otherwise stated. EcR293p40His cells stably expressing the pIND(SP1)p40His plasmid (12, 28) were cultivated in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS) purchased from Biosera (East Sussex, UK), 2 mm l-glutamine, 400 μg/ml zeocin, and 600 μg/ml G418 from Sigma (Poole, UK). HT29 cells were grown in minimum essential media (MEM) supplemented with 10% FBS and 2 mm l-glutamine. LNCaP cells were grown in RPMI1630, 10% FBS, 0.5% glucose, and 1% HEPES. Cells were grown at 37 °C and 5% CO2.
Small Molecule Compounds
Celecoxib and rofecoxib were obtained from Hefei Scenery Chemicals (Anhui, China). The celecoxib analogue TFM-C was synthesized by Onyx Scientific (Sunderland, UK). The geldanamycin analogue 17-(allylamino)geldanamycin (17AAG), and the ER ATPase inhibitor thapsigargin were both purchased from Sigma. All were dissolved in DMSO-based stock solutions before use.
Transient Transfection of the p19 Subunit of IL-23 into p40His-producing Cell Lines
The development of the ponasterone A-inducible HEK293-based EcR293p40His stable cell line has been described previously (28). This comprises HEK293 cells stably expressing both the pVRxR ecdysone receptor (EcR) plasmid and the pIND(SP1)p40His plasmid. To facilitate IL-23 expression, pIND(SP1)p19FLAG or pIND(SP1)p19His (as indicated) were transiently transfected into EcR293p40His cells to produce the IL-23 heterodimer p40/p19. All vectors were produced in our laboratory.
Purification of His-tagged p19, p40, p80, and IL-23 through Ni2+-NTA-Agarose
Purification of secreted individual subunits or subunit dimers was performed using Ni2+-nitrilotriacetic acid (Ni2+-NTA)-agarose (Invitrogen) as described previously (28).
Direct and Indirect Purification of p19FLAG
EcR293p40His cells transfected with 0.5 μg of pIND(SP1)p19FLAG were left for 24 h to recover. Drug treatment by celecoxib or TFM-C was initiated 2 h before induction with 5 μm ponasterone A to allow interaction of inhibitors with their targets before cytokine production. After 20 h of induction, cultured medium was removed and subjected to hexahistidine Ni2+-NTA-based purification to purify p19FLAG via p40His of IL-23. Alternatively, cells were washed with PBS and lysed in lysis buffer containing 50 mm Tris-HCl, 20 mm EDTA, and 1% nonidet P-40 for 15 min on ice. Samples were centrifuged for 15 min at 14,500 rpm and the supernatant retained. 0.25 μg of monoclonal α-M2-FLAG (Sigma) was added to lysis supernatant and turned on end for 16 h at 4 °C. Protein A-Sepharose (Sigma) was added, and samples were turned on end for 2 h at room temperature. Sepharose beads were pelleted gently at 1,000 rpm and washed three times with 0.1% Triton X-100 PBS, pH 7.2 with the resulting washed pellet treated with reducing SDS-PAGE loading buffer.
Effect on Secretion of IgG
Mouse IgG1 secreting hybridoma cells (SP2-derived) were plated in 6-well plates at a cell density of 4 × 105 cells per well and allowed to rest for 24 h. Cells were treated with 50 μm CE, 50 μm TFM-C, 5 μm thapsigargin, or DMSO only control. After 2 h, drug-containing medium was removed, and cells washed in copious PBS. Medium containing identical concentrations of ER-targeting compounds was re-added. After 8 h, medium was analyzed by reducing SDS-PAGE. Secreted IgG heavy chain was visualized by Western blot using goat anti-mouse horseradish peroxidase (Bio-Rad).
Primers for RT-PCR
Real-time PCR of ER chaperones was performed using the QuantiTect SYBR Green RT-PCR kit from Qiagen and the following Qiagen functionally validated primer sets for gene expression assays. GAPDH (QT01192646), CALR (QT00089215), GRP94 (QT00046963), GRP78 (QT00096404), ERp72 (QT00015883), ERp57 (QT00048776), ERp44 (QT00025263), GRP170 (QT00046214), DANJ (QT00197043), ERdj4 (QT00002716), ERdj5 (QT00088529), ERdj1 (QT00070182), ERdj3 (QT00042560), BAP (QT00073262), STCH (QT00045325), FKBP10 (QT00075229), SEC63 (QT00089719), ERp29 (QT00013153), LMAN1 (QT00065583), TXNDC12 (QT00079177), ERo1α (QT00096551), ERo1β (QT00050456), RAMP4 (QT00089327), HERP (QT00026418), and PDIA6 (QT00037086).
Quantification of ER Chaperone Gene Expression by RT- PCR
Samples for analysis of chaperone gene up-regulation in EcR293p40His, HT29, and LNCaP cells were obtained following treatment with 0.1 μm 17AAG, 50 μm TFM-C, 5 μm thapsigargin, or 100 μm rofecoxib for 6 h. Untreated cells acted as control. Total cellular RNA was extracted using the Absolutely RNA Miniprep kit from Stratagene. RNA was quantified using the Quant-iT Ribogreen RNA Assay kit from Invitrogen following the standard protocol. RT-PCR was performed on a DNA Engine Opticon 2 (MJ Research) using the recommended protocol of 30 min at 50 °C, 15 min at 95 °C, followed by 45 cycles of 15 s at 94 °C, 30 s at 56 °C, and 30 s at 76 °C. Reactions contained 2 ng of total cellular RNA, and all primer reactions were tested in the absence of template as negative control.
Optimization of HERP Membrane Protein Isolation
HERP purification was optimized to ensure compatibility with subsequent Ni2+-NTA enrichment of p80 or IL-23 folding complexes. Three protocols were compared. Approximately 30 × 106 EcR293p40His cells were added to three T175 flasks. Cells were subsequently treated with 1 μm thapsigargin for 16 h. Cultured medium was then removed, and cells were washed with PBS, scraped, and pelleted at 1000 rpm for 5 min. Protocol 1 was based on Oka et al. (29). The cell pellet was resuspended in 1 ml of sample buffer containing 10 mm Hepes-KOH pH 7.4, 250 mm sucrose, 10 mm KCl, 15 mm MgCl2, 1 mm EDTA, and 1 mm EGTA. Samples were homogenized by syringing through a 26-gauge needle 30 times at 4 °C. Samples were centrifuged at 1,000 × g for 5 min at 4 °C, and the subsequent pellet labeled as plasma membrane fraction. The supernatant was centrifuged again at 10,000 × g for 5 min with the resulting supernatant named the cytosolic fraction and the second pellet the ER membrane fraction. Protocol 2 was based on Lehner et al. (30). Cell pellets were resuspended in 40 mm Tris with 150 units/ml benzonase. Cells were vortexed for 5 min at 4 °C then sonicated 20 times in 1-s pulses. Vortexing and sonication steps were repeated. Samples were centrifuged at 12,000 × g for 10 min at room temperature. The supernatant was removed and labeled cytosolic fraction. The pellet was washed twice with 40 mm Tris with 150 units/ml benzonase buffer and resuspended in buffer containing 8 m urea, 2% w/v CHAPS, 2% w/v Triton X-100, 40 mm Tris, and 100 mm dithiothreitol. Vortexing, sonication, and centrifugation steps were performed as before with the resultant pellet labeled as membrane fraction and the supernatant labeled solubilized membrane proteins. For protocol 3, cells were resuspended in 8 m urea and protease inhibitors and left on ice for 90 min. Samples were centrifuged at 12,000 × g for 10 min at room temperature. The pellet was labeled membrane pellet whereas the supernatant was labeled soluble proteins. All labeled fractions were treated with reducing SDS-PAGE loading buffer and subsequently analyzed for the presence of HERP.
HERP Up-regulation at Protein Level
4 × 106 EcR293p40His cells were added to T75 flasks and left for 24 h. Cells were left untreated or treated with 50 μm TFM-C, 75 μm TFM-C, or 1 μm thapsigargin. At specific time intervals, medium was removed, cells were washed with PBS, and HERP was solubilized using a modified version of protocol 2 described above. The initial 40 mm Tris with 150 units/ml benzonase, vortexing, sonication, and centrifugation step was removed due to excessive loss of HERP at this step. Cells were resuspended in 8 m urea, 10 mm Tris-HCl, pH 8.0 containing generic protease inhibitors. Vortexing, sonication, centrifugation, and treatment with reducing loading buffer proceeded as before.
Cross-linking and Immunoprecipitation of p80-HERP and IL-23-HERP Complexes
Approximately 30 × 106 EcR293p40His cells were added to three T175 flasks each. Two were induced for 8 h with 5 μm ponasterone A. One flask was left uninduced as a control. One induced flask was treated with 50 μm TFM-C and all were left for 24 h. After this time, culture medium was discarded and cells washed with ice-cold PBS before being scraped and pelleted at 1000 rpm for 5 min. Cell pellets were treated with dithiobis(succinimidyl-propionate) (DSP) thiol-containing cross-linker to a final concentration of 100 μg/ml and immediately vortexed before incubation for 30 min at room temperature. Tris-HCl, pH 7.5 was added to a final concentration of 50 mm, vortexed, and incubated at room temperature for 15 min to quench the cross-linking reaction. Cross-linked cell pellets were lysed using a modified version of protocol 2 based on membrane protein solubilization in 8 m urea and 1% protease inhibitors, as outlined earlier. Samples were processed for purification on Ni2+-NTA agarose as described before (12). The procedure used for isolation of IL-23-HERP complexes was similar with the following exception: EcR293p40His cells were transfected with pIND(SP1)p19His 48 h prior to induction with ponasterone A.
Western Blotting and Immunodetection
Ni2+-NTA-agarose-purified media samples were separated through either reduced or non-reduced 4–12% Bis-Tris NuPAGE gels using NuPAGE MOPS running buffer (Invitrogen). After blotting, polyvinylidene difluoride membranes (Amersham Biosciences/GE Healthcare) were blocked with 5% nonfat dry milk in PBS for 1 h and probed with the following primary antibodies; mouse α-p40 antibody C8.6 (BD Biosciences), goat α-p19 antibody (R&D Systems), mouse α-HERP HT2 (29), and mouse α-M2-FLAG antibody (Sigma). For immunoblot detection of p40His and p19His following reducing SDS-PAGE we used an α-His6 mouse IgG1 mAb (Clone BMG-His-1; Roche). The secondary antibodies used were horseradish peroxidase (HRP)-conjugated goat α-mouse (Jackson Immunoresearch), and HRP-conjugated cross-reactive donkey α-sheep/goat (Serotec). Immobilon Western Substrate (Millipore) chemiluminescent detection reagent and Kodak autoradiographic film (Amersham Biosciences/GE Healthcare) were used to visualize protein bands present.
HERP siRNA Knockdown and CHX Chase Experiment
5 × 105 EcR293p40His cells were plated in 24-well plates and left for 16 h. Cells were treated with a final concentration of 50 nm validated AllStars control siRNA or 50 nm validated HERP siRNA both from Qiagen, using Lipofectamine RNAiMax siRNA transfection reagent (Invitrogen), based on the manufacturer's recommended protocol. After 48 h, cell medium was changed, and siRNA re-added. Cells were induced with 5 μm ponasterone A 2 h after siRNA re-addition. After a further 22 h, cells were treated with 50 μm TFM-C. Two hours later, cells were treated with 10 μg/ml CHX. At 0, 2, 4, and 8 h after CHX addition, cells were washed in PBS and lysed on ice in buffer containing 50 mm NaH2PO4, 300 mm NaCl, 15 mm imidazole, 0.5% Triton X-100, and 1% protease inhibitors, pH 8.0. Lysates were subjected to Ni2+-NTA-agarose purification. The resulting eluate was mixed with non-reducing SDS-PAGE loading buffer and subjected to SDS-PAGE and Western blot.
Statistics
Statistical analysis was performed using SPSS software (SPSS, Chicago). Data were analyzed for homogeneity of variance by Levene's test. One-way ANOVA and univariate ANOVA tests were carried out where applicable and are indicated in figure legends. Post-hoc Bonferroni correction was applied where appropriate. Data are plotted as mean ± S.E. with significance indicated as p < 0.05.
RESULTS
Celecoxib and TFM-C Inhibit IL-23 Secretion by Causing Intracellular Cytokine Retention
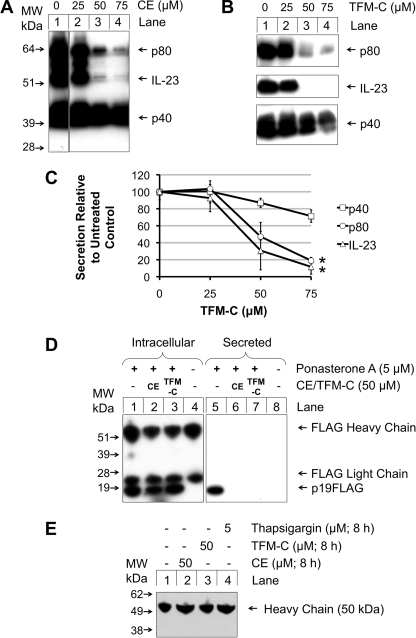
We have previously shown that secretion of IL-12 and p80 is blocked by both celecoxib and TFM-C (12). To further characterize the properties of TFM-C on members of the IL-12 dimer cytokine family, EcR293p40His cell lines previously described (12, 31) were transiently transfected with pIND(SP1)p19FLAG to facilitate secretion of the p40/p19 heterodimer IL-23. Secretion of both p80 and IL-23 was largely inhibited at a concentration of 50 μm celecoxib (Fig. 1A), whereas that of p40 was relatively unaffected. Similar effects were seen with TFM-C, which exhibits a 205-fold higher IC50 for inhibition of COX-2 (12) (Fig. 1B). In a semi-quantitative analysis of 3 experiments by densitometry (Fig. 1C), a minor decrease in p40 monomer secretion is seen at 50 μm TFM-C, while at this concentration IL-23 and p80 secretion show a significant 50–75% drop. At 75 μm TFM-C, p40 monomer levels have dropped by 25% while p80 and IL-23 secretion has decreased by 80–90%. In Fig. 1D, 50 μm celecoxib or TFM-C were added to cells secreting IL-23, and both secreted and intracellular p19 levels were analyzed. Intracellular levels of the IL-23 subunit p40 have previously been shown to be unaffected and were therefore not examined (12). Celecoxib or TFM-C did not reduce intracellular levels of p19, immunoprecipitated, and detected via the C-terminal FLAG peptide (Fig. 1D, left panel). In the right panel of Fig. 1D, secreted IL-23 was captured on Ni2+-NTA via the His tag on p40, and the cytokine was detected in Western blot via the FLAG tag on p19 subunit. This revealed virtually complete suppression of IL-23 secretion upon drug treatment. Thus, in agreement with our earlier data on IL-12 and p80 (12), celecoxib and TFM-C seem to block secretion of IL-23 through a COX-2-independent process involving intracellular retention of the cytokine.
FIGURE 1.
Celecoxib and TFM-C inhibit secretion of recombinantly expressed IL-23 and p80 but only to a lesser extent of monomeric p40. A and B, EcR293p40His cells were transfected with 0.5 μg of pIND(SP1)p19FLAG. After 24 h, cells were treated with a range of concentrations of celecoxib (A) or TFM-C (B) (lanes 2–4) or left as untreated control (lane 1). After 2 h of celecoxib treatment, cells were induced with 5 μm ponasterone A, and 20 h later, secreted His-tagged proteins in the medium were purified using Ni2+-NTA-agarose and subjected to non-reducing SDS-PAGE and Western blot. Blots were probed with α-p40 C8.6, which detects p40, p80, and IL-23. C, effects of TFM-C on secretion of p40, p80, and IL-23 were quantified by densitometric scanning. Values are expressed as the mean of three experiments ± S.E. Statistical analysis for differences between p40 and either p80 or IL-23 at corresponding concentrations was carried out by univariate ANOVA with Bonferroni correction, *, p < 0.05. D, EcR293p40His cells were transfected with pIND(SP1)p19FLAG. 50 μm celecoxib (lane 2) or 50 μm TFM-C (lane 3) were added. After 2 h, cells corresponding to lanes 1–3 were induced with 5 μm ponasterone A with lane 1 acting as induced but drug-free control and lane 4 as uninduced drug-free control. After 20 h, intracellular p19FLAG in cell lysates was purified by α-M2 FLAG protein A-Sepharose immunoprecipitation and resolved on reducing-PAGE and Western blot. Corresponding culture medium (lanes 5–8) was Ni2+-NTA purified to capture IL-23 via p40His, resolved by reducing-PAGE and Western blot. Blots were probed with α-M2 FLAG against p19FLAG of IL-23. E, IgG1-secreting hybridoma cells were pretreated with 50 μm CE (lane 2), 50 μm TFM-C (lane 3), or 5 μm thapsigargin (lane 4), or left untreated (lane 1). After 2 h, medium was removed, cells washed with PBS, and medium containing each compound at identical concentrations was re-added. After 8 h, medium was subjected to reducing SDS-PAGE and probed for IgG heavy chain secretion by Western blot.
To test whether TFM-C would similarly inhibit secretion of other oligomeric proteins, its effect was assessed on an IgG1 secreting hybridoma cell line (Fig. 1E). The α-mouse antibody utilized detected only the heavy chain of the secreted IgG. Because the heavy chain of IgG is only secreted in complex with the light chain (32), this can be taken as a measure of whole IgG secretion. In both cases, 50 μm celecoxib or TFM-C had no effect on IgG secretion. The ER Ca2+-ATPase inhibitor thapsigargin has previously been shown to inhibit p80 secretion at as low a concentration as 0.05 μm (12). Even when used at the high concentration of 5 μm, thapsigargin did not inhibit IgG secretion. Thus, similar to thapsigargin, TFM-C appears to display a certain level of selectivity in the type of oligomeric proteins of which it blocks secretion. All further experiments were performed with TFM-C only.
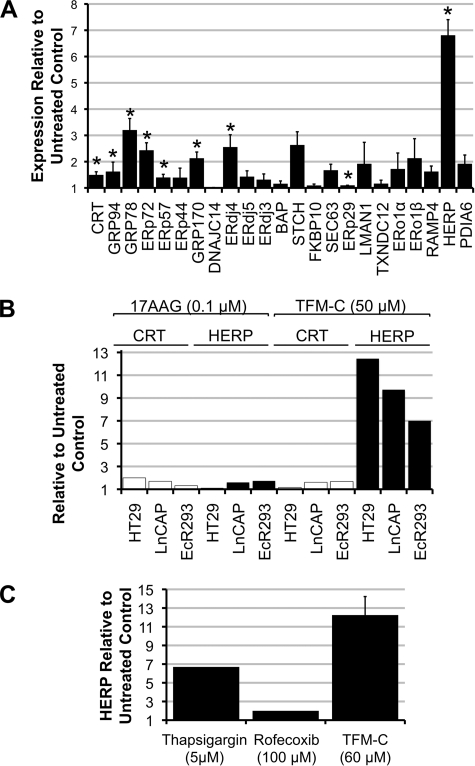
TFM-C Significantly Up-regulates the ERAD-linked Protein HERP
To study the fate of intracellularly retained IL-12 family subunits, gene expression changes for a panel of ER-resident proteins were examined upon treatment with TFM-C. This was achieved through RT-PCR of 23 ER-resident proteins linked to ER quality control, chaperone function, and ERAD (Fig. 2A). 50 μm TFM-C was used, as this was the lowest concentration of TFM-C capable of quantitative inhibition of p80, IL-12, and IL-23 secretion. In this screen, CRT, GRP94, GRP78, ERp72, ERp57, GRP170, ERdj4, ERp29, and HERP were all shown to undergo up-regulation of statistical significance. HERP, an ER membrane protein, previously identified to be involved in degradation of ERAD-targeted proteins (25, 26), was up-regulated by TFM-C by 700% in comparison with untreated control samples. To determine if this was a cell type- and/or drug-specific effect, up-regulation of HERP and CRT (as a control) was examined in HT29 and LNCaP cell lines alongside EcR293p40His cells treated with TFM-C or the GRP94 inhibitor 17-(allylamino)geldanamycin (17AAG) (Fig. 2B). In all cell lines tested, TFM-C was found to significantly upregulate HERP gene expression with little effect on CRT. 17AAG caused only minor increases in CRT and HERP expression levels. We also compared TFM-C to the selective COX-2 inhibitor rofecoxib and the ER Ca2+-ATPase inhibitor thapsigargin in HT29 cells (Fig. 2C). 100 μm rofecoxib showed a negligible effect on HERP expression. 5 μm thapsigargin induced a 7-fold increase in HERP expression levels, whereas 50 μm TFM-C induced a 12-fold increase. Thus, the dramatic up-regulation of HERP seen with TFM-C seems to be associated with ER Ca2+ perturbance because it can be reciprocated with thapsigargin.
FIGURE 2.
TFM-C dramatically up-regulates HERP gene expression in both EcR293p40His cells and cancer cell lines. A, RT-PCR screen for the effects of 50 μm TFM-C on gene expression of 23 ER factors. EcR293p40His cells were treated with 50 μm TFM-C for 6 h after which total RNA was extracted and quantified. mRNA levels for 23 ER-related proteins were analyzed by real time-PCR for changes in expression. Results are calculated relative to untreated control cells and corrected for changes in GAPDH expression. Values are expressed as the mean of three independent experiments ± S.E. Statistical analysis for significance compared with untreated control was performed using the 2-tailed Student's t test, *, p < 0.05. CRT, calreticulin; GRP, glucose-regulated protein; ERdj, ER-associated DNAJ; BAP, BiP-associated protein; STCH, stress 70 protein chaperone; FKBP10, FK506-binding protein 10; SEC63, ER protein translocation subcomplex subunit Sec63; LMAN1, lectin mannose-binding 1; TXNDC12, thioredoxin domain-containing 12; ERo, ER oxidoreductase; RAMP4, ribosome-associated ER protein 4; PDIA6, protein-disulfide isomerase A6. B, HERP up-regulation by 50 μm TFM-C is shared by HT29, LNCaP, and EcR293 cell lines. HT29, LNCaP, and HEK293 (EcR293p40His) cells were treated with 0.1 μm 17AAG or 50 μm TFM-C for 6 h. Total cellular RNA was extracted, quantified, and assayed by real-time PCR for up-regulation HERP and the calcium-binding chaperone CRT. Results are calculated relative to untreated control and corrected for changes in GAPDH expression. All values are expressed as the average of two or three experiments, whereas LNCaP data for 17AAG up-regulation of HERP is expressed as a single representative experiment. C, HERP up-regulation is induced by both thapsigargin and TFM-C, but not by the COX-2 inhibitor rofecoxib. HT29 cells were treated for 6 h as indicated, after which total cellular RNA was extracted and quantified. HERP gene expression was assayed by RT- PCR. Values for TFM-C are expressed as the average of two experiments ± S.D. Values for thapsigargin and rofecoxib are shown as a single representative experiment.
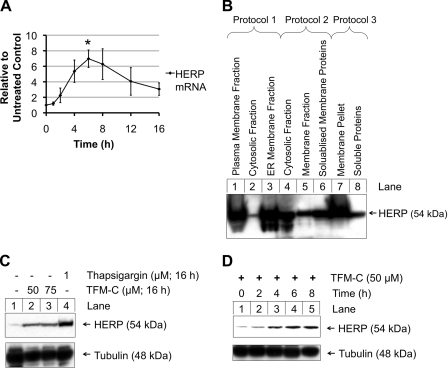
Kinetics of HERP mRNA and Protein Induction by TFM-C and Optimization of HERP Purification from ER Membranes
The up-regulation of HERP at the mRNA level upon TFM-C treatment (50 μm) was further analyzed in a time course assay (Fig. 3A), revealing that HERP gene expression was rapidly induced, peaking in the 4–8-h time period, after which mRNA levels gradually decreased until 16 h. We sought to determine if p80 homodimers retained in TFM-C-treated cells interacted with HERP. This required confirmation that HERP was up-regulated at the protein level. HERP being a membrane protein, considerable effort was dedicated to identification of a suitable solubilization approach facilitating release of HERP from the ER membrane in a form compatible with its capture via His-tagged p40 on Ni2+-NTA-agarose. Three isolation protocols were tested starting from cells treated with thapsigargin, previously shown to significantly up-regulate HERP (33) (Fig. 3, B and C). Full details of these protocols are provided under “Experimental Procedures.” With protocol 2 resulting in significant amounts of solubilized HERP (Fig. 3B, lane 6) and being compatible with Ni2+-NTA chromatography, further experiments were performed using this protocol. Up-regulation of HERP at the protein level by TFM-C, and thapsigargin was demonstrated (Fig. 3C). In time curve analysis, up-regulation of HERP protein levels began from 4 h onwards (Fig. 3D). Levels of tubulin were unaffected by TFM-C treatment (Fig. 3, C and D).
FIGURE 3.
TFM-C up-regulates mRNA and protein levels of HERP. A, kinetics of HERP mRNA up-regulation by TFM-C. EcR293p40His cells were treated with 50 μm TFM-C. At 0, 2, 4, 8, 12, and 16 h, total cellular RNA was extracted, quantified, and assayed by RT-PCR for HERP gene expression. Values were calculated relative to a corresponding untreated control sample and corrected for changes in GAPDH. The 6-h value is expressed as the mean of six experiments ± S.E. All other time points are expressed as the mean of three experiments ± S.E. Statistical analysis was performed by univariate ANOVA with Bonferroni correction, *, p < 0.05. B, HERP immunoblot of cell fractions obtained through three different protocols for purification of HERP compatible with Ni2+-NTA purification (see “Experimental Procedures”) for a full description. C, EcR293p40His cells were treated with 50 μm TFM-C (lane 2), 75 μm TFM-C (lane 3), or 1 μm thapsigargin (lane 4), or were left untreated (lane 1). After 16 h, cells were harvested and subjected to HERP solubilization via a simplified version of protocol 2 (see “Experimental Procedures”). Samples were subjected to reducing SDS-PAGE and Western blot using α-HERP HT2. D, EcR293p40His cells were treated with 50 μm TFM-C and harvested after 0, 2, 4, 6, and 8 h of treatment. Samples were subjected to HERP solubilization using the simplified protocol 2 and resolved using reducing-PAGE and Western blot with α-HERP HT2.
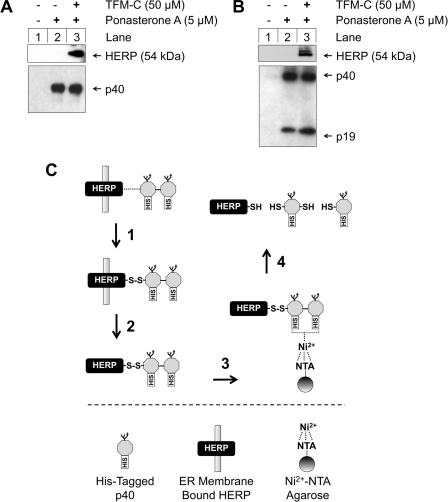
TFM-C Induces Interaction of HERP with p40/p80 and IL-23
We attempted to clarify whether direct protein interactions occur between HERP and p40/p80 or IL-23 in TFM-C-treated cells (Fig. 4). A schematic diagram outlining the successive steps that formed the basis of this experiment, comprising p40-HERP complex cross-linking with DSP, HERP solubilization, and p40His Ni2+-NTA-agarose affinity purification, is shown in Fig. 4C. Cross-linking by the thiol-cleavable cross-linker DSP was essential to maintain the noncovalent interaction between p40 and HERP following membrane protein solubilization with 8 m urea. Probing for co-captured HERP identified HERP co-purified with p40/p80 (Fig. 4A) or IL-23 (Fig. 4B) only under the conditions of both cytokine induction and TFM-C treatment. No HERP was shown to be co-captured in the absence of cytokine expression or without TFM-C treatment. As p80 and IL-23 are retained in the ER upon TFM-C treatment (but less so the p40 monomer), the cytokine population associated with HERP is likely to represent p80 and IL-23 rather than a single-standing p40 monomer.
FIGURE 4.
HERP interacts with p80 and IL-23 in TFM-C-treated but not in untreated cells. A, EcR293p40His cells were induced with 5 μm ponasterone A (lanes 2 and 3) to produce p40His. After 8 h, 50 μm TFM-C was added (lane 3). After another 16 h, cells were harvested, and incubated with DSP. The modified protocol 2, compatible with Ni2+-NTA purification was used to solubilize HERP from the ER membrane. p40His-containing cross-linked complexes were purified through Ni2+-NTA. Eluted p40 and co-captured HERP were resolved by reducing SDS-PAGE and Western blot probed with α-His6 Ab (for detection of p40) or α-HERP HT2. B, similar experiment using EcR293p40His cells transfected with pIND(SP1)p19His. p40 and p19 were detected using the α-His6 mAb. Eluates were quantified to ensure equal protein loading on SDS-PAGE. C, schematic representation of the procedure used to demonstrate HERP-p40His interaction. After induction of p40 and treatment with TFM-C: 1) cell pellets were resuspended and cross-linked using the thiol-cleavable cross-linker DSP to convert any non-covalent into covalent protein interactions; 2) HERP was solubilized from the ER membrane, thus facilitating 3) affinity capture of HERP-containing p40His complexes on Ni2+-NTA; subsequently, His-tagged proteins were eluted from Ni2+-NTA-agarose using 50 mm EDTA; 4) this eluate was then reduced to break the DSP cross-linker and to allow components co-captured with p40His to be identified as individual bands on reducing SDS-PAGE/immunoblot.
siRNA Knockdown Coupled to CHX Chase Experiments Reveal That HERP Is Required for p80 Degradation in TFM-C-treated Cells
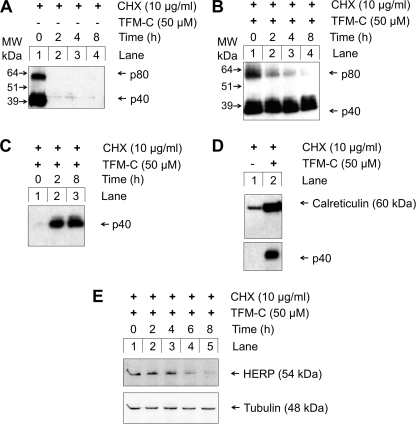
To investigate whether the interaction between p40/p80 and HERP is functionally relevant and leads ultimately to degradation of p80 homodimers, we investigated the effect of siRNA knockdown of HERP on intracellular p80 levels in CHX chase experiments.
CHX used for a short time blocks translation of p40 and thus indirectly entry of newly formed cytokines into the ER, whereas TFM-C treatment prevents exit of p80 from the secretory pathway. The combination of both treatments would act to create an intracellularly trapped population of p80 dimers, which is stable over time, unless it is subject to degradative mechanisms. Fig. 5A shows that following addition of CHX, as expected, cells empty their p40 and p80 content. The same experiment, performed this time in the presence of TFM-C, shows that at 2 h of CHX chase, intracellularly trapped p80 homodimers are largely degraded and at 4–8 h are practically absent (Fig. 5B). However, under these conditions, intracellular p40 persisted over the duration of the CHX chase experiment. Nonetheless, a significant fraction of monomer p40 was secreted under these conditions (Fig. 5C), as anticipated (see Fig. 1, A and B). The intracellularly retained undegraded p40 population present at 6 h of CHX chase was further analyzed and appeared to be associated with CRT (Fig. 5D). Thus, it is likely that this population may not be the secreted monomeric form, but p40 committed to the assembly of p80, prior to disulfide bond formation. As such, this population would be in the process of folding, recognized by the calreticulin ER quality control system, and intercepted. We have previously demonstrated a role for CRT in IL-12 and p80 folding (12, 28). Disulfide bond formation has previously been identified as an intermediate or final event following initial subunit association, preceding successful secretion from the ER (34). At any rate, as the fate of disulfide-bonded p80 was the main focus of this experiment, we have not further investigated this retained yet undegraded p40 population in more detail.
FIGURE 5.
Divergent effects of cycloheximide chase on p80 and p40 in TFM-C-treated cells. A and B, EcR293p40His cells were induced to produce p40/p80. Following 24 h, cells were treated with 50 μm TFM-C for 2 h (B only) after which 10 μg/ml CHX was added. At 0, 2, 4, and 8 h after CHX addition, cells were harvested and lysed. p40/p80 was purified using Ni2+-NTA-agarose, resolved on non-reducing SDS-PAGE and detected using α-p40 C8.6 by Western blot. C, secreted p40 was purified from the culture medium of TFM-C-treated cells at 0, 2, or 8 h after addition of CHX. D, EcR293p40His cells were treated with TFM-C for 2 h before treatment with CHX for a further 6 h. Intracellular p40His was isolated by Ni2+-NTA chromatography and probed for associated calreticulin. Lane 1, uninduced drug-free control. E, CHX chase experiments were carried out to determine the kinetics of degradation of HERP. EcR293p40His cells were pretreated with 50 μm TFM-C before addition of CHX. Cells were lysed in 8 m urea, 10 mm Tris-HCl, pH 8.0, and probed with the α-HERP Ab HT2.
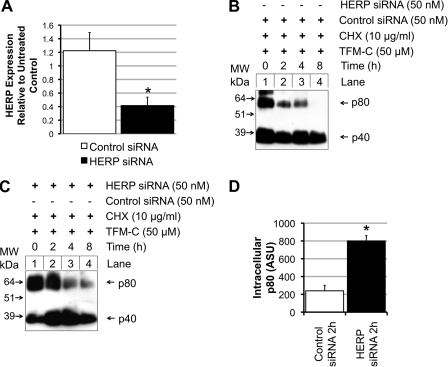
Fig. 5E shows the degradation kinetics of HERP following CHX addition, which coincide with those published before (23). HERP levels remained roughly constant for up to 4 h of CHX chase, as opposed to the much faster disappearing p80 population (Fig. 5B). Introduction of HERP siRNA was capable of reducing HERP mRNA by 60% in RT-PCR experiments (Fig. 6A). In Fig. 6B, the CHX chase experiment was repeated, this time using non-silencing control siRNA, confirming rapid degradation of p80 at 2 h. When the experiment was performed using HERP siRNA, p80 levels were stabilized against degradation at the 2-h time point (Fig. 6C, compare lanes 2 in B and C). Semi-quantitative approximation of p80 levels by densitometry at the 2-h time point over three independent experiments (Fig. 6D), showed that HERP siRNA caused a 4-fold increase in intracellular p80 levels at 2 h of CHX chase when compared with non-silencing siRNA. The stabilization of the trapped intracellular p80 population in response to HERP knockdown appeared only sustainable over the initial 2-h period of the experiment with degradation returning at the 4- and 8-h time points. This lack of effect at the 4- and 8-h time points (compare Fig. 6, B and C, lanes 3 and 4), is probably due to ineffectiveness of the siRNA over a longer time period, or to engagement of alternative HERP-independent degradation pathways.
FIGURE 6.
HERP siRNA knockdown blocks intracellular degradation of p80 in TFM-C-treated cells. A, HERP siRNA represses HERP mRNA expression levels. 50 nm control siRNA and 50 nm HERP siRNA were transfected into EcR293p40His cells. After 48 h, the medium was changed, and new siRNA was added. After a further 24 h, cells were harvested, and total RNA extracted and quantified. The effects of control siRNA and HERP siRNA on HERP mRNA levels were assayed by RT-PCR. Results are expressed relative to untreated control cells and corrected for changes in GAPDH. Values are stated as the mean of three experiments ± S.E. Statistical analysis was performed by one-way ANOVA, *, p < 0.05. B+C, EcR293p40His cells were transfected with 50 nm control siRNA (B) or 50 nm HERP siRNA (C) for 48 h, after which the medium was changed and siRNA replaced. After 24 h, 50 μm TFM-C were added, and 2 h later 10 μg/ml CHX. At 0, 2, 4, and 8 h after CHX addition, cells were harvested and lysed. p40/p80 was purified using Ni2+-NTA-agarose, resolved on non-reducing SDS-PAGE and detected using the α-p40 C8.6 by Western blot. D, effects of HERP siRNA relative to control siRNA on intracellular p80 levels at 2 h of CHX treatment were quantified by densitometric scanning. Values are expressed as the mean of three experiments ± S.E. Statistical analysis was performed by one-way ANOVA, *, p < 0.05.
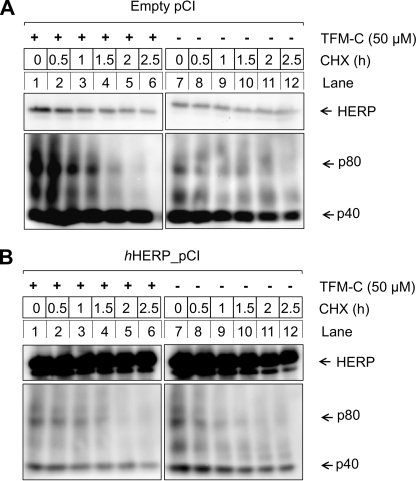
Overexpression of HERP Enhances Degradation of p80 in TFM-C-treated but Not in Untreated Cells
A complemental experiment was performed in which we examined the effect of overexpression of full-length human HERP on p80 degradation in TFM-C-treated or untreated cells (Fig. 7). Overexpression of HERP greatly enhanced degradation of p80 in TFM-C-treated cells, but had no effect on the level of p80 in untreated cells. Thus, high levels of HERP in the absence of ER stress do not seem to interfere with the productive folding of p80.
FIGURE 7.
Overexpression of HERP enhances degradation of p80 in TFM-C-treated but not in untreated cells. EcR293p40His cells were transfected with empty pCI vector (A) or pCI expressing full-length human HERP from the CMV promoter (B). At 30 h after transfection, cells were induced with ponasterone A, and at 48 h, cells were treated with 50 μm TFM-C or a similar volume of DMSO. Two hours later, 30 μg/ml CHX was added. At the times indicated, cells were harvested and p40/p80 was purified from the soluble lysate on Ni2+-NTA-agarose, and subjected to SDS-PAGE and Western blot (antibody used was C8.6). Insoluble cell pellets were dissolved in 8 m urea, 10 mm Tris-HCl, pH 8.0 and sonicated. Detection of HERP was performed with the HT2 antibody.
DISCUSSION
In this study we have demonstrated the ability of celecoxib and the non-coxib analogue TFM-C to inhibit the secretion of IL-23, as well as of IL-12 (p35/p40) and p80 (p40/p40) (12), whereas leaving secretion of monomeric p40 largely unaffected. TFM-C strongly up-regulates the expression of the ER transmembrane protein HERP, and by use of co-immunoprecipitation, CHX chase, and siRNA/overexpression assays, HERP is involved in delivery of p80 and IL-23 for degradation in TFM-C-treated cells.
In conjunction with our earlier work (12), we can now present a more detailed rationale for the putative mechanism by which TFM-C inhibits folding and secretion, and induces degradation of IL-12-type cytokines. Whereas TFM-C was designed to have no COX-2 inhibitory activity (IC50 for inhibition of COX-2 of 8.2 μm for TFM-C versus 0.04 μm for celecoxib; see Ref. 12), both celecoxib and TFM-C perturb Ca2+ homeostasis, most likely through blockage of ER Ca2+- ATPases (12, 16). TFM-C selectively inhibits secretion of dimeric polypeptide conformers of the IL-12 family (IL-12, p80, and IL-23), and to a much lesser extent of p40 monomers. Analysis of IgG1 secretion (Fig. 1) demonstrates that TFM-C-mediated perturbation of ER function does not induce a generic secretion block of oligomeric proteins and that TFM-C has a certain degree of selectivity for an as yet undefined group of susceptible secreted proteins that includes IL-12/p80/IL-23. Proteomic analysis of the secretome of cells treated with TFM-C is currently being undertaken to address the diversity and nature of susceptible secretory proteins. ER-targeting compounds, such as tunicamycin, have previously been shown to have little effect on the successful secretion of monoclonal antibodies (35).
At the lowest concentration of celecoxib and TFM-C required for inducing IL-12 and p80 retention, there are no cytotoxic effects or significant alterations in the level of p40 or p35 subunit transcription (12). The comparison between intracellular and secreted levels of p19 (Fig. 1D) indicates that in celecoxib- and TFM-C-treated cells, p19 is not secreted as subunit of IL-23, yet it is still present intracellularly at levels similar to untreated control. In TFM-C-treated cells, p80 shows increased interaction with calreticulin, a luminal ER chaperone involved in ER quality control (ERQC) of folding and retention of misfolded proteins, as well as decreased interaction with the chaperone ERp44 (12). Thus, TFM-C seems to induce, either through ER Ca2+ perturbation or an as yet unknown mechanism, compositional changes in the ER environment making it more hostile to sustenance of folding of IL-12-type proteins, thus leading to abrogation of ensuing secretion. Misfolded or non-native proteins are normally intercepted by the ERQC system prior to retrotranslocation and delivery for ERAD (36). We wondered whether this holds true for the non-secreted p80 dimers produced in TFM-C-treated cells. We addressed this question in first instance by analyzing the effect of TFM-C on transcriptional regulation of 23 ER-resident proteins. Out of 23 ER chaperones and associated factors analyzed, HERP was identified as the factor showing the most dramatic, i.e. 7-fold, up-regulation by TFM-C. To a lesser extent, the mRNA levels of CRT, GRP78, GRP94, GRP170, ERp72, ERp57, ERdj4, and ERp29 showed statistically significant differences as well. Of these, celecoxib has already been shown to increase the expression of the ER chaperones GRP78 and GRP94 in a wider proteomics analysis of its effects on intracellular proteins (13), and of ORP150/GRP170, GRP78, ERdj3, and ERdj4 in human gastric carcinoma cell lines (18, 37). The celecoxib analogue 2,5-dimethylcelecoxib (DMC) has also been shown to up-regulate GRP78 in glioblastoma cell lines (38). Thus, the modulation of ER chaperone expression in response to celecoxib is shared with DMC and TFM-C despite the structural modifications in the latter compounds, and is reminiscent of the unfolded protein response (UPR) (39). Part of the induction of the UPR is a decrease in general protein synthesis as biological means toward decreasing the protein folding load within the ER (40). This has shown to be the case with celecoxib treatment, but only at higher concentrations approaching 100 μm, whereas general protein synthesis remains relatively unaffected below this concentration (41). It would appear that 50 μm TFM-C induces a pre-apoptotic state in the ER, which causes retention of IL-12 family cytokines. Mild TFM-C-induced ER calcium perturbation may result in ineffective Ca2+-dependent chaperone function. A link between GRP78 and GRP94 up-regulation because of ER calcium depletion has long been identified (42). The low cytotoxicity of both celecoxib and TFM-C in EcR293p40His cells (12), coupled to little apoptosis in 50 μm celecoxib-treated cells (18) suggests at this level, that cells can successfully compensate for calcium perturbation through increased chaperone expression. This appears sufficient to prevent terminal protein misfolding on a scale, which would lead to overwhelming of the unfolded protein response and apoptosis, a tipping point which seems to be reached at concentrations around 100 μm celecoxib or TFM-C, and much less so at 50 μm.
Mechanistically of relevance to the cellular tolerance for 50 μm TFM-C, was the discovery that HERP was by some distance the ER factor showing the highest level of inducibility. HERP is ubiquitously expressed (19), whereas analysis in HT29, LNCaP, and EcR293 (HEK293) cells (Fig. 2B) suggests that TFM-C-induced up-regulation is also ubiquitous. HERP is also strongly up-regulated by thapsigargin, but only marginally by the selective COX-2 inhibitor rofecoxib or the geldanamycin analogue 17AAG (Fig. 2C), in line with an earlier report showing HERP up-regulation by thapsigargin and the Ca2+ ionophore A23187 (19). HERP resides normally in the trans-Golgi network, but relocates mainly to the ER upon calcium perturbation with thapsigargin (43). In neuronal cells, HERP is capable of preventing ER stress-induced apoptosis by stabilizing ER Ca2+ levels (33). Taken together with the data presented here, it can be inferred with some degree of certainty that the HERP-inducing properties of TFM-C are a COX-2-independent function of celecoxib and are likely associated with perturbation of ER calcium levels.
HERP is part of high molecular mass protein complex involved in ERAD (26) and is regulated by the ER stress-specific branch of the UPR (21–23), making it a prime candidate for a role in degradation of secretion-incompetent IL-12-type cytokines retained within TFM-C-treated cells. In immunoprecipitation experiments under DSP cross-linking conditions, HERP was co-captured with p80 and IL-23 in TFM-C-treated cells (Fig. 4). Because of the reducing nature of the experiment, it cannot conclusively be proven that HERP is co-captured with the cytokine subunits in monomeric or dimeric form, or indeed both. Because TFM-C and celecoxib have consistently been shown to leave p40 monomer secretion largely intact (Fig. 1, A and B), it can be inferred that the co-captured HERP is likely to be associated with retained p80 or IL-23.
A role for HERP in degradation of secretion-incompetent p80 was further substantiated in CHX chase experiments using cells transfected with HERP siRNA and treated with TFM-C (Fig. 6). Whereas in cells transfected with control siRNA, p80 is virtually completely degraded at 2 h of CHX chase, a dramatic accumulation of p80 was evident in cells treated with HERP siRNA at this time point. In a complementary experiment, ectopic expression of HERP effectuated rapid and enhanced clearance of p80 in TFM-C-stressed cells compared with cells transfected with empty vector, while no such effect was apparent in untreated cells (Fig. 7). Thus, unlike other proteins degraded in a HERP-dependent manner, such as connexin 43 (24) and CD3-delta (26), both of which are native substrates of ERAD, p80 does not appear to be targeted for degradation by HERP in the absence of an ER stress response. It is therefore possible that the p80 dimers intercepted by and degraded through HERP under conditions of TFM-C treatment occur in a non-native/misfolded conformation. Our earlier observation of enhanced interaction between p80 and the ERQC factor CRT in TFM-C-treated cells (12), lends support to this hypothesis.
At this point, it is unknown whether p80 and IL-23 interact directly with HERP or whether HERP is simply co-captured as part of the larger HERP-containing retrotranslocation complex, because of DSP cross-linking used in this experiment (Fig. 4). HERP has also been identified to bind directly to the ER transmembrane E3 ligase HRD1. HRD1 in turn binds to the ER membrane protein Derlin-1 and the cytosolic p97 (26, 27). The membrane protein VIMP is subsequently recruited to the complex by p97. Studies of the GRP78 substrates, non-secreted Igκ LC, mutant Igγ LC and truncated Igγ HC, show all three interact with HERP (25). Furthermore, US11 from cytomegalovirus induces MHC I complex retrotranslocation resulting in a direct association with Derlin-1 and VIMP. Thus, HERP and p80/IL-23 may directly interact, but as Derlin-1 is the putative translocation pore (27), it is also possible that these cytokines interact with Derlin-1 and that HERP is co-purified through its interaction with Derlin-1 via HRD1. Further elucidation of the composition of the cytokine-associated heteromeric HERP retrotranslocation complex is being addressed through diagonal electrophoresis.
In conclusion, the intracellular retention of IL-12, IL-23, and p80 by celecoxib and TFM-C represents a novel anti-inflammatory property of celecoxib, one independent of COX-2 inhibition. This is likely to contribute significantly to the overall spectrum of anti-inflammatory properties of celecoxib, because of inhibition of differentiation of IL-12-based IFN-γ producing Th1 cells and IL-23-based IL-17 producing Th17 cells; as well as of the neutrophil chemo-attractant properties of p80 (44).
Acknowledgments
We thank members of the Neurogenomiks Laboratory for helpful discussions. The kind provision by Dr. Kochi Kokame of the full-length human HERP pCI expression vector is gratefully acknowledged.
This work was supported by Grants RRG11.5 RSG/1726 (to K. V.) from the Northern Ireland R&D Office for Health and Personal Social Services, Ikerbasque, the Basque Foundation for Science, Bilbao, Spain, and the Ministerio de Ciencia e Innovación, Madrid, Spain (MEC-2008; SAF2008-00433).
- IL
- interleukin
- CE
- celecoxib
- CHX
- cycloheximide
- CRT
- calreticulin
- DSP
- dithiobis[succinimidylpropionate]
- ER
- endoplasmic reticulum
- ERAD
- endoplasmic reticulum-associated protein degradation
- ERQC
- endoplasmic reticulum-associated quality control
- HERP
- homocysteine-induced endoplasmic reticulum protein
- TFM-C
- 4-[5-(4- trifluoromethylphenyl)-3-(trifluoromethyl)-1H-pyrazol-1-yl]benzenesulfonamide (short name: 4-trifluoromethyl-celecoxib)
- UPR
- unfolded protein response
- PBS
- phosphate-buffered saline
- NTA
- nitrilotriacetic acid
- ANOVA
- analysis of variance
- MOPS
- 4-morpholinepropanesulfonic acid
- COX
- cyclooxygenase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
REFERENCES
- 1.Collison L. W., Vignali D. A. (2008) Immunol. Rev. 226, 248–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kroenke M. A., Carlson T. J., Andjelkovic A. V., Segal B. M. (2008) J. Exp. Med. 205, 1535–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinman L. (2008) J. Exp. Med. 205, 1517–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torti D. C., Feldman S. R. (2007) J. Am. Acad. Dermatol. 57, 1059–1068 [DOI] [PubMed] [Google Scholar]
- 5.Mannon P. J., Fuss I. J., Mayer L., Elson C. O., Sandborn W. J., Present D., Dolin B., Goodman N., Groden C., Hornung R. L., Quezado M., Yang Z., Neurath M. F., Salfeld J., Veldman G. M., Schwertschlag U., Strober W. (2004) N. Engl. J. Med. 351, 2069–2079 [DOI] [PubMed] [Google Scholar]
- 6.Cooper A. M., Khader S. A. (2007) Trends Immunol. 28, 33–38 [DOI] [PubMed] [Google Scholar]
- 7.Mikols C. L., Yan L., Norris J. Y., Russell T. D., Khalifah A. P., Hachem R. R., Chakinala M. M., Yusen R. D., Castro M., Kuo E., Patterson G. A., Mohanakumar T., Trulock E. P., Walter M. J. (2006) Am. J. Respir. Crit. Care Med. 174, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell T. D., Yan Q., Fan G., Khalifah A. P., Bishop D. K., Brody S. L., Walter M. J. (2003) J. Immunol. 171, 6866–6874 [DOI] [PubMed] [Google Scholar]
- 9.Segal B. M., Constantinescu C. S., Raychaudhuri A., Kim L., Fidelus-Gort R., Kasper L. H. (2008) Lancet Neurol. 7, 796–804 [DOI] [PubMed] [Google Scholar]
- 10.Leonardi C. L., Kimball A. B., Papp K. A., Yeilding N., Guzzo C., Wang Y., Li S., Dooley L. T., Gordon K. B. (2008) Lancet 371, 1665–1674 [DOI] [PubMed] [Google Scholar]
- 11.Vandenbroeck K., Alloza I., Gadina M., Matthys P. (2004) J. Pharm. Pharmacol. 56, 145–160 [DOI] [PubMed] [Google Scholar]
- 12.Alloza I., Baxter A., Chen Q., Matthiesen R., Vandenbroeck K. (2006) Mol. Pharmacol. 69, 1579–1587 [DOI] [PubMed] [Google Scholar]
- 13.Lou J., Fatima N., Xiao Z., Stauffer S., Smythers G., Greenwald P., Ali I. U. (2006) Cancer Epidemiol. Biomarkers Prev. 15, 1598–1606 [DOI] [PubMed] [Google Scholar]
- 14.Grösch S., Maier T. J., Schiffmann S., Geisslinger G. (2006) J. Natl. Cancer Inst. 98, 736–747 [DOI] [PubMed] [Google Scholar]
- 15.Schönthal A. H. (2007) Br. J. Cancer 97, 1465–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson A. J., Hsu A. L., Lin H. P., Song X., Chen C. S. (2002) Biochem. J. 366, 831–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pyrko P., Kardosh A., Liu Y. T., Soriano N., Xiong W., Chow R. H., Uddin J., Petasis N. A., Mircheff A. K., Farley R. A., Louie S. G., Chen T. C., Schönthal A. H. (2007) Mol. Cancer Ther. 6, 1262–1275 [DOI] [PubMed] [Google Scholar]
- 18.Tsutsumi S., Namba T., Tanaka K., Arai Y., Ishihara T., Aburaya M., Mima S., Hoshino T., Mizushima T. (2006) Oncogene 25, 1018–1029 [DOI] [PubMed] [Google Scholar]
- 19.Kokame K., Agarwala K. L., Kato H., Miyata T. (2000) J. Biol. Chem. 275, 32846–32853 [DOI] [PubMed] [Google Scholar]
- 20.Lenz B., Bleich S., Beutler S., Schlierf B., Schwager K., Reulbach U., Kornhuber J., Bönsch D. (2006) Exp. Cell Res. 312, 4049–4055 [DOI] [PubMed] [Google Scholar]
- 21.Ma Y., Hendershot L. M. (2004) J. Biol. Chem. 279, 13792–13799 [DOI] [PubMed] [Google Scholar]
- 22.Liang G., Audas T. E., Li Y., Cockram G. P., Dean J. D., Martyn A. C., Kokame K., Lu R. (2006) Mol. Cell Biol. 26, 7999–8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sai X., Kokame K., Shiraishi H., Kawamura Y., Miyata T., Yanagisawa K., Komano H. (2003) FEBS Lett. 553, 151–156 [DOI] [PubMed] [Google Scholar]
- 24.Hori O., Ichinoda F., Yamaguchi A., Tamatani T., Taniguchi M., Koyama Y., Katayama T., Tohyama M., Stern D. M., Ozawa K., Kitao Y., Ogawa S. (2004) Genes Cells 9, 457–469 [DOI] [PubMed] [Google Scholar]
- 25.Okuda-Shimizu Y., Hendershot L. M. (2007) Mol. Cell 28, 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulze A., Standera S., Buerger E., Kikkert M., van Voorden S., Wiertz E., Koning F., Kloetzel P. M., Seeger M. (2005) J. Mol. Biol. 354, 1021–1027 [DOI] [PubMed] [Google Scholar]
- 27.Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. (2004) Nature 429, 841–847 [DOI] [PubMed] [Google Scholar]
- 28.Alloza I., Martens E., Hawthorne S., Vandenbroeck K. (2004) Anal. Biochem. 324, 137–142 [DOI] [PubMed] [Google Scholar]
- 29.Oka Y., Hirabayashi Y., Ishii T., Takahashi R., Sasaki T. (2007) Tohoku J. Exp. Med. 212, 431–437 [DOI] [PubMed] [Google Scholar]
- 30.Lehner I., Niehof M., Borlak J. (2003) Electrophoresis 24, 1795–1808 [DOI] [PubMed] [Google Scholar]
- 31.Alloza I., Vandenbroeck K. (2005) J. Pharm. Pharmacol. 57, 213–218 [DOI] [PubMed] [Google Scholar]
- 32.Lee Y. K., Brewer J. W., Hellman R., Hendershot L. M. (1999) Mol. Biol. Cell 10, 2209–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan S. L., Fu W., Zhang P., Cheng A., Lee J., Kokame K., Mattson M. P. (2004) J. Biol. Chem. 279, 28733–28743 [DOI] [PubMed] [Google Scholar]
- 34.Leitzgen K., Knittler M. R., Haas I. G. (1997) J. Biol. Chem. 272, 3117–3123 [DOI] [PubMed] [Google Scholar]
- 35.Barnabé N., Butler M. (1998) J. Biotechnol. 60, 67–80 [DOI] [PubMed] [Google Scholar]
- 36.Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Namba T., Hoshino T., Tanaka K., Tsutsumi S., Ishihara T., Mima S., Suzuki K., Ogawa S., Mizushima T. (2007) Mol. Pharmacol. 71, 860–870 [DOI] [PubMed] [Google Scholar]
- 38.Kardosh A., Golden E. B., Pyrko P., Uddin J., Hofman F. M., Chen T. C., Louie S. G., Petasis N. A., Schönthal A. H. (2008) Cancer Res. 68, 843–851 [DOI] [PubMed] [Google Scholar]
- 39.Lai E., Teodoro T., Volchuk A. (2007) Physiology 22, 193–201 [DOI] [PubMed] [Google Scholar]
- 40.Malhotra J. D., Kaufman R. J. (2007) Semin. Cell Dev. Biol. 18, 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pyrko P., Kardosh A., Schönthal A. H. (2008) Biochem. Pharmacol. 75, 395–404 [DOI] [PubMed] [Google Scholar]
- 42.Li W. W., Alexandre S., Cao X., Lee A. S. (1993) J. Biol. Chem. 268, 12003–12009 [PubMed] [Google Scholar]
- 43.Tuvia S., Taglicht D., Erez O., Alroy I., Alchanati I., Bicoviski V., Dori-Bachash M., Ben-Avraham D., Reiss Y. (2007) J. Cell Biol. 177, 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paunovic V., Carroll H. P., Vandenbroeck K., Gadina M. (2008) Rheumatology 47, 771–776 [DOI] [PubMed] [Google Scholar]