Abstract
Previously we showed that the inactive form of p90 ribosomal S6 kinase 1 (RSK1) interacts with the regulatory subunit, PKARIα, of protein kinase A (PKA), whereas the active RSK1 interacts with the catalytic subunit (PKAc) of PKA. Herein, we demonstrate that the N-terminal kinase domain (NTK) of RSK1 is necessary for interactions with PKARIα. Substitution of the activation loop phosphorylation site (Ser-221) in the NTK with the negatively charged Asp residue abrogated the association between RSK1 and PKARIα. This explains the lack of an interaction between active RSK1 and PKARIα. Full-length RSK1 bound to PKARIα with an affinity of 0.8 nm. The NTK domain of RSK1 competed with PKAc for binding to the pseudosubstrate region (amino acids 93–99) of PKARIα. Overexpressed RSK1 dissociated PKAc from PKARIα, increasing PKAc activity, whereas silencing of RSK1 increased PKAc/PKARIα interactions and decreased PKAc activity. Unlike PKAc, which requires Arg-95 and -96 in the pseudosubstrate region of PKARIα for their interactions, RSK1/PKARIα association requires all four Arg residues (Arg-93–96) in the pseudosubstrate site of PKARIα. A peptide (Wt-PS) corresponding to residues 91–99 of PKARIα competed for binding of RSK1 with PKARIα both in vitro and in intact cells. Furthermore, peptide Wt-PS (but not control peptide Mut-PS), by dissociating RSK1 from PKARIα, activated RSK1 in the absence of any growth factors and protected cells from apoptosis. Thus, by competing for binding to the pseudosubstrate region of PKARIα, RSK1 regulates PKAc activity in a cAMP-independent manner, and PKARIα by associating with RSK1 regulates its activation and its biological functions.
Keywords: Bioenergetics, Calcium, Channels/Potassium, Diseases/Neurodegeneration, Organisms/Rat, Subcellular Organelles/Mitochondria, Tissue/Organ Systems/Brain, Mitochondrial Permeability Transition
Introduction
The cyclic AMP-dependent protein kinase (PKA)2 represents one of the best studied kinases that is involved in regulating numerous physiological processes. PKA is a heterotetramer that consists of two regulatory and two catalytic (PKAc) subunits (1). Depending upon which of the two forms of the regulatory subunit, PKARI or PKARII, PKAc binds to, the PKA holoenzyme is classified as type I or II, respectively (1). PKARI and PKARII themselves exist in two forms, α and β; thus, there are essentially four forms of the regulatory subunits (1). Likewise, four forms of the catalytic subunits may be expressed in cells and tissues (2). The heterotetrameric form of the PKA holoenzyme is inactive, and the binding of two cAMP molecules to each of the regulatory subunits causes dissociation of PKAc, resulting in its activation (3, 4). Both PKARI and PKARII, via binding of their pseudosubstrate and inhibitory regions, respectively, to the catalytic cleft of PKAc, inhibit its activity (5–8). However, other regions of PKARIα also interact with the large lobe of PKAc (7).
The four isoforms of the p90 ribosomal S6 kinases (RSK1-RSK4) are members of a family with two kinase domains, an N-terminal kinase domain (NTK) and a C-terminal kinase domain (CTK). Although RSK1–3 share considerable sequence homology, RSK4 is larger and may also function differently (9, 10). The CTK domain of RSK1–3 is activated by Erk1/2-mediated phosphorylation of a Thr residue (Thr-573 in rat RSK1) in their activation loop. The activated CTK then autophosphorylates the RSKs to permit docking of PDK1 that then phosphorylates a Ser residue (Ser-221 in rat RSK1) in the activation loop of the NTK (11). The fully active RSKs, via their NTK, then phosphorylate their respective substrates (12). The fully active RSK1 and RSK2 are also translocated to the nucleus where they phosphorylate several transcription factors to mediate their biological actions (13). RSK1 and RSK2 play a pivotal role in regulating a variety of biological functions including inhibition of apoptosis (14, 15), activation of the mTOR pathway (16), and cell proliferation (17, 18). RSK1 and RSK2 have also been shown to play an important role in the proliferation of breast and prostate tumor cells (17, 18) and in augmenting cardiac hypertrophy (19). Previous studies from our laboratory have shown that the inactive form of RSK1 associates with PKARIα, and upon its activation, RSK1 binds PKAc (20). Thus, independent of its activation state, RSK1 is associated with PKA and thereby also with protein kinase A-anchoring proteins (AKAPs) (20). These interactions are specific for RSK1, since RSK2 and RSK3 do not associate with either PKARIα or PKAc (20). The indirect (via PKA subunits) interaction of RSK1 with AKAPs is necessary for the nuclear localization of RSK1, and perturbations in this interaction with either PKARIα or AKAPs alters the biological outcome (e.g. anti-apoptotic actions) of RSK1 (20). Recently, we have also shown that the indirect association of RSK1 with D-AKAP1 brings the RSK1 in proximity of the catalytic subunit of protein phosphatase 2A (PP2Ac) that is bound to D-AKAP1, and this permits PP2Ac to dephosphorylate and regulate RSK1 activation (21). Moreover, we have recently shown that the last 13 residues on RSK1 form the PKAc binding site and that autophosphorylation of Ser-732 by NTK of RSK1 within this region is required for interactions with PKAc (22). The PKAc binding region on RSK1 is also the Erk1/2 binding site, except that autophosphorylation of Ser-732 results in dissociation of Erk1/2 (22, 23). Thus, the phosphorylation status of Ser-732 determines whether RSK1 associates with Erk1/2 or PKAc (22).
The purpose of the studies described in this report was to identify the regions on PKARIα and inactive RSK1 that interact with each other and to further understand the functional implications of this interaction. Our findings demonstrate that the NTK domain of RSK1 is the PKARIα binding site, and consistent with our previous reports that inactive RSK1 interacts with PKARIα (20, 21), substitution of Ser-221 in the activation loop of RSK1 with negatively charged Asp residue abrogated this interaction. Additionally, our data show that the pseudosubstrate domain of PKARIα comprising the sequence (93RRRRGAI99) is necessary for association with RSK1; Ala-98 represents the phosphorylation site (P site) if it were Thr or Ser. Substitution of Arg-93/94 with Ala, which abrogates the RSK1 consensus phosphorylation sequence, also abolished the binding of RSK1 with PKARIα without altering its PKARIα/PKAc interactions. Moreover, RSK1 competed with PKAc for binding to PKARIα and modulated endogenous PKAc activity. Akin to our recent report showing that silencing of PKARIα activates RSK1 (21), the dissociation of the PKARIα/RSK1 interaction by a cell-permeable peptide corresponding to the pseudosubstrate region of PKARIα mimicked the silencing of PKARIα and, by increasing the amount of active RSK1, augmented the anti-apoptotic actions of RSK1. These data demonstrate that the catalytic cleft of the inactive NTK interacts with the pseudosubstrate region of PKARIα and that this mode of association is necessary for regulating the activation of RSK1 and its biological actions as well as regulating endogenous PKAc activity.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
cDNA sequences corresponding to the different regions of RSK1 were PCR synthesized using rat RSK1 as the template (provided by Dr. Warner Greene, University of California, San Francisco) and inserted in vector pHM6 or pGEX-4T-3 at EcoRI and NotI sites to express the HA-tagged or GST-tagged RSK1 polypeptides. Bovine PKARIα cDNA (provided by Dr. Susan S. Taylor, Univ. of California, San Diego) was inserted in-frame with EYFP in vector pEYFP-N1 at NheI and SalI sites to express PKARIα-EYFP. Site-directed mutagenesis of this construct was performed using mutagenic primers for universal PCR.
Cell Culture and Transfection
Both B82L cells (mouse lung fibroblasts) and HEK293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine, and penicillin/streptomycin. Methotrexate (5 μg/ml) was added for maintenance of B82L cells. For transfection, HEK293T cells were plated in 60-mm dishes at 4 × 105 cells/dish. The next day, cells were transfected with plasmids using FuGENE 6 transfection reagent (Roche Diagnostics).
Pulldown of PKARIα with cAMP-Agarose
The experimental procedures detailed in our previous report were used (20). Essentially, lysates (500 μg of protein) of HEK293T cells that were serum-starved were mixed with cAMP-agarose (Sigma) for 2 h at 4 °C in the presence and absence of 50 mm cAMP (Sigma). The resin was then washed twice with high salt buffer (10 mm HEPES, pH 7.4, 1.5 mm MgCl2, 10 mm KCl, 0.5 m NaCl, 0.1% Igepal CA-630, 1 mm dithiothreitol, 100 μm phenylmethylsulfonyl fluoride, and 1 μg/ml each pepstatin A, aprotinin, and leupeptin) followed by 2 washes with no-salt buffer (as described above but without NaCl). Proteins in the complex were identified by Western analyses using the following antibodies: anti-PKARIα (BD Biosciences) and anti-HA horseradish peroxidase (Roche Diagnostics).
GST Pulldown Assays
Bacteria (TOP10 competent cells; Invitrogen) transformed with vector alone or pGEX-4T-3-RSK1-(1–317) were grown in Luria broth containing ampicillin (100 μg/ml) until the A600 reached 0.6 and then induced to express GST or GST-RSK1-(1–317) with isopropyl β-d-1-thiogalactopyranoside (0.5 mm) at 20 °C for 3 h. Bacterial lysates were incubated with glutathione-Sepharose to purify GST or GST-tagged RSK1-(1–317). Thereafter, glutathione-Sepharose beads (15 μl) with prebound GST or GST-RSK1-(1–317) (5 μg) were incubated in the pulldown buffer (20 mm Hepes, pH 7.4, 150 mm NaCl, 5 mm MgCl2, 2 mm MnCl2, 0.1% Triton X-100, 1 mm dithiothreitol, 0.1 mm ATP, and 0.1 mg/ml bovine serum albumin) with purified PKARIα (10 pmol) or its mutant PKARIαΔ91 with the deletion of amino acids 1–91 (provided by Dr. Susan Taylor, University of California, San Diego) at 4 °C for 2 h in a final volume of 400 μl. After three thorough washes with the same buffer, the complex was eluted with Laemmli sample buffer and subjected to SDS-PAGE for Western analyses. To investigate the competition between RSK1 and PKAc for binding to PKARIα before the GST pulldown assays, PKARIα (10 pmol) was incubated with different indicated amounts of PKAc to form the holoenzyme as described before (22). Thereafter, the mixture was added to GST-RSK1-(1–317) (5 μg) bound to glutathione-Sepharose. To examine the ability of the short peptides corresponding to the pseudosubstrate region of PKARIα to dissociate PKARIα/RSK1 interactions, synthetic peptides Wt-PS (sequence: KGRRRRGAI) and Mut-PS (sequence: KGAARRGAI) corresponding to the pseudosubstrate region 91–99 were obtained from New England Peptides (Gardner, MA). These peptides were preincubated on ice with GST-RSK1-(1–317) (5 μg) for 15 min before being mixed with PKARIα (10 pmol).
Immunoprecipitations
After overnight serum starvation, HEK293T cells were washed twice with ice-cold PBS and scraped into lysis buffer (20 mm Hepes, pH 7.4, 150 mm NaCl, 1 mm EDTA, 0.5% Triton X-100, 2.5 mm MgCl2, 0.1 mm ATP, 1 mm dithiothreitol, 0.1 mm phenylmethylsulfonyl fluoride, and proteinase inhibitor mixture (Roche Diagnostics). Cell lysates were cleared by centrifugation at 20,000 × g for 15 min. The supernatants (500 μg of protein except for immunoprecipitation with anti-PKARIα antibody, where 1 mg protein was used) were incubated for 2 h at 4 °C with 0.4 μg of anti-RSK1 antibody or 2 μg of anti-PKAc antibody (both from Santa Cruz Biotechnology, Santa Cruz, CA) or 2 μg of PKARIα antibody (BD Biosciences) together with 15 μl of protein G-conjugated agarose beads. After three thorough washes with lysis buffer, the immunoprecipitates were eluted with Laemmli sample buffer and subjected to SDS-PAGE for Western analyses. To detect changes in the phosphorylation status of RSK1, B82L cells that had been serum-starved were treated with 2 μm each of palmitoylated, cell-permeable peptides Wt-PS or Mut-PS for 10 min. Thereafter, cells were lysed in a buffer containing 50 mm Hepes, pH 7.5, 1% Triton X-100, 0.5% CHAPS (ICN Biomedicals Inc., Aurora, OH), 150 mm NaCl, 1 mm dithiothreitol, 1 mm sodium orthovanadate, 50 mm NaF, 5 mm sodium pyrophosphate, 10 mm β-glycerophosphate, 100 μm phenylmethylsulfonyl fluoride, 1 μm microcystin, and 1 μg/ml each pepstatin A, aprotinin, and leupeptin. The immunoprecipitations were performed with anti-RSK1 antibody as above, and the phosphorylation status of RSK1 was monitored by anti-phospho-Ser-380 RSK antibody (Epitomics, Burlingame, CA).
Surface Plasmon Resonance
All measurements were performed using the Bio-Rad ProteOn XPR36 instrument. The PKARIα was immobilized on GLC sensor chips using Proteon amine coupling kit. After activating the surface of the chip, PKARIα (200 pm) in 10 mm sodium acetate, pH 4.00, was injected at the rate of 30 μl/min for 300 s. A total of ∼5000 resonance units of PKARIα were immobilized on the chip. Residual N-hydroxysulfosuccinimide esters on the chip surface were reacted with ethanolamine. The binding of PKAc and RSK1 with PKARIα was performed at 25 °C in buffer containing 20 mm Mops, pH 7.0, 150 mm KCl, 1 mm Tris(2-carboxyethyl)phosphine, 0.005% of polysorbate 20, 1 mm MnCl2, and 0.2 mm AMP-PNP. Different concentrations of PKA catalytic subunit (200, 100, 50, 25, and 12.5 nm), GST (100, 50, 25, 12.5, 6.25 nm), GST full-length RSK1 (GST-FL-RSK1) (25, 12.5, 6.25, 3.15 and 1.56 nm), or GST-RSK1-(1–317) (100, 50, 25, 12.5, 6.25 nm) were injected over immobilized PKARIα at the flow rate of 30 μl/min for 600 s. After monitoring the association and dissociation of proteins, the chip surface was regenerated either with glycine pH 2.0 or 10 mm NaOH for 18 s at a flow rate of 100 μl/min. GST protein at concentrations similar to those shown for GST-RSK1-(1–317) was used as a reference. GST alone did not bind to the immobilized PKARIα. Kinetic constants were calculated using the Biacore pseudo-first-order rate equation, and affinity constants (KD) were calculated from the equation KD = kdissociation/kassociation. The GST and GST fusion proteins, prepared as described above, were eluted with GSH and concentrated in 30 mm MES, pH 6.5, 1 mm EDTA, 50 mm KCl, 5 mm β-mercaptoethanol, and protease inhibitor mixture (Roche Diagnostics) using centrifugal concentrators (QMWL, 10 kDa).
Silencing of Endogenous RSK1
Endogenous RSK1 was silenced by the two siRNAs as described in our previous publication (22). The first siRNA sequence was: sense, GGA CCA AGA UGG AGA GAG ACA UCC T; antisense, AGG AUG UCU CUC UCC AUC UUG GUC CGA. The second siRNA sequence was: sense, CCU CUA UGU GGA UGA GUC UGG GAA C; antisense, GUU CCC AGA CUC AUC CAC AUA GAG GAU. The control siRNA sequence was: sense, GGA UGA UAU UCC UCC UUG UGU CUG UCC; antisense, GGA CAG ACA CAA GGA GGA AUA UCA UCC.
In Vitro Kinase Activity Assays
After treatment of cells with 2 μm each of the palmitoylated, cell-permeable, peptides Wt-PS or Mut-PS or with EGF (50 nm), triplicate immunoprecipitates of RSK1, performed as described above for detection of RSK1 phosphorylation, were resuspended in 20 mm Hepes, pH 7.5, 1 mm sodium orthovanadate, 1 mm NaF, 1 mm dithiothreitol, 25 mm β-glycerophosphate, 5 mm MgSO4, 200 μm Kemptide (Leu-Arg-Arg-Ala-Ser-Leu-Gly), 125 μm ATP, and 10 μCi of [γ-32P]ATP (final volume, 120 μl) for 10 min at room temperature. Where indicated, the RSK1/2 inhibitor, SL-0101 (1 μm), was added to the kinase activity reaction buffer. Reactions were terminated by the addition of equal volume of 20% trichloroacetic acid, and after centrifugation (16,000 × g, 5 min), aliquots (100 μl) of each supernatant were spotted onto P81 paper (Whatman), air-dried, and washed 3 times with 0.5% phosphoric acid. The filters were then dried and counted in a liquid scintillation counter. One parallel set of immunoprecipitated proteins was denatured in Laemmli sample buffer, separated on 10% polyacrylamide gels, and analyzed by immunoblotting to ensure that equal amount of RSK1 was immunoprecipitated under different conditions.
Kinase assays with purified PKAc, PKARIα, and GST-FL-RSK1 were performed as described previously (22). Essentially, PKAc (2 nm final concentration) was mixed with different concentrations of GST or GST-FL-RSK1 in buffer containing 20 mm Hepes, pH 7.4, 5 mm MgCl2, 2 mm MnCl2, 1 mm dithiothreitol, 5 mm NaF, 0.1 mg/ml bovine serum albumin, 0.1 mm ATP, and then PKARIα (3 nm) was added. This mixture was incubated for 1 h at room temperature. Kinase activity reactions (50 μl final volume) were initiated by the addition of 0.2 mm Kemptide and 2.5 μCi of [γ-32P]ATP. The reactions were stopped after 15 min with 50 μl of 20% trichloroacetic acid.
Apoptosis Assay
B82L cells (50,000 cells/well) in a 24-well dish were transfected with control or RSK1-specific siRNAs as described before (22). Forty-eight hours later cells were serum-starved overnight. Wherever indicated, cells were treated with 2 μm Wt-PS or Mut-PS peptide for 10 min followed by stimulation with 50 nm EGF for 10 min, as indicated. The cells were then treated with or without 20 ng/ml TNF-α plus 25 μg/ml cycloheximide (CHX) to induce apoptosis. One hour after TNF/CHX treatment, DNA fragmentation was measured using the Cell Death detection kit (Roche Applied Science).
RESULTS
NTK of RSK1 Interacts with PKARIα
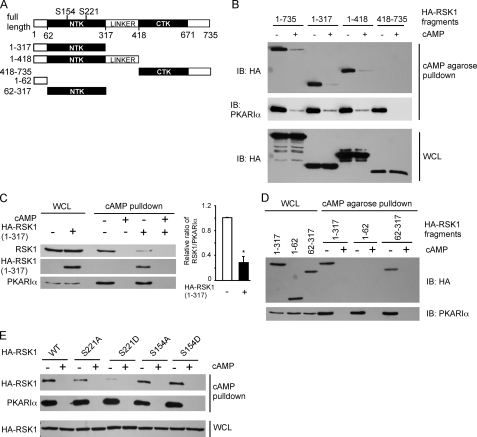
The initial aim of our study was to identify the region on inactive RSK1 that interacts with PKARIα. Therefore, we transfected HEK293T cells with HA-tagged full-length (1–735) RSK1 or the different constructs shown in Fig. 1A that include the N-terminal 317 residues (1–317) containing the NTK or its N-terminal 418 residues that contain both the NTK and the linker region as well as the C-terminal 318 amino acids (418–735) containing the CTK and the C-terminal tail. After serum-starving the cells, the endogenous PKARIα from the cell lysates was pulled down with cAMP-agarose, and proteins in the pulldown assays were analyzed for HA-RSK1 or its fragments. As shown in Fig. 1B, the HA-tagged full-length (1–735) as well as 1–317 and 1–418 fragments of RSK1 were pulled down along with endogenous PKARIα. However, the HA-(418–735) fragment of RSK1 was not pulled down with PKARIα by the cAMP-agarose. Specificity of the interactions between RSK1 and its fragments with PKARIα is shown by the lack of these proteins when exogenous free cAMP was added to dissociate PKARIα from the cAMP-agarose. Next, we determined if overexpression of the N-terminal 317 residues of RSK1 competed with endogenous RSK1 for association with endogenous PKARIα. Fig. 1C shows that overexpression of the HA-(1–317) fragment of RSK1 decreased the amount of endogenous RSK1 that was pulled down with PKARIα. These data (Fig. 1, B and C) demonstrate that the N-terminal 317 amino acids of RSK1 form the PKARIα binding site. To determine whether the NTK or the extreme N-terminal 62 amino acids of RSK1 (Fig. 1A) form the PKARIα binding site, HEK293T cells were transfected to express shorter HA-tagged fragments of RSK1 comprising the N-terminal 62 residues (1–62) and residues encompassing the NTK-(62–317) as well as HA-RSK1-(1–317). The cAMP-agarose pulldown assays of PKARIα from cell lysates contained HA-tagged N-terminal 317 residues of RSK1 and the HA-NTK-(62–317) but not the HA-1–62 form of RSK1 (Fig. 1D). These data suggest that PKARIα binds the NTK domain of RSK1.
FIGURE 1.
RSK1, via its N-terminal kinase domain, interacts with PKARIα. A, shown is a schematic of rat RSK1 and HA-tagged RSK1 constructs used; numbers represent the amino acids in rat RSK1. B–E, shown is pulldown of HA-tagged RSK1 or its polypeptides with cAMP-agarose. HEK293T cells were transfected to express HA-tagged RSK1 or its polypeptides. After depriving of serum overnight, cells were lysed and incubated with cAMP-agarose to pull down PKARIα in the presence and absence of 50 mm cAMP as indicated. The proteins in the complex were monitored using anti-HA, anti-PKARIα, or anti-RSK1 antibodies. IB, immunoblot. B, PKARIα binds to RSK1 fragments containing the N-terminal part of RSK1. C, HA-RSK1-(1–317) competes with endogenous RSK1 for binding to PKARIα. The panel on the right shows quantification of band intensities of RSK1 as a ratio of band intensities of PKARIα (mean ± S.E.) from three experiments. *, p < 0.05 as compared with the control. D, the NTK of RSK1 binds to PKARIα. E, substitution of Ser-221 of RSK1 with a negatively charged residue abrogates the interaction between RSK1 and PKARIα. Representatives of three similar experiments are shown for all panels. WCL, whole cell lysates.
We have previously shown that the inactive form of RSK1 binds PKARIα, and the phosphorylated, active form of RSK1 binds PKAc (20–22). The NTK of RSK1 contains two phosphorylation sites, a Ser-221 in its activation loop that is phosphorylated by PDK1 and a Ser-154 that is phosphorylated by an as yet unidentified kinase (11, 24). To determine whether the phosphorylation-mediated negative charge at these two sites altered the association of RSK1 with PKARIα, HEK293T cells were transfected to express the HA-tagged wild-type RSK1 or its S221A, S221D, S154A, and S154D mutants. Interestingly, except for the S221D mutant, all forms of RSK1 (wild type, S221A, S154A, and S154D) were pulled down with PKARIα. Substitution of Ser with Asp at position 221 can interfere with PKARIα binding by either the negative charge or the bulk that is introduced. However, because active, phospho-RSK1 does not interact with PKARIα (20, 22), our data suggest that the Asp substitution acts as a phospho-mimic and that phosphorylation of the activation loop residue (Ser-221) in the NTK of RSK1 determines its interactions with PKARIα.
Pseudosubstrate Region of PKARIα Interacts with the NTK Domain of RSK1
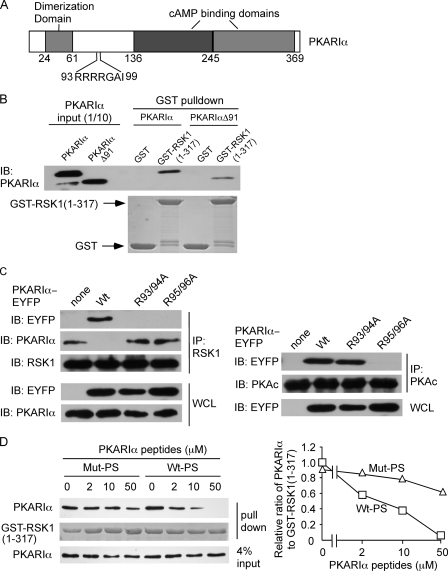
Next we focused on identifying the region on PKARIα that is necessary for interactions with RSK1. For this purpose, pure, full-length PKARIα or PKARIαΔ91 with deletions of the first 91 amino acids were incubated with GST or GST-RSK-(1–317). After pulling down GST or its fusion protein with glutathione-Sepharose, the presence of PKARIα forms was monitored. Deletion of the N-terminal 91 residues of PKARIα that contains the AKAP binding region and the PKARIα dimerization domain (Fig. 2A (25–27)) did not alter its interactions with the N-terminal 317 residues of RSK1 (Fig. 2B). Because the RSK1 consensus phosphorylation sequence comprises RXRXX(S/T) and because the pseudosubstrate region of PKARIα contains 93RRRRGAI99 with Ala-98 being the P-site (Fig. 2A (7, 28)), we investigated whether this region constitutes the binding site for the NTK of RSK1. HEK293T cells were transfected to express PKARIα-EYFP or its mutants in which either Arg residues at positions 93 and 94 or Arg-95 and -96 in the pseudosubstrate region were substituted with Ala. Immunoprecipitation of RSK1 from these cells demonstrated that although the wild-type PKARIα interacted with RSK1, PKARIα R93A/R94A and PKARIα R95A/R96A did not associate with RSK1 (Fig. 2C). As expected from the structure of the type I PKA holoenzyme (7), immunoprecipitation of PKAc from the same cells demonstrated that the interactions between PKAR1α and PKAc were abolished when Arg-95 and -96 were substituted with Ala but not when R93 and R94 were replaced with Ala (Fig. 2C). Thus, the Arg residues on PKARIα required for its association with PKAc and RSK1 are those that form the consensus phosphorylation sequences for the two respective kinases. Because disruption of the RSK1 consensus phosphorylation sequence in the pseudosubstrate region of PKARIα abrogated the interaction between these proteins, we reasoned that a peptide corresponding to the pseudosubstrate region of PKARIα should compete for the binding of RSK1-(1–317) with PKARIα. Indeed, the addition of a peptide (Wt-PS; sequence KGRRRRGAI) corresponding to the pseudosubstrate region of PKARIα in a concentration-dependent manner, attenuated the association of purified PKARIα with GST-RSK-(1–317) (Fig. 2D). On the other hand, the control peptide (Mut-PS; sequence KGAARRGAI), in which the underlined residues corresponding to Arg-93 and -94 in the PKARIα pseudosubstrate region were substituted with Ala, did not compete for interactions between purified PKARIα and GST-RSK-(1–317) (Fig. 2D). These data (Fig. 2, C and D) demonstrate that RSK1 binds the pseudosubstrate region of PKARIα.
FIGURE 2.
RSK1 binds to the pseudosubstrate region of PKARIα. A, shown is a schematic of bovine PKARIα; numbers represent the amino acids in bovine PKARIα. B, the truncated PKARIα (deletion of amino acids 1–91, PKARIαΔ91) retains the ability to interact with RSK1. Pure PKARIα or PKARIαΔ91 (10 pmol each) was incubated with glutathione-Sepharose prebound with GST or GST-RSK1-(1–317) (5 μg). The amounts of PKARIα or PKARIαΔ91 in the pulldown complex were detected with anti-PKARIα antibody. GST or GST-RSK1-(1–317) was stained with Coomassie Blue. IB, immunoblot. C, substitution of Arg-93/94 or Arg-95/96 on PKARIα to Ala selectively abrogates the interactions of PKARIα with RSK1 and PKAc. HEK293T cells were transfected with plasmids expressing C-terminal fusion of Wt-PKARIα, PKARIα (R93A/R94A), or PKARIα (R95A/R96A) with enhanced yellow fluorescent protein (wt-PKARIα-EYFP, PKARIα (R93A/R94A)-EYFP, and PKARIα (R95A/R96A)-EYFP, respectively). Cell lysates were immunoprecipitated (IP) with anti-RSK1 or PKAc antibody. The immune complex was probed with anti-EYFP, PKARIα, RSK1, or PKAc antibodies. WCL, whole cell lysate. D, the peptide corresponding to the PKARIα pseudosubstrate region (Wt-PS) competes for the interaction of PKARIα with RSK1. GST-RSK1-(1–317) (5 μg) prebound to glutathione-Sepharose was incubated with PKARIα peptides, Wt-PS, or Mut-PS at the concentrations indicated at 4 °C for 15 min before being mixed with PKARIα (10 pmol). GST-RSK1-(1–317) was stained with Coomassie Blue. The panel on the right shows the quantification of relative intensities of PKARIα bands as a ratio of GST-RSK1 bands from two identical experiments. Wt-PS, peptide with wild-type PKARIα pseudosubstrate sequence KGRRRRGAI). Mut-PS, peptide with mutated PKARIα pseudosubstrate sequence (KGAARRGAI).
Affinities of RSK1 and PKAc for PKARIα
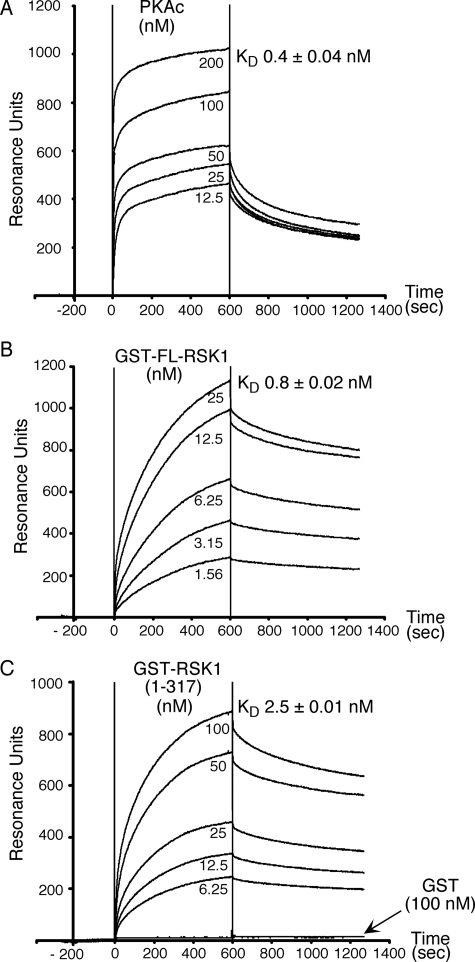
Next, using surface plasmon resonance, we determined the affinities of GST-tagged full-length (FL) and N-terminal 317 residues (1–317) of RSK1 as well as PKAc for PKARIα. As shown in Fig. 3, PKAc bound PKARIα with an affinity of 0.4 ± 0.04 nm (Fig. 3A), a value similar (0.7 nm) to that previously reported for binding of PKAc to immobilized PKARIα (29). Under identical conditions, GST-FL-RSK1 and GST-RSK1-(1–317) bound to immobilized PKARIα with affinities of 0.8 ± 0.02 and 2.5 ± 0.01 nm, respectively (Fig. 3, B and C). Controls with GST alone did not show any binding (Fig. 3C). These data demonstrate that the affinities of PKAc and full-length RSK1 for PKARIα are very similar, with the difference that PKAc associates with PKARIα at a faster rate than RSK1 (Kassoc for PKAc = 1.53 ± 0.04 × 106 m−1 s−1 versus Kassoc for GST-FL-RSK1 = 2.91 ± 0.1 × 105 m−1 s−1), and RSK1 dissociates from PKARIα at a slower rate than PKAc (Kdissoc for GST-FL-RSK1 = 2.32 ± 0.01 × 10−4 s−1 versus Kdissoc for PKAc = 7.04 ± 0.14 × 10−4 s−1). The differences in the affinities of FL-RSK1 and its N terminus RSK1-(1–317) for PKARIα could be due to some steric hindrance from the GST tag in the context of the short RSK1-(1–317) protein versus the full-length RSK1.
FIGURE 3.
Affinities of PKAc and RSK1 for PKARIα. PKARIα was immobilized on GLC sensor chips of the Bio-Rad Proteon XPR36 instrument, and binding isotherms were monitored by infusing the different concentrations of PKAc (A), GST-full-length RSK1 (GST-FL-RSK1) (B), or GST-RSK1-(1–317) (C) for the 600-s periods (denoted by vertical lines) as described under “Experimental Procedures.” The amount of immobilized PKARIα corresponded to 5000 resonance units. Controls with GST alone (C) were also performed. Representatives of six similar independent experiments are shown. KD values presented are the mean ± S.E. from six different experiments.
PKAc and RSK1 Compete for Binding to PKARIα, and This Competition Regulates PKAc Activity
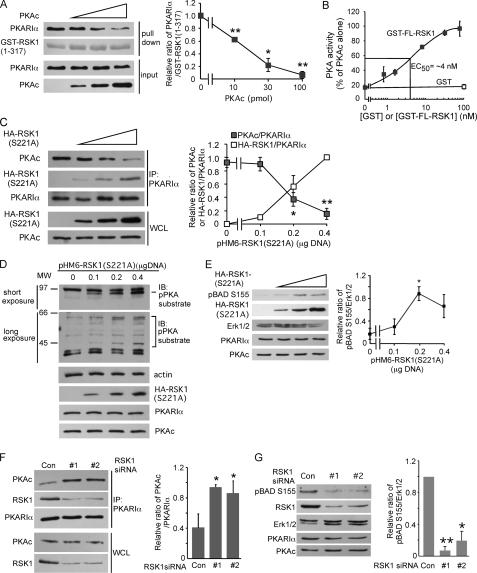
Because both RSK1 (Fig. 2) and PKAc (7, 8, 28) bind the pseudosubstrate region of PKARIα, we reasoned that PKAc and RSK1 might compete for binding to PKARIα. In support of this notion, we had previously observed that the association of RSK1 with PKARIα decreased the interactions between PKAc and PKARIα (20). Nonetheless, to more directly demonstrate that PKAc and the NTK of RSK1 compete for binding with PKARIα, GST-RSK1-(1–317) was incubated with purified PKARIα and increasing concentrations of PKAc. The amount of PKARIα associated with the GST-RSK1-(1–317) was then determined by glutathione-Sepharose pulldown assays. Fig. 4A shows that increasing concentrations of PKAc decreased the amount of PKARIα that was associated with the GST-RSK1-(1–317) fragment that contains the NTK. Consistent with its ability to compete with PKAc for binding to PKARIα, increasing amounts of GST-FL-RSK1 decreased the formation of the PKA holoenzyme as assessed by measurements of PKAc activity in the mixture of PKARIα and PKAc with different concentrations of GST-FL-RSK1 (Fig. 4B). Notably, the EC50 concentration of GST-FL-RSK1 required to compete with holoenzyme formation in these experiments was 2-fold greater than the concentration of PKAc (2 nm) in the assay (Fig. 4B). This is similar to the 2-fold difference in the affinities of PKAc and GST-FL-RSK1 for PKARIα (Fig. 3).
FIGURE 4.
Competition between PKAc and RSK1 for association with PKARIα regulates PKA activity. A, PKAc competes with RSK1-(1–317) for binding to PKARIα. PKARIα (10 pmol) was preincubated with different indicated amounts of PKAc to form the holoenzyme before mixing with glutathione resin pre-bound to GST-RSK1-(1–317) (5 μg) for the pulldown assay. GST-RSK1-(1–317) was stained with Coomassie Blue. The panel on the right is quantification of the ratio of the band intensities of PKARIα relative to GST-RSK1 from three similar experiments. *, p < 0.05; ** p < 0.01, Student's unpaired t test analysis. B, GST full-length RSK1 (GST-FL-RSK1) competes with PKAc for PKARIα and decreases the formation of the PKA holoenzyme. PKAc (2 nm final concentration) and increasing indicated concentrations of GST-FL-RSK1 (or GST 100 nm) were added to PKARIα (3 nm). After incubation of the mixture for 1 h, formation of the PKA holoenzyme was monitored by measuring PKAc activity as described under “Experimental Procedures.” C, increasing expression of HA-RSK1 (S221A) decreases the association of endogenous PKAc with PKARIα. HEK293T cells were transfected with the indicated different amounts of plasmid expressing HA-RSK1 (S221A). Cell lysates were immunoprecipitated (IP) with anti-PKARIα antibody. The right hand panel shows the quantification of band intensities of PKAc or HA-RSK1 as a ratio of the band intensities of PKARIα from 3 similar experiments. *p < 0.05; **, p < 0.01. D–E, overexpression of HA-RSK1 (S221A) activates PKA. Experiments were the same as in panel C, except that the cell lysates were probed with anti-phospho-PKA substrate (D) or anti- phospho-BAD Ser-155 antibodies (E). In D, a representative of three similar experiments is shown. IB, immunoblot. In E, the right-hand panel shows quantification of band intensities of phospho-BAD-Ser-155 and Erk1/2 from three experiments; *, p < 0.05. F, silencing of RSK1 increases the interactions between endogenous PKAc and PKARIα. B82L cells were transfected with RSK1-specific siRNA #1 (20 nm) or #2 (40 nm) for 56 h and then serum-starved overnight. The cell lysates were immunoprecipitated with anti-PKARIα antibody. The right-hand panel shows quantification of band intensities of PKAc as a ratio of PKARIα band intensities from three experiments. *, p < 0.05, as compared with control. G, silencing of RSK1 decreases phosphorylation of BAD on Ser-155. RSK1 was silenced as in F. Quantified band intensities of phospho-BAD-Ser-155 and Erk1/2 from three experiments are shown on the right. *, p < 0.05; **, p < 0.01, as compared with control siRNA (Con). WCL, whole cell lysate.
To assess the competition between RSK1 and PKAc for binding to endogenous PKARIα in intact cells, we transfected HEK293T cells with increasing amounts of plasmid expressing HA-RSK1 (S221A) and examined the amounts of PKAc and HA-RSK1 in the PKARIα pulldown assays. As the expression of HA-RSK1(S221A) in HEK293T cells increased, more HA-RSK1(S221A) was associated with PKARIα, and this was accompanied by decreasing amounts of PKAc in the PKARIα pulldown assays (Fig. 4C). Moreover, by competing for PKARIα binding and dissociating PKAc from PKARIα, increasing the expression of HA-RSK1(S221A) in intact cells increased PKAc activity as assessed by augmented immunoreactivity of cellular proteins to anti-phospho-PKA substrate antibody (Fig. 4D) and an increase in phosphorylation of Ser-155, the PKAc site (30, 31), on BAD (Fig. 4E). Because PKARIα-free PKAc is unstable, in the experiments shown in Fig. 4, D and E, which involve longer term expression of HA-RSK1 (S221A), if anything, the activity of the PKAc monitored may be an underestimate. To determine whether endogenous RSK1 regulated PKAc activity by competing with PKAc for binding to PKARIα, RSK1 was silenced using two different RSK1-specific siRNAs. As shown in Fig. 4F, after silencing of endogenous RSK1, immunoprecipitates of endogenous PKARIα contained greater amounts of PKAc, indicating that endogenous RSK1 and PKAc compete for association with endogenous PKARIα; the amounts of PKARIα or PKAc were not altered by silencing of RSK1 (Fig. 4, F and G). Additionally, consistent with increased PKAc/PKARIα interactions, when RSK1 was silenced, the endogenous PKA activity was decreased as monitored by phosphorylation of BAD on Ser-155, the PKAc site (Fig. 4G). Together these data (Fig. 4) show that RSK1 competes with PKAc for binding to PKARIα and that, via this competition, RSK1 can modulate the activity of PKAc in vitro as well as in intact cells.
RSK1/PKARIα Interaction Regulates Activation of RSK1 and Its Ability to Modulate Cellular Apoptosis
The next aim of our studies was to determine the functional consequences of the interactions between inactive RSK1 and PKARIα on the activation of RSK1 and its ability to inhibit apoptosis. Recently, using a variety of approaches including silencing of PKARIα, we have shown that the PKARIα/RSK1 interaction is necessary to bring RSK1 in proximity of PP2Ac on AKAPs such as D-AKAP1 (21). By bringing RSK1 in proximity of PP2Ac, PKARIα ensures that the RSK1 is dephosphorylated and maintained in its inactive form in the absence of any growth factors (21). Therefore, we reasoned that the cell-permeable, palmitoylated peptide Wt-PS that competes with PKARIα for association with RSK1 (Fig. 2D) may augment the activation state of the RSK1 in unstimulated cells and also regulate its ability to phosphorylate its cellular substrates and function. The palmitoylated peptide Mut-PS, in which the Arg residues corresponding to Arg-93 and -94 in the pseudosubstrate site substituted with Ala, was used as the control. These experiments were performed in B82L cells that contain endogenous PKARIα but not PKARII (20). Peptide Wt-PS, but not Mut-PS, decreased the association of endogenous PKARIα with endogenous RSK1 (Fig. 5A). Because PKARIα, via its interactions with AKAPs, also brings RSK1 in proximity with the AKAP-associated catalytic subunit of PP2Ac (21), peptide Wt-PS-mediated dissociation of PKARIα and RSK1 was also accompanied by a decrease in PP2Ac in the immunoprecipitations of RSK1 (Fig. 5A). As expected from its ability to dissociate RSK1 from PKARIα and, therefore, also from the complex containing PP2Ac (Fig. 5A and Ref. 21), peptide Wt-PS also activated endogenous RSK1 under basal conditions (i.e. in the absence of growth factors). Thus, as assessed by its phosphorylation on Ser-380 (Fig. 5A) and in vitro kinase activity of immunoprecipitated RSK1 (supplemental Fig. S1), RSK1 was active in peptide Wt-PS-treated cells but not in cells that had been treated with Mut-PS. That the activity in the immunoprecipitates represents RSK1 activity and not any other associated kinase(s) is shown by the finding that SL-0101, a selective RSK1/2 inhibitor (18), inhibited the activity (supplemental Fig. S1).
FIGURE 5.
PKARIα/RSK1 interactions regulate RSK1 activation and apoptosis. A, peptide (Wt-PS) corresponding to the pseudosubstrate region of PKARIα disrupts the interaction of PKARIα and RSK1 and increases RSK1 phosphorylation in unstimulated cells. B82L cells were serum-starved overnight and then were treated with 2 μm each of Wt-PS or Mut-PS (for sequences, see the legend to Fig. 2) for 10 min. RSK1 from the cell lysates was immunoprecipitated (IP) with anti-RSK1 antibody. The proteins in the immune complex were detected with Western analysis. The quantitative data using band intensities of the indicated proteins from three experiments are shown in the panels on the right. B, Wt-PS, but not Mut-PS, disrupts the association of RSK1 with PKARIα, increases the binding of endogenous PKAc to PKARIα, and decreases phosphorylation of BAD on Ser-155. Cell lysates were prepared as in A, and PKARIα was immunoprecipitated with anti-PKARIα antibody. The proteins in the immune complexes or whole cell lysates (WCL) were subjected to Western analysis. Quantification of data using band intensities of the indicated proteins (n ≥ 3 experiments) are shown in the panels on the right. C, Wt-PS, but not Mut-PS, increases basal RSK1 activity and decreases apoptosis. After overnight deprivation of serum, B82L cells were treated with PKARIα peptides Wt-PS or Mut-PS (2 μm each) for 10 min followed by stimulation with 50 nm EGF for 10 min. Cell lysates were examined for phosphorylation of Ser-380 on RSK1 and Ser-112 on BAD. The panels on the right represent quantification of band intensities of the indicated proteins from three experiments. For the apoptosis assay, after treatments with or without EGF for 10 min, cells were treated with TNF-α (20 ng/ml) plus cycloheximide (25 μg/ml) (TNF/CHX) for 1 h, and DNA fragmentation was monitored. Data are the mean ± S.E. of A405 per μg of protein (n = 3). D, silencing of RSK1 abrogates the ability of Wt-PS to inhibit apoptosis. Procedures were as in C, except that cells were transfected with RSK1 siRNAs for 56 h before experimentation. The inset shows silencing of RSK1 by the two siRNAs. Data are the mean ± S.E. of A405 per μg of protein (n = 3).
As observed with silencing of RSK1 (Fig. 4F), the dissociation of RSK1 from PKARIα by peptide Wt-PS was accompanied by an increase in the association between PKAc and PKARIα (Fig. 5B) with a resultant decrease in PKA activity as monitored by phosphorylation of BAD on Ser-155, the PKA site (30, 31). These findings further corroborate our observations that endogenous RSK1 and PKAc compete for association with PKARIα. Note that although the pseudosubstrate region of PKARIα is one of the binding sites for PKAc, PKARIα also interacts with the large lobe of PKAc (7, 32). Therefore, the inability of peptide Wt-PS to compete the PKARIα/PKAc interactions may reflect differences in the affinity of the peptide versus the pseudosubstrate domain of full-length PKARIα for the catalytic cleft of PKAc. Alternatively, because the concerted interactions of the pseudosubstrate region and cAMP binding domains of PKARIα with PKAc are required for binding and inhibiting the activity of PKAc (32), the loss of interaction only with the pseudosubstrate region is insufficient to release PKAc from PKARIα. Whatever, the mechanism, it is clear from our data that peptide Wt-PS releases RSK1 from PKAR1α and thereby increases PKAc/PKARIα interactions and also decreases PKAc activity in intact cells.
Next, we investigated whether peptide PS-elicited RSK1 activation altered the phosphorylation of its substrate BAD on Ser-112, the RSK1 site (33), and modulated cell survival. Peptide Wt-PS, but not the mutant peptide (Mut-PS), increased RSK1 activation in the absence of growth factor and also augmented phosphorylation of BAD on Ser-112 (Fig. 5C). The basal apoptosis in B82L cells was low and not altered either by EGF or any of the two peptides. In contrast, TNF-α and cycloheximide (TNF-α/CHX)-induced apoptosis was inhibited by peptide Wt-PS to the same extent as by EGF; the control peptide (Mut-PS) was ineffective (Fig. 5C). The inhibition of apoptosis by peptide Wt-PS and EGF was not additive (Fig. 5C). That the effects of peptide PS were mediated by RSK1 is shown by our findings that silencing of RSK1 using two different siRNAs against it obliterated the ability of peptide Wt-PS to protect against TNF-α/CHX-induced apoptosis (Fig. 5D). Interestingly, siRNA-mediated silencing of RSK1 increased basal apoptosis in the absence of TNF-α and CHX, suggesting that low levels of RSK1 activity under basal conditions contribute toward cell survival.
DISCUSSION
Our finding that the pseudosubstrate region of PKARIα is involved in interactions with the NTK of RSK1 when the Ser-221 in RSK1 is replaced by a residue (Ala) that cannot be phosphorylated but not when Ser-221 is substituted by a negatively charged residue explains why inactive, and not the active RSK1 associates with PKARIα. The finding that the NTK domain of RSK1 interacts with the pseudosubstrate region of PKARIα and that Arg residues 93 and 94 are necessary for these interactions explains certain earlier reported results from our laboratory. Thus, the lack of an interaction of RSK1 with PKARII (20) is explained by the differences in the pseudosubstrate region of PKARIα (RRRRGAI) and the inhibitory site on PKARII (FNRRVSV); the pseudosubstrate region of PKARIα represents a RSK1 consensus phosphorylation sequence, whereas the cognate region on PKARII does not. That the intact RSK1 consensus phosphorylation sequence on the pseudosubstrate region of PKARIα is required for its interaction with the NTK of RSK1 suggests that the catalytic cleft of the NTK is involved in the interactions. The catalytic cleft residues in PKAc also contact the pseudosubstrate site on PKARIα and the cognate inhibitory region on PKARII with a resultant inhibition of PKAc activity (7, 8, 28). In the case of RSK1, however, the inactive, but not the active form of RSK1 associates with PKARIα (20, 21), and substitution of Ser-221 with Asp abrogates this interaction, suggesting that the Ser-221 phosphorylation or a negative charge at this site alters the orientation of the residues in the NTK catalytic cleft, which interacts with the pseudosubstrate site on PKARIα. This explains why active RSK1 does not interact with PKARIα and also explains the inability of PKARIα to inhibit the catalytic activity of active RSK1 (see the supplemental data in Chaturvedi et al. (20)).
Although the NTK domains of RSK1, RSK2, and RSK3 are similar, only RSK1 associates with PKARIα (20, 21). Thus, the small differences between the catalytic cleft residues of RSK1 and the other RSK isoforms are most likely the determinants of the specificity of binding with PKARIα. The structure of the NTK of RSK1 has been solved (34). Unfortunately, however, residues in the activation loop surrounding Ser-221 were not ordered and, therefore, were not resolved in the structure (34). Hence, it is difficult to predict the precise residues in the RSK1 NTK catalytic cleft that interact with the Arg residues (Arg-93–96) in PKARIα, whose substitutions also abrogate PKARIα/RSK1 interactions. On the other hand, the structure of the type I PKA holoenzyme clearly shows the interactions of Arg-95 and -96 with Glu-127, -170, and -230 in the catalytic cleft of PKAc including Thr-197 in the activation loop that is phosphorylated (7). Consistent with the structural data, the R95A/R96A mutant of PKARIα does not interact with PKAc, whereas the R93A/R94A mutant associates with PKAc (Fig. 2C), as these residues are not in direct contact with PKAc (7). Notably, although R95A/R96A PKARIα does not associate with PKAc (Fig. 2C), suggesting that this site as an important determinant of PKAc/PKARIα interaction, peptide Wt-PS, which contains the PKARIα pseudosubstrate region, did not compete for PKARIα/PKAc association, indicating that once the PKAc is bound to PKARIα, competition at the pseudosubstrate region of PKARIα alone is not sufficient to dissociate PKAc (Fig. 5B) but, rather, increases PKARIα/PKAc interactions by competing RSK1 from PKARIα. This may either be reflective of the relative affinities of peptide Wt-PS and pseudosubstrate region of full-length PKARIα for PKAc or the inability of competition at the pseudosubstrate site alone to overcome the concerted interactions of PKAc with different binding sites on PKARIα (7, 32).
Our findings that the two kinases have fairly similar (0.4 and 0.8 nm) affinities for PKARIα and compete for interactions with PKARIα in vitro as well as in intact cells (Figs. 3–5) imply that the basal activity of PKAc in cells may be regulated by the amount of RSK1 that is expressed. To this end, the ability of overexpressed RSK1 to decrease the PKARIα/PKAc association and increase PKAc activity in cells combined with the observation that silencing of endogenous RSK1 increases PKAc/PKARIα interactions and decreases PKAc activity demonstrate the physiological role of this RSK1/PKARIα association in regulating PKAc activity without involving changes in cAMP levels. In this context it should also be noted that active RSK1, by associating with PKAc, decreases the ability of cAMP to activate type I PKA activity (22). Hence, depending upon the abundance of active and inactive RSK1, the PKA activity can either be inhibited or increased, respectively. These findings from our laboratory (Ref. 22 and this report) underscore the ability of RSK1 to modulate PKA activity.
Although the NTK of RSK1 is sufficient to bind PKARIα (Fig. 1), the affinity of GST-RSK-(1–317) was 3-fold lower than that of GST-FL-RSK1 (Fig. 3). This difference could be due to either some steric hindrance from the GST tag in the context of the small region (1–317) of RSK1 or that there is another site on RSK1 that is involved in the regulation of the association between RSK1 and PKARIα. Although we have not observed interactions of PKARIα with any other region on RSK1 beside the NTK, future co-crystallization and structural studies may reveal further subtleties in the interactions between these proteins.
Besides regulating PKA activity, the other physiological function of the PKARIα/RSK1 interaction concerns regulation of RSK1 activation. Consistent with our recent report that PKARIα, by associating with RSK1, brings it in the proximity of PP2Ac that then dephosphorylates RSK1 to maintain it in an inactive state (21), dissociation of RSK1 from PKARIα with the Wt-PS peptide increased RSK1 activity in cells without the addition of any growth factors. This increase in RSK1 activity was also reflected in the elevated phosphorylation of the proapoptotic protein BAD and protection of cells against apoptosis. These findings underscore how the PKARIα/RSK1 interactions regulate both the cellular activity of PKA as well as RSK1.
One of the implications of our finding that PKAc and RSK1 compete for binding PKARIα is that in the context of the PKARIα dimer, one of the monomers may be associated with PKAc, whereas the other may be associated with RSK1. The other implication of the competitive binding to PKARIα concerns the Carney complex, an inherited autosomal disease in which one of the alleles of the PKAR1A gene is mutated, resulting in haploinsufficiency of PKARIα (35, 36). Decreased PKARIα levels in Carney complex have been attributed to elevated PKAc activity. This activation of PKAc may be further augmented by the competition between PKAc and RSK1 for binding the lower amounts of PKARIα in this condition. Moreover, the decreased amounts of PKARIα in Carney complex, by decreasing the association of RSK1 with AKAPs and PP2Ac, may also increase RSK1 activity that then leads to augmented survival and growth of tumors in these patients.
In summary, in this communication we have identified RSK1 as another kinase that interacts with the pseudosubstrate site of PKARIα and competes with PKAc for this binding. Although a number of proteins have been reported to interact with PKARIα, only RSK1 competes with PKAc for the pseudosubstrate domain on PKARIα. We also show that the PKARIα/RSK1 interaction has important roles in regulating both PKAc activity independent of cAMP and the activation state of RSK1 and its biological actions.
Supplementary Material
Acknowledgments
We thank Dr. Susan S. Taylor (Univ. of California, San Diego) for the purified PKAc and PKARIα. We also thank Dr. Warner Greene (Univ. of California, San Francisco) for the rat RSK1 cDNA.
This work was supported, in whole or in part, by National Institutes of Health Grant GM079226.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PKA
- protein kinase A
- PKAc
- catalytic subunit of PKA
- PKAR
- PKA regulatory subunit
- RSK
- p90 ribosomal S6 kinase
- AKAP
- PKA-anchoring proteins
- NTK
- N-terminal kinase domain
- CTK
- C-terminal kinase domain
- PP2Ac
- catalytic subunit of protein phosphatase 2A
- CHX
- cycloheximide
- FL
- full-length
- EYFP
- enhanced yellow fluorescent protein
- Erk
- extracellular signal-regulated kinase
- HA
- hemagglutinin
- GST
- glutathione S-transferase
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- Mops
- 4-morpholinepropanesulfonic acid
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate
- MES
- 4-morpholineethanesulfonic acid
- siRNA
- small interfering RNA
- EGF
- epidermal growth factor
- TNF
- tumor necrosis factor
- BAD
- Bcl-xL/Bcl-2-associated death promoter
- D-AKAP1
- dual specific AKAP1.
REFERENCES
- 1.Døskeland S. O., Maronde E., Gjertsen B. T. (1993) Biochim. Biophys. Acta 1178, 249–258 [DOI] [PubMed] [Google Scholar]
- 2.Skålhegg B. S., Taskén K. (1997) Front. Biosci. 2, d331–342 [DOI] [PubMed] [Google Scholar]
- 3.Saraswat L. D., Filutowics M., Taylor S. (1988) Methods Enzymol. 159, 325–336 [DOI] [PubMed] [Google Scholar]
- 4.Taylor S. S., Buechler J. A., Yonemoto W. (1990) Annu. Rev. Biochem. 59, 971–1005 [DOI] [PubMed] [Google Scholar]
- 5.Boucher M. J., Duchesne C., Lainé J., Morisset J., Rivard N. (2001) Biochem. Biophys. Res. Commun. 285, 207–216 [DOI] [PubMed] [Google Scholar]
- 6.Knighton D. R., Zheng J. H., Ten Eyck L. F., Ashford V. A., Xuong N. H., Taylor S. S., Sowadski J. M. (1991) Science 253, 407–414 [DOI] [PubMed] [Google Scholar]
- 7.Kim C., Xuong N. H., Taylor S. S. (2005) Science 307, 690–696 [DOI] [PubMed] [Google Scholar]
- 8.Taylor S. S., Kim C., Vigil D., Haste N. M., Yang J., Wu J., Anand G. S. (2005) Biochim. Biophys. Acta 1754, 25–37 [DOI] [PubMed] [Google Scholar]
- 9.Carriere A., Ray H., Blenis J., Roux P. P. (2008) Front. Biosci. 13, 4258–4275 [DOI] [PubMed] [Google Scholar]
- 10.Dümmler B. A., Hauge C., Silber J., Yntema H. G., Kruse L. S., Kofoed B., Hemmings B. A., Alessi D. R., Frödin M. (2005) J. Biol. Chem. 280, 13304–13314 [DOI] [PubMed] [Google Scholar]
- 11.Jensen C. J., Buch M. B., Krag T. O., Hemmings B. A., Gammeltoft S., Frödin M. (1999) J. Biol. Chem. 274, 27168–27176 [DOI] [PubMed] [Google Scholar]
- 12.Richards S. A., Fu J., Romanelli A., Shimamura A., Blenis J. (1999) Curr. Biol. 9, 810–820 [DOI] [PubMed] [Google Scholar]
- 13.Chen R. H., Sarnecki C., Blenis J. (1992) Mol. Cell. Biol. 12, 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertolotto C., Maulon L., Filippa N., Baier G., Auberger P. (2000) J. Biol. Chem. 275, 37246–37250 [DOI] [PubMed] [Google Scholar]
- 15.Tan Y., Ruan H., Demeter M. R., Comb M. J. (1999) J. Biol. Chem. 274, 34859–34867 [DOI] [PubMed] [Google Scholar]
- 16.Roux P. P., Ballif B. A., Anjum R., Gygi S. P., Blenis J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13489–13494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark D. E., Errington T. M., Smith J. A., Frierson H. F., Jr., Weber M. J., Lannigan D. A. (2005) Cancer Res. 65, 3108–3116 [DOI] [PubMed] [Google Scholar]
- 18.Smith J. A., Poteet-Smith C. E., Xu Y., Errington T. M., Hecht S. M., Lannigan D. A. (2005) Cancer Res. 65, 1027–1034 [PubMed] [Google Scholar]
- 19.Rolfe M., McLeod L. E., Pratt P. F., Proud C. G. (2005) Biochem. J. 388, 973–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaturvedi D., Poppleton H. M., Stringfield T., Barbier A., Patel T. B. (2006) Mol. Cell. Biol. 26, 4586–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaturvedi D., Cohen M. S., Taunton J., Patel T. B. (2009) J. Biol. Chem. 284, 23670–23681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao X., Patel T. B. (2009) J. Biol. Chem. 284, 33070–33078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roux P. P., Richards S. A., Blenis J. (2003) Mol. Cell. Biol. 23, 4796–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cavet M. E., Lehoux S., Berk B. C. (2003) J. Biol. Chem. 278, 18376–18383 [DOI] [PubMed] [Google Scholar]
- 25.Newlon M. G., Roy M., Morikis D., Carr D. W., Westphal R., Scott J. D., Jennings P. A. (2001) EMBO J. 20, 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newlon M. G., Roy M., Morikis D., Hausken Z. E., Coghlan V., Scott J. D., Jennings P. A. (1999) Nat. Struct. Biol. 6, 222–227 [DOI] [PubMed] [Google Scholar]
- 27.Banky P., Newlon M. G., Roy M., Garrod S., Taylor S. S., Jennings P. A. (2000) J. Biol. Chem. 275, 35146–35152 [DOI] [PubMed] [Google Scholar]
- 28.Buechler Y. J., Taylor S. S. (1991) J. Biol. Chem. 266, 3491–3497 [PubMed] [Google Scholar]
- 29.Herberg F. W., Dostmann W. R., Zorn M., Davis S. J., Taylor S. S. (1994) Biochemistry 33, 7485–7494 [DOI] [PubMed] [Google Scholar]
- 30.Virdee K., Parone P. A., Tolkovsky A. M. (2000) Curr. Biol. 10, 1151–1154 [DOI] [PubMed] [Google Scholar]
- 31.Lizcano J. M., Morrice N., Cohen P. (2000) Biochem. J. 349, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim C., Cheng C. Y., Saldanha S. A., Taylor S. S. (2007) Cell 130, 1032–1043 [DOI] [PubMed] [Google Scholar]
- 33.Shimamura A., Ballif B. A., Richards S. A., Blenis J. (2000) Curr. Biol. 10, 127–135 [DOI] [PubMed] [Google Scholar]
- 34.Ikuta M., Kornienko M., Byrne N., Reid J. C., Mizuarai S., Kotani H., Munshi S. K. (2007) Protein Sci. 16, 2626–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirschner L. S., Carney J. A., Pack S. D., Taymans S. E., Giatzakis C., Cho Y. S., Cho-Chung Y. S., Stratakis C. A. (2000) Nat. Genet. 26, 89–92 [DOI] [PubMed] [Google Scholar]
- 36.Kirschner L. S., Sandrini F., Monbo J., Lin J. P., Carney J. A., Stratakis C. A. (2000) Hum. Mol. Genet 9, 3037–3046 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.