Abstract
Her4 (ErbB-4) and Her2/neu (ErbB-2) are receptor-tyrosine kinases belonging to the epidermal growth factor receptor (EGFR) family. Crystal structures of EGFR and Her4 kinase domains demonstrate kinase dimerization and activation through an allosteric mechanism. The kinase domains form an asymmetric dimer, where the C-lobe surface of one monomer contacts the N-lobe of the other monomer. EGFR kinase dimerization and activation in vitro was previously reported using a nickel-chelating lipid-liposome system, and we now apply this system to all other members of the EGFR family. Polyhistidine-tagged Her4, Her2/neu, and Her3 kinase domains are bound to these nickel-liposomes and are brought to high local concentration, mimicking what happens to full-length receptors in vivo following ligand binding. Addition of nickel-liposomes to Her4 kinase domain results in 40-fold activation in kinase activity and marked enhancement of C-terminal tail autophosphorylation. Activation of Her4 shows a sigmoidal dependence on kinase concentration, consistent with a cooperative process requiring kinase dimerization. Her2/neu kinase activity is also activated by nickel-liposomes, and is increased further by heterodimerization with Her3 or Her4. The ability of Her3 and Her4 to heterodimerize and activate other family members is studied in vitro. Her3 kinase domain readily activates Her2/neu but is a poor activator of Her4, which differs from the prediction made by the asymmetric dimer model. Mutation of Her3 residues 952ENI954 to the corresponding sequence in Her4 enhanced the ability of Her3 to activate Her4, demonstrating that sequence differences on the C-lobe surface influence the heterodimerization and activation of ErbB kinase domains.
Keywords: Breast Cancer, Growth Factors, Liposomes, Receptor-Tyrosine Kinase, Signal Transduction
Introduction
Her4 (ErbB-4) and Her2/neu (ErbB-2) are members of the ErbB family of receptor-tyrosine kinases, with the founding member of this family being epidermal growth factor receptor (EGFR)3 (1). The ErbB receptor-tyrosine kinases have an extracellular domain that is involved in ligand binding and receptor homo- or heterodimerization. Their intracellular domain consists of the juxtamembrane region, tyrosine kinase domain, and C-terminal tail. EGFR, Her2/neu, and Her4 contain catalytically active kinase domains, whereas Her3 (ErbB-3) has an inactive kinase domain and must heterodimerize with another ErbB receptor to signal (2). Several studies on ErbB heterodimerization have shown that Her2/neu is the preferred heterodimerization partner for the other three ErbB receptors (3–5). These studies also provide evidence for formation of EGFR-Her3 and EGFR-Her4 heterodimers in response to ligands that bind to Her3 or Her4 (3, 6).
Both Her4 and Her2/neu play important roles in the development and the normal physiology of the cardiovascular and nervous systems (7, 8). Her4 and Her2/neu knock-out mice both die around embryonic day 10.5 because of malformation of the cardiac trabeculae, and additionally, they are found to have abnormalities in the hindbrain or in sensory ganglia and motor nerves (9, 10). In human breast cancers, Her2/neu gene amplification is found in 20–30% of patients, resulting in unregulated tyrosine kinase signaling that increases cancer cell proliferation, migration, and propensity to metastasize (11). Treatment of Her2/neu-amplified breast cancers with the monoclonal antibody, trastuzumab (Herceptin), forms an essential part of state of the art, breast cancer treatment (12).
Multiple crystal structures of the extracellular domain of all four ErbB receptor-tyrosine kinases have been solved and provide a detailed understanding of ligand binding and receptor dimerization (reviewed in Refs. 13, 14). However, our structural understanding of the intracellular domain of the ErbB receptors is less complete. Multiple crystal structures of the EGFR tyrosine kinase domain have been solved (reviewed in Ref. 15), and in 2008, two structures of the Her4 tyrosine kinase domain were published (16, 17). An important advance in the understanding of how ErbB receptor dimerization activates the tyrosine kinase domain came from the crystal structure of an EGFR kinase domain dimer (18). The contact surfaces of the observed dimer are located on the C-lobe of one monomer (called the “donor” monomer) and the N-lobe of the other monomer (the “acceptor” monomer), and therefore, this dimer was named the asymmetric dimer. Comparison of this EGFR kinase domain dimer to the structure of the cyclin-cyclin-dependent kinase complex showed similarities in that the “donor” EGFR kinase domain monomer acted as a “cyclin-like” activator for the “acceptor” EGFR monomer (18). This asymmetric dimer model was supported by biochemical studies on recombinant EGFR kinase domain protein and by mutational studies on the full-length EGFR protein. The crystal structure of Her4 kinase domain shows that it also adopts a very similar, asymmetric dimer structure (16).
The biochemical studies on EGFR kinase domain which supported the asymmetric dimer model involved the use of a reconstituted in vitro system. In this system, the EGFR kinase domain was anchored to the surface of a liposome and brought to a high local concentration, thereby mimicking what happens in vivo on the cell membrane when the full-length EGFR protein binds its ligand and dimerizes (18). The anchoring of the EGFR kinase domain to liposome was achieved by incorporating a polyhistidine tag on the kinase and a nickel-chelating lipid into the liposome. This in vitro system was first developed by Weis and co-workers (19, 20) in their studies of a prokaryotic signal transduction system that mediates bacterial chemotaxis. Small unilamellar vesicles (SUVs or liposomes) containing this nickel-chelating lipid were used to anchor and cluster the polyhistidine-tagged Tar chemotaxis protein and form a 3-protein complex containing Tar, an adaptor protein, and the CheA protein kinase. This complex resulted in a 180-fold increase in CheA protein kinase activity. Application of this system to EGFR allowed for in vitro studies of EGFR kinase domain dimerization and discovery of the mechanism of action of Mig6/RALT, an endogenous inhibitory protein of EGFR (21).
In this report, we show that this nickel-chelating lipid-liposome system is broadly applicable to the ErbB receptor-tyrosine kinases. Her4 binds to and dimerizes on the surface of these liposomes, and this results in a marked enhancement of its tyrosine kinase activity. Likewise, Her2/neu is also activated by addition of these liposomes. This system offers the unique advantage that ErbB kinase domain heterodimerization can now be studied in vitro, and we utilize this to test predictions made by the EGFR asymmetric dimer model. Specifically, we observe that the ability of the Her3 kinase domain to activate Her4 does not follow the model predictions and that this difference is due to specific residues on the Her3 kinase domain C-lobe surface.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Dioleoyl-phosphatidylcholine (DOPC) and nickel-1,2-dioleoyl-sn-glycero-3-([N-(5-amino-1-carboxypentyl)iminodi acetic acid]succinyl)-nickel salt (Ni-NTA-DOGS) were purchased from Avanti Polar Lipids (Alabaster, AL). Antibodies: Her2/neu phospho-Tyr-877 antibody was from Abcam (catalogue no. ab47262, Cambridge, MA) and anti-phosphotyrosine, clone 4G10 was from Millipore (Billerica, MA). Substrate peptide, biotin-GGMEDIYFEFMGGKKK, was synthesized by W. M. Keck Biotechnology Resource Laboratory at Yale University (New Haven, CT). [γ-32P]ATP was from PerkinElmer (Waltham, MA). Avidin and ECL reagents were from Pierce. All other chemicals and reagents were from Sigma-Aldrich.
DNA Constructs
The Her4 kinase domain (residue 702–1029) and kinase domain plus C terminus (residues 702–1308) were cloned into the pFastBacHT vector (Invitrogen) with NotI, XbaI ends. Her4 amino acid numbering used here is based on the Her4 JM-a/Cyt-1 isoform (NCBI Refseq accession number NP_005226.1) with its signal peptide sequence included. The Her3 kinase domain (residues 693–1021) was cloned into pFastBacHT vector using EcoRI, KpnI sites. Site-directed mutagenesis was done using the QuikChange kit (Stratagene). Each construct was verified by DNA sequence analysis. Recombinant baculoviruses were prepared following the Bac-to-Bac protocol (Invitrogen). P2 viral stocks were titered using the FastPlax kit (Novagen, Madison, WI). The Her2/neu kinase domain (residues 705–1029) in pFastBacHT was a gift from Drs. Daniel Leahy and Aruna Sathyamurthy (Johns Hopkins University). Her3 and Her2/neu NCBI Refseq accession numbers are NP_001973.2 and NP_004439.2, respectively. Full-length Her3 and Her4 JM-a/Cyt1 isoform cDNAs were kindly provided by Dr. Graham Carpenter (Vanderbilt University).
Protein Expression
Sf9 cells were grown in shaker flasks at 27 °C and 120 rpm in IPL-41 medium (Invitrogen) supplemented with 10% bovine calf serum supplemented (Hyclone), yeastolate extract (Invitrogen), penicillin, streptomycin, and Pluronic F-68. Cultures at a density of 1 × 106 cell/ml were infected at 1 MOI and incubated for 48 h. Cells were harvested by centrifugation then washed once with resuspension buffer (20 mm Hepes, pH 7.5, 4 mm NaHCO3, 55 mm KCl, 5 mm CaCl2, 78 mm sucrose). The pellets were snap-frozen in dry ice/ethanol bath and stored at −80 °C. Her3 and Her4 cell pellets were lysed by sonication in Buffer A (25 mm Tris-HCl, pH 8, 300 mm NaCl, 10 mm imidazole, 10% glycerol, 1 mm DTT) supplemented with phenylmethylsulfonyl fluoride, benzamidine, DNase, and protease inhibitor mixture tablets lacking EDTA (Roche). Lysates were centrifuged for 45 min at 20,000 × g. 0.2–0.5 ml of Ni-NTA beads (Qiagen) were added to the supernatants and incubated on a rotator for 1 h at 4 °C. The beads were poured into a 10-ml disposable column (Bio-Rad) and the flow-through collected. The column was washed with 10 column volumes of Buffer A + 20 mm imidazole, and the proteins were eluted with Buffer A + 50–125 mm imidazole. Her2/neu cell pellets were processed the same way except the lysis buffer was supplemented with 1% Tween-20, and the protein was eluted with Buffer A + 50–125 mm imidazole + 0.01% Tween-20. Her2/neu was concentrated using an Amicon 10K spin-concentrator (Millipore) and buffer exchanged in 20 mm Tris-HCl, pH 8, 150 mm NaCl, 0.01% Tween-20, 1 mm DTT. Aliquots were snap-frozen then stored at −80 °C. Her4 and Her3 was further purified by gel filtration chromatography on Superdex 200 GL 10/300 column (GE Healthcare) using 20 mm Tris-HCl, pH 8, 150 mm NaCl, 1 mm DTT buffer. Pooled fractions were concentrated, as indicated above, snap frozen, and then stored −80 °C.
Liposome Preparation
DOPC and the nickel-chelating lipid, Ni-NTA-DOGS, were aliquoted (total lipid = 1 mg/ml) into glass tubes and dried under a gentle stream of N2. The dried lipid films were placed under high vacuum for 2 h to remove residual solvents, then stored under N2 at −20 °C for no more than 2 weeks before use. Liposomes were prepared by a modification of the reverse phase method of Papahadjopoulos et al. (22). One volume of diethyl ether was added to the dried lipid film followed by 1 volume of liposome buffer (20 mm Tris-HCl, pH 7.5, 100 mm NaCl). The solution was then sonicated for about 20 s in a bath sonicator (power delivered to the bath is 80 kHz and 80 watts, Laboratory Supplies Co., Hicksville, NY) to form an emulsion. The emulsion was then transferred to a 50 ml round bottom flask and rotary-evaporated to remove the ether. The resulting multi-lamellar vesicles were then used to make small unilamellar vesicles (SUV) by the extrusion method using a mini-extruder (Avanti Polar Lipids) with 200-nm polycarbonate membranes. The diameter of these SUV was determined to be 180–190 nm by dynamic light scattering. Total phospholipid concentration was determined by the method of Stewart (23).
Kinase Assays
Radiometric kinase assays in solution were carried out as described in Ref. 16. Briefly Her4 or Her2/neu kinase domain proteins were incubated in 35 mm Tris, pH 7.5, 10 mm MgCl2, 100 μm Na3VO4, 2 mm DTT, 100 μm ATP, 1 μCi of [γ-32P]ATP, 75 mm NaCl, 5% glycerol, 0.005% Tween-20 with 100 μm biotin-GGMEDIYFEFMGGKKK as the peptide substrate in a 25-μl reaction volume. This peptide sequence was developed as an optimized peptide substrate for recombinant Her2/neu kinase domain (24), and the Km for this peptide was previously measured to be 170 μm for Her4 kinase domain and 60 μm for Her2/neu kinase domain (16). Reactions were initiated by the addition of 0.25–2.5 μm kinase, carried out for 6–8 min at 30 °C, and stopped by the addition of 10 μl of 100 mm EDTA. To each sample, 10 μl of 10 mg/ml avidin was added, and 25-μl samples were transferred to centrifugal filtration units with 30,000 NMWL membranes (Millipore) and washed three times with 100 μl of wash solution (0.5 m sodium phosphate, 0.5 m NaCl, pH 8.5). Retained 32P was measured by scintillation counting.
To determine the kinase activity on the liposomes, kinases were incubated with liposomes on ice for 15∼25 min before the reaction. Total lipid concentration was 0.5 mg/ml. No glycerol or Tween-20 was present when assaying Her4 kinase activity with liposomes, and the kinase was diluted to 25 nm immediately prior to addition of assay mixture noted above. Activities proved to be linear with time in the ranges used, and the limiting substrate turnover was less than 10% (20% for Her4 with liposomes) for all rate measurements. Duplicate measurements were generally within 15%. Mixing or heterodimerization experiments were performed by adding the two different kinase monomers together at a concentration of 50–100 ng/μl prior to liposome addition.
Binding Studies of Kinase Domains to Liposomes
Aliquots were removed from the binding reactions and diluted to 150 μl in liposome buffer. Liposome-bound proteins were separated from free proteins by ultracentrifugation for 20 min at 120,000 × g in a Beckman ultracentrifuge in a TLA 100.1 rotor. An aliquot was removed prior to centrifuging for the total protein, and another aliquot of the supernatant was taken after centrifugation for the free protein. The protein samples were separated on a 10% Tris-glycine SDS-PAGE, stained with SimplyBlue (Invitrogen), and analyzed by densitometry. The bound fraction was calculated as the (ODtotal − ODfree)/ODtotal. Calculation of Kd was performed by fitting to a single site binding equation using SigmaPlot 2001 software (Windows version 7.0).
Measurement of Autophosphorylation
Her2/neu kinase domain or Her4 kinase domain plus C-terminal (residues 702–1308) proteins were incubated in 30 mm Tris, pH 7.5, 10 mm MgCl2, 100 μm Na3VO4, 2 mm DTT, 100 μm ATP, 1 μCi of [γ-32P]ATP, 60 mm NaCl, 5% glycerol, 0.005% Tween-20 in the presence or absence of liposomes containing 5 mol% Ni-NTA-DOGS (total lipid concentration = 0.5 mg/ml). Reactions were initiated by the addition of 1.25 μm kinase, carried out at room temperature, and stopped at the indicated time points using SDS-PAGE loading buffer with 50 mm EDTA. Reactions were run on a 10% SDS-polyacrylamide gel, and visualized using phosphor imaging. Autophosphorylation of Her2/neu Tyr-877 was measured by conducting the same reaction in the absence of [γ-32P]ATP, and performing a Western blot using the Her2/neu phospho-Tyr-877 antibody (Abcam, catalogue no. ab47262, dilution 1:10,000).
RESULTS
Her4 Kinase Domain Is Activated by Dimerizing on the Surface of Nickel-liposomes
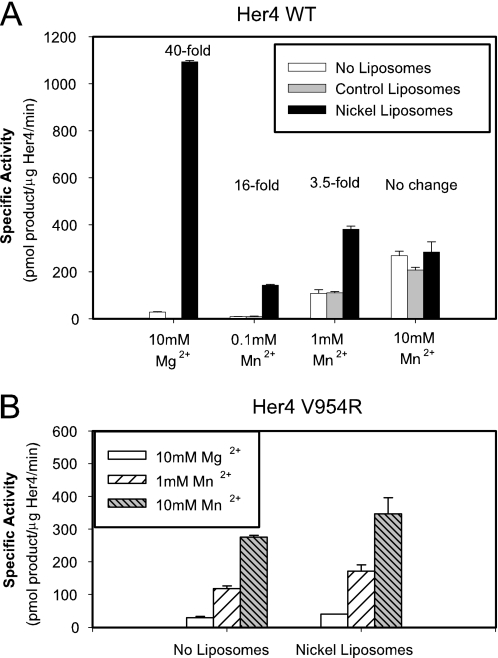
The kinase domain of Her4 was made using a baculoviral protein expression system and purified using nickel-NTA beads followed by gel filtration chromatography. Protein purity of greater than 95% was achieved (supplemental Fig. S1). Small unilamellar vesicles containing 5 mol% Ni-NTA-DOGS and 95 mol% DOPC (which we will term “nickel-liposomes”) or 100% DOPC (control liposomes) were prepared by extrusion. The kinase activity of this Her4 kinase domain protein was tested in the absence of liposomes and in the presence of control liposomes or nickel-liposomes. Her4 kinase activity was increased up to 40-fold upon short preincubation with the nickel-liposome (Fig. 1A). This effect requires both the presence of the nickel-chelating lipid on the liposomes and the His6 tag on the protein as neither control liposomes nor Her4 kinase domain with the His6 tag removed by TEV protease treatment showed this activation (Fig. 1A). The activity of the Her4 kinase domain in the presence of control liposomes was the same as activity in the absence of liposomes (Fig. 1A). Further evidence of the specificity of this effect is shown by competition with imidazole, which will disrupt binding of the His6 tag to the nickel-chelating lipid. Addition of 50–100 mm imidazole abrogated the increase in Her4 kinase activity observed upon addition of nickel-liposomes, but had no significant effect on kinase activity measured in the presence of control liposomes (Fig. 1B).
FIGURE 1.
Activation of the Her4 kinase domain by nickel-liposomes. A, Her4 kinase domain protein containing the N-terminal polyhistidine tag or Her4 having the His6 tag removed by TEV protease treatment was incubated with liposomes for 15–20 min on ice. The kinase domain-liposome complex was incubated at 30 °C with a kinase reaction mixture containing 100 μm ATP, 1 μCi of [γ-32P]ATP, 10 mm MgCl2, and 100 μm biotin-GGMEDIYFEFMGGKKK peptide as in “Experimental Procedures.” The phosphorylated peptide was measured by liquid scintillating counting. B, His6-tagged Her4 kinase domain was incubated with liposomes in the presence of 0–100 mm imidazole, pH 7.5. Kinase assays were performed as above. C, Her4 kinase domain proteins were incubated with liposomes, and kinase assays performed as above. Mixing of Her4 I712Q and V954R proteins at 1:1 molar ratio was done prior to addition of liposomes. Fold change is calculated relative to the activity with control liposomes. The Her4 amino acid numbering scheme used here is based on the Her4 JM-a/Cyt-1 isoform with its signal peptide included. Data are representative of two (A and B) or four (C) independent experiments. D, predicted effects of the Her4 mutations used in C on kinase dimerization and activation.
Based on the similarities between the crystallographic studies of the EGFR and Her4 kinase domains (16), activation of the Her4 kinase domain is also expected to depend on formation of an asymmetric dimer. Mutation of either the N-lobe surface of this dimer interface, Her4 kinase domain mutant I712Q, or the C-lobe surface, Her4 kinase domain mutant V954R, abrogates the increase in kinase activity observed upon addition of nickel-liposomes (Fig. 1C). By mixing these two mutant kinase domain proteins at a 1:1 molar ratio, the N-lobe mutant, Her4 I712Q, which has an intact C-lobe dimerization surface, is able to act as a donor monomer for the C-lobe mutant, Her4 V954R (Fig. 1D). The mixing of these two mutant Her4 proteins reconstructs the asymmetric dimer interface and is predicted to restore kinase activation. In keeping with this prediction, mixing the N-lobe and C-lobe mutants (Her4 I712Q + V954R) stimulates Her4 kinase activity by 8.4-fold (Fig. 1C).
Activation of Her4 Kinase Domain on the Surface of Nickel-liposomes Shows Cooperativity
The binding of the Her4 kinase domain protein to the nickel-liposomes was measured by incubating varying amounts of Her4 kinase domain with a fixed amount of nickel-liposome and then separating the bound and free protein by ultracentrifugation, which pellets the liposomes. The binding of the His6-tagged Her4 kinase domain to liposomes was well fit by a hyperbolic binding curve (R2 = 0.974). The Kd for His6-tagged Her4 binding to nickel-liposomes was calculated to be ∼3 μm, as expected for the His6-nickel interaction (Fig. 2A). In contrast, the activation of Her4 kinase domain on the nickel-liposome showed a clearly sigmoidal dependence on Her4 kinase domain concentration, implying a cooperative process (Fig. 2B). This complex kinase activation concentration dependence produced a Hill coefficient of 2.1 ± 0.2 (Fig. 2B, inset). One explanation for this sigmoidal kinetics and Hill coefficient is that kinase activation requires the formation of a kinase domain dimer.
FIGURE 2.
Binding and activation of Her4 on the nickel-liposomes. A, binding of His6-tagged Her4 kinase domain to nickel-liposomes was measured by incubating 0–17.3 μm Her4 with nickel-liposome (8 μm bulk concentration of accessible Ni-NTA-DOGS lipid) and then separating bound from free Her4 by ultracentrifugation at 120,000 × g. Data were fit to a single site binding equation, y = ax/(b + x). The data are representative of four experiments. B, cooperativity of Her4 kinase domain activation on nickel-liposomes. Her4 kinase domain concentration during binding was varied and kinase activity measured as in Fig. 1 and the “Experimental Procedures.” Data points represent mean ± S.D. of n = 4. Data were fit to the Hill equation of y = y0 + [axb/(cb + xb)], where b represents the Hill coefficient. The inset shows transformation of the data so that the slope = −Hill coefficient. Linear regression of this transformed data demonstrates a Hill coefficient of 2.1 ± 0.2. Data are representative of three experiments. Her4 kinase domain concentrations reported in A and B represent bulk solution concentrations of the kinase.
Effect of Divalent Cations
Divalent cations, such as the physiological cation Mg2+ or non-physiological cations Mn2+ and Co2+, are essential cofactors for protein kinase activity, but their effect on kinase activity is complex. Two cation binding sites in the active site of kinases have been defined, and binding of cations to the second, lower affinity site can either be stimulatory or inhibitory (25–27). It was previously reported that Her4 kinase activity is higher in the presence of Mn2+ than with Mg2+, but that Mn2+ concentrations >0.5 mm resulted in a reduction in Her4 kinase activity (28).
Because Mn2+ is widely used in the in vitro assay of tyrosine kinases to increase kinase activity, we sought to determine if the choice of cation influences the activation of Her4 by the nickel-liposomes. The increase in Her4 kinase activity upon addition of nickel-liposomes was seen in the presence of 0.1 or 1 mm Mn2+, though both the relative and absolute increase in kinase activity is less than with 10 mm Mg2+ (Fig. 3A). In the presence of 10 mm Mn2+, the activation of Her4 kinase activity by the nickel-liposomes was lost. The effect of Mn2+ is independent of Her4 kinase domain dimerization because the Her4 C-lobe mutant (V954R), which does not dimerize, shows the same response to varying the divalent cation in the absence or presence of nickel-liposomes (Fig. 3B). This suggests that the effects of Mn2+ occur directly on the kinase monomers themselves, most likely at the active site of the kinase. Because Mg2+ is the likely physiological cation utilized by protein kinases and because the activity of Her4 was greatest with 10 mm Mg2+ when bound to the nickel-liposome (Fig. 3A), we used 10 mm Mg2+ for all further experiments.
FIGURE 3.
Effect of divalent cations, Mg2+ and Mn2+, on Her4 kinase domain activation. A, Her4 WT kinase domain was incubated in the presence or absence of the indicated liposomes and the kinase reaction conducted in the presence of the indicated concentration of Mg2+ or Mn2+. Fold change is calculated relative to the activity in the absence of liposomes. B, Her4 V954R kinase domain was incubated in the absence or presence of nickel-liposomes and the kinase reaction conducted in the presence of the indicated concentration of Mg2+ or Mn2+. Data are representative of three (A) or two (B) independent experiments.
Enhancement of Her4 and Her2/neu Autophosphorylation by Dimerization on the Nickel-liposomes
To study the effect of the nickel-liposomes on ErbB receptor autophosphorylation, we expressed a recombinant Her4 protein containing the kinase domain plus the entire C-terminal tail (residues 702 to C-terminal end). This longer Her4 protein, which we will call Her4 kd+tail, contains all of the tyrosine phosphorylation sites identified in a recent mass spectrometry-based study of Her4 (29). Autophosphorylation of Her4 kd+tail protein was markedly enhanced by binding to the nickel-liposomes as compared with that in the absence of liposomes (Fig. 4A). Further, a shift in electrophoretic mobility of the autophosphorylated Her4 kd+tail protein was seen, suggesting that a higher stoichiometry of phosphorylation was achieved by the addition of nickel-liposomes. The time scale of autophosphorylation shown here, 20 s to 5 min, matches that of the signal transduction events seen upon addition of the Her4 ligands to cells expressing full-length Her4 (5, 30).
FIGURE 4.
Autophosphorylation of Her4 and Her2/neu. A, Her4 kd+tail was incubated with 100 μm ATP, 1 μCi of [γ-32P]ATP, and either with or without nickel-liposomes, as in the “Experimental Procedures.” 32P incorporation into Her4 kd+tail was visualized by phosphorimaging. B, upper panel, Her2/neu kinase domain autophosphorylation was measured as in A. B, lower panel, autophosphorylation of Her2/neu was conducted in a non-radioactive reaction. Phosphorylation at Tyr-877 was measured using a phosphospecific Ab to this site.
We also wanted to determine how other ErbB family members would behave when bound to the surface of the nickel-liposomes. The kinase domain of Her2/neu (residues 705–1029) was expressed using the baculoviral system. Similar to Her4, incubation of Her2/neu kinase domain with nickel-liposomes increased Her2/neu kinase activity (Fig. 5A), though this effect was more modest than the activation seen with Her4. Her2/neu kinase domain autophosphorylation was tested upon binding to the nickel-liposomes, and this effect also was markedly increased by the addition of nickel-liposomes (Fig. 4B). We previously described a phosphorylation site in the kinase domain of Her2/neu at Tyr-877 (31). Measurement of Her2/neu Tyr-877 phosphorylation, using a phosphospecific antibody to this site, demonstrates that Tyr-877 phosphorylation is readily detected after a 10-s reaction (Fig. 4B, lower panel). The degree of Her2/neu Tyr-877 phosphorylation seen without liposomes is essentially the same as background (Fig. 4B).
FIGURE 5.
Heterodimerization and activation of Her2/neu kinase domain by Her3 and kinase dead Her4. A, activation of Her2/neu kinase domain by Her3. B, activation of Her2/neu kinase domain by kinase-dead Her4 (Her4 D843N). For both A and B, kinase domain proteins were mixed at 1:1 molar ratio immediately prior to nickel-liposome addition, and kinase assay was measured as in Fig. 1. Fold change is calculated relative to Her2/neu kinase activity in the absence of liposomes. Data are representative of three experiments.
Activation of Her2/neu by Heterodimerization
Previous studies on Her2/neu have suggested that it is the preferred heterodimerization partner for the other ErbB receptor-tyrosine kinases and that the Her2/neu-Her3 heterodimer, in particular, forms an “oncogenic unit” that contributes to breast cancer proliferation (3, 32). We expressed the kinase domain of Her3 and mixed it with the Her2/neu kinase domain and nickel-liposomes to determine if this produced further activation of Her2/neu kinase activity. Consistent with prior reports (33), the Her3 kinase domain on its own showed no significant kinase activity, either in the presence of nickel-liposomes or in the absence of liposomes (supplemental Table S1). As described above, incubation of Her2/neu kinase domain with nickel-liposomes resulted in a 3.5-fold increase in kinase activity (Fig. 5A). Addition of Her3 kinase domain in a 1:1 molar ratio to Her2/neu, followed by incubation with nickel-liposomes, resulted in a 5.5-fold increase in Her2/neu kinase activity compared with Her2/neu in the absence of liposomes (Fig. 5A).
We generated a kinase-dead mutant of the Her4 kinase domain by mutating the catalytic base Asp-843 to Asn (D843N). Mixing Her2/neu kinase domain with kinase-dead Her4 in 1:1 molar ratio in the presence of nickel-liposomes resulted in a 5.9-fold increase in Her2/neu kinase activity (Fig. 5B). In contrast, performing this same mixing experiment in the absence of liposomes demonstrated no change in Her2/neu kinase activity. Under these conditions, Her2/neu kinase domain was activated to essentially the same extent by Her3 and by Her4 D843N kinase domains (Fig. 5, A and B).
Heterodimerization of Her4 and Her3
The asymmetric dimer model was proposed in 2006 based on the crystal structure and biochemical experiments on EGFR kinase domain (18). This model is further supported by the finding that the Her4 kinase domain crystallized in a very similar, asymmetric dimer (16). Based on the primary sequence alignment between the four ErbB family members, it was proposed that all four ErbB kinase domains, including Her3, can act as donor monomers in the dimer pair, because the amino acid residues at their C-lobe contact surface are conserved (18). To test this prediction, we used the C-lobe mutant, Her4 V954R kinase domain protein, which is unable to dimerize and activate on its own on the surface of the nickel-liposome, but can form an activated kinase dimer when mixed with other Her4 protein constructs (Figs. 1C and 6). The consequences of this model are shown schematically in Fig. 6A. When mixed with either the N-lobe mutant, Her4 I712Q, or the kinase-dead Her4 mutant, Her4 D843N, along with nickel-liposomes, Her4 V954R showed a 15-fold increase in kinase activity (Fig. 6B).
FIGURE 6.
Heterodimerization of Her4 with Her3. A, predicted effects of the Her4 mutations and of Her3 on Her4 kinase activity. The mutations shown here are in the Her4 kinase domain. The prediction made by the asymmetric dimer model in the case of Her4 V954R mixed with Her3 is highlighted because the model prediction is not borne out by the observed experimental results (below). B, Her4 and Her3 kinase domain proteins were mixed at 1:1 molar ratio immediately prior to nickel-liposome addition, and the kinase assay was measured as in Fig. 1. Specific activity was corrected for the amount of active Her4 kinase present in each pairwise mixture. Fold change is calculated relative to kinase activity in the absence of liposomes. C, kinase-dead Her4 (D843N) or Her3 WT was mixed with WT Her4 kinase domain at the indicated molar ratio, nickel-liposomes were added, and kinase assays were performed as in Fig. 1. Data are representative of three (B) or two (C) experiments.
The asymmetric dimer model predicts that Her3 kinase domain and the kinase-dead Her4 D843N kinase domain should be comparable activators of other ErbB kinase domain monomers (18). We observed that Her3 kinase domain and kinase-dead Her4 D843N proteins are able to activate Her2 to the same extent (Fig. 5, A and B). By contrast, there is a striking difference in the ability of these two proteins to activate the C-lobe mutant, Her4 V954R. Mixing Her4 V954R and Her4 D843N kinase domains in the presence of nickel-liposomes resulted in a 15-fold increase in Her4 kinase activity, whereas mixing Her4 V954R with Her3 kinase domain resulted only in a 2.7-fold increase in Her4 kinase activity (Fig. 6B). This suggests that there is a functional difference between Her3 kinase domain and Her4 D843N. To confirm that this functional difference observed between Her3 and kinase-dead Her4 was not limited to this mixing experiment with Her4 V954R, we repeated this experiment using Her4 WT kinase domain (Fig. 6C). Mixing WT Her4 and kinase dead Her4 (D843N) at a 1:1 ratio had minimal effect on Her4 activation. In contrast, mixing Her3 with WT Her4 decreased Her4 activation by 50%. When we increased the ratio of inactive kinase to WT Her4 to 2:1, a similar difference between kinase-dead Her4 and Her3 is seen, though the overall activity measured in these conditions is less than the 1:1 mixing because of the higher amounts of inactive kinases present. Unlike WT Her4, Her4 V954R cannot activate itself (as shown in Figs. 6B and 1C), and therefore it is a useful mutation to use in heterodimerization experiments. As demonstrated here, the results obtained both with Her4 V954R and WT Her4 kinase domains support the same conclusion.
The asymmetric dimer model further predicts that the N-lobe mutant, Her4 I712Q cannot be activated by a kinase-dead ErbB monomer, because the Her4 I712Q mutation disrupts the surface where a donor monomer would bind to it (Fig. 6A). Consistent with this prediction, Her4 I712Q kinase domain shows no activation with either the Her3 or Her4 D843N kinase domains (Fig. 6B). The 1.2–1.3-fold increase in Her4 I712Q kinase activity seen here upon addition of nickel-liposomes is observed with Her4 I712Q kinase domain on its own (Fig. 1C).
Mutation of Predicted Her3 C-lobe Surface Residues
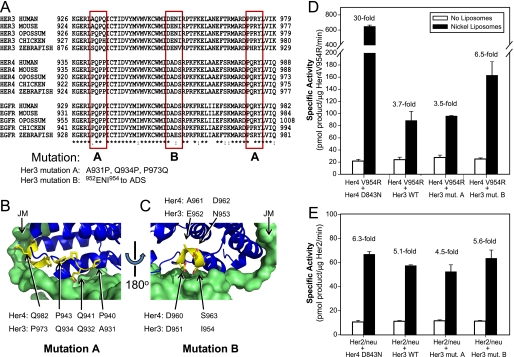
To address the mechanism of this difference between Her4 and Her3, we examined the primary amino acid sequence of the C-lobe surface of Her3 as compared with Her4 and EGFR kinase domains (Fig. 7A). We used the protein structure of Her4 kinase domain + juxtamembrane segment (PDB accession number 2R4B) to predict which residues in Her3 may be surface residues and involved in the heterodimerization interface (Fig. 7, B and C). Using this information, we hypothesized that Her3 residues 952ENI954 or residues Ala-931, Gln-934, Pro-973, which are largely conserved in Her3 from human to zebrafish, but are different in Her4 and EGFR, may be responsible for this functional difference between Her3 and Her4. We mutated these residues to directly test this prediction. For mutation A, we mutated Her3 at three residues: A931P, Q934P, and P973Q and for mutation B, we changed Her3 residues 952ENI954 to ADS. Both of these mutations are intended to make Her3 more “Her4-like”. We then tested the effects of these two mutations using kinase assays with nickel-liposomes (Fig. 7D). We observed that Her3 mutation B has an increased ability to activate Her4 as compared with WT Her3 kinase domain (6.5-fold versus 3.7-fold) whereas it causes no change in Her3 ability to activate Her2/neu (Fig. 7, D and E). Similar to wild-type Her3, Her3 mutation B lacks kinase activity (supplemental Table S1) and therefore, this increased ability to activate Her4 is mediated through heterodimerization. Unlike Her3 mutation B, Her3 mutation A did not increase Her3 ability to activate Her4 kinase domain in this assay. This assay uses the kinase domains only, and therefore, this assay cannot rule out that Her3 mutation A has some effect on the full-length protein. The ability of Her3 mutation B to increase activation of Her4 suggests that the amino acid sequence differences on the C-lobe surface of the donor monomer affects the degree of heterodimerization and activation between the kinase domains of the ErbB receptors.
FIGURE 7.
Mutation of Her3 to enhance its ability to activate Her4. A, multisequence alignment of the intracellular portions of Her3, Her4, and EGFR across five species, using ClustalW2 (47). Shown by red boxes are three regions on the C-lobe dimerization surface of the kinase domain where contact residues are divergent across this family. To make these contact sites in Her3 more “Her4-like,” the following mutations were created. Her3 mutation A: A931P/Q943P/P973Q. Her3 mutation B: 952ENI954 to ADS. B, contacts made by residues in mutation A. The Her4 donor monomer is shown in blue, and the PQPP and PQRY sequences in it are highlighted in yellow (structure shown is PDB ID: 2R4B) (17). The Her4 acceptor monomer is shown as a green surface. This crystal structure includes the juxtamembrane (JM) segment, which is seen on the left side of the image and contacts Her4 residue Gln-982 through Tyr-984 in the donor monomer. C, contacts made by residues in mutation B. The Her4 asymmetric dimer is rotated 180° relative to B, and the DADS sequence in the donor monomer is highlighted in yellow. Panels B and C are generated with PyMol version 1.1 (48). D, Her4 V954R was mixed with either kinase-dead Her4, WT Her3 or mutant Her3 at 1:1 molar ratio immediately prior to nickel-liposome addition and kinase assay was measured as in Fig. 1. Specific activity was corrected for the amount of active Her4 kinase present in each pairwise mixture. Fold change is calculated relative to kinase activity in the absence of liposomes. E, mixing of Her2/neu with the specified Her4 or Her3 kinase domain proteins was performed as in D. Data in D and E are representative of two independent experiments each.
DISCUSSION
Use of nickel-chelating lipids and liposomes is potentially applicable to the study of many membrane-associated proteins and protein domains. In this study, we demonstrate that binding of Her4 and Her2/neu to nickel-liposomes increases their kinase-specific activity 40-fold and 3.5-fold, respectively. Her2/neu can be further activated by heterodimerizing with either Her3, which is intrinsically kinase-dead, or with a kinase-dead mutant of Her4. This activation of the ErbB kinase domains by nickel-liposomes can be disrupted through competition of the nickel-polyhistidine interaction with imidazole. Further, while the binding of ErbB kinase domains to the nickel-liposomes follows a single-site binding model, kinase activation is found to be a cooperative process, with a Hill coefficient of close to 2, consistent with formation of a kinase dimer. Mutation of the asymmetric dimer interface, either Her4 mutation I712Q or V954R, eliminates Her4 kinase activation. Mixing of these two mutant kinases allows for reconstitution of a functional asymmetric dimer interface and restoration of activation, providing additional evidence for the formation of an activated kinase domain dimer. We observe that this activation on the surface of the nickel-liposomes is greatest in the presence of Mg2+, rather than with Mn2+, and this difference is likely due to direct effects of Mn2+ in the kinase catalytic site rather than an effect on kinase dimerization.
We used this nickel-liposome-based system to probe predictions made by the asymmetric dimer model. Based on the crystal structure of the EGFR kinase domain asymmetric dimer and multisequence alignment of the four ErbB kinase domains, it was proposed that all ErbB kinase domains should be effective donor monomers (18). Her3 is intrinsically kinase dead, and one of reasons for this effect is the catalytic base Asp residue, which is conserved among kinases, is an Asn residue in Her3. By making the same amino acid change in Her4 (Her4 D843N), we render Her4 kinase dead and generate a protein that we can directly compare with Her3. We observe that both Her3 and kinase dead Her4 are equally effective in their ability to activate Her2/neu. In contrast, there is a striking difference in the ability of Her3 and kinase-dead Her4 to activate Her4. Mutation of Her3 residues 952–954 to the corresponding sequence in Her4 (mutation B) increased the ability of Her3 to activate Her4. This demonstrates that differences in the predicted C-lobe surface amino acids influence the degree of activation between different ErbB kinase heterodimers. We note that Her3 mutation B does not reproduce the full activation seen with kinase-dead Her4, and it is likely that additional amino acid residues distant from the C-lobe contact surface also influence how the Her3 kinase domain heterodimerizes.
We observe that the Her2/neu kinase domain showed a smaller degree of activation upon binding to nickel-liposomes than the Her4 kinase domain. While Her2/neu kinase specific activity is further increased upon heterodimerization with Her3 or Her4 (up to 5.9-fold), it still did not achieve the 40-fold activation observed with Her4 bound to nickel-liposomes. Prior reports have suggested that the recombinant Her2/neu kinase domain is an intrinsically less active kinase than EGFR or Her4 kinase domains (28, 34). Her2/neu residues in the loop between the αC helix and the β4 sheet were found to play a major role in controlling the kinase activity of Her2/neu (34). Mutating residues in this loop to match EGFR (Her2/neu G776S, G778D) or a cancer-associated insertion in this loop (Her2/neu G776YVMA) significantly increased Her2/neu kinase activity (34–36). We speculate that this may be an evolutionary adaptation as Her2/neu is the only member of the ErbB family that does not bind a ligand. The crystal structure of the Her2/neu extracellular domain has shown that it adopts an extended conformation with its dimerization surfaces exposed (37, 38). Given that the Her2/neu extracellular domain appears to be “poised” to interact with other EGFR family members, limiting the ability of the Her2/neu kinase domain to be activated by homo- or heterodimerization could represent an important physiologic regulatory mechanism.
An important consideration in the interpretation of these experiments is that they measure the dimerization of the kinase domains only. The extracellular domain, transmembrane domain, and the intracellular juxtamembrane region also contribute significantly to homo- and heterodimerization of the ErbB receptor-tyrosine kinases (13, 14, 39, 40), and they are not present in this reconstituted in vitro system. After the initial submission of this report, further biochemical and structural studies on the EGFR intracellular juxtamembrane region were published (41, 42). These studies clearly demonstrate that the juxtamembrane region contains an activation domain, also called the juxtamembrane latch. The crystal structure of this region demonstrates that the juxtamembrane activation domain of the acceptor monomer cradles the donor monomer (41). Kinase assays were performed comparing the activity of EGFR constructs containing the juxtamembrane region and the kinase domain in solution to kinase domain only constructs attached to nickel-liposomes (42). We anticipate that these juxtamembrane containing protein constructs can also be tested with the nickel-liposome system. In vivo, the transmembrane domain and the lipids on the inner leaflet of the plasma membrane will affect how the juxtamembrane region interacts with the kinase domain, and this can be studied with this in vitro system.
Assaying membrane-associated proteins, such as the kinase domains of receptor-tyrosine kinases, in the presence of nickel-liposome has multiple advantages. First, this assay method can be broadly applied to multiple receptor-tyrosine kinases. Zhang et al. (18) demonstrated its utility for studies on EGFR. We demonstrate its general applicability to the remainder of the ErbB family. Weis and co-workers (43) recently reported activation of insulin receptor, Tie2, and the Ephrin receptor, EphB2, kinase activity by a nickel-chelating lipid containing nanoparticle. Additionally, this method permits the generation of membrane associated protein complexes in vitro and opens up important research approaches and experimental designs that are not possible in intact cells. The ability to generate a four protein prokaryotic signaling complex on the nickel-liposomes has been demonstrated (19). Given that this system permits rapid autophosphorylation of the Her4 C-terminal tail tyrosine sites, testing SH2-containing proteins for their ability to complex in vitro with Her4 and mediate subsequent signaling steps should be feasible.
Nickel-liposomes can also be used to study disease associated mutations in vitro. An activating mutation in EGFR, L858R, has been found in lung cancer patients who are never-smokers, and this mutation is associated with a markedly enhanced response to EGFR-targeted tyrosine kinase inhibitor drugs (44–46). Assaying this mutation using nickel-liposomes demonstrated that EGFR L858R kinase domain has an ∼20-fold increase in catalytic efficiency (kcat/Km) as compared with WT EGFR kinase (18). In addition, using the dimeric and activated form of tyrosine kinases in small molecule inhibitor screens opens up many important pharmacological and pharmaceutical possibilities. The affinity and detailed structure-activity relationship of inhibitors for the dimeric state of the kinases could be directly probed. Screens for inhibitors of kinase dimerization can also be constructed. Of course, adequate controls to evaluate for effects of lipophilic, small molecules partitioning into liposomes would need to be included.
In conclusion, this nickel-chelating lipid-liposome system is potentially applicable to the study of many membrane-associated proteins and protein domains. We demonstrate that Her4 and Her2/neu kinase domains dimerize and become activated upon binding to the nickel-liposomes. This system is used to induce the formation of ErbB kinase domain heterodimers in vitro. The Her3 kinase domain is found to readily activate Her2/neu but to be a poor activator of Her4, which differs from the prediction made by the asymmetric dimer model, and we observed that mutating Her3 residues 952–954 to the corresponding sequence in Her4 increased the ability of Her3 to activate Her4. This ability to study ErbB heterodimerization in vitro is a unique feature of the nickel-liposome system and offers significant future potential for analyzing the function of the entire intracellular half of the EGFR-ErbB family of receptor-tyrosine kinases.
Supplementary Material
Acknowledgments
We thank Linda Pike, Daniel J. Leahy, Philip A. Cole, and Aruna Sathyamurthy for critical reading of the manuscript.
Note Added in Proof
A crystal structure of the Her3 kinase domain has been recently published (Jura, N., Shan, Y., Cao, X., Shaw, D. E., and Kuriyan, J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21608–21613). Review of this crystal structure (Protein Data Bank code 3KEX) shows that Her3 residues Asp-951, Asn-953, and Ile-954 are surface-exposed.
This work was supported, in whole or in part, by National Institutes of Health Grant K22CA128951 (NCI Transition Career Development Award, K22) (to R. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
- EGFR
- epidermal growth factor receptor
- DOPC
- dioleoylphosphatidylcholine
- Ni-NTA-DOGS
- nickel-1,2-dioleoyl-sn-glycero-3-([N-(5-amino-1-carboxypentyl)iminodiacetic acid]succinyl)-nickel salt
- SUV
- small unilamellar vesicles
- DTT
- dithiothreitol
- WT
- wild type.
REFERENCES
- 1.Carpenter G. (2003) Exp. Cell Res. 284, 66–77 [DOI] [PubMed] [Google Scholar]
- 2.Citri A., Skaria K. B., Yarden Y. (2003) Exp. Cell Res. 284, 54–65 [DOI] [PubMed] [Google Scholar]
- 3.Tzahar E., Waterman H., Chen X., Levkowitz G., Karunagaran D., Lavi S., Ratzkin B. J., Yarden Y. (1996) Mol. Cell Biol. 16, 5276–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graus-Porta D., Beerli R. R., Daly J. M., Hynes N. E. (1997) EMBO J. 16, 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karunagaran D., Tzahar E., Beerli R. R., Chen X., Graus-Porta D., Ratzkin B. J., Seger R., Hynes N. E., Yarden Y. (1996) EMBO J. 15, 254–264 [PMC free article] [PubMed] [Google Scholar]
- 6.Olayioye M. A., Graus-Porta D., Beerli R. R., Rohrer J., Gay B., Hynes N. E. (1998) Mol. Cell Biol. 18, 5042–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birchmeier C. (2009) Exp. Cell Res. 315, 611–618 [DOI] [PubMed] [Google Scholar]
- 8.Pentassuglia L., Sawyer D. B. (2009) Exp. Cell Res. 315, 627–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gassmann M., Casagranda F., Orioli D., Simon H., Lai C., Klein R., Lemke G. (1995) Nature 378, 390–394 [DOI] [PubMed] [Google Scholar]
- 10.Lee K. F., Simon H., Chen H., Bates B., Hung M. C., Hauser C. (1995) Nature 378, 394–398 [DOI] [PubMed] [Google Scholar]
- 11.Hynes N. E., MacDonald G. (2009) Curr. Opin. Cell Biol. 21, 177–184 [DOI] [PubMed] [Google Scholar]
- 12.Romond E. H., Perez E. A., Bryant J., Suman V. J., Geyer C. E., Jr., Davidson N. E., Tan-Chiu E., Martino S., Paik S., Kaufman P. A., Swain S. M., Pisansky T. M., Fehrenbacher L., Kutteh L. A., Vogel V. G., Visscher D. W., Yothers G., Jenkins R. B., Brown A. M., Dakhil S. R., Mamounas E. P., Lingle W. L., Klein P. M., Ingle J. N., Wolmark N. (2005) N. Engl. J. Med. 353, 1673–1684 [DOI] [PubMed] [Google Scholar]
- 13.Burgess A. W., Cho H. S., Eigenbrot C., Ferguson K. M., Garrett T. P., Leahy D. J., Lemmon M. A., Sliwkowski M. X., Ward C. W., Yokoyama S. (2003) Mol. Cell 12, 541–552 [DOI] [PubMed] [Google Scholar]
- 14.Lemmon M. A. (2009) Exp. Cell Res. 315, 638–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bose R., Zhang X. (2009) Exp. Cell Res. 315, 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu C., Tarrant M. K., Choi S. H., Sathyamurthy A., Bose R., Banjade S., Pal A., Bornmann W. G., Lemmon M. A., Cole P. A., Leahy D. J. (2008) Structure 16, 460–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood E. R., Shewchuk L. M., Ellis B., Brignola P., Brashear R. L., Caferro T. R., Dickerson S. H., Dickson H. D., Donaldson K. H., Gaul M., Griffin R. J., Hassell A. M., Keith B., Mullin R., Petrov K. G., Reno M. J., Rusnak D. W., Tadepalli S. M., Ulrich J. C., Wagner C. D., Vanderwall D. E., Waterson A. G., Williams J. D., White W. L., Uehling D. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2773–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X., Gureasko J., Shen K., Cole P. A., Kuriyan J. (2006) Cell 125, 1137–1149 [DOI] [PubMed] [Google Scholar]
- 19.Shrout A. L., Montefusco D. J., Weis R. M. (2003) Biochemistry 42, 13379–13385 [DOI] [PubMed] [Google Scholar]
- 20.Montefusco D. J., Asinas A. E., Weis R. M. (2007) Methods Enzymol. 423, 267–298 [DOI] [PubMed] [Google Scholar]
- 21.Zhang X., Pickin K. A., Bose R., Jura N., Cole P. A., Kuriyan J. (2007) Nature 450, 741–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szoka F., Olson F., Heath T., Vail W., Mayhew E., Papahadjopoulos D. (1980) Biochim. Biophys. Acta 601, 559–571 [DOI] [PubMed] [Google Scholar]
- 23.Stewart J. C. (1980) Anal. Biochem. 104, 10–14 [DOI] [PubMed] [Google Scholar]
- 24.Jan A. Y., Johnson E. F., Diamonti A. J., Carraway K. L., III, Anderson K. S. (2000) Biochemistry 39, 9786–9803 [DOI] [PubMed] [Google Scholar]
- 25.Armstrong R. N., Kondo H., Granot J., Kaiser E. T., Mildvan A. S. (1979) Biochemistry 18, 1230–1238 [DOI] [PubMed] [Google Scholar]
- 26.Zheng J., Trafny E. A., Knighton D. R., Xuong N. H., Taylor S. S., Ten Eyck L. F., Sowadski J. M. (1993) Acta Crystallogr. D Biol. Crystallogr. 49, 362–365 [DOI] [PubMed] [Google Scholar]
- 27.Sun G., Budde R. J. (1999) Biochemistry 38, 5659–5665 [DOI] [PubMed] [Google Scholar]
- 28.Brignola P. S., Lackey K., Kadwell S. H., Hoffman C., Horne E., Carter H. L., Stuart J. D., Blackburn K., Moyer M. B., Alligood K. J., Knight W. B., Wood E. R. (2002) J. Biol. Chem. 277, 1576–1585 [DOI] [PubMed] [Google Scholar]
- 29.Kaushansky A., Gordus A., Budnik B. A., Lane W. S., Rush J., MacBeath G. (2008) Chem. Biol. 15, 808–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plowman G. D., Green J. M., Culouscou J. M., Carlton G. W., Rothwell V. M., Buckley S. (1993) Nature 366, 473–475 [DOI] [PubMed] [Google Scholar]
- 31.Bose R., Molina H., Patterson A. S., Bitok J. K., Periaswamy B., Bader J. S., Pandey A., Cole P. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9773–9778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holbro T., Beerli R. R., Maurer F., Koziczak M., Barbas C. F., 3rd, Hynes N. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8933–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guy P. M., Platko J. V., Cantley L. C., Cerione R. A., Carraway K. L., 3rd (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8132–8136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan Y. X., Wong L., Ding J., Spiridonov N. A., Johnson R. C., Johnson G. R. (2008) J. Biol. Chem. 283, 1588–1596 [DOI] [PubMed] [Google Scholar]
- 35.Shigematsu H., Takahashi T., Nomura M., Majmudar K., Suzuki M., Lee H., Wistuba, Fong K. M., Toyooka S., Shimizu N., Fujisawa T., Minna J. D., Gazdar A. F. (2005) Cancer Res. 65, 1642–1646 [DOI] [PubMed] [Google Scholar]
- 36.Wang S. E., Narasanna A., Perez-Torres M., Xiang B., Wu F. Y., Yang S., Carpenter G., Gazdar A. F., Muthuswamy S. K., Arteaga C. L. (2006) Cancer Cell 10, 25–38 [DOI] [PubMed] [Google Scholar]
- 37.Cho H. S., Mason K., Ramyar K. X., Stanley A. M., Gabelli S. B., Denney D. W., Jr., Leahy D. J. (2003) Nature 421, 756–760 [DOI] [PubMed] [Google Scholar]
- 38.Garrett T. P., McKern N. M., Lou M., Elleman T. C., Adams T. E., Lovrecz G. O., Kofler M., Jorissen R. N., Nice E. C., Burgess A. W., Ward C. W. (2003) Mol. Cell 11, 495–505 [DOI] [PubMed] [Google Scholar]
- 39.Thiel K. W., Carpenter G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19238–19243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macdonald-Obermann J. L., Pike L. J. (2009) J. Biol. Chem. 284, 13570–13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Red Brewer M., Choi S. H., Alvarado D., Moravcevic K., Pozzi A., Lemmon M. A., Carpenter G. (2009) Mol. Cell 34, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jura N., Endres N. F., Engel K., Deindl S., Das R., Lamers M. H., Wemmer D. E., Zhang X., Kuriyan J. (2009) Cell 137, 1293–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esposito E. A., Shrout A. L., Weis R. M. (2008) J. Biomol. Screen 13, 810–816 [DOI] [PubMed] [Google Scholar]
- 44.Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Fulton L., Mardis E., Kupfer D., Wilson R., Kris M., Varmus H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13306–13311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paez J. G., Jänne P. A., Lee J. C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F. J., Lindeman N., Boggon T. J., Naoki K., Sasaki H., Fujii Y., Eck M. J., Sellers W. R., Johnson B. E., Meyerson M. (2004) Science 304, 1497–1500 [DOI] [PubMed] [Google Scholar]
- 46.Lynch T. J., Bell D. W., Sordella R., Gurubhagavatula S., Okimoto R. A., Brannigan B. W., Harris P. L., Haserlat S. M., Supko J. G., Haluska F. G., Louis D. N., Christiani D. C., Settleman J., Haber D. A. (2004) N. Engl. J. Med. 350, 2129–2139 [DOI] [PubMed] [Google Scholar]
- 47.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 48.DeLano W. L. (2002) The PyMol Molecular Graphics System, DeLano Scientific [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.