Abstract
SIRT1 (Sirtuin type 1), a mammalian orthologue of yeast SIR2 (silent information regulator 2), has been shown to mediate a variety of calorie restriction (CR)-induced physiological events, such as cell fate regulation via deacetylation of the substrate proteins. However, whether SIRT1 deacetylates activator protein-1 (AP-1) to influence its transcriptional activity and target gene expression is still unknown. Here we demonstrate that SIRT1 directly interacts with the basic leucine zipper domains of c-Fos and c-Jun, the major components of AP-1, by which SIRT1 suppressed the transcriptional activity of AP-1. This process requires the deacetylase activity of SIRT1. Notably, SIRT1 reduced the expression of COX-2, a typical AP-1 target gene, and decreased prostaglandin E2 (PGE2) production of peritoneal macrophages (pMΦs). pMΦs with SIRT1 overexpression displayed improved phagocytosis and tumoricidal functions, which are associated with depressed PGE2. Furthermore, SIRT1 protein level was up-regulated in CR mouse pMΦs, whereas elevated SIRT1 decreased COX-2 expression and improved PGE2-related macrophage functions that were reversed following inhibition of SIRT1 deacetylase activity. Thus, our results indicate that SIRT1 may be a mediator of CR-induced macrophage regulation, and its deacetylase activity contributes to the inhibition of AP-1 transcriptional activity and COX-2 expression leading to amelioration of macrophage function.
Keywords: Immunology/Cellular Response, Protein/Post-translational Modification, Protein/Protein-Protein Interactions, Transcription/AP-1, Calorie Restriction, Cyclooxygenase-2 (COX-2), Macrophage, Sirtuin Type 1 (SIRT1)
Introduction
Transcription factor activator protein-1 (AP-1)3 represents homodimers or heterodimers of the Jun and Fos families. c-Fos and c-Jun are the subunits predominantly expressed in mammalian cells. In response to growth factors, cytokines, oxidative stress, or pharmacological stimuli, such as phorbol 12-myrisatate 13-acetate (PMA), AP-1 binds to the promoters of target genes to modulate their expression, which is in turn involved in cell proliferation, differentiation, and inflammation (1). Fos and Jun belong to the basic leucine zipper (bZIP) family, which preferentially binds to 12-O-tetradecanoylphorbol-13-acetate response element sites and to the cAMP response element with slightly lower affinity (2). The basic fragment of bZIP is responsible for sequence-specific DNA binding, and the leucine zipper is critical for the dimerization of ZIP proteins. In addition to direct binding to cis-acting elements, AP-1 could also interact with other transcriptional regulators, participating in transcriptional regulation, and the bZIP domain contributes to the association with co-activator (3) or co-repressor (4). The transcriptional activity of AP-1 is regulated by the post-translational modification, including acetylation (5). The acetylation modification of transcriptional factors changes the protein-protein or protein-DNA interaction as an important regulatory mechanism of transcription (6).
Cyclooxygenase-2 (COX-2), one of the classic AP-1 targets, is the rate-limiting enzyme for prostaglandin (PG) production. Macrophage-derived PGs regulate functions of both macrophage itself and the neighboring cells as well. Excessive PGE2 inhibits the phagocytosis activity and the bacterial killing function of macrophages (7, 8). Increased PGE2 compromises the tumoricidal activity of macrophages and natural killer cells (9–11). The expression and activity of COX-2 are induced by various stimuli such as PMA, inflammatory factors (lipopolysaccharide), growth factors, and cytokines. COX-2 expression is increased in cells from aged animals, whereas calorie restriction (CR), a regimen extending the life span of organisms and maintaining many physiological processes in a youthful state, suppresses the age-related COX-2 increase (12).
The Sir2 gene family regulates transcriptional silencing and extends the lifespan of Saccharomyces cerevisiae and Caenorhabditis elegans. SIRT1, the mammalian orthologue of Sir2, is a NAD+-dependent deacetylase of numerous substrates. SIRT1 has been shown to mediate a variety of physiological events such as cell fate regulation, increased metabolic rate, and higher oxygen consumption by deacetylation of the substrate targets. For example, SIRT1 deacetylates p53 protein at lysine 382 and represses its transcriptional function to protect cells from apoptosis (13). Deacetylation of FOXO3 and FOXO4 by SIRT1 switches cells from apoptosis to cell cycle arrest under stress (14, 15). SIRT1 deacetylates liver X-activated receptors and positively regulates its activity, promoting cholesterol efflux from cells (16). In macrophage cell line MonoMac6, SIRT1 has been reported to inhibit proinflammatory mediator release including interleukin-8 and tumor necrosis factor-α through deacetylating RelA/p65 subunit and inhibiting NF-κB (17).
CR extends the life span of organisms such as yeast, nematodes, fruit flies, and mice (13). In mammals, CR up-regulates expression of SIRT1 in a variety of tissues including kidney, liver, and fat, and SIRT1 transgenic mice display some phenotypes similar to mice on a calorie-restricted diet (18, 19). SIRT1 is implicated in CR-induced physiological events such as cell survival and senescence effects by regulating its substrates of SIRT1, such as p53 and FOXOs (20). CR has also been reported to influence macrophage functions (21, 22), delay the age-related chronic inflammatory diseases, and decrease AP-1 activity (23) and COX-2 expression (12). Based on these observations, we hypothesized that SIRT1 may decrease AP-1 activity and COX-2 expression to modulate macrophage function and that up-regulation of SIRT1 in macrophages may mimic the beneficial effect of CR on macrophages. In this report, we identified the direct interaction between SIRT1 and the bZIP domain of c-Fos/c-Jun and found that the deacetylase activity of SIRT1 is required for repression of AP-1 transcriptional activity. Furthermore, SIRT1 inhibited COX-2 expression and PGE2 production in macrophages, resulting in improved macrophage phagocytosis and tumoricidal functions. Finally, our results demonstrated that SIRT1 expression is increased in macrophages from CR mice, leading to decreased COX-2 expression and PGE2 production, and improves PGE2-related macrophage functions.
EXPERIMENTAL PROCEDURES
Mice and pMΦ Preparations
Specific pathogen-free male C57BL/6 mice were obtained from the Laboratory Animal Center of the Chinese Academy of Military Medical Sciences (Beijing, China). Mice aged 2–3 months were housed singly in cages at a constant temperature (22–25 °C) with a 12:12-h light-dark cycle. The mice were given ad libitum access to water and feed for a 1-week acclimation period and randomly assigned to two groups. A group of mice was given ad libitum (AL) access to water and feed as control group, and another group of mice was given 60% of daily calorie intake of control mice as CR group. Composition of the diet and the feeding protocol was described elsewhere in detail (24). All of the animal experiments were performed in accordance with institutional guidelines. Thioglycollate-elicited pMΦs were obtained from mice that had been injected with 2 ml of 3% (w/v) thioglycollate 5 days before sacrifice. The isolated cells were incubated in RPMI 1640 medium containing 10% fetal bovine serum for 3 h followed by washing three times with phosphate-buffered saline to remove nonadherent cells before treatment.
Reagents and Antibodies
The reagents, including PMA, thioglycollate, resveratrol (RSV), and Sirtinol were purchased from Sigma-Aldrich. Anti-HA and anti-β-actin monoclonal antibodies were from Sigma-Aldrich, and anti-Fos, -Jun, and -hSIRT1 (human SIRT1) rabbit polyclonal antibodies were from Santa Cruz Biotechnology. Anti-mSIRT1 (mouse SIRT1) and anti-acetyl-lysine antibody were from Upstate, and anti-COX-2 antibody was from Cayman Chemicals.
Plasmid and Virus Production
The AP-1-luc plasmid was obtained by the insertion of a double-stranded oligonucleotide representing seven AP-1 binding sites into the pTK-luc plasmid. The full-length and deletion mutant c-Fos and c-Jun expression vectors were constructed by inserting the human c-Fos or c-Jun cDNA in the frame of pcDNA3.1 vector. SIRT1 expression vectors were gifts from Dr. Ishikawa (25). The mouse COX-2 promoter plasmid containing a 1068-bp fragment, −1003 to +65 relative to the transcription start site, was subcloned into pGL3 vector (COX-2-luc). The wild type COX-2 promoter was modified using a site-directed mutagenesis kit (Promega). The AP-1 binding element (5′-ACA GAG TCA C-3′ from −95 to −86) was modified to 5′-ACA TTA TAA C-3′. The underlined sequences denote the mutated site, and it was confirmed by DNA sequencing. GST fusion proteins were expressed in Escherichia coli strain BL-21 by using the pET42a vector system. Replication-defective adenoviral vectors expressing SIRT1 (Ad-SIRT1) or control green fluorescent protein (Ad-GFP) were prepared using the AdEasy vector kit (Quantum Biotechnologies) as described in the supplier's protocol. The adenovirus-mediated knockdown of SIRT1 (Ad-SIRT1 RNAi) and control vector (Ad-U6) were generated using the same system. The RNAi sequence was reported previously (26). pMΦs were infected with the above adenovirus for 36 h and were cultured in fresh RPMI 1640 for further treatment.
Transfections and Luciferase Assays
HEK293 cells and RAW264.7 were grown as recommended by ATCC and were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Luciferase assays were performed using a dual luciferase reporter assay system (Promega). Luciferase activity was normalized by transfection efficiency using pRL-TK reporter (Promega) as an internal control (30 ng). The results are expressed as percentages of relative luciferase activity of the control group, which was set as 100%.
Co-immunoprecipitation, AP-1 Acetylation Assays, GST Pulldown Assays, and Western Blotting
The cells were collected, and the proteins were solubilized in IP buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 1% protease inhibitor mixture) at 4 °C. One milligram of lysated protein was incubated with specific antibodies and precipitated with protein A- or G-agarose (Upstate Biotechnology). Purification of GST fusions (GST-Fos-HA, GST-Jun-HA, and control GST-HA) and maltose-binding protein-tagged SIRT1 from bacterial lysates was performed, and the binding assays in vitro were carried out using Glutathione-Sepharose (GE Healthcare) according to the manufacture's instructions. Precipitated proteins or total lysates were separated in 12% PAGE followed by immunoblotting with specific antibodies. The blots were then incubated with horseradish peroxidase-conjugated secondary antibody and exposed to ECL reagent for detection of protein expression.
DNA-Protein Binding Assays
The probe is a biotinylated, double-stranded oligonucleotide corresponding to human cox-2 promoter sequence −72/−18, which contains the binding site of AP-1 (5′-ACA GAG TCA C-3′). Four micrograms of probe were incubated with 400 μg of nuclear protein extract and 30 μl of streptavidin-agarose bead slurry at room temperature for 2 h with shaking. The mixture were pelleted and washed with phosphate-buffered saline for three times. The bound proteins were analyzed by consequent Western blotting.
Chromatin IP
pMΦs stimulated with PMA (50 ng/ml) or DMSO for 2 h were treated with 1% formaldehyde for 15 min. The cross-linked chromatin was sheared by sonication, and the sonicated complex was used for IP with specific antibodies or nonimmune rabbit IgG as control. Immunoprecipitated complexes were collected by using protein A-Sepharose beads (Upstate Biotechnology). The immunoprecipitated chromatin was amplified by PCR using primers for COX-2 promoter. The primers were as follows: COX-2/F-302, 5′-CAG ACT CAG CGA ACC ACA GGG-3′; and COX-2/R-17, 5′-TGA CAA CTG GCT GCT AAT GGG G-3′. The resulting PCR products (286 bp) were separated by 2% agarose gel electrophoresis.
PGE2 Assays
The PGE2 levels were measured with a commercially available enzyme-linked immunosorbent assay kit (Cayman Chemicals) according to the manufacturer's specifications.
Phagocytosis Assays and Tumoricidal Function Assays
Yeasts were deactivated and labeled with FITC in Na2HPO4 (50 mm, pH 9.2) and were incubated with macrophages at effector to target ratio of 3:1. After incubating for 45 min, the unattached extracellular yeast cells were removed, and trypan blue was used to quench the FITC fluorescence of outside yeasts. The cells and fluorescent yeasts internalized by cells were counted under fluorescent microscope. For all of the experiments, 100 cells/high power field were counted, and three high power fields/section were analyzed. The phagocytic percentage was calculated as the count of cells engulfed yeasts/the total cell number of 100, and the phagocytic index was calculated as the engulfed yeasts/the total 100 cells. Four times 105 macrophages in a well on 24-well plate were overlaid with 4 × 104 suspended target cells, K562 cells, or Jurkat cells. After co-incubation at 37 °C for 24 h, the remaining suspended cells were measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide. The absorbance was read at 540 nm on a Bio-Rad microplate reader. The percentage of growth inhibition of leukemia cells was calculated by (1 − Atarget remained/Atarget standard × 100%).
Statistical Analysis
All of the numerical results are expressed as the means ± S.D. derived from three independent experiments unless otherwise stated. The statistical analyses were performed using one-factor analysis of variance, and statistically significant differences were established as p < 0.05.
RESULTS
SIRT1 Interacts with bZIP Domain of c-Fos and c-Jun
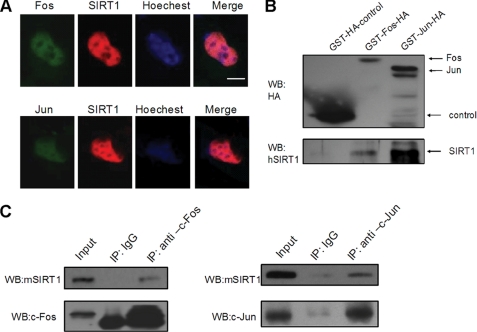
To investigate the potential relationships between SIRT1 and AP-1, we first investigated whether SIRT1 co-localizes with AP-1 in vivo. GFP-fused human c-Fos or c-Jun and red fluorescent protein (Cherry)-fused human SIRT1 were co-transfected into HEK293 cells. Subcellular localization of c-Fos/c-Jun and SIRT1 was visualized by monitoring green or red fluorescence. The GFP-Fos, GFP-Jun, and Cherry-SIRT1 were found primarily co-localized in the nuclear (Fig. 1A).
FIGURE 1.
SIRT1 interacts with c-Fos and c-Jun. A, HEK293 cells were transfected with plasmids coding for SIRT1 fused to Cherry and c-Fos or c-Jun fused to GFP. The nuclei were stained with Hoechst 33342 (blue). The cells were analyzed by fluorescent microscopy. The scale bar represents 10 μm. B, GST pulldown assay examines the direct interaction between SIRT1 and AP-1 in vitro. C, pMΦs were treated with PMA (50 ng/ml) for 3 h and were lysed for IP and Western blotting (WB) as indicated. The images are representative of three independent experiments.
Then we performed glutathione S-transferase pulldown assays with GST-tagged Fos-HA or Jun-HA and maltose-binding protein-tagged SIRT1 to address whether SIRT1 directly interacts with c-Fos/c-Jun. We found obvious binding of SIRT1 to c-Fos or c-Jun in this in vitro system (Fig. 1B), which demonstrated that the SIRT1-AP-1 interaction is physically direct.
We next examined whether SIRT1 interacts with c-Fos/c-Jun in macrophages. Mouse primary pMΦs were treated with PMA and were lysed for IP with c-Fos/c-Jun antibody and blotting with anti-SIRT1 antibody. We found obvious binding of SIRT1 to c-Fos or c-Jun in microphages (Fig. 1C), which indicates that the SIRT1-AP-1 interaction may have biological significance.
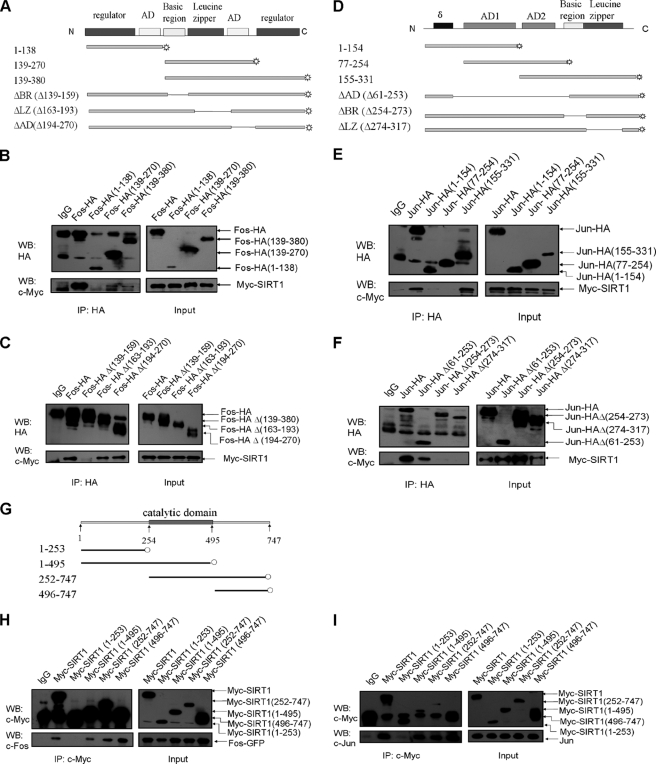
To map the domain of c-Fos that is required for the interaction with SIRT1, we generated expression vectors containing HA-tagged full-length or truncated c-Fos (Fig. 2A). The full-length or truncated c-Fos expression vectors were co-transfected to HEK293 cells with Myc-tagged SIRT1. The cell lysates were immunoprecipitated with anti-HA antibody and then analyzed by Western blotting using the anti-Myc antibody. The results showed that c-Fos interacted with SIRT1, and the C terminus of c-Fos was responsible for the interaction (Fig. 2B). Additional experiments showed that deletion of c-Fos residues 139–159 (basic region) completely abolished the interaction between SIRT1 and c-Fos (Fig. 2C). A reciprocal IP assay with anti-Myc antibody and blotting with anti-HA antibody revealed the similar interaction (supplemental Fig. S1, A and B). These data strongly suggest that the basic region of c-Fos is necessary for its binding to SIRT1.
FIGURE 2.
Identification of the binding domain on AP-1 and SIRT1. A, D, and G, schematic diagram of human c-Fos (A), c-Jun (D), and SIRT1 (G) is shown. AD, activation domain; BR, basic region; LZ, leucine zipper. B, C, E, F, H, and I, HEK293 cells were transfected with the indicated expression vectors for 36 h and were lysed for IP and Western blotting (WB). Plasmids encoding HA-tagged full-length or deletion mutants of c-Fos were co-transfected with plasmids encoding Myc-tagged SIRT1 into cells (B and C). Similar studies were performed in cells transfected with plasmids encoding HA-tagged c-Jun and Myc-tagged SIRT1 (E and F). The cells expressing Myc-tagged deletion mutants of SIRT1 and GFP-fused c-Fos or HA-tagged c-Jun were analyzed using IP and Western blotting (H and I). The images are representative of two independent experiments.
Similarly, we generated expression vectors containing HA-tagged full-length or truncated c-Jun (Fig. 2D). To determine which region of c-Jun is important for the interaction, we co-transfected cells with HA-tagged c-Jun and Myc-tagged SIRT1 and performed IP with anti-HA antibody followed by blotting with anti-Myc antibody. As is obvious in Fig. 2E, the C terminus of c-Jun was found to be responsible for its interaction with SIRT1 (Fig. 2E). Deletion of c-Jun residues 254–273 (basic region) and 274–317 (leucine zipper) completely abrogated the SIRT1-c-Jun interaction (Fig. 2F). IP with anti-Myc antibody and blotting with anti-HA antibody confirmed the above results (supplemental Fig. S1, C and D). These results suggested that the bZIP domain of c-Jun is required for the steady association between c-Jun and SIRT1.
Finally, to examine which regions of SIRT1 are responsible for the SIRT1-AP-1 interaction, we constructed Myc-tagged deletion mutants of SIRT1 (Fig. 2G). HEK293 cells were co-transfected with the full-length SIRT1 vector or each of the truncated SIRT1 vectors and c-Fos or c-Jun expression vectors. IP with anti-Myc antibody and blotting with anti-c-Fos or anti-c-Jun antibody revealed that deletion residues 254–747 completely abolished the SIRT1-AP-1 interaction (Fig. 2, H and I). We found that either the catalytic domain (residues 254–495) or the C terminus (residues 496–747) of SIRT1 is independently sufficient for the binding to AP-1 (c-Fos or c-Jun).
SIRT1 Suppresses AP-1 Transcriptional Activity
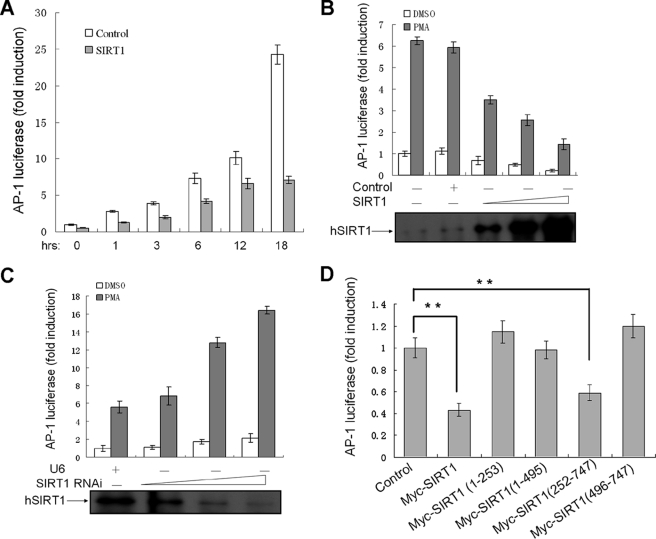
To investigate whether SIRT1 regulates the transcriptional activity of AP-1, we performed luciferase assays using AP-1-luc reporter in HEK293 cells. The cells were also transfected with SIRT1 expression vector or control vector pcDNA3.1 and then treated with PMA. PMA stimulated AP-1 reporter activity in a time-dependent manner, which was inhibited by SIRT1 for all time points (Fig. 3A).
FIGURE 3.
SIRT1 suppresses AP-1 transcriptional activity. A, HEK293 cells were transiently transfected with 0.1 μg of AP-1 luciferase reporter (AP-1-luc), 30 ng of pRL-TK, and SIRT1 expression vectors or control (pcDNA3.1) for 24 h and were co-cultured with PMA (50 ng/ml) or vehicle DMSO. The relative luciferase activities were measured at the different time points. The relative luciferase activities are presented as the means ± standard deviation (S.D.) of triplicate samples and are representative of three independent experiments. B and C, AP-1-luc and pRL-TK were co-transfected into HEK293 cells with increasing amounts of the SIRT1 expression (0.1, 0.3, and 0.9 μg) or SIRT1 RNAi plasmid (0.3, 1.5, and 3 μg) for 24 h. The cells were stimulated with PMA (50 ng/ml) for 3 h, and the relative luciferase activities were measured. Proteins from the DMSO-treated samples were separated by SDS-PAGE and probed to detect hSIRT1. D, HEK293 cells transfected with 0.1 μg of AP-1-luc, 30 ng of pRL-TK, and 1 μg of SIRT1 expression vectors or control (pcDNA4-Myc) were stimulated with PMA (50 ng/ml) for 3 h, and the luciferase activities were examined. **, p < 0.01 versus control.
We further transfected HEK293 cells with increasing amounts of SIRT1 expression construct or SIRT1 RNAi vector. The PMA-induced AP-1 luciferase activities declined with increasing levels of SIRT1 in a dose-dependent manner (Fig. 3B). Conversely, the AP-1 activity was elevated by siRNA-mediated knockdown of SIRT1 (Fig. 3C). Taken together, these data demonstrated that SIRT1 is a negative regulator of AP-1 transcriptional activity.
We wondered which regions of SIRT1 are crucial for AP-1 activity regulation and found that the full-length and mutant (252–747) SIRT1 suppressed AP-1 transcriptional activity, but other mutants lost the effect (Fig. 3D). These results demonstrated that inhibition of AP-1 by SIRT1 is dependent on both the catalytic domain and the C terminus, although both domains bound to AP-1 independently as described above (Fig. 2, H and I).
SIRT1 Deacetylase Activity Contributes to Repression of AP-1 Transcriptional Activity
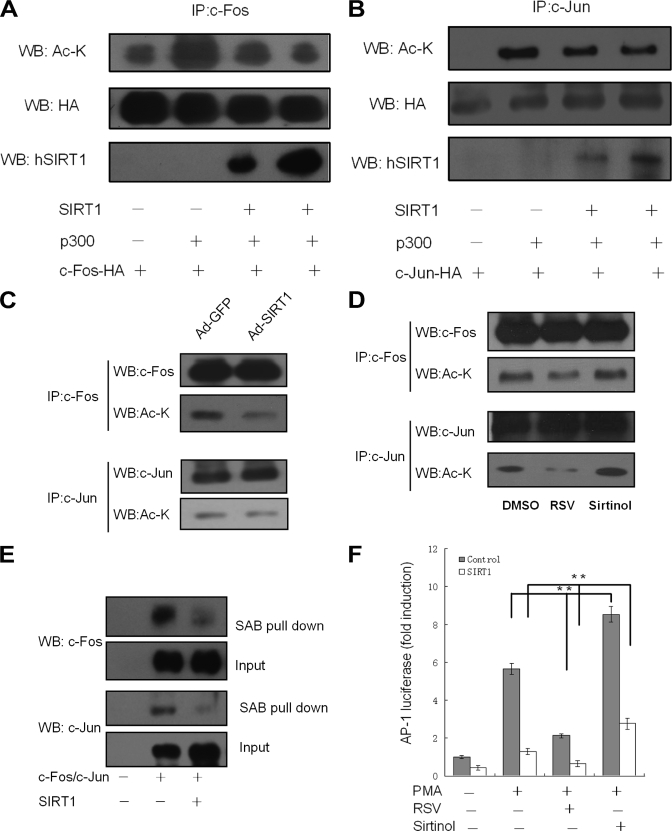
Because SIRT1 usually functions through deacetylating target proteins, we wondered whether SIRT1 influences the AP-1 acetylation status. Vector encoding HA-tagged c-Fos was transfected to cells with expression plasmids for p300 and SIRT1 as indicated (Fig. 4A). c-Fos was immunoprecipitated and analyzed by blotting with anti-acetylated lysine antibody. The precipitates were also blotted with anti-HA antibody to standardize the total amount of c-Fos. c-Fos was observed to be acetylated without overexpression of p300, an acetyltransferase, whereas exogenous expression of p300 enhanced the c-Fos acetylation. Overexpression of SIRT1 nearly abolished the p300-induced acetylation. To test whether acetylation of c-Jun is regulated by SIRT1, we performed similar experiments (Fig. 4B). Acetylated c-Jun was detected in cells overexpressing p300 and c-Jun-HA but was not observed in cells with c-Jun-HA alone. SIRT1 obviously reduced c-Jun acetylation, although not completely.
FIGURE 4.
SIRT1 deacetylase activity is important for suppression of AP-1. A and B, HEK293 cells were co-transfected with p300, SIRT1 expression vector and HA tagged c-Fos (A) or HA tagged c-Jun (B) as indicated. The cell extracts were immunoprecipitated with anti-HA antibody and probed with the different antibodies. C, pMΦs were infected with Ad-GFP or Ad-SIRT1 for 36 h and treated with PMA (50 ng/ml) for 3 h. D, pMΦs were treated with RSV (50 μm) and Sirtinol, respectively, (25 μm) for 1 h followed by PMA (50 ng/ml) for 3 h. The cell extracts were immunoprecipitated and probed with indicated antibodies. The figures are representative of three independent experiments. E, nuclear extracts were incubated with biotinylated oligonucleotides containing the AP-1 binding sequence of COX-2 promoter and streptavidin-agarose beads (SAB), and the precipitated complex was analyzed by Western blotting (WB). Input was used as a control. The images are representative of two independent experiments. F, HEK293 cells were co-transfected with 0.1 μg of AP-1-luc and 0.3 μg of indicated expression vectors. The cells were further treated with RSV (50 μm) and Sirtinol (25 μm), respectively, for 1 h followed by PMA (50 ng/ml) for 3 h. The luciferase activities are presented as the means ± S.D. of triplicate samples and are representative of three independent experiments. **, p < 0.01.
We next examined whether SIRT1 deacetylates endogenous c-Fos/c-Jun in macrophages. Mouse primary pMΦs with adenovirus-mediated expression of SIRT1 or control GFP were treated with PMA. c-Jun or c-Fos was immunoprecipitated and analyzed by blotting with anti-acetylated lysine antibody. We found that overexpression of SIRT1 obviously reduced the c-Fos/c-Jun acetylation level in macrophages (Fig. 4C). We further examined the influence of SIRT1 activator RSV and inhibitor Sirtinol on the acetylation of c-Fos/c-Jun. RSV reduced the c-Fos/c-Jun acetylation level, which was, however, increased by Sirtinol (Fig. 4D). These results suggested that SIRT1-mediated deacetylation of AP-1 is dependent on its deacetylase activity.
The acetylation level could contribute to transcriptional factor activity by influencing its DNA binding (6). Next, we asked the question of whether AP-1 DNA binding activity is also influenced by SIRT1. We transfected HEK293 cells with expression vectors of c-Fos/c-Jun and SIRT1 as described earlier and performed a DNA binding assay. The cell nuclear extracts were incubated with biotinylated double-strand DNA oligonucleotide, which is a region of human COX-2 promoter containing the AP-1 binding site. The oligonucleotides were pulled down by streptavidin-agarose beads, and the captured proteins were analyzed by Western blotting using anti-c-Fos or anti-c-Jun antibody. As expected, SIRT1 reduced the c-Fos and c-Jun binding to the COX-2 promoter probe. The nuclear extracts were also directly blotted by anti-c-Fos or anti-c-Jun antibody as input (Fig. 4E). These data proved that SIRT1 inhibits the recruitment of AP-1 to DNA.
We further examined the effects of SIRT1 activator RSV and inhibitor Sirtinol on the transactivation of AP-1 (Fig. 4F). HEK 293 cells were transfected with the expression plasmid of SIRT1 or control and were pretreated with each compound for 1 h followed by stimulation with PMA for another 3 h. SIRT1 suppressed basal AP-1 transactivation and reduced AP-1 activity to a lower level in the treatment of RSV, whereas Sirtinol increased AP-1 activity. These results demonstrated that the deacetylase activity of SIRT1 mediates suppression of AP-1 activity.
Furthermore, the expression vectors for SIRT1, plus c-Fos or c-Jun, were also co-transfected to cells (Fig. S2). SIRT1 suppressed the basal activity of AP-1. Overexpression of c-Fos or c-Jun enhanced AP-1 luciferase activity, which was decreased by co-transfection of SIRT1 expression plasmid, which further proved the SIRT1 to be a suppressor of AP-1.
SIRT1 Inhibits COX-2 Expression in Macrophages
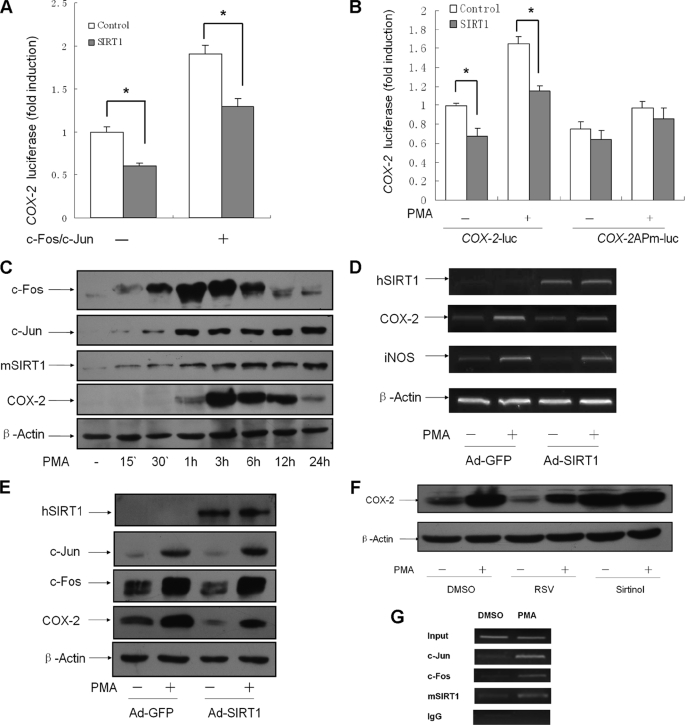
It is well established that AP-1 is important in transcriptional activation of COX-2 gene (28). As described earlier, SIRT1 suppressed the AP-1 DNA affinity to the COX-2 promoter (Fig. 4E). Next, to determine whether the suppressive effect of SIRT1 on AP-1 activity results in down-regulation of COX-2 gene transcription, we constructed a luciferase reporter plasmid (COX-2-luc) containing the mouse COX-2 promoter (−1003/+65) and examined the COX-2 promoter luciferase activity. HEK293 cells were transfected with the reporter construct, the c-Fos/c-Jun expression vector, and the SIRT1 expression vector. c-Fos/c-Jun caused a 2-fold increase in COX-2 promoter activity, and SIRT1 squelched the activation of COX-2 promoter, either at the basal level or in the presence of c-Fos/c-Jun overexpression (Fig. 5A). We further generated another luciferase reporter construct from COX-2-luc by site-directed mutation of a putative AP-1 binding site, which is at position −95/−86 in the COX-2 promoter. The cells were transfected with the wild type or mutant COX-2 promoter reporter and were treated with PMA. Luciferase activity assay showed that mutation of the AP-1 site resulted in a failure to respond to PMA stimulation (Fig. 5B). The inhibitory effect of SIRT1 on COX-2 promoter was destroyed by the AP-1 site mutation (Fig. 5B).
FIGURE 5.
SIRT1 inhibits COX-2 expression in macrophages. A, c-Fos/c-Jun expression plasmids (0.1 μg of each) and 0.3 μg of COX-2-luc were co-transfected with 0.3 μg of SIRT1 expression vector or control (pcDNA3.1) into HEK293 cells. Luciferase activities are presented as the means ± S.D. of triplicate samples and are representative of three independent experiments. *, p < 0.05 versus control. B, HEK293 cells transfected with 0.3 μg of COX-2-luc or COX-2APm-luc (AP-1 site mutant) and 0.3 μg of SIRT1 or control vector were treated with PMA (50 ng/ml) or vehicle for 3 h. The data present the means ± S.D. of triplicate samples and are representative of three independent experiments. *, p < 0.05 versus control. C, pMΦs were treated with PMA (50 ng/ml) for the indicated time. The protein expression of c-Fos, c-Jun, mSIRT1, and COX-2 was analyzed by Western blotting (WB). D and E, pMΦs infected with Ad-SIRT1 or control Ad-GFP were treated with PMA (50 ng/ml) for 3 h. COX-2 expression levels at RNA and protein were analyzed by reverse transcription-PCR (D) and WB (E). The images are representative of three experiments with similar results. F, pMΦs were pretreated with RSV (50 μm), Sirtinol (25 μm), and vehicle DMSO, respectively, for 1 h and then incubated with PMA (50 ng/ml) for another 3 h. Western blotting was performed using extracted protein. The images are representative of three experiments with similar results. G, chromatin was prepared from pMΦs treated with PMA (50 ng/ml) or DMSO for 3 h. Chromatin IP analysis was carried out with primers for mouse COX-2 promoter. The images are representative of three independent experiments with similar results.
We next examined whether SIRT1 influences COX-2 expression in macrophages. Mouse primary pMΦs with adenovirus-mediated expression of SIRT1 or control GFP were treated with PMA, and the expressions of hSIRT1, c-Fos, c-Jun, and COX-2 were analyzed by Western blotting. PMA significantly increased in the protein level of c-Fos, c-Jun, and COX-2 in pMΦs, and the COX-2 expression reached maximum levels at 3 h after PMA stimulation (Fig. 5C). Semiquantitative RT-PCR showed that exposure of pMΦs to PMA for 3 h led to an induction in COX-2 mRNA, which was depressed by overexpression of SIRT1 (Fig. 5D). SIRT1 decreased COX-2 expression in cells with or without PMA treatment, whereas overexpression of SIRT1 did not significantly affect protein expression of c-Fos or c-Jun in this cell model (Fig. 5E). In agreement with the result in pMΦs, macrophage-like RAW264.7 cells transiently transfected with SIRT1 expression vector had a lower level of COX-2 compared with the cells transfected with control vector (supplemental Fig. S3). RSV significantly reduced basal or PMA-stimulated COX-2 expression in pMΦs, whereas Sirtinol dramatically promoted COX-2 protein expression (Fig. 4F). We further performed a chromatin immunoprecipitation assay and found that endogenous SIRT1 in pMΦs was recruited to the AP-1 binding site of COX-2 promoter (Fig. 5G).
SIRT1 Regulates PGE2-related Macrophage Functions
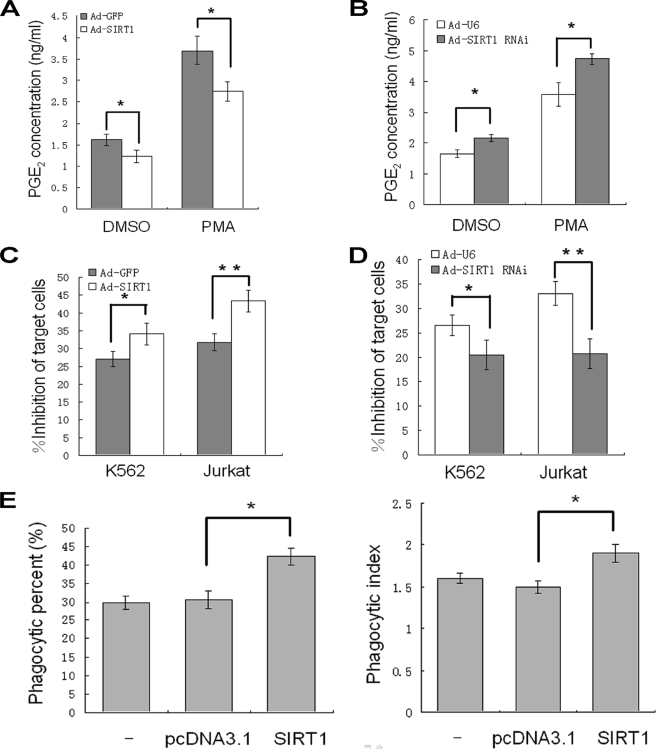
Because COX-2 is the rate-limiting synthase in the biosynthesis of PGs, we wondered whether the inhibition of COX-2 expression by SIRT1 affects PGE2 synthesis. Indeed, adenovirus-mediated overexpression of SIRT1 decreased PGE2 concentration in the supernatant of pMΦs (Fig. 6A). Reciprocally, the knockdown of SIRT1 increased PGE2 production from pMΦs in the presence or absence of PMA (Fig. 6B).
FIGURE 6.
SIRT1 improves macrophage functions. A and B, pMΦs were infected with Ad-GFP or Ad-SIRT1 (A) or Ad-U6 or Ad-SIRT1 RNAi (B), respectively, for 36 h and treated with PMA (50 ng/ml) or DMSO as control for 3 h. The PGE2 production are presented as the means ± S.D. of three independent experiments. *, p < 0.05. C and D, adenovirus-infected pMΦs were co-cultured with K562 or Jurkat cells for 24 h. Leukemia growth inhibition by macrophages is presented as the means ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01. E, RAW264.7 cells transfected with SIRT1 expression plasmid or vector control (pcDNA3.1) or not transfected were co-cultured with FITC-labeled yeast. The phagocytic percentage and phagocytic index were calculated, and the data are presented as the means ± S.D. of three independent experiments. *, p < 0.05.
One of the most important characteristics of macrophages is the ability to selectively attack against neoplastic cells, which is impaired by excessive PGE2 (11). To examine the potential effects of SIRT1 on macrophage tumoricidal activity, pMΦs were co-incubated with target cells, the suspended erythroleukemic K562 cells, or Jurkat cells for 24 h. The resultant cytotoxicity of macrophage to target tumor cells was evaluated as a percentage of inhibition of the target cells. Compared with cells expressing control GFP, the pMΦs with overexpression of SIRT1 had higher cytotoxic effect against K562 or Jurkat cells (Fig. 6C). Furthermore, knockdown of endogenous SIRT1 by RNAi significantly impaired the antitumoral activity of macrophages (Fig. 6D).
Macrophages excel at capturing and engulfing particles, including apoptotic cells and microbial invaders, which are crucial for nonspecific immune responses (29). Overproduction of PGE2 inhibits macrophages phagocytosis (8). To address whether SIRT1 affects macrophage uptake of particles, we transiently transfected SIRT1 expression vector and control pcDNA3.1 vector to RAW264.7 cells. Transfection of SIRT1 expression vector resulted in increased macrophage phagocytic percentage and phagocytic index, indicating the improved phagocytosis function (Fig. 6E).
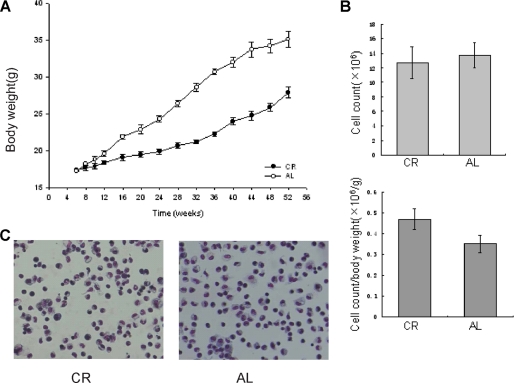
CR Does Not Influence Macrophage Formation
SIRT1 is increased in tissues of CR mice, mediating various CR-induced physical and cellular events (18, 26). However, it is unclear whether CR affects macrophage functions. We wondered whether any potential effects of CR on macrophages are mediated by SIRT1. To address this question we utilized a previously published CR model (24). pMΦs from mice fed ad libitum (AL group) or with 60% of the caloric base line of diet (CR group) for 1 year were extracted. The number of exudate pMΦs obtained by lavage exhibited no difference between AL and CR groups (Fig. 7A). However, CR significantly increased the ratio of macrophage population to body weight, because the mice fed with the calorie-restricted diet had lower body weights (Fig. 7, A and B). By Wright-Giemsa staining, we did not find any obvious split of nuclei or morphological abnormality in pMΦs in both AL and CR mice. In addition, the ability of pMΦs to adhere to and spread on the plate surface is similar between the two groups (Fig. 7C). Because the overall morphology and cell populations looked similar in pMΦs from CR and AL mice, CR may not significantly influence macrophage formation in the present model.
FIGURE 7.
CR does not influence macrophage formation. A and B, mean weekly body weights (A) and the number of pMΦs (B) for mice of AL and CR group were examined. The data are the means ± S.D. derived from 10 mice of each group. C, pMΦs were stained with May-Grünwald Giemsa.
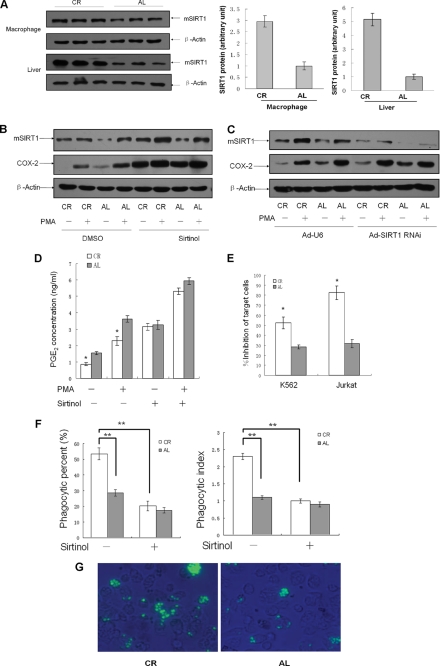
Elevated SIRT1 Decreases COX-2 Expression and Improves PGE2-related Macrophage Functions in Macrophages of CR Mouse
Our results further showed that SIRT1 protein was up-regulated in liver as well as in pMΦs from CR mice (Fig. 8A). We treated pMΦs with Sirtinol or infected pMΦs with Ad-SIRT1 RNAi to knock down endogenous SIRT1 activity. By Western blotting assay, we found that CR-induced down-regulation of COX-2 expression was reversed following knockdown of SIRT1 or Sirtinol treatment (Fig. 8, B and C). We finally examined the modulation of macrophage functions by CR. The PGE2 synthesis by pMΦs from CR mice is lower than that of AL mice, and Sirtinol distinctly antagonized CR-mediated repression of PGE2 production (Fig. 8D). CR also meliorated the macrophage cytotoxicity to tumor cells, including K562 and Jurkat cells (Fig. 8E). We next evaluated phagocytosis of FITC-labeled yeasts by pMΦs from CR mice and found that the phagocytic percentage and phagocytic index of pMΦs derived from CR mice were increased, compared with the AL group. Sirtinol dramatically eliminated the improved phagocytosis activity of pMΦs isolated from CR mice (Fig. 8, F and G).
FIGURE 8.
Elevated SIRT1 in macrophages of CR mouse decreases COX-2 expression and improves PGE2-related macrophage functions. A, SIRT1 expression in pMΦs and liver from mice of each group was analyzed by Western blotting. Each lane represents an individual mouse. The data are presented as the means ± S.D. of three samples. B and C, Western blotting analysis of COX-2 expression in pMΦs from AL and CR mice. pMΦs were treated with Sirtinol (25 μm) for 3 h (B) or were infected with SIRT1 RNAi adenovirus for 36 h (C) followed by treatment with PMA (50 ng/ml) or vehicle for another 3 h. The images are representative of six experiments with similar results. D, pMΦs were treated with Sirtinol (25 μm) or vehicle DMSO for 3 h and PMA (50 ng/ml) for another 3 h, and PGE2 production is presented as the means ± S.D. of three independent experiments. **, p < 0.01 versus the AL group. E–G, the tumoricidal activity (E) and phagocytosis of pMΦs (F and G) from each group were analyzed. The data are presented as the means ± S.D. of six independent experiments. **, p < 0.01.
DISCUSSION
SIRT1 is a NAD+-dependent histone deacetylase that catalyzes the removal of acetyl groups from a number of histone or non-histone targets, belonging to class III deacetylase (30, 31). The non-histone substrates of SIRT1 include a variety of important transcription factors and some transcription co-factors, such as p53, FOXOs, and PGC-1α (13, 32). The downstream effects of target deacetylation include changes in cellular metabolism, cell survival, and senescence. The biological functions of SIRT1 are thought to be mediated by its deacetylase activity (33). In the present study we found that SIRT1 decreased c-Fos/c-Jun acetylation induced by p300 and inhibited the transcriptional activity of AP-1. Moreover, RSV, which mimics the effect of SIRT1 (34, 35), decreased AP-1 transcriptional activity synergistically with SIRT1. Sirtuin inhibitor (36) Sirtinol, however, abolished the inhibitory effect of SIRT1 on AP-1 transcription. Furthermore, we found that SIRT1 inhibited AP-1 DNA binding to the COX-2 promoter in accordance with the previous report that acetylation modification is crucial for the DNA binding activity of transcriptional factors (6). In addition, our experiments also identified the specific AP-1-binding regions of SIRT1, namely the C terminus and the catalytic domain. The inhibitory effects on AP-1 transcriptional activity of constructs containing both domains were similar to full-length SIRT1. The C terminus of SIRT1 itself could bind AP-1, although it alone did not suppress AP-1 luciferase activity, implicating that the catalytic domain is necessary for the SIRT1-mediated inhibitory effects on AP-1. Taken together, these results indicate that SIRT1 deacetylase activity plays important roles in inducing AP-1 transcriptional inhibition.
For AP-1, the conserved bZIP domain is crucial for transcriptional regulation because it is responsible not only for dimerization and DNA binding but also for the interaction between the bZIP domain and other transcriptional factors or co-regulators. Earlier studies have shown that the basic region of c-Fos and c-Jun is involved in the interaction with general transcriptional factors, including TFIIE and TFIIF (37). The bZIP domain has also been shown to interact with TATA-binding protein association factor-1 (38) and the multiprotein bridging factor 1 (3). More recently, two groups reported binding between c-Jun and SIRT1 (39, 40). In line with these observations and extended to c-Fos, we found that SIRT1 directly interacted with the bZIP domain of both c-Fos and c-Jun, resulting in a reduction in the p300-induced acetylation of c-Fos and c-Jun. c-Jun has been reported to be acetylated by CBP/p300 at 271 lysine residues within the basic region of bZIP domain (5), which promoted us to investigate the relationships between the acetylation status of the basic region and SIRT1-mediated transcriptional inhibition. Surprisingly, none of the lysine mutants within the basic region of c-Jun affected AP-1 transcriptional activity in our experiments (data not shown). Recent reports showed that the N terminus of SIRT1, which lacks the deacetylase activity, was sufficient to stimulate the methyltransferase activity of SUV39H1. It may be because the N terminus of SIRT1 could alter conformation of SUV39H1 by direct interaction (41). Indeed, SIRT1 can utilize a noncatalytic mechanism to protect neuron survival (42). Further studies are needed to elucidate whether the binding of SIRT1 to bZIP domain of c-Fos and c-Jun participates in AP-1 transcriptional regulation in a deacetylase-independent manner.
CR decreases AP-1 DNA binding in mouse epidermis (43) and attenuates the aging-related AP-1 activation (23, 44). Numerous studies have linked the beneficial effects of mammalian CR with increased cellular SIRT1 expression (18, 26). Our results proved that overexpression of SIRT1 suppressed AP-1 activity, which seems to explain AP-1 inhibition by CR. Furthermore, CR has also been shown to decrease COX-2 expression (12) and plasma PGE2 levels (45). Excessive PGE2 destroys the macrophage-induced cytotoxicity of tumor cells (11) and the phagocytosis activity of macrophages (7, 8). In the present study, macrophages with adenovirus-mediated SIRT1 overexpression displayed similar effects on COX-2 expression, PGE2 production, and macrophage functions, compared with CR mice macrophages where higher endogenous SIRT1 was expressed. Because CR has been reported to broadly suppress carcinogenesis and counteract aging-related chronic inflammatory diseases, such as neural degeneration diseases and atherosclerosis (23, 24, 46, 47), the up-regulated SIRT1 in macrophages may represent a novel mechanism whereby CR is beneficial.
Sirtinol or knockdown of SIRT1 by RNAi similarly promoted AP-1 functions with promoting AP-1 transcriptional activity and COX-2 expression. Moreover, Sirtinol also abolished the CR-induced suppression of PGE2 production and improvement of macrophage functions, which implicate a potential role for deacetylase activity in the modulation of macrophage functions.
In summary, SIRT1 interacts with the bZIP domain of c-Fos/c-Jun and inhibits AP-1 transcriptional activity. The deacetylase activity of SIRT1 contributes to the decreased AP-1 transcriptional activity, reduced COX-2 expression, decreased PGE2 production, and improved PGE2-related macrophage phagocytosis and tumoricidal functions. Furthermore, SIRT1 may mediate CR-induced amelioration of macrophage functions through its deacetylase activity. These findings suggest that reinforcing SIRT1 activity in macrophages may be an effective strategy to prevent inflammatory diseases.
Supplementary Material
Acknowledgments
We thank Wei Zheng; Yun-Biao Lu for the Myc-SIRT1 and maltose-binding protein-SIRT1 construct; Qing-Jun Zhang for SIRT1 RNAi vector and p300 expression plasmid; and Li Li, Shuang Zhou, Hui-Na Zhang, Guang Liu, and Zhi-Gang She for technical assistance. We are grateful to Dr. Hua Cai University of California Los Angeles (UCLA) for revision on the manuscript.
This work was supported by National Basic Research Program of China Grants 2005CB522402 and 2006CB503801, National 863 Project Grant 2006AA02A406, and National Natural Science Foundation of China Grant 30721063.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- AP-1
- activator protein-1
- CR
- calorie restriction
- bZIP
- basic leucine zipper
- COX-2
- cyclooxygenase-2
- RNAi
- RNA interference
- PG
- prostaglandin
- pMΦ
- peritoneal macrophages
- PMA
- phorbol 12-myrisatate 13-acetate
- AL
- ad libitum
- RSV
- resveratrol
- HA
- hemagglutinin
- GST
- glutathione S-transferase
- GFP
- green fluorescent protein
- IP
- immunoprecipitation
- DMSO
- dimethyl sulfoxide
- FITC
- fluorescein isothiocyanate.
REFERENCES
- 1.Eferl R., Wagner E. F. (2003) Nat. Rev. Cancer 3, 859–868 [DOI] [PubMed] [Google Scholar]
- 2.Angel P., Imagawa M., Chiu R., Stein B., Imbra R. J., Rahmsdorf H. J., Jonat C., Herrlich P., Karin M. (1987) Cell 49, 729–739 [DOI] [PubMed] [Google Scholar]
- 3.Miotto B., Struhl K. (2006) Mol. Cell Biol. 26, 5969–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S. K., Kim J. H., Lee Y. C., Cheong J., Lee J. W. (2000) J. Biol. Chem. 275, 12470–12474 [DOI] [PubMed] [Google Scholar]
- 5.Vries R. G., Prudenziati M., Zwartjes C., Verlaan M., Kalkhoven E., Zantema A. (2001) EMBO J. 20, 6095–6103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo J., Li M., Tang Y., Laszkowska M., Roeder R. G., Gu W. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serezani C. H., Chung J., Ballinger M. N., Moore B. B., Aronoff D. M., Peters-Golden M. (2007) Am. J. Respir. Cell Mol. Biol. 37, 562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aronoff D. M., Canetti C., Peters-Golden M. (2004) J. Immunol. 173, 559–565 [DOI] [PubMed] [Google Scholar]
- 9.Schultz R. M., Pavlidis N. A., Stylos W. A., Chirigos M. A. (1978) Science 202, 320–321 [DOI] [PubMed] [Google Scholar]
- 10.Yakar I., Melamed R., Shakhar G., Shakhar K., Rosenne E., Abudarham N., Page G. G., Ben-Eliyahu S. (2003) Ann. Surg. Oncol. 10, 469–479 [DOI] [PubMed] [Google Scholar]
- 11.Yamada H., Kuroda E., Matsumoto S., Matsumoto T., Yamada T., Yamashita U. (2002) Clin. Exp. Immunol. 128, 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y. J., Kim H. J., No J. K., Chung H. Y., Fernandes G. (2006) Life Sci. 78, 2523–2532 [DOI] [PubMed] [Google Scholar]
- 13.Guarente L., Picard F. (2005) Cell 120, 473–482 [DOI] [PubMed] [Google Scholar]
- 14.Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L. (2004) Cell 116, 551–563 [DOI] [PubMed] [Google Scholar]
- 15.Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 16.Li X., Zhang S., Blander G., Tse J. G., Krieger M., Guarente L. (2007) Mol. Cell 28, 91–106 [DOI] [PubMed] [Google Scholar]
- 17.Yang S. R., Wright J., Bauter M., Seweryniak K., Kode A., Rahman I. (2007) Am. J. Physiol. Lung Cell Mol. Physiol. 292, L567–L576 [DOI] [PubMed] [Google Scholar]
- 18.Cohen H. Y., Miller C., Bitterman K. J., Wall N. R., Hekking B., Kessler B., Howitz K. T., Gorospe M., de Cabo R., Sinclair D. A. (2004) Science 305, 390–392 [DOI] [PubMed] [Google Scholar]
- 19.Bordone L., Cohen D., Robinson A., Motta M. C., van Veen E., Czopik A., Steele A. D., Crowe H., Marmor S., Luo J., Gu W., Guarente L. (2007) Aging Cell 6, 759–767 [DOI] [PubMed] [Google Scholar]
- 20.Rogina B., Helfand S. L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15998–16003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stapleton P. P., Fujita J., Murphy E. M., Naama H. A., Daly J. M. (2001) Nutrition 17, 41–45 [DOI] [PubMed] [Google Scholar]
- 22.Sun D., Muthukumar A. R., Lawrence R. A., Fernandes G. (2001) Clin. Diagn. Lab. Immunol. 8, 1003–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castello L., Froio T., Cavallini G., Biasi F., Sapino A., Leonarduzzi G., Bergamini E., Poli G., Chiarpotto E. (2005) FASEB J. 19, 1863–1865 [DOI] [PubMed] [Google Scholar]
- 24.Birt D. F., Pelling J. C., White L. T., Dimitroff K., Barnett T. (1991) Cancer Res. 51, 1851–1854 [PubMed] [Google Scholar]
- 25.Takata T., Ishikawa F. (2003) Biochem. Biophys. Res. Commun. 301, 250–257 [DOI] [PubMed] [Google Scholar]
- 26.Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M. W., Guarente L. (2004) Nature 429, 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deleted in proof
- 28.Guo Y. S., Hellmich M. R., Wen X. D., Townsend C. M., Jr. (2001) J. Biol. Chem. 276, 22941–22947 [DOI] [PubMed] [Google Scholar]
- 29.Aderem A., Underhill D. M. (1999) Annu. Rev. Immunol. 17, 593–623 [DOI] [PubMed] [Google Scholar]
- 30.Blander G., Guarente L. (2004) Annu. Rev. Biochem. 73, 417–435 [DOI] [PubMed] [Google Scholar]
- 31.Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000) Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 32.Gasser S. M., Cockell M. M. (2001) Gene 279, 1–16 [DOI] [PubMed] [Google Scholar]
- 33.Brooks C. L., Gu W. (2009) Nat. Rev. Cancer 9, 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borra M. T., Smith B. C., Denu J. M. (2005) J. Biol. Chem. 280, 17187–17195 [DOI] [PubMed] [Google Scholar]
- 35.Howitz K. T., Bitterman K. J., Cohen H. Y., Lamming D. W., Lavu S., Wood J. G., Zipkin R. E., Chung P., Kisielewski A., Zhang L. L., Scherer B., Sinclair D. A. (2003) Nature 425, 191–196 [DOI] [PubMed] [Google Scholar]
- 36.Grubisha O., Smith B. C., Denu J. M. (2005) FEBS J. 272, 4607–4616 [DOI] [PubMed] [Google Scholar]
- 37.Martin M. L., Lieberman P. M., Curran T. (1996) Mol. Cell. Biol. 16, 2110–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lively T. N., Nguyen T. N., Galasinski S. K., Goodrich J. A. (2004) J. Biol. Chem. 279, 26257–26265 [DOI] [PubMed] [Google Scholar]
- 39.Dey S., Bakthavatchalu V., Tseng M. T., Wu P., Florence R., Grulke E. A., Yokel R., Dhar S. K., Yang H. S., Chen Y., St Clair D. K. (2008) Carcinogenesis 29, 1920–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao Z., Ye J. (2008) Biochem. Biophys. Res. Commun. 376, 793–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaquero A., Scher M., Erdjument-Bromage H., Tempst P., Serrano L., Reinberg D. (2007) Nature 450, 440–444 [DOI] [PubMed] [Google Scholar]
- 42.Pfister J. A., Ma C., Morrison B. E., D'Mello S. R. (2008) PLoS ONE 3, e4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Przybyszewski J., Yaktine A. L., Duysen E., Blackwood D., Wang W., Au A., Birt D. F. (2001) Carcinogenesis 22, 1421–1427 [DOI] [PubMed] [Google Scholar]
- 44.Kim H. J., Jung K. J., Seo A. Y., Choi J. S., Yu B. P., Chung H. C. (2002) Age 25, 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalani R., Judge S., Carter C., Pahor M., Leeuwenburgh C. (2006) J. Gerontol. A Biol. Sci. Med. Sci. 61, 211–217 [DOI] [PubMed] [Google Scholar]
- 46.Mai V., Colbert L. H., Berrigan D., Perkins S. N., Pfeiffer R., Lavigne J. A., Lanza E., Haines D. C., Schatzkin A., Hursting S. D. (2003) Cancer Res. 63, 1752–1755 [PubMed] [Google Scholar]
- 47.Lee C. K., Weindruch R., Prolla T. A. (2000) Nat. Genet. 25, 294–297 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.