Abstract
The apoB RNA-editing enzyme, catalytic polypeptide-like (APOBEC) family of proteins includes APOBEC1, APOBEC3, and activation-induced deaminase, all of which are zinc-dependent cytidine deaminases active on polynucleotides and involved in RNA editing or DNA mutation. In contrast, the biochemical and physiological functions of APOBEC2, a muscle-specific member of the family, are unknown, although it has been speculated, like APOBEC1, to be an RNA-editing enzyme. Here, we show that, although expressed widely in striated muscle (with levels peaking late during myoblast differentiation), APOBEC2 is preferentially associated with slow-twitch muscle, with its abundance being considerably greater in soleus compared with gastrocnemius muscle and, within soleus muscle, in slow as opposed to fast muscle fibers. Its abundance also decreases following muscle denervation. We further show that APOBEC2-deficient mice harbor a markedly increased ratio of slow to fast fibers in soleus muscle and exhibit an ∼15–20% reduction in body mass from birth onwards, with elderly mutant animals revealing clear histological evidence of a mild myopathy. Thus, APOBEC2 is essential for normal muscle development and maintenance of fiber-type ratios; although its molecular function remains to be identified, biochemical analyses do not especially argue for any role in RNA editing.
Keywords: Development Differentiation, Development Differentiation/Muscle, Diseases/Muscular, Evolution/Protein, Gene/Knock-out, Tissue/Organ Systems/Muscle/Skeletal, AID/APOBEC Family, Deaminases
Introduction
The AID/APOBEC5 proteins form a subgroup of a superfamily of zinc-dependent deaminases, with AID, APOBEC1, and APOBEC3 being able to deaminate cytosine to uracil in the context of polynucleotide substrates (1–3). APOBEC1 is an RNA-editing enzyme that acts on apolipoprotein B RNA in the small intestine to yield a premature stop codon, thereby yielding the shorter form of apolipoprotein B polypeptide. AID catalyzes targeted deamination of deoxycytidines within the context of the immunoglobulin locus in B lymphocytes, where this mutagenic action underpins antibody gene diversification. The APOBEC3 proteins are able to deaminate deoxycytidine in the replication intermediates of various retroelements or retroviruses, with human APOBEC3F and APOBEC3G acting as host factors that restrict and hypermutate human immunodeficiency virus-1.
In contrast, APOBEC2 (which, along with AID, is likely the oldest member of the AID/APOBEC family (1)) is exclusively expressed in cardiac and skeletal muscle (4, 5). The crystallographic structure of APOBEC2 was the first structure of an AID/APOBEC protein to have been determined and indeed has been used as a paradigm on which to model the structures of the other polynucleotide deaminase family members (6). The function of APOBEC2 is, however, wholly unknown. On the basis of its homology to APOBEC1, it has widely been speculated to be an RNA-editing enzyme, although it has not yet been demonstrated to exhibit any biochemical or physiological activity.
Here, to gain insight into the biology of APOBEC2, we have analyzed its pattern of expression within skeletal muscle and have also identified phenotypic consequences of APOBEC2 deficiency. Skeletal muscle comprises several fiber types that exhibit different metabolic features for energy production (7). Fast-twitch fibers have a high potential for producing ATP by anaerobic pathways and high enzymatic activities of glycolytic enzymes, whereas slow-twitch fibers have higher aerobic potential and higher activities of enzymes related to oxidative metabolism (8). We found that APOBEC2 is especially associated with slow-twitch fibers and is particularly abundant in soleus muscle (which has a high proportion of slow fibers) but that APOBEC2 deficiency actually causes a shift from fast to slow fibers in soleus muscle, which is evident even in young mice. Once the mice age to >6 months, a mild myopathy develops. Thus, APOBEC2 is required for normal muscle development, affecting the ratio of slow to fast myofibers, although our investigations into the biochemical properties of the protein do not provide especial support to the idea that it fulfills its function through RNA editing.
EXPERIMENTAL PROCEDURES
Mice and Cell Culture
APOBEC2-deficient mice (9) bred onto a C57BL/6 background for at least nine generations were maintained in the barrier facility in Cambridge, UK, under UK Home Office Project License 80/2226 and in Fukuoka, Japan, under Kyusyu University's rules for animal welfare, with wild-type littermates from interheterozygous crosses or inbred C57BL/6 mice providing controls. For sciatic nerve denervation, 15-week-old mice were anesthetized with pentobarbital (1 mg/kg), a 1-cm incision was made in the skin along the axis of the femur in the right hind limb, and a 3–5-mm section of sciatic nerve was excised to prevent re-innervation.
The mouse C2C12 myoblast cell line (10) was maintained at a low density in Dulbecco's modified Eagle's medium supplemented with 20% in FBS. Cultures of mouse primary myoblasts (11) were established by digesting minced limb muscles from 2–5-day-old mice with Liberase Blendzyme 3 (0.14 units/ml; Roche Diagnostics, West Sussex, UK) in Ham's F-10 medium (Invitrogen, Paisley, UK) without FBS for 30–45 min. After passing through a nylon mesh and washing in Ham's F-10 + 20% FBS, cells were resuspended in growth medium (Ham's F-10 and 20% FBS containing 2.5 ng/ml basic fibroblast growth factor (Roche Diagnostics)). Myoblasts were enriched as the cells that did not adhere to plastic dishes during two rounds of preplating for 45 min each and cultured on collagen-coated dishes for at least 2 weeks, repeating the preplating procedure twice weekly. To induce myoblast differentiation, the growth medium was replaced with Dulbecco's modified Eagle's medium supplemented with 2% horse serum. Cell fusion index was determined as the ratio of the number of nuclei in myotubes (cells with two or more nuclei) to the number of nuclei in solitary cells. Six randomly chosen fields from each dish were counted at a magnification of ×100.
Histology and Immunohistochemistry
For histology, serial transverse 5-μm sections of frozen or paraffin-embedded muscle were stained with hematoxylin and eosin. For immunohistochemistry, freshly prepared tissue samples were fixed in 4% paraformaldehyde and embedded in paraffin, and sections (5–8 μm) were cut and mounted. After blocking for 30 min with 10% FBS and 2% goat serum in phosphate-buffered saline with 0.1% Triton X-100 (PBST), myosin HC isoform expression was determined by incubating sections with mAb (Sigma) MY-32 or A4.951, specific for slow myosin HC, and mAb NOQ7.5.4D, specific for fast myosin, and developing with biotinylated secondary antibody (goat anti-mouse IgG), followed by streptavidin/peroxidase reagents (Vector Laboratories, Burlingame, CA).
To stain for APOBEC2, sections were subjected to antigen retrieval in which deparaffinized and rehydrated slides were boiled (2× 5 min in 0.1 m Tris (pH 9) for APOBEC2 staining and in 0.1 m citrate (pH 6) for myosin HC staining) and, after cooling, treated with 0.3% H2O2 in methanol for 10 min, followed by two 5-min washes with phosphate-buffered saline and blocking for 30 min with 2% goat serum in PBST. Detection of APOBEC2 was performed using an affinity-purified anti-APOBEC2 polyclonal antiserum (diluted 1:1000 in PBST) prepared from rabbits that had been hyperimmunized with a fusion of MBP to mouse APOBEC2. The serum was absorbed against boiled MBP-expressing Escherichia coli, and APOBEC2-specific antibodies were then purified by binding to MBP-APOBEC2 fusion protein that had been coupled to a HiTrap-NHS-Sepharose column (GE Healthcare, Buckinghamshire, UK) and elution (after extensive washing) with 0.1 m glycine (pH 2.8). The staining was developed using biotinylated goat anti-rabbit antiserum and a streptavidin/peroxidase/substrate kit (VECTASTAIN, Vector Laboratories).
Western Blot Analysis
For Western blots, myosin HC was detected using mAb A4.1025 (1:200 dilution; Alexis, London, UK), whereas histone H3 was detected using mAb 6002 (1:10,000 dilution; Abcam, Cambridge, UK). APOBEC2 was detected either with a polyclonal antiserum (either generated from rabbits immunized with MBP-APOBEC2 or obtained commercially (Abcam)) or with anti-APOBEC2 mAb hA2-33-1 (IgG2bκ; 0.5 μg/ml). This mAb was generated by fusion of spleen cells from A/J mice that had been hyperimmunized with an MBP-human APOBEC2 fusion protein with cells of the mouse NS0 plasmacytoma. mAb hA2-33-1 was obtained following screening of hybridoma supernatants for antibodies recognizing both mouse and human APOBEC2.6 Western blots were developed using goat anti-mouse horseradish peroxidase (Jackson ImmunoResearch Laboratories, Suffolk, UK, or Vector Laboratories), and bands were visualized using ECL.
Analysis of APOBEC2 RNA
Oligo(dT)-primed cDNA was generated from aliquots (3 μg) of total RNA that had been extracted from muscle samples using TRIzol reagent (Invitrogen, Carlsbad, CA). Real-time quantitative reverse transcription-PCR was performed in triplicate using the LightCycler system (Roche Diagnostics, Penzberg, Germany) according to the manufacturer's protocol. The expression levels of APOBEC2 mRNA were determined by normalizing relative to hypoxanthine-guanine phosphoribosyltransferase expression. The forward and reverse primers were 5′-GACCGGTTCTGTCATGTCG and 5′-ACCTGGTTCATCATCACTAATCAC for hypoxanthine-guanine phosphoribosyltransferase and 5′-TCCTGAAGTAGGCAACAGAGC and 5′-GCCATCCTGGTCATTGCT for APOBEC2, respectively. Northern blot analyses were performed on total RNA purified and DNase I-treated using an RNeasy® fibrous tissue kit (Qiagen GmbH) and probed with APOBEC2 and β-actin cDNAs labeled with [α-32P]dCTP.
Biochemical Analysis of APOBEC2
To determine the native molecular mass of APOBEC2, extracts of mouse gastrocnemius muscle were prepared by disruption under liquid N2 and homogenization in lysis buffer (10 mm Tris (pH 7.4), 15 mm NaCl, 5 mm MgCl2, 0.1 mm ZnCl2, complete protease inhibitor (Roche Diagnostics), and 0.2 mm phenylmethylsulfonyl fluoride). They were then clarified by centrifugation (twice at 350 × g for 5 min), treated (or not) with RNase A (100 μg/ml; Sigma), adjusted to 150 mm in NaCl, and subjected to gel filtration on a HiLoad 26/60 Superdex 200 column (GE Healthcare) that had been calibrated using a calibration kit for molecular weights of 12,000–200,000 (Sigma). Fractions containing APOBEC2 were identified by Western blotting. Myosin HC isoforms in extracts of snap-frozen muscle samples were also distinguished by virtue of their differential mobility in high resolution, low cross-linked 8% acrylamide/glycerol gels as described previously (12).
Cross-linking studies were performed to screen for possible RNA binding essentially as described (14). The binding site for APOBEC1 in apoB mRNA (A-I in Ref. 14), as well as a sequence of 32 tandem AU repeats, was transcribed in vitro with T7 polymerase from PCR products in the presence of 80 μCi of [α-32P]UTP (3000 Ci/mmol). Labeled RNA (5 ng, ∼2 × 108 cpm/μg) was incubated with 1 μg of MBP-APOBEC1 or MBP-APOBEC2 fusion protein that had been purified from E. coli in 15 μl of buffer (20 mm Hepes (pH 7.5), 1 mm dithiothreitol, and 100 mm NaCl) for 30 min at room temperature in microtiter plates. The plates were put on ice and irradiated with 250 mJ/cm2 UV light in a Stratalinker (Stratagene, La Jolla, CA). Samples were treated with 0.5 mg/ml RNase A for 15 min at 37 °C, separated on a 12% BisTris gel, stained with Coomassie Blue, dried, and analyzed by autoradiography using a PhosphorImager.
RESULTS
APOBEC2 Is Widely Expressed in Muscle but with Stronger Staining of Slow Fibers
Previous studies (4, 5, 9) have revealed that APOBEC2 expression is largely restricted to striated muscle. To ascertain whether APOBEC2 is present in similar abundance in all myofibers, we developed conditions that allowed an immunohistochemical analysis. Although APOBEC2 was readily detected in all hind limb muscle fibers examined, the intensity of staining differed between individual fibers (Fig. 1A). Analysis of both serial and longitudinal sections revealed that the observed variation in staining intensity reflected a difference between individual fibers rather than simply variation in intensity along the fiber length. Staining of serial sections with antibodies against different myosin HC isoforms revealed that stronger APOBEC2 staining was much more frequently observed in slow rather than fast myofibers (Fig. 1, B and C).
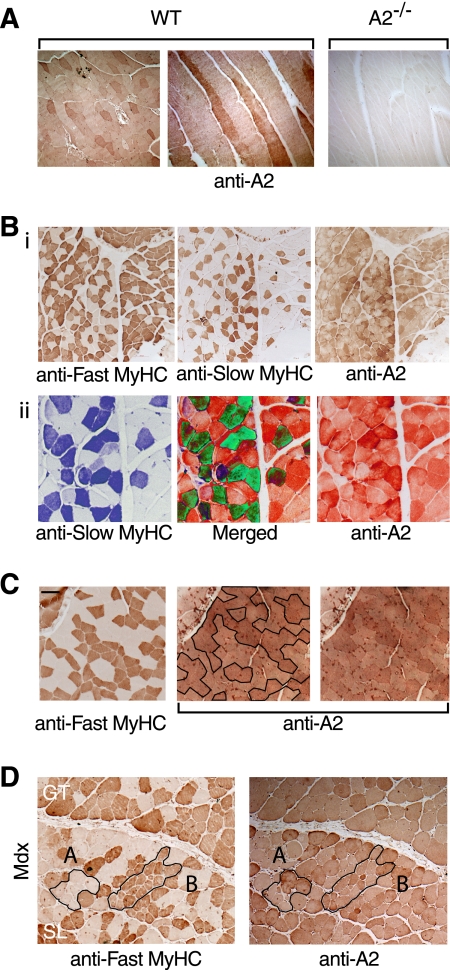
FIGURE 1.
APOBEC2 is highly expressed in slow myofibers. A, lateral and longitudinal sections of hind limb muscle from a 3-month-old C57BL/6 mouse (left and middle panels) and parallel staining of longitudinal sections of hind limb muscle from an APOBEC2-deficient (A2−/−) mouse to demonstrate the specificity of staining (right panel). WT, wild-type. B, panel i, staining of serial sections of hind limb muscle from 3-month-old wild-type mice for fast and slow myosin HC (MyHC) isoforms and for APOBEC2; panel ii, regions of the sections depicted in panel i shown in higher magnification but with pseudo-coloring such that fibers staining strongly for both slow myosin HC and APOBEC2 are seen as green in the merged panel. C, comparison of serial sections of soleus muscle from 6-month-old wild-type mice stained for the fast myosin HC isoform and APOBEC2. The two panels stained for APOBEC2 are identical except that, in the one on the left, slow fibers (which lack fast myosin HC) are enclosed by a black line. Scale bar = 200 μm (left panel). D, comparison of serial sections of soleus muscle from 6-month-old male mdx mice stained for the fast myosin HC isoform and APOBEC2. Examples of damaged/regenerating fibers are outlined in black, with region A comprising slow fibers and region B comprising fast fibers. GT, gastrocnemius muscle; SL, soleus muscle.
To determine whether APOBEC2 expression levels change during myoblast differentiation, we used Western blot analysis to monitor APOBEC2 abundance during in vitro differentiation of neonatal mouse myoblasts. Expression of APOBEC2 (like that of myosin HC) dramatically increased during late differentiation, in contrast to the relatively constant level of histone H3 expression (Fig. 2). Similar results were obtained during in vitro differentiation of the mouse C2C12 myoblast cell line (data not shown). However, APOBEC2 levels do not appear to be significantly different in newly formed (as opposed to well established) myofibers. In mice lacking dystrophin (mdx mice), significant muscle pathology develops with age and is manifested by an increased proportion of regenerating fibers (small fibers and fibers with centrally located nuclei) (15). We did not detect evidence of differential APOBEC2 staining of such newly formed fibers (Fig. 1D).
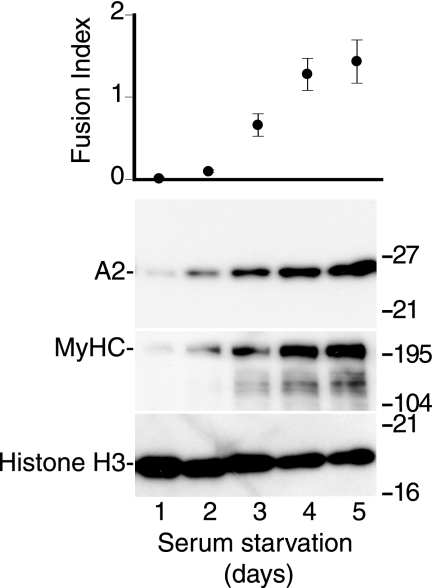
FIGURE 2.
APOBEC2 expression increases during myoblast differentiation in vitro. Primary mouse myoblasts were cultured under low serum conditions to induce muscle cell differentiation. At days 1–5 after the start of serum starvation, the ratio of multinucleated myotubes to mononucleated myoblasts (fusion index) was determined, and cell extracts were analyzed by Western blotting for expression of APOBEC2 (A2), myosin HC (MyHC), and histone H3.
If APOBEC2 is generally more highly expressed in slow as opposed to fast fibers, one would expect higher abundance of APOBEC2 in soleus compared with gastrocnemius muscle. Using both Northern and Western blot analyses (because these provide more quantitative information on abundance than immunohistochemistry), it is evident that (in both young and old mice) APOBEC2 is indeed much more highly expressed in soleus muscle than in other muscles examined (Fig. 3). Thus, APOBEC2 expression increases late during myoblast differentiation and, although widely expressed in striated muscle, is much more abundant in soleus compared with gastrocnemius muscle and in slow as opposed to fast myofibers.
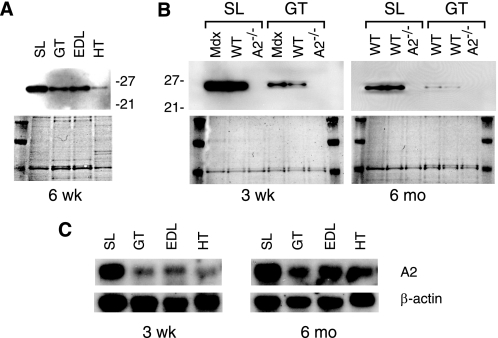
FIGURE 3.
APOBEC2 is highly expressed in soleus muscle. A, abundance of APOBEC2 in extracts of different muscles from a 6-week (wk)-old male C57BL/6 mouse as determined by Western blot analysis (upper panel), controlling for loading by staining for total protein with Ponceau reagent (lower panel). SL, soleus muscle; GT, gastrocnemius muscle; HT, heart. B, Western blot comparison of APOBEC2 abundance in extracts of soleus and gastrocnemius muscle from 3-week-old or 6-month (mo)-old wild-type (WT) and mdx mice, with APOBEC2-deficient (A2−/−) mice serving as a negative control. C, Northern blot analysis of APOBEC2 RNA in various muscles, with β-actin serving as a control.
APOBEC2-deficient Mice Weigh Less than Controls
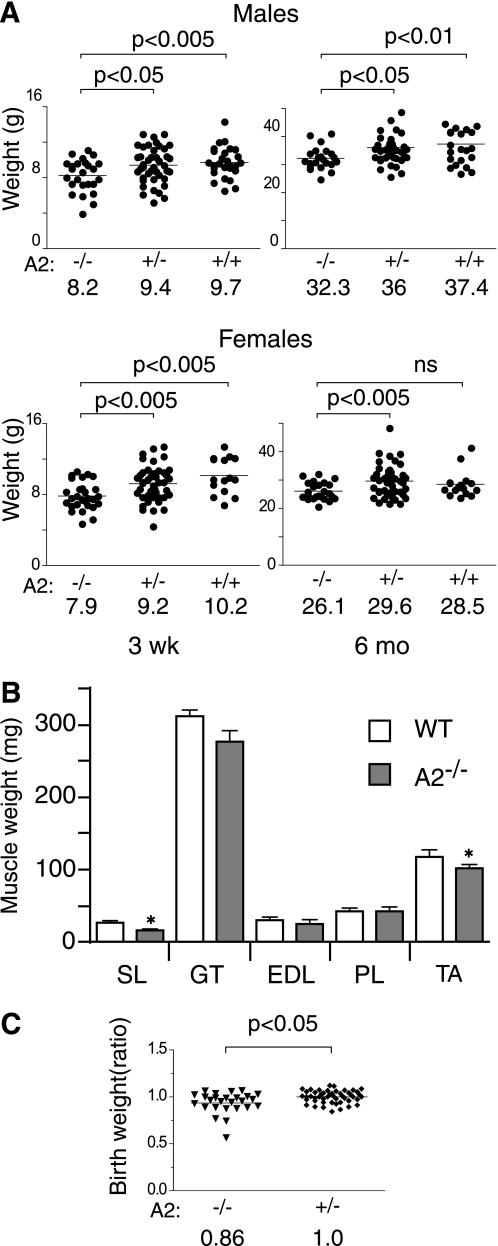
Previous studies revealed that APOBEC2-deficient mice are viable and fertile and reach adulthood (9). However, to screen for the possibility of more subtle developmental defects, we compared overall body mass in a cohort of APOBEC2-deficient and control littermates that had been generated by interbreeding APOBEC2+/− heterozygotes. The results revealed that, at the time of weaning, APOBEC2-deficient mice of both genders had a total body mass that was significantly (15–20%) lower than that of control wild-type or heterozygous littermates (Fig. 4A). The reduction in body weight persisted as the mice grew older, with there still being a substantial body weight reduction in 6-month-old mice. The overall body mass difference appeared to broadly correlate with a general reduction of the mass of individually dissected skeletal muscles (Fig. 4B), although we could not exclude the possibility that reduced mass of other body components might not contribute disproportionately to the overall reduction in body weight of APOBEC2-deficient mice. A body mass difference was in fact evident from birth: APOBEC2-deficient pups born to male APOBEC2-deficient fathers and heterozygous mothers were 14% lighter at 24 h than their heterozygous littermates (Fig. 4C). The weight reduction was not a result of competition between APOBEC2-deficient and wild-type littermates because it was also observed when comparing offspring of homozygous APOBEC2−/− breedings with control inbreedings (data not shown).
FIGURE 4.
APOBEC2-deficient mice weigh less than control siblings. A, the body weights of APOBEC2 (A2)-deficient (−/−), heterozygous (+/−), and wild-type (+/+) mice from heterozygous breedings were determined at the time of weaning (21 days after birth) and at 6 months. The mean weights for the different groups are indicated. Data were analyzed using Student's t test, with the significance for each comparison shown. ns, not significant. B, the weights of individual muscles from groups of five male 15-week-old wild-type (WT) and five male 15-week-old APOBEC2-deficient mice are shown. SL, soleus muscle; GT, gastrocnemius muscle; PT, plantaris muscle; TA, tibialis anterior muscle. Error bars are means ± S.E., with asterisks denoting differences significant at p < 0.05 (Student's t test). C, the body weights of newborn mice born to breedings of an APOBEC2-deficient father and a heterozygous mother are presented, with the individual weights given as a ratio of the absolute weight of an individual animal to that of the mean of APOBEC2+/− heterozygotes within the same litter.
Increased Proportion of Slow Fibers in Soleus Muscle from APOBEC2−/− Mice
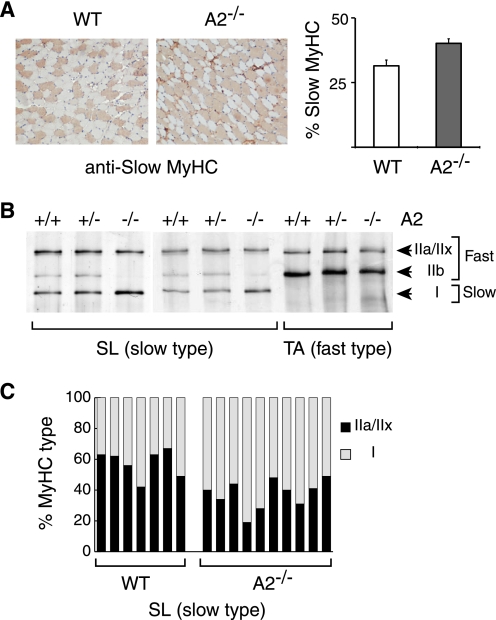
Given the differential abundance of APOBEC2 in fast and slow muscle fibers, we wondered whether APOBEC2 deficiency would affect the development of these fiber forms. Immunohistochemical analysis did indeed reveal a significant change in the ratio of fiber forms in soleus muscle of APOBEC2−/− mice: the proportion of fibers staining with an antibody against slow myosin HC increased from 31% in 15-week-old wild-type mice to 40% in knock-out mice (Fig. 5A). Comparison of soleus muscle extracts from 3-month-old APOBEC2-deficient and control mice by SDS-PAGE revealed that APOBEC2 deficiency was indeed accompanied by an increase in the ratio of slow to fast myosin HC isoforms as demonstrated by an increase in the proportion of type I (slow) and concomitant disappearance of type IIb isoforms. This change was already evident at 3 weeks of age (Fig. 5, B and C).
FIGURE 5.
Increased proportion of slow fibers in APOBEC2−/− mouse soleus. A, immunohistochemical staining with an anti-slow myosin HC (MyHC) mAb of transverse sections of soleus muscle from 15-week-old wild-type (WT) and APOBEC2−/− (A2−/−) mice. Analysis of five pairs of 15-week-old mice revealed that the proportion of soleus fibers staining with the anti-slow myosin HC mAb increased from 31% in control mice to 40% in APOBEC2-deficient mice. B, comparison of the relative abundance of slow and fast myosin HC isoforms in soleus muscle (SL; left and middle panels) and tibialis anterior muscle (TA; right panel) from 3-month-old litter-matched wild-type, APOBEC2+/−, and APOBEC2−/− mice analyzed by silver staining of muscle extracts subjected to SDS-PAGE in low cross-linked gels. C, histogram showing the ratios of fast (MyHCIIa+IIx) to slow (MyHCI) myosins in soleus muscle of seven wild-type and nine APOBEC2-deficient mice as judged by densitometry of silver-stained SDS-polyacrylamide gels.
Changes in APOBEC2 Expression and Muscle Fiber Type following Denervation
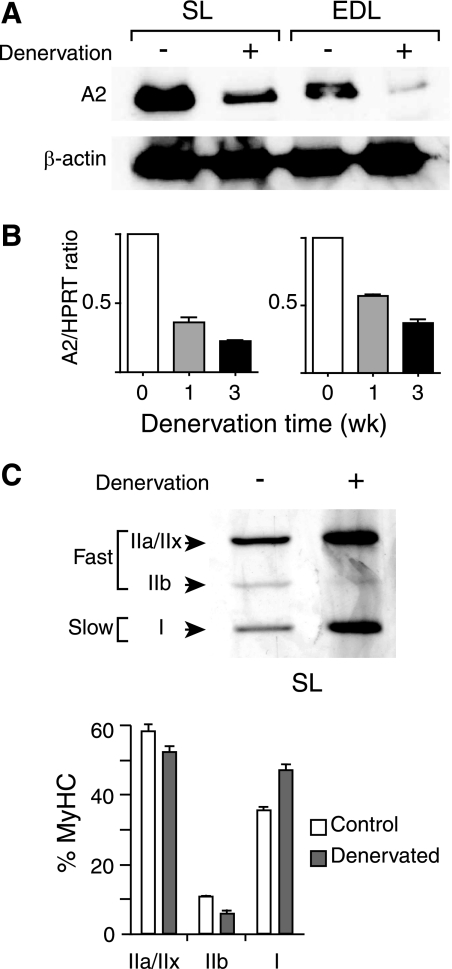
We were somewhat surprised that, although APOBEC2 was preferentially expressed in slow muscle, APOBEC2 deficiency actually led to an increase in the proportion of slow fibers in soleus muscle. However, a similar correlation between reduced APOBEC2 expression and a fast-to-slow fiber-type switch was seen following muscle denervation. Thus, removal of a section of the sciatic nerve provides an experimentally useful model for muscle atrophy and fiber-type shift because it yields a quick and pronounced change in skeletal muscle (16, 17). Denervation of both mouse and rat soleus muscle has recently been found to be associated with a switch from fast to slow fiber type (18, 19), and at least in the case of rat, a proteomic analysis suggested an accompanying reduction in APOBEC2 (19). We therefore performed a denervation analysis in mouse and found that, in both soleus and EDL muscle, this led to a substantial decrease in APOBEC2 expression, as judged by both Western and Northern blotting, as well as an increase in the ratio of slow to fast myosin HC isoforms (Fig. 6).
FIGURE 6.
Reduced APOBEC2 expression and fiber-type switch following denervation. A, Western blot analysis of APOBEC2 expression in soleus (SL) and EDL muscle from 15-week-old male mice that had or had not been subjected to sciatic nerve denervation 2 weeks previously. Protein loading was controlled by reprobing for β-actin. B, reverse transcription-PCR analysis of APOBEC2 (A2) expression in soleus and EDL muscle from 15-week-old male mice that had or had not been subjected to sciatic nerve denervation 1 or 3 weeks (wk) previously. The reverse transcription-PCR signal was calculated relative to that given by hypoxanthine-guanine phosphoribosyltransferase (HPRT). C, analysis of change in myosin HC (MyHC) isoform expression in soleus muscle 3 weeks following denervation in male C57BL/6 mice by high resolution SDS-PAGE. Histograms present the results of the analysis of five mice.
APOBEC2-deficient Mice Develop Myopathy with Age
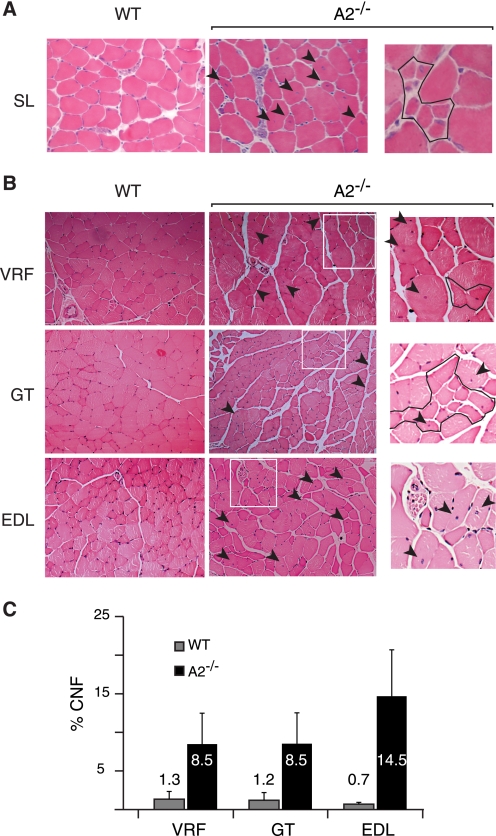
Although APOBEC2-deficient mice are smaller than controls and exhibit an increased proportion of slow fibers in soleus muscle, they appear healthy and do not have a significantly reduced life span. We nevertheless examined muscle from both 8-week-old and 8–10-month-old APOBEC2-deficient mice to see whether there was any histological evidence of myopathy. Whereas the younger mice showed no increase in the proportion of regenerating fibers, myopathic changes were evident in all muscle types examined from the older animals. Not only was there increased variability in the size of myofibers in all muscles examined, but there was also an increased proportion of myofibers harboring centrally (as opposed to peripherally) located nuclei (Fig. 7 and Table 1), classic signs of regenerating muscle and postnatal myofibers associated with myopathy. Although striking, the myopathy evident in 8–10-month-old APOBEC2-deficient mice was significantly milder than that evident in 6-month-old mdx mice, which carry a mutated dystrophin gene (see Fig. 1D). Furthermore, no change was observed in the localization of laminin or dystrophin in the muscle tissue of APOBEC2-deficient mice (data not shown).
FIGURE 7.
Histological evidence of myopathy in 8–10-month-old APOBEC2−/− mice. A, examples of a region of soleus muscle (SL) in 15-week-old wild-type (WT) or APOBEC2-deficient (A2−/−) mice. The arrowheads indicate fibers with centrally located nuclei, and the right panel shows a region with immature myotubes. B, comparison of transverse sections of vastus and rectus femoris (VRL), gastrocnemius (GT), and EDL muscle from 9-month-old APOBEC2-deficient and wild-type mice. The boxed areas in the middle panels are shown in higher magnification in the right panels, where outlined regions contain smaller fibers. C, histogram comparing the percentage of centrally nucleated fibers (CNF) in different muscles from 9-month-old APOBEC2-deficient and control mice.
TABLE 1.
Quantification of central nucleated myofibers in hematoxylin/eosin sections of control and APOBEC2-deficient hind limb muscles
Samples are from 9–10-month-old control and APOBEC2-deficient (A2−/−) mice (n = 3 for each group).
| Vastus and rectus femoris | |||||
|---|---|---|---|---|---|
| Control |
A2−/− |
||||
| No. fibers in section | No. central nuclei | % | No. fibers in section | No. central nuclei | % |
| 229 | 8 | 3.49 | 242 | 5 | 2.07 |
| 105 | 0 | 0.00 | 148 | 7 | 4.73 |
| 250 | 2 | 0.80 | 156 | 16 | 10.26 |
| 127 | 2 | 1.57 | 160 | 22 | 13.75 |
| 212 | 0 | 0.00 | 174 | 26 | 14.94 |
| 179 | 4 | 2.23 | 126 | 5 | 3.97 |
| 214 | 17 | 7.94 | |||
| 212 | 21 | 9.91 | |||
| 183 | 16 | 8.74 | |||
| Total: 1118 | 1.35 (±1.37)a | 1750 | 8.48 (±4.34)a | ||
| Gastrocnemius | |||||
| 172 | 4 | 2.33 | 333 | 39 | 11.71 |
| 134 | 1 | 0.75 | 389 | 30 | 7.71 |
| 232 | 3 | 1.29 | 266 | 15 | 5.64 |
| 270 | 2 | 0.74 | 207 | 24 | 11.59 |
| 293 | 5 | 1.71 | 162 | 19 | 11.73 |
| 263 | 1 | 0.38 | 252 | 7 | 2.78 |
| Total: 1380 | 1.2 (±0.72)a | Total 1743 | 8.53 (±3.79)a | ||
| EDL | |||||
| 324 | 2 | 0.62 | 289 | 27 | 9.34 |
| 335 | 1 | 0.30 | 270 | 30 | 11.11 |
| 146 | 1 | 0.68 | 156 | 36 | 23.08 |
| 241 | 2 | 0.83 | 236 | 57 | 24.15 |
| 338 | 2 | 0.59 | 298 | 35 | 11.74 |
| 305 | 3 | 0.98 | 404 | 37 | 9.16 |
| 334 | 3 | 0.90 | 237 | 32 | 13.50 |
| 234 | 36 | 15.38 | |||
| Total: 2037 | 0.70 (±0.23)a | 14.68 (±5.89)a | |||
a Mean ± S.D. as in Fig. 6C.
Thus, APOBEC2 deficiency does yield an evident myopathy, but it is not especially severe, and unlike the reduction in muscle mass and fiber-type switch, the myopathy develops only in older mice. Interestingly, whereas APOBEC2-deficient mice already show a marked reduction in body mass without evident myopathy at the time of weaning, a similar early body mass reduction is not as evident in mdx mice despite the fact that mdx mice go on to develop much more severe pathology. This presumably reflects a compensatory hypertrophy that develops in the mdx mice. Thus, mdx mice that are also deficient in APOBEC2 exhibit the early body mass reduction characteristic of APOBEC2 deficiency along with the severe pathology attributable to the mdx mutation (supplemental Table S1).
APOBEC2 Is a Free Tetramer with No Evidence of High Affinity for RNA
The molecular function of APOBEC2 is not known, but based on sequence homology to other members of the AID/APOBEC family, it has been proposed to be a nucleic acid-editing enzyme. The AID and APOBEC3 proteins function physiologically as DNA editors, and their DNA deaminase activity can be sensitively monitored in a bacterial mutation assay (20, 21). Previous studies have failed to identify any DNA mutator or deaminase activity associated with APOBEC2 (9, 21); it is unlikely therefore to be a DNA-editing enzyme. Testing for a possible RNA-editing activity is more difficult, especially in the absence of any informed speculation concerning the possible target RNA. However, all RNA-editing enzymes identified to date either bind RNA or associate with an RNA-binding partner; we therefore asked whether APOBEC2 shares either of these attributes.
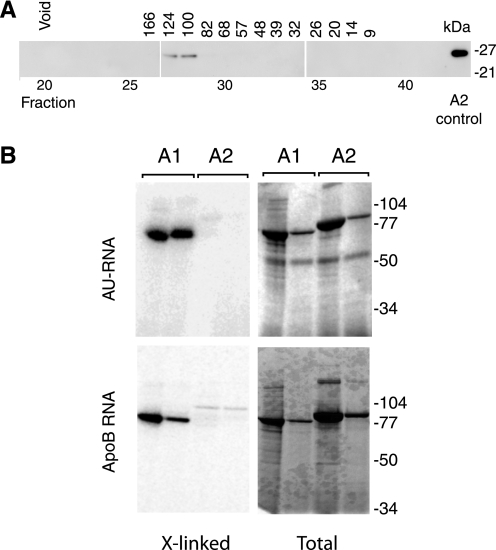
Analysis of APOBEC2 that had been modified by the addition of a One-STrEP peptide tag (13) and pulled down from transfected mouse C2C12 myoblast cells did not reveal any high stoichiometry-associated protein: the most abundantly co-purified protein (present at 5% of the APOBEC2 band) was revealed by mass spectrometric analysis to be actin, which was also found in the pulldown from the untransfected control (data not shown). Western blot analysis of extracts prepared from mouse gastrocnemius muscle revealed that APOBEC2 was present in the soluble fraction; gel filtration of the extract on a calibrated fast protein liquid chromatography column yielded a single peak of APOBEC2 protein, eluting with a molecular mass of 100–124 kDa (Fig. 8A). Extensive pretreatment with RNase A did not affect the APOBEC2 gel filtration profile. Thus, it appears most likely that APOBEC2 is present in muscle as a soluble homotetramer without any tightly high stoichiometry-associated protein. This is consistent with the fact that N-terminally truncated recombinant APOBEC2 purified from E. coli crystallizes as a homotetramer (6).
FIGURE 8.
APOBEC2 is a soluble homotetramer showing little evidence of RNA binding. A, APOBEC2 from gastrocnemius muscle extracts was eluted from a calibrated Sephadex gel filtration column and detected by Western blotting following SDS-PAGE. Preincubation of the extract with RNase did not affect the APOBEC2 elution profile (not shown). B, MBP-APOBEC1 (A1) and MBP-APOBEC2 (A2) were compared for binding to AU repeat RNA and apolipoprotein B targets. The MBP fusion proteins produced from E. coli were incubated at two different concentration (differing 10-fold) with 32P-labeled RNA. After UV cross-linking and digestion with RNase, samples were subjected to SDS-PAGE, and proteins were identified by staining (Total), whereas cross-linked RNA (X-linked) was detected by autoradiography.
We were interested in determining whether APOBEC2, like APOBEC1 and APOBEC3 family members (14, 22), could bind to RNA. Recombinant MBP-APOBEC1 and MBP-APOBEC2 were compared in their ability to bind radiolabeled oligonucleotide RNA substrates (both the minimal APOBEC1-binding sequence of apoB RNA and AU repeat RNA) in UV cross-linking experiments. Although strong cross-linking of both the AU repeat sequence and the apoB sequence could be detected with MBP-APOBEC1, no such strong binding could be detected with MBP-APOBEC2 (Fig. 8B).
DISCUSSION
The APOBEC2 gene is found in all vertebrates so far investigated, with its expression being largely restricted to striated muscle. Here, we have shown that, although found in all skeletal muscle types and myofibers examined, high levels of APOBEC2 are especially associated with slow muscle and, within soleus muscle, with slow as opposed to fast muscle myofibers. Curiously, however, we found that a deficiency in APOBEC2 actually results in an increase (rather than a decrease) in the proportion of slow fibers in soleus muscle, an observation that is consistent with the finding that denervation (which causes a fast-to-slow myosin HC shift) is also accompanied by a reduction in APOBEC2 expression. These results suggest that, although APOBEC2 is likely to largely function in slow muscle (because that is where it is predominantly expressed), it cannot be essential for the specification of slow fibers (because the proportion of slow fibers is actually increased in the soleus muscle of APOBEC2-deficient mice).
An obvious conundrum presented by our findings is why, if APOBEC2 is largely expressed in slow muscle, should APOBEC2 deficiency then result in a fast-to-slow fiber-type shift in soleus muscle. Several possible explanations can be envisioned. One is that APOBEC2 deficiency leads to an inadequacy/defect in slow muscle and that the fiber-type shift is then triggered to achieve some form of compensation. Another possibility is that APOBEC2 is somehow implicated in the turnover of slow myofibers. Indeed, skeletal muscle mass is determined by dynamic balance of synthesis and degradation of muscle proteins, with muscle mass, fiber size, and fiber composition being normally regulated in response to changes in physical stimuli, nutrition, and pathological conditions (19, 23).
Although reduced muscle mass and a fiber-type switch are already evident in young mice, histological evidence of myopathy (an increased proportion of newly generated fibers as judged by fiber size and the presence of centrally located nuclei) only becomes evident in older mice. Interestingly, others have noted increased APOBEC2 abundance in the muscles of aging rats (24). Maybe this increase in APOBEC2 expression with age could reflect other changes in muscle characteristics such as turnover of muscle proteins or shift in fiber type, although this remains to be determined.
Based on it structural homology to APOBEC1, APOBEC2 has widely been presumed to be an RNA-editing enzyme. Similar considerations led others to assume that the lymphocyte-specific AID protein (another member of the AID/APOBEC family) would also prove to be an RNA-editing enzyme (25). However, the discovery that AID instead functions as a DNA deaminase (26) contributed to more recent speculation that APOBEC2 might act on DNA (27). However, we remain somewhat skeptical as to whether APOBEC2 functions as a polynucleotide deaminase at all. We have previously demonstrated that a cytidine deaminase activity that was previously assigned to recombinant APOBEC2 is most likely attributable to bacterial deaminases contaminating the recombinant APOBEC2 preparation (9). Not only did we find no mutator activity associated with APOBEC2 (such activity might be expected for a DNA deaminase), we also found no evidence of an affinity for RNA or high stoichiometry association with a partner, either of which is usually associated with RNA-editing enzymes.
The proposition that APOBEC2 does not function as a polynucleotide deaminase is consistent with the APOBEC2 amino acid sequence because it lacks conserved regions that have been shown to be important for deaminase activity in other APOBEC family members, e.g. the tryptophan residue adjacent to the zinc coordination motif (28, 29) and the cluster of basic amino acid residues in the N-terminal portion, which are likely to be implicated in nucleic acid binding (29–31). Furthermore, comparison of the fragment crystal structures of APOBEC2 and APOBEC3G highlights differences surrounding the active site that may account for differences in substrate interactions (29).
Thus, APOBEC2 may deaminate a substrate other than a polynucleotide, or the possibility even remains that it might fulfill some structural role without displaying any deaminase activity at all. Such a structural role has indeed been proposed for some of the individual APOBEC domains found within multidomain APOBEC3 deaminases (reviewed in Ref. 3). However, if APOBEC2 acts as a deaminase for a non-polynucleotide substrate, identification of its specific physiological function is most likely to come from the identification of such a substrate.
Supplementary Material
Acknowledgments
We are grateful to Sara Pruzina for help with analysis of mice, Marie Mikl and Meng Wang for helpful discussions, and Mason Lu for provision of anti-APOBEC2 mAbs (supported by Medical Research Council U.1051.03.008).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
M. Lu and M. S. Neuberger, unpublished data.
- AID
- activation-induced deaminase
- APOBEC
- apolipoprotein B RNA-editing enzyme, catalytic polypeptide-like
- FBS
- fetal bovine serum
- HC
- heavy chain
- mAb
- monoclonal antibody
- MBP
- maltose-binding protein
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- EDL
- extensorum digitorum longus.
REFERENCES
- 1.Conticello S. G., Thomas C. J., Petersen-Mahrt S. K., Neuberger M. S. (2005) Mol. Biol. Evol. 22, 367–377 [DOI] [PubMed] [Google Scholar]
- 2.Navaratnam N., Sarwar R. (2006) Int. J. Hematol. 83, 195–200 [DOI] [PubMed] [Google Scholar]
- 3.Conticello S. G. (2008) Genome Biol. 9, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liao W., Hong S. H., Chan B. H., Rudolph F. B., Clark S. C., Chan L. (1999) Biochem. Biophys. Res. Commun. 260, 398–404 [DOI] [PubMed] [Google Scholar]
- 5.Anant S., Mukhopadhyay D., Sankaranand V., Kennedy S., Henderson J. O., Davidson N. O. (2001) Am. J. Physiol. Cell Physiol. 281, C1904–C1916 [DOI] [PubMed] [Google Scholar]
- 6.Prochnow C., Bransteitter R., Klein M. G., Goodman M. F., Chen X. S. (2007) Nature 445, 447–451 [DOI] [PubMed] [Google Scholar]
- 7.Berchtold M. W., Brinkmeier H., Müntener M. (2000) Physiol. Rev. 80, 1215–1265 [DOI] [PubMed] [Google Scholar]
- 8.Takekura H., Yoshioka T. (1987) J. Muscle Res. Cell Motil. 8, 342–348 [DOI] [PubMed] [Google Scholar]
- 9.Mikl M. C., Watt I. N., Lu M., Reik W., Davies S. L., Neuberger M. S., Rada C. (2005) Mol. Cell. Biol. 25, 7270–7277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yaffe D., Saxel O. (1977) Differentiation 7, 159–166 [DOI] [PubMed] [Google Scholar]
- 11.Rando T. A., Blau H. M. (1994) J. Cell Biol. 125, 1275–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizunoya W., Wakamatsu J., Tatsumi R., Ikeuchi Y. (2008) Anal. Biochem. 377, 111–113 [DOI] [PubMed] [Google Scholar]
- 13.Junttila M. R., Saarinen S., Schmidt T., Kast J., Westermarck J. (2005) Proteomics 5, 1199–1203 [DOI] [PubMed] [Google Scholar]
- 14.Navaratnam N., Shah R., Patel D., Fay V., Scott J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spencer M. J., Mellgren R. L. (2002) Hum. Mol. Genet. 11, 2645–2655 [DOI] [PubMed] [Google Scholar]
- 16.Nwoye L., Mommaerts W. F., Simpson D. R., Seraydarian K., Marusich M. (1982) Am. J. Physiol. 242, 401–408 [DOI] [PubMed] [Google Scholar]
- 17.Midrio M., Danieli-Betto D., Megighian A., Velussi C., Catani C., Carraro U. (1992) Pflugers Arch. 420, 446–450 [DOI] [PubMed] [Google Scholar]
- 18.Agbulut O., Vignaud A., Hourde C., Mouisel E., Fougerousse F., Butler-Browne G. S., Ferry A. (2009) Am. J. Physiol. Cell Physiol. 296, C205–C214 [DOI] [PubMed] [Google Scholar]
- 19.Sato Y., Shimizu M., Mizunoya W., Wariishi H., Tatsumi R., Buchman V. L., Ikeuchi Y. (2009) Biosci. Biotechnol. Biochem. 73, 1749–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petersen-Mahrt S. K., Harris R. S., Neuberger M. S. (2002) Nature 418, 99–103 [DOI] [PubMed] [Google Scholar]
- 21.Harris R. S., Petersen-Mahrt S. K., Neuberger M. S. (2002) Mol. Cell 10, 1247–1253 [DOI] [PubMed] [Google Scholar]
- 22.Jarmuz A., Chester A., Bayliss J., Gisbourne J., Dunham I., Scott J., Navaratnam N. (2002) Genomics 79, 285–296 [DOI] [PubMed] [Google Scholar]
- 23.Arany Z. (2008) Curr. Opin. Genet. Dev. 18, 426–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piec I., Listrat A., Alliot J., Chambon C., Taylor R. G., Bechet D. (2005) FASEB J. 19, 1143–1145 [DOI] [PubMed] [Google Scholar]
- 25.Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. (2000) Cell 102, 553–563 [DOI] [PubMed] [Google Scholar]
- 26.Neuberger M. S., Harris R. S., Di Noia J., Petersen-Mahrt S. K. (2003) Trends Biochem. Sci. 28, 305–312 [DOI] [PubMed] [Google Scholar]
- 27.Rai K., Huggins I. J., James S. R., Karpf A. R., Jones D. A., Cairns B. R. (2008) Cell 135, 1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen K. M., Martemyanova N., Lu Y., Shindo K., Matsuo H., Harris R. S. (2007) FEBS Lett. 581, 4761–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holden L. G., Prochnow C., Chang Y. P., Bransteitter R., Chelico L., Sen U., Stevens R. C., Goodman M. F., Chen X. S. (2008) Nature 456, 121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bransteitter R., Pham P., Scharff M. D., Goodman M. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4102–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen K. M, Harjes E., Gross P. J., Fahmy A., Lu Y., Shindo K., Harris R. S., Matsuo H. (2008) Nature 452, 116–119 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.