Abstract
Cold is a limiting environmental factor that adversely affects plant growth and productivity. Calcium/calmodulin-mediated signaling is believed to play a pivotal role in plant response to cold stress, but its exact role is not clearly understood. Here, we report that CRLK1, a novel calcium/calmodulin-regulated receptor-like kinase, is crucial for cold tolerance in plants. CRLK1 has two calmodulin-binding sites with different affinities as follows: one located at residues 369–390 with a Kd of 25 nm, and the other located at residues 28–112 with a Kd of 160 nm. Calcium/calmodulin stimulated the kinase activity, but the addition of chlorpromazine, a calmodulin antagonist, blocked its stimulation. CRLK1 is mainly localized in the plasma membrane, and its expression is stimulated by cold and hydrogen peroxide treatments. Under normal growth conditions, there is no noticeable phenotypic difference between wild-type and crlk1 knock-out mutant plants. However, as compared with wild-type plants, the crlk1 knock-out mutants exhibited an increased sensitivity to chilling and freezing temperatures. Northern analysis showed that the induction of cold-responsive genes, including CBF1, RD29A, COR15a, and KIN1 in crlk1 mutants, is delayed as compared with wild-type plants. These results indicate that CRLK1 is a positive regulator of cold tolerance in plants. Furthermore, our results suggest that CRLK1 plays a role in bridging calcium/calmodulin signaling and cold signaling.
Keywords: Arabidopsis, Calcium, Calcium/Calmodulin, Calcium/Cellular Regulation, Phosphorylation, Phosphorylation/Kinases/Serine-Threonine, Signal Transduction/Calcium
Introduction
Cold, a major environmental factor, affects plant growth and reduces productivity (1). It has been well documented that numerous cold-induced physiological and biochemical responses exist, including accumulation of reactive oxygen species, changes in membrane lipid composition, and the accumulation of compatible osmolytes (1, 2). Cold acclimation also induces the expression of certain COR genes such as RD29A, COR15a, and KIN1, which function to stabilize membranes against freezing-induced injury (1). The CBF/DREB1- or CRT/DRE-binding factors, a family of closely related AP2/ERF transcription factors, play a major role in controlling the COR gene expression (3), and CBFs themselves are activated by cold through ICE1 (4, 5).
Calcium, a universal second messenger, acts as a mediator of stimulus-response coupling in the regulation of plant growth, development, and responses to environmental stimuli (6–10). A variety of stress signals, such as cold, high salt, and hydrogen peroxide, triggers rapid increases in Ca2+ levels in plant cells (11–13). Intracellular free Ca2+ changes are recognized by Ca2+ receptors such as calcium-dependent protein kinases (10), calcineurin-B-like proteins (14), and calmodulin (CaM).4 CaM is a ubiquitous Ca2+-binding protein in animals and plants (15, 16). The presence of CaM-dependent protein phosphorylation in plants has been reported (17–19). It has been suggested that Ca2+/CaM-mediated signaling via the phosphorylation cascade is crucial for regulating COR gene expression and cold acclimation (20–22). However, little is known about the identity of the CaM-regulated protein kinase(s) involved in cold signal transduction.
In the Arabidopsis genome, there are at least 600 receptor-like kinases (RLKs) representing nearly 2.5% of the annotated protein-coding genes. The RLKs resemble animal receptor kinases in that they have the general structure of an extracellular domain, a single transmembrane sequence, and an intracellular kinase domain. The function of most of the plant RLKs is not well understood (23). It has been reported that CaM interacts with Brassica oleracea S locus receptor kinase (24) and Arabidopsis AtCaMRLK1 (25), but CaM has no effect on the activity of these kinases. Earlier, we characterized a CaM up-regulated cytoplasmic receptor-like kinase, but its function is not yet known (18). Here, we report the isolation of a plant-specific, plasma membrane-anchored Ca2+/CaM-regulated RLK (CRLK1) from an Arabidopsis cold-stressed cDNA library. Characterization of the crlk1 null mutants and complementation lines suggests that CRLK1 confers freezing tolerance by acting as a positive regulator of COR gene expressions.
EXPERIMENTAL PROCEDURES
Plant Materials
Arabidopsis thaliana ecotype Columbia and mutant line were grown in a 1:1 mixture of soil mix and vermiculite under 14-h light/10-h dark at 22 °C with light intensity of 220 μmol quanta m−2 s−1 if not otherwise specified.
Petri Dish Chilling and Freezing Tolerance Assays
Chilling sensitivity was tested as described previously (4). After 2 days of stratification at 4 °C, the seeds were germinated at 22 °C for 7 days on MS solid medium with 1% sucrose. Chilling stress was imposed by incubating the seedlings at 6 ± 1 °C with 50 ± 2 μmol quanta m−2 s−1 light. To measure the chlorophyll content, 2-week-old plants at 20–22 °C under 220 μmol quanta m−2 s−1 light were transferred to a growth chamber at 4 ± 1 °C at the same light intensity. Chlorophyll extraction was performed as described previously (26). Freezing tolerance was essentially assayed as described previously (27). Seeds were sterilized and scattered on Petri dishes containing Gamborg basal salts solidified with 0.9% agar. The plates were kept at 4 °C for 2 days and moved to 22 °C under continuous light. Ten days after germination, the plates with seedlings were transferred to a chamber at −1 °C without light for 16 h. Then the chamber was set to cool by decreasing the temperature 1 °C per h. The plates were removed at desired temperatures, thawed at 4 °C for 12 h in the dark, and then returned to the original growth conditions. After 2 days, the surviving plants were counted. Three repeats for each line were tested for the freezing tolerance and used for statistical analysis.
Cloning of CRLK1
The cold-treated Arabidopsis seedling cDNA expression library was screened using 35S-CaM (potato PCM1 and PCM6) as described previously (18). The positive cDNA clones were sequenced on both strands, and CRLK1 was one of the positive clones. The full length of CRLK1 was cloned with the partial cDNA as a probe.
Construction of DNA Templates Coding CRLK1 Proteins
The templates coding for various CRLK1 mutants were produced by PCR amplification from their cDNAs with the gene-specific primers and cloned downstream of the His6 tag into the BamHI and SalI sites in pET-32 (Novagen). The primers used are as follows: ggaggatccttctgctttaggtatcatag/gtcgtcgacttacaaaatcacgtcttcag (M28–440); ggaggatccttctgctttaggtatcatag/gtcgtcgacattcttctgccgctttc (M28–392); ggaggatccttctgctttaggtatcatag/gtcgtcgaccatgttaggacgttttctag (M28–369); ggaggatccttctgctttaggtatcatag/gtcgtcgaccccatcatgaagatactctag (M28–228); ggaggatccacgtgtaatttcacgactttg/gtcgtcgaccatgttaggacgttttctag (M112–369); and ggaggatccatgagggacattgttcagg/gtcgtcgacttacaaaatcacgtcttcag (M28–440). The underlined letters are the BamHI and SalI sites. All constructs were verified by sequencing. The constructs were transformed into E. coli strain BL21(DE3) pLysS and purified by nickel-nitrilotriacetic acid (Novagen).
CaM Binding Assay
CaM binding overlay assay was carried out as described previously (18) using 35S-labeled potato CaM PCM1. The synthetic peptides were prepared in the Laboratory of Bioanalysis and Biotechnology, Washington State University. Gel mobility assay was carried out as described previously (18) using bovine CaM (Sigma). The purified recombinant M369-440 and M28–228 were used for CaM-binding affinity assay as described previously (18).
Transient Transformation Assays with GFP Fusion Constructs
The full-length CRKL1 or M28–440 was translationally fused at the C terminus with GFP in the BamHI site of psmGFP. Plasmid DNA was introduced into the onion epidermal cells by particle bombardment. The onion peels were examined using argon laser (488 nm) for green fluorescence. Plasmolysis of the onion epidermal cells was induced upon addition of 0.8 m mannitol.
RNA Isolation, Northern Analysis, and Reverse Transcription-PCR
Total RNA was isolated from frozen tissue following the manufacturer's instructions (TRIzol Reagent, Invitrogen). Northern analysis was performed using the full-length cDNA of CRLK1, RD29A (At5g52310), COR15a (At2g42540), KIN1 (At5g15960) and CBF1 (At4g25490) and 18 S rDNA as probes.
Antibody Preparation, Plant Protein Isolation, and Western Analysis
The peptide corresponding to amino acids 385–400 of CRLK1 was conjugated with keyhole limpet hemocyanin. Antibody was raised in rabbits by GenMed (San Francisco, CA). The soluble and membrane proteins were fractionated as described previously (18). Plasma membrane fraction was prepared from microsomes using two-phase partitioning experiments (28). Specifically, the microsomes were separated in a buffer containing 6.2% (w/w) dextran T500 (GE Healthcare), 6.2% (w/w) polyethylene glycol (Sigma), 11% (w/v) sucrose, 5 mm potassium phosphate buffer, pH 7.8, and 1 and 5 mm KCl in the first and second phase system, respectively. Membrane proteins in the upper and lower phase were pelleted by ultracentrifugation. The pellets were resuspended in resuspension medium (10% (v/v) glycerol, 0.5 mm dithiothreitol, 1 mm MOPS-KOH, pH 7, and 5 μm leupeptin), immediately frozen, and kept at −80 °C until used. The purity of the plasma membrane fraction and other membrane fractions was tested by Western blotting using antibody against plasma membrane marker H+-ATPase (29) and tonoplast marker V-ATPase (Agrisera). Protein content was determined by Bradford reagent (Bio-Rad). Western blotting was carried out following the procedure suggested by the manufacturer (Roche Applied Science).
Immunoprecipitations and Protein Phosphorylation Assays
Immunoprecipitation of CRLK1 from wild-type or crlk1 mutant plants was performed using the anti-CRLK1 antibody as described previously (6, 18). Specifically, 1 g of rosette leaves was harvested, cut into ∼5-mm2 pieces, and soaked in ice-cold lysis buffer (10 mm HEPES-KOH, pH 7.4, 12.5% sucrose, 10 mm potassium acetate, 3 mm MgCl2, 1 mm CaCl2) supplemented with 0.01% detergent Silwet-77 and 1% formaldehyde for 30 min at 4 °C and subjected to two cycles of 5 min of vacuum treatment. The leaves were then washed two times for 20 min with 50 ml of lysis buffer supplemented with 0.3 m glycine at 4 °C and stored at −80 °C until further use. The total proteins or proteins from different fractions were prepared in lysis buffer supplemented with NaN3 and 1× Roche Applied Science complete protease inhibitors. Immunoprecipitation was carried out using anti-CRLK1 antibody. The immunoprecipitants were fractionated in SDS-PAGE and subjected to Western analysis. Calmodulin was detected using anti-calmodulin antibody. Phosphorylation of the recombinant proteins and phosphoamino acid analysis were carried out as described previously (30). One μm purified CRLK1 and mutant proteins were used in the autophosphorylation and substrate phosphorylation assays. All the CPZ experiments were done in the presence of Ca2+/CaM, unless stated otherwise. The experiments were repeated at least three times.
Isolation of T-DNA Insertion Knock-out Plants
The crlk1 insertion mutant was identified from SalK_016240 by PCR using two gene-specific primer pairs, TGATGGGGTAAGCTGCTTGCTA/CCCAAAAGAATTGAAAAACAAAATCA and a T-DNA left border primer (31). The amplified fragments were sequenced, and the T-DNA insertion site was determined. The mutant plants were further verified by Northern analysis using full-length cDNA as a probe and Western blotting analysis.
Genetic Complementation Analysis
Full length and deletions of the CRLK1 cDNA coding region were amplified by reverse transcription-PCR with gene-specific primers and fused with 35S promoter in pCAMBIA 1302 (Cambia, Canberra, Australia). These constructs were transformed into the crlk1 mutants by the floral dip method. Transgenic lines were selected on MS medium containing hygromycin. Homozygous transgenic lines were isolated using PCR. The expression levels of transgenes were studied using reverse transcription-PCR using the same primer pair (GCTGAGCTGGGATTTGAGAG/GAGAGGCCAAAGTCAGCAAC). A representative plant with the comparable transgene expression level in each complementation group was selected for further studies.
RESULTS
CRLK1 Is a CaM-binding Protein
Using 35S-labeled CaM screening, we isolated CRLK1 (At5g54590.2) from a cold-stressed Arabidopsis library. It encodes a 440-amino acid protein with a predicted molecular mass of ∼49 kDa. Bioinformatic analysis indicated that CRLK1 contained all 11 subdomains of the catalytic domain of the kinase, a transmembrane domain but no extracellular domain, suggesting that it is a membrane-anchored protein kinase (supplemental Fig. S1). CRLK1 has high homology with another Arabidopsis gene (At5g15730; 50% identity) and other plant RLKs such as the rice gene Os08g04420 (74% similarity, 66.9% identity) and tobacco NPK15 (66.5% similarity, 55.9% identity).
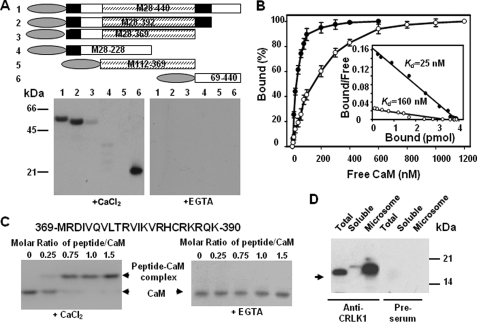
To verify that CRLK1 is a CaM-binding protein, a bacterially expressed mutant construct (M28-440) lacking the putative transmembrane domain was tested in a CaM binding assay. As shown in Fig. 1A, CaM bound CRLK1 in the presence of CaCl2 but not in the presence of EGTA, a Ca2+ chelator, suggesting that CaM binding to CRLK1 is Ca2+-dependent. To map the CaM-binding site(s), several deletion mutants lacking different parts of the CRLK (Fig. 1A) were tested for CaM binding. These analyses showed that CRLK1 has two Ca2+-dependent CaM-binding sites, one located in 369–390 amino acids, just downstream of the catalytic domain, and another in the N-terminal 28–112 amino acids (Fig. 1A). The dissociation constant (Kd) of CaM for the C- and N-terminal CaM-binding domains was ∼25 and 160 nm, respectively (Fig. 1B). Furthermore, a synthetic peptide corresponding to 369–390 amino acids was used for gel mobility shift assay. This peptide bound CaM in the presence of CaCl2 but not in the presence of EGTA (Fig. 1C). Similarly, a 20-mer peptide corresponding to the N-terminal 30–49 amino acids was also able to bind CaM in a Ca2+-dependent manner (supplemental Fig. S2). However, about four times peptide:CaM molar ratio were needed for CaM to fully form the complex with the peptide, suggesting the low CaM affinity to this peptide. Furthermore, co-immunoprecipitation was carried out to pull down the proteins prepared from cold-treated wild-type plants. The interaction was fixed using a prior in vivo cross-linking strategy (6). CaM was detected in the immunoprecipitant by the anti-CaM antibody. However, there is no CaM detected in the preimmune precipitant. These results support that CaM interacts with CRLK1 in vivo (Fig. 1D).
FIGURE 1.
CRLK1 is a Ca2+-dependent calmodulin-binding protein. A, CaM binds to CRLK1. CRLK1 deletion mutants (upper, domain structure is shown in the longest mutant, CaMBD is in a filled box, and kinase domain is in a hatched box) translationally fused with a His tag (oval box) were produced in E. coli. Purified recombinant proteins (2 μg) were subjected to SDS-PAGE and transferred onto a nitrocellulose membrane. The membrane was incubated with 50 nm 35S-PCM1 in a buffer containing either 2 mm EGTA or 0.1 mm CaCl2 (lower). B, saturation curve of 35S-CaM binding to purified M369-440 (solid circle) and M28–228 (open circle). Four pmol of recombinant proteins were spotted on a membrane and incubated with different amounts of 35S-PCM1. The amount of bound CaM at each point was represented as a percentage of the maximal binding. The average of three independent experiments was presented. The inset shows Scatchard plots of the binding data. C, gel mobility shift assay showing CaM binding to a synthetic peptide (amino acids 369–390 in CRLK1). D, CaM is co-immunoprecipitated with CRLK1 in planta. The total proteins or proteins from different fractions were prepared from wild-type plants treated at 4 °C for 4 h. Immunoprecipitation was carried out using anti-CRLK1 antibody. The immunoprecipitants were fractionated in SDS-PAGE and subjected to Western analysis. Calmodulin was detected using anti-calmodulin antibody.
Ca2+/CaM Regulates the Kinase Activity of CRLK1
To investigate the kinase activity of CRLK1, the recombinant deletion mutants were purified to homogeneity. Three proteins, M28–440, M28–392, and M28–369, were shown to autophosphorylate in the presence of 10 mm Mg2+ but not in the presence of Ca2+ alone (data not shown). To further study the role of Ca2+/CaM in regulating the kinase activity, all three proteins were subjected to autophosphorylation in the presence of CaM. The kinase activity of both M28–440 and M28–392 was enhanced with the increasing concentrations of bovine CaM. Fig. 2A shows that Ca2+/CaM stimulates the kinase activity of M28–392 and calcium alone, or CaM alone did not show any measurable effect on the kinase activity of M28–392 (supplemental Fig. S2B). Increasing concentrations of CaM antagonist, CPZ, abolished the Ca2+/CaM-stimulated kinase activity (Fig. 2A). CPZ alone did not show obvious effect on kinase activity of M28–292 (supplemental Fig. S2B). On the other hand, Ca2+/CaM showed no significant effect on kinase activity of M28–369, which lacks the C-terminal CaM-binding site, under both low and high concentrations of CaM (Fig. 2, A and B). We further studied the effects of two plant CaM isoforms, PCM1 and PCM6, on the kinase activity. PCM1 enhanced the kinase activity of CRLK1 like bovine CaM; however, PCM6 did not show such a stimulation (Fig. 2C), suggesting that the kinase activity of CRLK1 is regulated by a specific CaM isoform(s). When M28–392 was used to phosphorylate casein, in the presence of increasing concentrations of Ca2+/CaM, there was an increase in substrate phosphorylation. Addition of CPZ showed inhibition of Ca2+/CaM-stimulated activity. However, M28–389 did not show the significant substrate phosphorylation changes by increasing CaM concentration (Fig. 2, D and E). These results suggest that the CaM-binding site (residues 369–390) is responsible for the Ca2+/CaM stimulation of kinase activity, and the CaM-binding site in the N-terminal region has no pronounced effect on kinase activity. Phosphoamino acid analysis revealed that CRLK1 autophosphorylated at both serine and threonine residues (Fig. 2F), suggesting that CRLK1 is a serine/threonine kinase.
FIGURE 2.
Ca2+/calmodulin regulates the kinase activity of CRLK1. A, kinase activity of M28–392 and M28–369 at different concentrations of CaM and CaM antagonist CPZ. Inhibition of Ca2+/CaM-stimulated CRLK1 activity by CPZ was measured in the presence of 200 nm Ca2+/CaM. B, CaM does not stimulate the kinase activity of M28–369. C, plant CaM isoforms differentially regulate CRLK activity. Bovine serum albumin (BSA) was used as a negative control. D and E, substrate phosphorylaton for M28–392 and M28–369 at different concentrations of CaM and CaM antagonist CPZ. F, phosphoamino acid analysis showing that M28–440 autophosphorylates at serine and threonine residues.
CRLK1 Is Primarily a Plasma Membrane-associated Protein
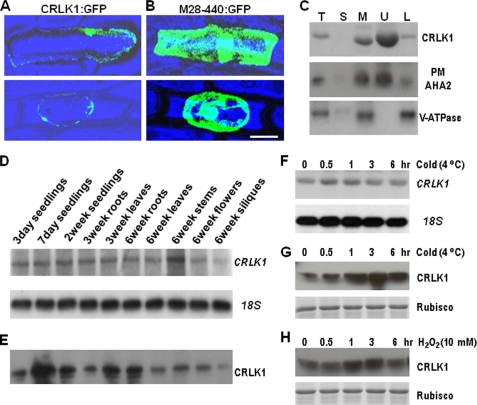
The existence of a transmembrane domain in the CRLK1 N terminus indicates it is a membrane protein. To verify this, full-length CRLK1:GFP fusion and M28–440:GFP fusion, which lacked the putative transmembrane domain, were transiently expressed in onion inner epidermal cells. The CRLK1:GFP predominantly localized on the periphery of the cell (Fig. 3A, upper panel). However, M28–440:GFP was detected throughout the cytoplasm and nucleus (Fig. 3B, upper panel). To further differentiate the location of CRLK1, the cells were plasmolyzed using 0.8 m mannitol. The green fluorescence for CRLK1:GFP was not in the cell wall but was mainly found in the periphery of the protoplast (Fig. 3A, lower panel). In addition, microsome preparation from wild-type (WT) plants was subjected to two-phase separation (28). The Western blot analysis using anti-CRLK1 antibody revealed that CRLK1 is located in the microsomes but not in the soluble fraction (Fig. 3C), confirming that CRLK1 is a membrane-associated protein. Furthermore, after the two-phase separation, CRLK1 was found predominantly in the plasma membrane-enriched upper fraction (Fig. 3C), a distribution pattern similar to that of a plasma membrane marker, proton-ATPase AHA2 (29, 32, 33). No CRLK1 enrichment was observed in the lower fraction where non-plasma membrane proteins such as tonoplast V-ATPase were enriched (Fig. 3C). Thus CRLK1 is a membrane protein predominantly associated with the plasma membrane.
FIGURE 3.
Expression of CRLK1. A and B, subcellular distribution of green fluorescence in onion epidermis. The full-length CRLK1 and M28–440 were inserted between cauliflower mosaic virus 35S promoter and GFP in psmGFP, respectively. The confocal images were taken after 24 h of particle bombardment. The plasmolysis was observed after treating with 0.8 m mannitol (lower panel). Bar, 50 μm. C, Western analysis showing that CRLK1 is a plasma membrane (PM) protein. 20 μg of total protein or soluble fraction and 5 μg of microsomes or plasma membrane-enriched upper phase or other membrane-enriched lower phase fractions from the wild-type plants were loaded in each lane, respectively. The immunoblots were used against anti-CRLK1 antibody, anti-plasma membrane AHA2 antibody, and anti-tonoplast V-ATPase. T, total proteins; S, soluble fraction; M, microsomal fraction; U, upper phase; L, lower phase. D, Northern analysis showing CRLK1 expression pattern. The full length of CRLK1 was used as a probe. 18 S RNA shows equal loading of total RNA. E, differential expression of CRLK1 protein in tissues by Western blotting using the anti-CRLK1 antibody. The microsomal fraction (5 μg) was used for Western analysis. F and G, expression of CRLK1 mRNA and protein treated by cold stress. H, expression of CRLK1 protein is stimulated by H2O2. Three-week-old plants were subjected to cold treatment (4 °C) or sprayed with 10 mm H2O2 and harvested after various periods as indicated. The total protein (20 μg) was used for Western analysis. The Coomassie-stained ribulose-bisphosphate carboxylase/oxygenase (Rubisco) bands show equal loading of protein samples.
CRLK1 Is a Positive Regulator of Cold Tolerance
The expression pattern of CRLK1 was further investigated by Northern analysis. One band around 2 kb showed a similar expression pattern in all tested tissues except in siliques (Fig. 3D). However, CRLK1 protein was differentially expressed in these tissues, as revealed by Western analysis (Fig. 3E). Highest expression levels of the CRLK1 protein were detected in 7-day-old seedlings. The CRLK1 protein was also detected in roots, leaves, flowers, and siliques. Because CRLK1 was isolated from a cold-treated cDNA library, the effect of low temperature on CRLK1 expression was further studied. Although there was no major change in mRNA level (Fig. 3F), the protein level of CRLK1 rapidly increased after cold treatment (Fig. 3G). After 30 min of cold treatment, the CRLK1 protein level began to increase, reached a maximum level at 3 h, then started to decrease after 6 h. Because cold stress induces the accumulation of H2O2, we examined its effect on the expression of CRLK1. H2O2 treatment resulted in an increase in CRLK1 level (Fig. 3H), suggesting that it is involved in cold-related oxidative stress signal transduction pathway.
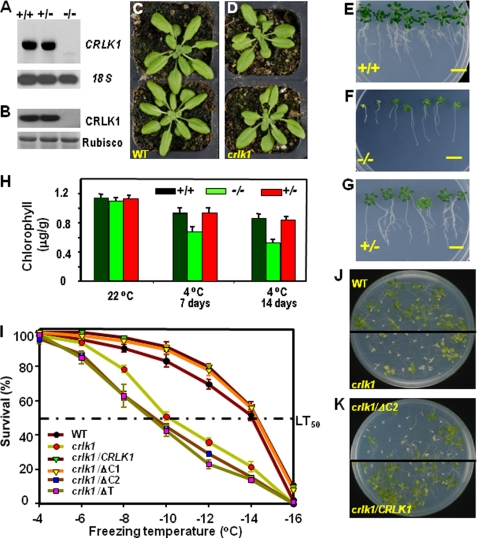
To gain insight into the functional significance of CRLK1, we isolated a crlk1 T-DNA insertion mutant. Northern analysis indicated that an ∼2-kb band corresponding to CRLK1 size was present in WT and heterozygous plants but not in homozygous plants. After a longer exposure, there was one weak band with a smaller size that appeared in homozygous plants (Fig. 4A). However, no CRLK1 proteins were detected in the crlk1 plants, whereas CRLK1 proteins were well detected in WT and heterozygous plants (Fig. 4B). Together the results indicate that crlk1 is a recessive null mutant at the protein level. The homozygous crlk1 and WT plants had no obvious phenotypic difference at 22 °C (Fig. 4, C and D). Plant response to chilling was tested at 6 °C, and chilling injury became apparent in the mutant plants after 3 weeks of cold treatment (supplemental Fig. S3D). Prolonged cold treatment up to 7 weeks produced significant phenotypic differences between the crlk1 and WT plants (Fig. 4, E and F). These differences included inhibited root growth, decreased number of lateral roots, and decreased shoot growth. In addition, the mutant leaves also showed yellowing, a sign of early senescence and the severity of chilling damage (Fig. 4, E and F). The F1 heterozygous plants obtained by backcrossing with WT plants showed similar chilling tolerance as WT (Fig. 4G). About 25% of the F2 population was sensitive to chilling stress, indicating that the crlk1 mutant has a single recessive mutation in crlk1 gene locus. The total chlorophyll content was tested to study the rate of senescence (Fig. 4H). At normal temperature, all plants had similar chlorophyll contents. The chlorophyll content in all plants decreased at 4 °C; however, it dropped rapidly in crlk1 homozygous plants, which contained around 65% chlorophyll as compared with WT and heterozygous plants treated at 4 °C for 14 days, supporting that crlk1 plants were more sensitive to the chilling treatment. Furthermore, the crlk1 plants were less freezing-tolerant than the WT plants at all freezing temperatures after cold acclimation. WT plants had a half-survival rate (LT50) at −14 °C, whereas crlk1 mutants had an LT50 at −10 °C (Fig. 4I). At −14 °C, about 50% of WT plants were still alive, whereas only 20% of the crlk1 plants survived (Fig. 4, I and J), which was comparable with hos9 and hos10 plants (26 and 28% survival rate, respectively) under the same condition. Most of the surviving crlk1 mutant seedlings also appeared yellow in color as compared with the WT. Without cold acclimation, both WT and crlk1 seedlings showed a similar LT50 of ∼5.8 °C (data not shown). This suggests that CRLK1 is required for cold acclimation.
FIGURE 4.
crlk1 knock-out mutants are sensitive to low temperature. A, expression of CRLK1 mRNA in crlk1 mutants. The total RNAs (10 μg) from wild-type (+/+), crlk1 heterozygous (+/−), and homozygous (−/−) seedlings were subjected to Northern analysis using the full-length CRLK1 as a probe. 18 S RNA shows equal loading of total RNA. B, expression of CRLK1 protein was abolished in homozygous crlk1 mutants. The total protein (20 μg) isolated from 3-week-old plants were used for Western analysis. Ribulose-bisphosphate carboxylase/oxygenase (Rubisco). C and D, crlk1 mutants had no obvious phenotypic difference as compared with wild-type plants at 22 °C. E–G, crlk1 homozygous plants (−/−) were more sensitive to chilling. The photos were taken after 7 weeks at 6 °C. Bar, 10 mm. H, chlorophyll content in WT and mutant plants. I, freezing survival rates following cold acclimation. The full-length and truncated forms of CRLK1 cDNA were inserted downstream of cauliflower mosaic virus 35S of pCAMBIA 1302. ΔC1 lacks the C-terminal 48 amino acids beyond the CaM-binding region; ΔC2 lacks the C-terminal 71 amino acids including the CaM-binding domain; ΔT lacks the N-terminal transmembrane domain. The average survival rates from three repeats of WT and complementation lines were used. J, crlk1 mutants are less tolerant to freezing than WT plants. Ten-day-old seedlings grown at 22 °C were incubated for 4 days at 4 °C before freezing treatment at −14 °C. The picture was taken 12 days after the freezing treatment. K, freezing tolerance test showing that complementation with truncated CRLK1 lacking the C-terminal CaM-binding site (crlk1/ΔC2) cannot rescue the freezing sensitivity.
We further prepared constructs of 35S::CRLK1 and ectopically expressed CRLK1 in crlk1 mutants (supplemental Fig. S3A). Four independently complemented plants (crlk1/CRLK1) showed a higher level of both CRLK1 mRNA and protein as compared with WT plants. There was no obvious change in CRLK1 expression in the control plants transformed with the empty vector. Under normal conditions, no significant difference in the phenotypes of the complemented lines and crlk1 plants was observed. However, after exposing to long term low temperature (6 °C), crlk1/CRLK1 plants rescued the cold sensitivity of crlk1 mutants and showed growth similar to WT plants. Furthermore, after cold acclimation, the freezing tolerance of crlk1/CRLK1 was enhanced as compared with crlk1 plants (Fig. 4, I and K). The LT50 value for crlk1/CRLK1 plants is around −14.3 °C, which was comparable with WT plants, suggesting that the level of CRLK1 in plants is correlated to freezing tolerance.
To understand the functional significance of CaM binding to CRLK1 in planta, we introduced the truncated CRLK1 (crlk1/ΔC2) lacking the C-terminal CaM-binding site into crlk1 plants. The crlk1/ΔC1, a truncated CRLK1 missing the C terminus beyond the CaM-binding region, was used as a control. Furthermore, crlk1/ΔT, a truncated CRLK1 without the N-terminal transmembrane domain, was introduced into crlk1 plants to study the function of membrane localization (supplemental Fig. S3A). Complementation plants carrying WT and mutated versions of CRLK1 with a comparable expression level of transgenes (supplemental Fig. S3B) were used for further studies. Without cold acclimation, no difference in freezing tolerance between the full-length and the deletion mutant lines was observed. However, after cold acclimation, crlk1/ΔC2 and crlk1/ΔT plants showed a higher sensitivity to freezing as compared with crlk1 plants (Fig. 4, I and K, and supplemental Fig. S3C). The LT50 value for these lines was comparable with crlk1 plants, indicating that the C-terminal CaM-binding site and N-terminal transmembrane domain are crucial for CRLK1 function in freezing tolerance.
CRLK1 Regulates COR Gene Expression
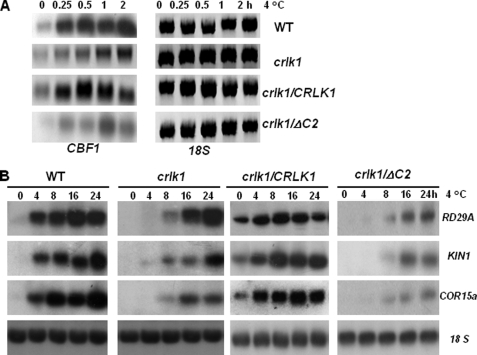
To investigate whether CRLK1 regulates cold acclimation by the CBF-dependent pathway, we studied the gene expression of CBF and COR genes RD29A, KIN1, and COR15a in the crlk1 null mutants and CRLK1 transgenic lines. The CBF family is the key cold-responsive genes controlling the expression of downstream COR genes (3). We used the full length of CBF1 cDNA as probe, which may also hybridize with CBF2 and CBF3 because CBF1, -2, and -3 share high homology in nucleotide sequences. Northern analysis revealed differences in the expression kinetics of CBF1 between WT and crlk1 plants in response to cold treatment (Fig. 5A). Significant cold-induced CBF1 expression was observed in WT after 0.25 h of cold treatment. However, the induction of CBF1 in crlk1 plants appeared at 1 h after cold treatment. We also studied the expression of COR genes RD29A, KIN1, and COR15a in the crlk1 mutants and CRLK1 complemented lines. In comparison with wild-type plants, the cold induction of RD29A, KIN1, and COR15a was delayed by more than 4 h in the crlk1 mutants, and the induction levels were also decreased for KIN1 and COR15a (Fig. 5B). In the complementation line with the full-length CRLK1(crlk1/CRLK1), cold induction of COR genes was also observed. Even under normal conditions, expression of COR genes was detected in these plants. Furthermore, the complementation line crlk1/ΔC2 carrying a truncated CRLK1 without the C-terminal CaM-binding site showed delayed expression of COR genes similar to crlk1 plants. This is in agreement with the results of the freezing tolerance tests. These results suggest that CRLK1 acts as a positive regulator of CBF1, RD29A, KIN1, and COR15.
FIGURE 5.
Altered expression of cold-responsive genes in crlk1 knock-out mutants and complementation lines. A, Northern analysis showing the altered expression of CBF in wild-type and crk1 mutant plants after cold treatment. The plants were treated at 4 °C for 0–2 h. 18 S RNA shows equal loading. B, Northern analysis showing expression of RD29A, KIN1, and COR15a genes in wild-type, crlk1 mutant plants, crlk1/CRLK1 (complementation lines with full-length CRLK1) and crlk1/ΔC2 (complementation lines with truncated CRLK1 lacking the C-terminal CaM-binding region) under cold treatment (4 °C) for various periods. 18 S RNA shows equal loading of total RNA. Ten μg of total RNA was loaded in each lane. The DNA fragments amplified with gene-specific primers were used as probes.
DISCUSSION
Our results document that CRLK1 is a calcium/CaM-regulated RLK anchored in plasma membrane. Calcium/CaM binding to CRLK1 regulates its kinase activity and plays a critical role in cold acclimation in planta. It has been well documented that the cold-induced changes in intracellular Ca2+ level is critical for plants to fully acclimate and for maximal cold induction of some COR genes (11). It has been suggested that calcium/CaM-mediated signaling is involved in the cold induction of COR genes because CaM genes are themselves induced by low temperature (34), and CaM inhibitors have been shown to inhibit cold-induced COR expression (20). Our results support that CaM acts as a positive regulator during cold acclimation. However, overexpression of CaM3 in Arabidopsis resulted in the inhibition of the COR expression, suggesting that this specific CaM gene may function as a negative regulator (21). There are 11 CaM genes in Arabidopsis, some of which show spatial and temporal regulation in the cell (15). Hence, it cannot be generalized that all CaMs show similar response to cold.
There are two CaM-binding sites in CRLK1. Although no significant role was detected for CaM binding to the N-terminal site on protein phosphorylation, we cannot exclude other roles for this binding. The mammalian epidermal growth factor receptor is a member of the CaM-binding receptor tyrosine kinase family with a CaM-binding site near the transmembrane domain. CaM regulates intracellular trafficking of epidermal growth factor receptor and mitogen-activated protein kinase (MAPK) signaling (35). It will be interesting to further dissect the function of the N-terminal CaM binding.
CRLK1 is localized in the plasma membrane, which is required for its function in planta because the truncated form of CRLK1 lacking the transmembrane domain did not rescue the phenotype of crlk1 null mutants. In the Arabidopsis genome, out of 600 RLK genes, more than 200 RLKs have no receptor configuration, and very few of them have known functions (36). Tomato Pto protein kinase, which confers resistance to the pathogen Pseudomonas syringae, is a cytoplasmic RLK without an extracellular domain and transmembrane domain. Pto is activated by avrPto, a small peptide secreted by P. syringae (37). Thus, even though CRLK1 has no receptor configuration, it could function directly in transducing an extracellular signal into cells in a manner similar to Pto, as well as playing a role in transducing the Ca2+/CaM-mediated signal triggered by stress. Alternatively, CRLK1 may form a dimer or oligomer with an RLK(s) with receptor configuration to recognize an extracellular ligand(s) or interact with other membrane protein(s) similar to TAK1. TAK1, a thylakoid membrane protein kinase without a receptor configuration, is required for light harvesting by phosphorylating the light-harvesting complex proteins in the thylakoid membrane (38). Isolation of CRLK1-interacting proteins/substrates will help in further dissecting this signal pathway. Accumulating evidence indicates that protein phosphorylation is involved in the pathway connecting the cold-triggered calcium changes and cold acclimation (22, 39). However, the protein kinase(s) responsible for inducing cold-regulated genes and activating freezing tolerance is elusive. Our results suggest that calcium/CaM/CRLK1-regulated phosphorylation can be an important signal node in the cold signal transduction pathway.
Identification of CRLK1 as a Ca2+/CaM-regulated protein kinase lays the groundwork for improving plant stress tolerance. In recent years, we have been able to define the structure/function relationships of Ca2+/CaM-binding proteins by manipulating their Ca2+ and/or CaM-binding sites (40). For example, altered Ca2+/CaM binding to DWF1 documented the possibility of producing size-engineered plants (6), although altered Ca2+ and Ca2+/CaM binding to chimeric calcium/calmodulin-dependent protein kinase produces spontaneous nodulation (7). Recently, we characterized the CaM-binding transcription factor AtSR1, which doses the salicylic acid level and regulates plant immunity (41). Redesigning CRLK1 orthologs in crop plants by manipulating their CaM-binding sites will help to enhance their ability to respond to the stress signal and to engineer stress-tolerant crops.
Supplementary Material
Acknowledgments
We thank J. Browse, C. Skidmore, and L. Barkan for their help in freezing tolerance tests; M. Palmgren for providing anti-AHA2 antibody; V. Franceschi, C. Davitt, and V. Lynch-Holm for microscopy; D. Takezawa for providing the cDNA library; Y. Chen and K. Kumar for their help in the laboratory. We also thank D. von Wettstein, J. Browse, and A. McCubbin for critically reading the manuscript.
This work was supported by the National Research Initiative Competitive Grant 2008-35100-04566 from the United States Department of Agriculture, the National Institute of Food and Agriculture, Grant ISO-0642146 from the National Science Foundation, and the Washington State University Agricultural Research Center.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- CaM
- calmodulin
- CPZ
- chlorpromazine
- GFP
- green fluorescent protein
- MOPS
- 3-(N-morpholino)propanesulfonic acid
- WT
- wild type
- COR
- cold-regulated gene
- CBF
- C-repeat-binding factor.
REFERENCES
- 1.Thomashow M. F. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599 [DOI] [PubMed] [Google Scholar]
- 2.Browse J., Xin Z. (2001) Curr. Opin. Plant Biol. 4, 241–246 [DOI] [PubMed] [Google Scholar]
- 3.Shinozaki K., Yamaguchi-Shinozaki K., Seki M. (2003) Curr. Opin. Plant Biol. 6, 410–417 [DOI] [PubMed] [Google Scholar]
- 4.Chinnusamy V., Ohta M., Kanrar S., Lee B. H., Hong X., Agarwal M., Zhu J. K. (2003) Genes Dev. 17, 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee B. H., Henderson D. A., Zhu J. K. (2005) Plant Cell 17, 3155–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du L., Poovaiah B. W. (2005) Nature 437, 741–745 [DOI] [PubMed] [Google Scholar]
- 7.Gleason C., Chaudhuri S., Yang T., Muñoz A., Poovaiah B. W., Oldroyd G. E. (2006) Nature 441, 1149–1152 [DOI] [PubMed] [Google Scholar]
- 8.Poovaiah B. W., Reddy A. S. (1987) CRC Crit. Rev. Plant Sci. 6, 47–103 [DOI] [PubMed] [Google Scholar]
- 9.Reddy A. S. (2001) Plant Sci. 160, 381–404 [DOI] [PubMed] [Google Scholar]
- 10.Sanders D., Pelloux J., Brownlee C., Harper J. F. (2002) Plant Cell 14, S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knight H., Trewavas A. J., Knight M. R. (1996) Plant Cell 8, 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pei Z. M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G. J., Grill E., Schroeder J. I. (2000) Nature 406, 731–734 [DOI] [PubMed] [Google Scholar]
- 13.Xiong L., Schumaker K. S., Zhu J. K. (2002) Plant Cell 14, S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luan S., Kudla J., Rodriguez-Concepcion M., Yalovsky S., Gruissem W. (2002) Plant Cell 14, S389–S400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang T., Poovaiah B. W. (2003) Trends Plant Sci. 8, 505–512 [DOI] [PubMed] [Google Scholar]
- 16.Bouche N., Yellin A., Snedden W., Fromm H. (2005) Annu. Rev. Plant Mol. Plant Physiol. 56, 435–466 [DOI] [PubMed] [Google Scholar]
- 17.Veluthambi K., Poovaiah B. W. (1984) Science 223, 167–169 [DOI] [PubMed] [Google Scholar]
- 18.Yang T., Chaudhuri S., Yang L., Chen Y., Poovaiah B. W. (2004) J. Biol. Chem. 279, 42552–42559 [DOI] [PubMed] [Google Scholar]
- 19.Zhang L., Lu Y. T. (2003) Trends Plant Sci. 8, 123–127 [DOI] [PubMed] [Google Scholar]
- 20.Tähtiharju S., Sangwan V., Monroy A. F., Dhindsa R. S., Borg M. (1997) Planta 203, 442–447 [DOI] [PubMed] [Google Scholar]
- 21.Townley H. E., Knight M. R. (2002) Plant Physiol. 128, 1169–1172 [DOI] [PubMed] [Google Scholar]
- 22.Monroy A. F., Sarhan F., Dhindsa R. S. (1993) Plant Physiol. 102, 1227–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiu S. H., Bleecker A. B. (2003) Plant Physiol. 132, 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanoosthuyse V., Tichtinsky G., Dumas C., Gaude T., Cock J. M. (2003) Plant Physiol. 133, 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charpenteau M., Jaworski K., Ramirez B. C., Tretyn A., Ranjeva R., Ranty B. (2004) Biochem. J. 379, 841–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poovaiah B. W., Leopold A. C. (1973) Plant Physiol. 52, 236–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xin Z., Browse J. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7799–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsson C., Widell S., Kjellbom P. (1987) Methods Enzymol. 148, 558–569 [Google Scholar]
- 29.Palmgren M. G., Sommarin M., Serrano R., Larsson C. (1991) J. Biol. Chem. 266, 20470–20475 [PubMed] [Google Scholar]
- 30.Takezawa D., Ramachandiran S., Paranjape V., Poovaiah B. W. (1996) J. Biol. Chem. 271, 8126–8132 [DOI] [PubMed] [Google Scholar]
- 31.Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., Gadrinab C., Heller C., Jeske A., Koesema E., Meyers C. C., Parker H., Prednis L., Ansari Y., Choy N., Deen H., Geralt M., Hazari N., Hom E., Karnes M., Mulholland C., Ndubaku R., Schmidt I., Guzman P., Aguilar-Henonin L., Schmid M., Weigel D., Carter D. E., Marchand T., Risseeuw E., Brogden D., Zeko A., Crosby W. L., Berry C. C., Ecker J. R. (2003) Science 301, 653–657 [DOI] [PubMed] [Google Scholar]
- 32.Fuglsang A. T., Guo Y., Cuin T. A., Qiu Q., Song C., Kristiansen K. A., Bych K., Schulz A., Shabala S., Schumaker K. S., Palmgren M. G., Zhu J. K. (2007) Plant Cell 19, 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jahn T., Fuglsang A. T., Olsson A., Brüntrup I. M., Collinge D. B., Volkmann D., Sommarin M., Palmgren M. G., Larsson C. (1997) Plant Cell 9, 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polisensky D. H., Braam J. (1996) Plant Physiol. 111, 1271–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tebar F., Villalonga P., Sorkina T., Agell N., Sorkin A., Enrich C. (2002) Mol. Biol. Cell 13, 2057–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiu S. H., Bleecker A. B. (2001) Sci. STKE 2001, re22. [DOI] [PubMed] [Google Scholar]
- 37.Tang X., Frederick R. D., Zhou J., Halterman D. A., Jia Y., Martin G. B. (1996) Science 274, 2060–2063 [DOI] [PubMed] [Google Scholar]
- 38.Snyders S., Kohorn B. D. (1999) J. Biol. Chem. 274, 9137–9140 [DOI] [PubMed] [Google Scholar]
- 39.Monroy A. F., Dhindsa R. S. (1995) Plant Cell 7, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang T., Du L., Poovaiah B. W. (2007) Funct. Plant Biol. 34, 343–352 [DOI] [PubMed] [Google Scholar]
- 41.Du L., Ali G. S., Simons K. A., Hou J., Yang T., Reddy A. S., Poovaiah B. W. (2009) Nature 457, 1154–1158 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.