Abstract
In the endoplasmic reticulum (ER), a number of thioredoxin (Trx) superfamily proteins are present to enable correct disulfide bond formation of secretory and membrane proteins via Trx-like domains. Here, we identified a novel transmembrane Trx-like protein 4 (TMX4), in the ER of mammalian cells. TMX4, a type I transmembrane protein, was localized to the ER and possessed a Trx-like domain that faced the ER lumen. A maleimide alkylation assay showed that a catalytic CXXC motif in the TMX4 Trx-like domain underwent changes in its redox state depending on cellular redox conditions, and, in the normal state, most of the endogenous TMX4 existed in the oxidized form. Using a purified recombinant protein containing the Trx-like domain of TMX4 (TMX4-Trx), we confirmed that this domain had reductase activity in vitro. The redox potential of this domain (−171.5 mV; 30 °C at pH 7.0) indicated that TMX4 could work as a reductase in the environment of the ER. TMX4 had no effect on the acceleration of ER-associated degradation. Because TMX4 interacted with calnexin and ERp57 by co-immunoprecipitation assay, the role of TMX4 may be to enable protein folding in cooperation with these proteins consisting of folding complex in the ER.
Keywords: Chaperones/Protein Folding, Protein/Disulfide, Protein/Folding, Subcellular Organelles/Endoplasmic Reticulum, Protein Folding, Protein Disulfide Isomerase, Redox, Thioredoxin
Introduction
Disulfide bond formation is a rate-limiting step in the correct folding of nascent polypeptides of many secretory and membrane proteins. Disulfide bonds are primarily produced co-translationally by oxidation of thiol groups between two cysteine residues in the lumen of the endoplasmic reticulum (ER)2 (1), where the redox environment is highly oxidative compared with that of cytosol (2). In addition, nascent polypeptides may need to be isomerized for correct disulfide bond formation to acquire a functional conformation (3–5).
In mammalian cells, ∼20 thioredoxin (Trx) superfamily oxidoreductases having Trx-like domain(s) have been identified in the ER. Trx superfamily proteins are thought to work as oxidoreductases via catalytic cysteine residues in the Trx-like domain and play an important role in proper disulfide bond formation (5, 6). One of the well known ER oxidoreductases is PDI (7), which possesses two active Trx-like domains, a and a′, with catalytic CXXC motifs, and two inactive domains, b and b′, for effective substrate recognition (8, 9). PDI catalyzes the formation of intra- and intermolecular disulfide bonds in collaboration with the FAD-binding oxidoreductase Ero1α/β, which obtains its oxidative potential from oxygen (10–12). ERp57, another ER oxidoreductase, forms folding complexes with calnexin and/or calreticulin, both of which are lectin-like molecular chaperones in the ER, and is involved in correct folding of nascent glycoproteins (13, 14). Oxidation by ERp57 is needed for modification of major histocompatibility complex class I (15) and CD1d (16). Interestingly, ERp57 also prevents one substrate, β-integrin, from becoming overoxidized, indicating that ERp57 can work as a reductase to reduce incorrectly formed disulfide bonds (17). Furthermore, the PDI-like protein ERdj5 was recently identified as a reductase that can cleave disulfide bonds of misfolded glycoproteins in the ER, facilitating ER-associated degradation (ERAD) (18). Although the functions of most Trx superfamily proteins remain unclear, a few well characterized proteins demonstrate that Trx superfamily proteins in the ER are important not only for productive folding of nascent polypeptides but also for elimination of misfolded proteins by ERAD (4, 19).
Four Trx superfamily proteins in the ER of mammalian cells have been reported to possess a transmembrane region (transmembrane Trx-like protein (TMX)) (5, 6). All four TMX family proteins (TMX1–TMX4) have an ER-targeting signal sequence, one Trx-like domain with a catalytic CXXC motif, and one transmembrane domain. TMX2 has an unusual catalytic motif, SNDC instead of CXXC (20) and is thought to be redox-inactive (6). TMX3 is most similar to PDI because of the presence of the b and b′ domains in addition to the catalytic Trx-like domain (21). TMX3 was suggested from in vitro analysis to have isomerase activity in the ER (21, 22). TMX (TMX1) has one Trx-like domain with oxidoreductase activity (23, 24), and a recent report demonstrated that TMX1 prevents an overexpressed major histocompatibility complex class I heavy chain from being degraded (25).
Here, we identified and characterized a TMX family protein, TMX4, as a novel reductase in the ER. TMX4 is a type I transmembrane protein, with a Trx-like domain facing the ER lumen and reductase activity in vitro. TMX4 interacts with calnexin, suggesting the possible involvement of TMX4 as a reductase in cooperation with TMX1 or ERp57 in the calnexin folding complex.
EXPERIMENTAL PROCEDURES
Cells and Antibodies
HeLa, human embryonic kidney (HEK293), and HepG2 cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and antibiotics (supplemented with nonessential amino acids for HepG2 cells). To induce ER stress, cells were treated with 300 nm thapsigargin (Sigma-Aldrich), 2 μg/ml tunicamycin (Sigma- Aldrich), and 5 mm dithiothreitol (DTT; Nacalai Tesque, Japan) for 6 h. The polyclonal anti-TMX4 C-terminal antibody (Sigma) and the antibody raised by immunizing rabbits against a recombinant C-terminal region (220–343) of the human TMX4 were used. The house-made rabbit antibody serum was affinity-purified using this antigen. The other antibodies used in this study were: anti-HA polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA), anti-PDI, anti-calnexin C terminus (Stressgen, British Columbia, Canada), anti-BiP, anti-calnexin N terminus (BD Transduction Laboratories), anti-β-tubulin (Tub-2.1) (ICN Biomedicals), anti-β-actin (Chemicon International), anti-calreticulin (MBL, Nagoya, Japan), anti-TMX1 (Abcam, Cambridge, UK), anti-ERp57 (Santa Cruz Biotechnology), anti-FLAG (Sigma-Aldrich), anti-α1-antitrypsin (DAKO), and anti-SEL1L (kindly supplied by Dr. Nobuko Hosokawa, Kyoto University, Japan).
Construction of Plasmids
The human TMX4 cDNA clone (DDBJ accession number AK075404) was supplied by the NBRC, NITE-DOB (Kisarazu, Japan). A human TMX4 cDNA with an HA tag sequence at the C terminus was subcloned into pT7blue (Novagen, Darmstadt, Germany) by TA cloning and finally subcloned into pCDNA3.1(+) (Invitrogen). The cDNA with HA tag was amplified using the following primer set: forward, 5′-GGATCCATGGCTGGTGGACGCTGCGGCCC-3′ and reverse, 5′-CTCGAGCTACAGTCCCTTGTCAGCAGCGTAATCTGGAACATCGTATGGTAATGCTGACTTTTACGCTGCC-3′. To express the recombinant Trx-like domain of TMX4, the Trx-like domain region (35–185) was subcloned into pCold-TF, which incorporates a His6 tag and trigger factor (TF) at the N terminus (TaKaRa, Japan).
Northern Blotting
Total RNA was obtained using an RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany). Total RNA (5 μg) was separated by electrophoresis using a 1% agarose, 2.2 m formaldehyde-denaturing gel, blotted onto nylon membranes (Gene Screen Plus, PerkinElmer Life Sciences), and then UV cross-linked. A human multiple-tissue Northern blot (Clontech) was used to analyze the tissue distribution of TMX4. The DNA probes used in this study were: the C-terminal fragment of human TMX4 (660–1050), a mouse BiP fragment (384–1391), the full-length glyceraldehyde-3-phosphate dehydrogenase (a gift from Dr. Koji Nagasawa, Kyoto University, Japan), and β-actin (Clontech). The mouse BiP fragment was amplified from pEYFP-C1-mBiP, which was a kind gift from Dr. Ikuo Wada (Fukushima Medical University School of Medicine, Japan). The probes were labeled with [32P]dCTP (GE Healthcare) using a random primer DNA labeling kit (Roche Applied Science) according to the manufacturer's instructions. Nonincorporated deoxynucleotides were removed using ProbeQuantTM G-50 Micro Columns (GE Healthcare). Membranes were hybridized in PerfectHYBTM hybridization solution (TOYOBO Co. Ltd., Osaka, Japan) with the probes at 68 °C for 16 h. The hybridized membranes were exposed to an imaging plate, and the radioactive signal was analyzed using a STORM PhosphorImager (GE Healthcare).
Cell Fractionation, Alkaline Extraction, and Trypsin Protection Assays
Cells were suspended in buffer I (50 mm Tris-HCl (pH 7.4), 5 mm EDTA), incubated on ice for 10 min, and then homogenized by passing 10 times through a 25-gauge needle. After homogenization, an equal volume of buffer II (buffer I with 880 mm sucrose) was added immediately. A postnuclear supernatant (PNS) was prepared by centrifuging the cell homogenate at 1,000 × g. The PNS was centrifuged at 12,000 × g to remove mitochondria, and a microsomal fraction was then obtained as a pellet by ultracentrifugation of the supernatant at 120,000 × g for 1 h. The supernatant was used as the cytosolic fraction. For alkaline extraction assays, the microsomal fraction was resuspended in alkaline buffer (100 mm Na2CO3 (pH 11.3)) on ice for 30 min and then centrifuged at 120,000 × g for 1 h to separate the alkaline-soluble fraction and the membrane-associated fraction. For trypsin protection assays, the PNS was treated with trypsin (Sigma) on ice for 15 min, and the reaction was then stopped by the addition of Laemmli SDS-PAGE sample buffer.
Endoglycosidase H (Endo H) Digestion
Cells were extracted in lysis buffer (50 mm HEPES (pH 7.5), 150 mm NaCl) containing 1% Nonidet P-40, 10 mm N-ethylmaleimide, and protease inhibitors. Cell extracts were centrifuged at 20,000 × g for 20 min at 4 °C. The supernatant was boiled for 10 min with 1% SDS and 1% 2-mercaptoethanol, adjusted to pH 5.4 by adding sodium acetate buffer (pH 5.4), and then digested with Endo H (Roche Applied Science) at 37 °C for 1 h.
Immunofluorescence
Cells overexpressing TMX4-HA were washed with phosphate-buffered saline and fixed with 4% paraformaldehyde for 20 min at room temperature. Cells were permeabilized with 0.2% Triton X-100 in phosphate-buffered saline at room temperature for 5 min followed by an incubation in 1% normal goat serum and 1% bovine serum albumin for 1 h. Cells were incubated with a rabbit anti-HA antibody, an anti-mouse PDI antibody for 1 h, and then with Alexa Fluor 488-conjugated goat anti-rabbit IgG and Alexa Flour 594-conjugated goat anti-mouse IgG (Invitrogen) as secondary antibodies for 1 h. Confocal images were obtained using a LSM 510 META confocal microscope (Carl Zeiss, Jena, Germany).
Transfection and Small Interfering RNA Sequences
Plasmids were transfected using Effectene (Qiagen), Lipofectamine 2000, or Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer's instructions. StealthTM RNA siRNAs targeting TMX4 were obtained from Invitrogen. The sequences were as follows: siTMX4-1, 5′-UUUACGCUGCCUCAAGGAGUCUUCC-3′, siTMX4-2, 5′-UGAUAUUACCACCAAGACCAGACCC-3′. Medium GC and StealthTM RNAi duplexes were used as negative controls.
Protein Expression, Purification, and Circular Dichroism (CD) Measurements
Escherichia coli BL21(DE3) cells (Novagen) carrying pCold-TF-TMX4-Trx (35A-185E) were cultured at 37 °C until the A600 reached 0.5. At this point, expression of the recombinant proteins was induced by cold shock at 15 °C for 24 h in the presence of 0.4 mm isopropyl-β-d-thiogalactoside. Harvested cells were sonicated in 20 mm sodium phosphate (pH 7.5), containing 50 mm imidazole and 0.5 m NaCl. The supernatant of the cell lysate was loaded onto a HisTrap column (GE Healthcare) equilibrated with the cell suspension buffer and eluted with the same buffer containing 0.5 m imidazole. To remove the trigger factor portion of TF-TMX4-Trx, the eluted fraction was treated with HRV3C protease (Novagen) using the buffer conditions described in manufacturer's instructions for 16 h at 4 °C. After digestion, the cleaved TF portion and uncleaved TF-TMX4-Trx, both of which have a His tag, were removed using a HisTrap column. The flow-through fraction was desalted and loaded onto a Resource Q column (GE Healthcare) equilibrated with 20 mm Tris-HCl (pH 8.0), and then fractions were eluted with a linear gradient of NaCl. TMX4-Trx-rich fractions were loaded onto a HiLoad 16/60 Superdex 75pg isofraction column. Purified recombinant TMX4-Trx was pooled and then stored at 4 °C. The CD spectrum of TMX4-Trx was measured using a 1-mm path length cell at 25 °C in phosphate-buffered saline (pH 7.4) containing 1 mm NDSB-201. The concentration of TMX4-Trx was 18 μm.
Insulin Reduction Assay
The insulin reduction assay was performed at 25 °C in 0.1 m sodium phosphate buffer (pH 7.0) and 8 mm GSH containing 150 mm NaCl, 2 mm EDTA, and 1 mm NDSB-201 using the method of Lambert and Freedman (26). In this assay, the enzyme-catalyzed reduction of the disulfide bonds in insulin is coupled to the reduction of GSSG to GSH by glutathione reductase (Sigma) and NADPH. The purified recombinant PDI a domain was kindly provided by Dr. Kenji Inaba (Kyushu University, Japan).
Oxidase and Isomerase Activity
Reduced and denatured RNase A was prepared using an overnight incubation of native bovine RNase A (Sigma) in 0.1 m Tris-HCl (pH 8.0) containing 6 m guanidine hydrochloride and 150 mm DTT. The reduced RNase A was exchanged into 0.1% acetic acid and then stored at −80 °C until use. Scrambled RNase A was obtained from Sigma. The oxidase and isomerase activities using RNase A as a substrate were measured by monitoring RNase A-catalyzed hydrolysis of cCMP spectrophotometrically, as described previously (27). Measurements were performed at 25 °C in 50 mm HEPES-NaOH (pH 7.5) containing 150 mm NaCl, 2 mm EDTA, 1 mm NDSB-201, 4.5 mm cCMP (as a substrate of RNase A), 0.4 mm GSSG, and 1.2 mm GSH as a redox buffer ([GSSG]:[GSH] = 1:3).
Measurement of Redox Equilibrium with Glutathione
The redox equilibrium between recombinant TMX4-Trx and glutathione was measured as follows. TMX4-Trx (1 μm) was incubated with 0.1 mm GSSG and various concentrations of GSH at 30 °C for 1 h in 0.1 m sodium phosphate buffer (pH 7.0) containing 1 mm EDTA and 1 mm NDSB-201 with ultracentrifugation at 120,000 × g to remove aggregates of TMX4-Trx. After incubation, trichloroacetic acid (10%) was added to prevent further thiol-disulfide exchange. The precipitated pellet was solubilized in 0.1 m sodium phosphate buffer (pH 7.0), containing 2% SDS and 1 mm 4-acetamido-4′-maleidylstilbene-2,2′-disulfonic acid (AMS; Invitrogen) or 1 mm methoxypolyethylene glycol (average molecular weight 2000)-maleimide (mPEG2K-mal) (Sunbright ME-020MA; NOF Corporation, Japan), followed by incubation at 25 °C for 1 h to alkylate free sulfhydryl groups of cysteines. The samples were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. The ratio of reduced form was quantified. The redox equilibrium constant (Keq) was calculated by fitting the fraction of the reduced form to the following equation: r = ([GSH]2/[GSSG])/(Keq+([GSH]2/[GSSG])), where r is the relative ratio of reduced TMX4-Trx.
Metabolic Labeling, Immunoprecipitation, Pulse-Chase Assays
Pulse-chase assays were performed 72 h after siRNA transfection and 24 h after NHK transfection. After preincubation in Dulbecco's modified Eagle's medium lacking methionine/cysteine (Invitrogen) for 1 h (for pulse-immunoprecipitation assay) or 25 min (for pulse-chase assay), HEK293 cells were pulse-labeled with 8.2 MBq/ml Express 35S protein labeling mix (PerkinElmer Life Sciences) for indicated periods. Cells were extracted in lysis buffer (150 mm NaCl, 50 mm HEPES (pH 7.5)) containing 1% Nonidet P-40 or 3% digitonin, 10 or 20 mm N-ethylmaleimide, and protease inhibitors. The cell extract was centrifuged at 20,000 × g for 20 min at 4 °C. The supernatant was used for immunoprecipitation with the indicated antibodies. Immune complexes were collected using protein A-Sepharose beads (GE Healthcare) or protein G-Sepharose beads (GE Healthcare). The eluted immune complexes were separated by SDS-PAGE, and the gels were then exposed to an imaging plate for 2 days. The radioactive bands were then detected using a FujiFilm PhosphorImager FLA7000IP. DNA plasmid of calnexin-HA was kind gift from Dr. Ikuo Wada (Fukushima Medical University School of Medicine, Japan).
Sucrose Density Gradient Centrifugation
Cells were lysed with lysis buffer containing 3% digitonin. The supernatant was applied to a linear 10–40% sucrose gradient and then centrifuged at 36,000 rpm in a swing bucket rotor for 14 h at 4 °C. Fractions (250 μl each) were collected from the top, separated by SDS-PAGE, and examined by Western blot analysis.
RESULTS
Expression of TMX4
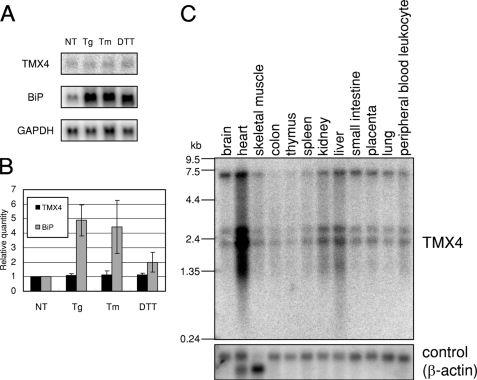
A data base search identified one putative ER membrane protein that has a Trx-like domain with a CPAC catalytic motif. Many ER resident molecular chaperones and ERAD components are induced or increase expression in response to ER stress (28). Following an ER stress, the expression of TMX4 in HeLa cells was examined by Northern blot analysis in the presence or absence of tunicamycin, thapsigargin, or DTT. As shown in Fig. 1, no induction of TMX4 expression was observed. Because mRNA of BiP was induced by the same treatment, we concluded that TMX4 is a protein that does not respond to ER stress (Fig. 1, A and B). This result is consistent with the observation that TMX4 does not contain an unfolded protein response element or an ER stress response element in the promoter region, as indicated by in silico analysis of genome sequences (28, 29).
FIGURE 1.
Expression profile of TMX4. A, expression of TMX4 mRNA before and after ER stress. Total mRNA from HeLa cells that were not treated (NT) or were treated with 300 nm thapsigargin (Tg), 2 μg/ml tunicamycin (Tm), and 5 mm DTT for 6 h were analyzed by Northern blotting. BiP was used as a positive control for induction by ER stress, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was an internal control. B, quantification of TMX4 mRNA before and after ER stress. Northern blot signals were quantified, as in A, and normalized to GAPDH mRNA levels. The results are means ± S.D. of three independent experiments. C, tissue distribution of mRNA expression of TMX4. A multiple human tissue blot was hybridized with TMX4 C-terminal cDNA (660–1050) and β-actin cDNA as a control.
The expression of TMX4 in various tissues was examined using a human MTN Blot (Clontech) on which poly(A) RNA from 12 different human tissues are blotted (Fig. 1C). Up to four bands hybridized with TMX4 cDNA, which may be due to the expression of TMX4 mRNAs with 3′ poly(A) tails of differing lengths. Although the strongest expression of TMX4 mRNA was observed in the heart, expression was ubiquitous in the various tissues examined thus far.
TMX4 Is a Type I Transmembrane Protein with a CXXC Motif in the ER Lumen
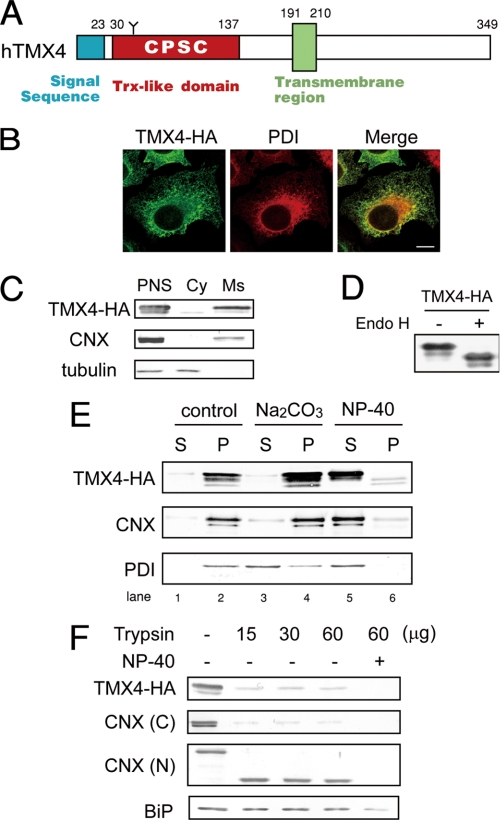
From the presence of a signal sequence predicted by the SignalP Server, TMX4 was predicted to localize to the ER (Fig. 2A and supplemental Fig. 1). Localization of TMX4 in the ER was confirmed by overexpression of the full-length C-terminal HA-tagged TMX4 (TMX4-HA) in HeLa cells. The staining pattern of TMX4-HA coincided with that of PDI by immunofluorescence microscopy (Fig. 2B). When cell lysates were biochemically separated into cytosolic and microsomal fractions, the microsomal fractions were found to contain both calnexin and TMX4-HA (Fig. 2C). Endo H treatment revealed that TMX4 contained an Endo H-sensitive N-glycan (Fig. 2D), consistent with the prediction by the NetNGlyc 1.0 Server that TMX4 contains one N-glycosylation site in the Trx-like domain (supplemental Fig. 1). The presence of an N-glycosylation site, in turn, suggests that the Trx-like domain of TMX4 faces the ER lumen.
FIGURE 2.
Subcellular localization of TMX4. A, domain structure of human TMX4. TMX4 has an ER targeting signal sequence, one Trx-like domain with a CXXC motif (CPSC), and one transmembrane region. One N-glycosylation site is present in the Trx-like domain. B, immunofluorescence analysis of HeLa cells expressing TMX4-HA. Cells were co-stained with anti-HA (green) and anti-PDI (red, ER marker). C, cellular fractionation assay. Cells expressing TMX4-HA were lysed, and the nuclear fraction was removed by centrifugation. The PNS was fractionated by sequential centrifugation into cytosolic (Cy) and microsomal (Ms) fractions. The fractions were analyzed by Western blotting using anti-HA, anti-calnexin (CNX, an ER marker protein), and anti-tubulin (a cytosolic marker protein). D, Endo H digestion. Cell lysates expressing TMX4-HA were incubated with or without Endo H and analyzed by Western blotting using an HA antibody. E, alkaline extraction of microsomal TMX4. A microsomal fraction prepared from cells transiently expressing TMX4-HA was fractionated by ultracentrifugation after no treatment (lanes 1 and 2) or treatment with alkaline buffer (Na2CO3, lanes 3 and 4) or 1% Nonidet P-40 (lanes 5 and 6). The fractions were analyzed by Western blotting using anti-calnexin (an ER membrane marker protein) and anti-PDI (an ER luminal marker protein). F, trypsin digestion of TMX4. A PNS fraction prepared from cell lysates expressing TMX4-HA was treated with the indicated concentration of trypsin and analyzed by Western blotting using anti-HA; anti-calnexin N terminus, which recognizes the luminal portion of calnexin; anti-calnexin C terminus, which recognizes the cytosolic portion of calnexin; and anti-BiP (an ER luminal marker protein).
From the primary sequence, TMX4 was predicted to be a transmembrane protein because of the existence of a putative transmembrane region. Treatment of the microsomal fraction with alkaline buffer, which removes peripheral membrane proteins, confirmed the transmembrane nature of TMX4. After alkaline treatment, TMX4 remains in the membrane fraction, similar to the well known ER transmembrane protein calnexin (Fig. 2E). When a PNS fraction was treated with trypsin, the C-terminal HA tag of TMX4 was quickly digested (Fig. 2F). This result is analogous to detection of the type I transmembrane protein calnexin by an antibody specific for its cytosolic C terminus. Taken together, these observations indicate that the C terminus of TMX4 is in the cytosol and that TMX4 is a type I transmembrane protein of the ER with a Trx-like domain facing into the ER lumen.
Redox State of TMX4 in Cells
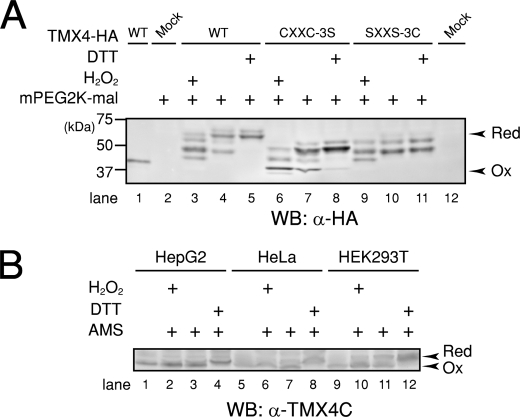
The redox state of the CXXC motif of TMX4 was monitored using a cysteine alkylation method with a maleimide reagent to determine whether the catalytic cysteines in the CXXC motif alternate between oxidized and reduced forms (30, 31). Proteins were trichloroacetic acid-precipitated from a suspension of HeLa cells overexpressing TMX4-HA and resuspended in HEPES buffer containing mPEG2K-mal, which can covalently bind to reduced cysteine residues. Immunoblotting analysis with an anti-HA antibody showed that mPEG2K-mal modified TMX4 migrated more slowly than the oxidized form in SDS-polyacrylamide gels. Although the actual molecular mass of mPEG2K-mal is around 2 kDa, the migration of reduced TMX4 was detected slower than expected because mPEG-mal simultaneously binds water (32).
Blots of TMX4-wild type samples showed multiple bands corresponding to the number of reduced cysteines (Fig. 3A, lane 4). If cells were treated with the oxidant H2O2 or the reductant DTT, the redox state of TMX4 was dramatically changed, as shown in Fig. 3A, lanes 3 and 5, respectively. These results suggest that TMX4 can change its redox state according to the redox environment in living cells.
FIGURE 3.
In vivo redox states of TMX4. A, redox state of TMX4-HA. TMX4-HA transfected cells were lysed by addition of trichloroacetic acid to prevent further disulfide exchange. Trichloroacetic acid precipitates were resolved in 0.1 m sodium phosphate buffer (pH 7.0), 2% SDS, with or without 1 mm mPEG2K-mal. The mPEG2K-mal modifications were performed at 25 °C for 1 h and analyzed by Western blotting (WB). Reduced (Red) and oxidized (Ox) TMX4 were prepared by pretreatment of cells with DTT and H2O2. Two cysteine mutants of TMX4, TMX4-CXXC-3S and TMX4-SXXS-3C, in which cysteines outside the CXXC motif or in the CXXC motif were mutated to serines, respectively, were used to clarify which cysteines are crucial for the effect of DTT or H2O2 treatment. B, redox state of endogenous TMX4. The redox state of endogenous TMX4 was analyzed by AMS modification.
Because human TMX4 has five (two catalytic and three noncatalytic) cysteine residues (supplemental Fig. 1), we constructed two TMX4 cysteine mutants: one has an intact CXXC motif with mutations in the other three cysteines to serine (CXXC-3S), and the other has the CXXC motif mutated to SXXS with the three noncatalytic cysteines intact (SXXS-3C). The noncatalytic cysteines were stable even when cells were treated with H2O2 or DTT (Fig. 3A, lanes 9–11). In contrast, the electrophoretic mobility of CXXC-3S was slower in the presence of DTT and faster in the presence of H2O2 (Fig. 3A, lanes 6–8), strongly suggesting that these cysteines in the CXXC motif were reduced or oxidized depending on the cellular redox environment.
We next analyzed the redox state of endogenous TMX4 in three different cell lines (Fig. 3B). The antibody against the TMX4 C terminus does not recognize endogenous TMX4 after mPEG2K-mal treatment; therefore, we modified free cysteines with AMS, which is a much smaller molecule than mPEG2K-mal. As predicted by the overexpression data, the redox state of endogenous TMX4 was modified depending on cellular redox environment. Under normal conditions, most TMX4 was oxidized, but still both reduced and oxidized bands of TMX4 were observed. Interestingly, the ratios of reduced to oxidized bands of TMX4 were different among the three cell lines, and the biological meaning of this phenomenon will be addressed in future studies (Fig. 3B, lanes 3, 7, and 11).
Oxidoreductase Activity of Recombinant TMX4
TMX4 has one Trx-like domain with a catalytic CXXC motif. To clarify whether this Trx-like domain has oxidoreductase activity, we constructed and expressed a recombinant protein containing the Trx domain (35A-185E) of TMX4 (TMX4-Trx) in bacteria. TMX4-Trx was expressed in bacteria, but was not recovered in the soluble fraction. Subsequently, we fused the bacterial TF to the N terminus of TMX4-Trx (TF-TMX4-Trx), which was successfully recovered in the soluble fraction (data not shown). After cleavage of TF with the HRV3C protease, we purified TMX4-Trx by affinity chromatography combined with biochemical purification (supplemental Fig. 2A). This recombinant TMX4-Trx was thought to be properly folded because it showed a typical α/β type signal in a far UV CD spectrum (supplemental Fig. 2B).
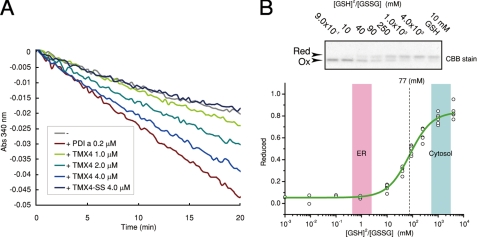
TMX4 oxidoreductase activity was examined using this recombinant TMX4-Trx. The reductase activity of TMX4-Trx was measured by insulin reduction coupled with glutathione reductase, where the decrease in NADPH detected by absorbance at 340 nm was a measure of the reductase activity, as described under “Experimental Procedures. ” As shown in Fig. 4A, TMX4 could catalyze the reduction of oxidized insulin by GSH in a dose-dependent manner (Fig. 4A). This activity was exerted through the CXXC motif in the Trx domain of TMX4 because the TMX4-SS mutant, in which the catalytic cysteines in the CXXC motif were mutated to serines, showed no enzyme activity (Fig. 4A).
FIGURE 4.
In vitro analysis of the Trx-like domain of TMX4. A, insulin reduction assay of recombinant TMX4-WT-Trx and TMX4-SS-Trx. Enzyme-catalyzed reduction of insulin disulfide bonds by GSH is coupled to the reduction of GSSG by glutathione reductase. The insulin reductase activity of the enzyme was measured with 8 mm GSH at 25 °C by spectrophotometrically monitoring NADPH consumption, which is concomitant with GSSG reduction by glutathione reductase. CBB, Coomassie Brilliant Blue. Red, reduced; Ox, oxidized. B, upper, redox equilibrium assay with glutathione at 30 °C. The free sulfhydryl groups of the cysteine residues were modified with AMS after incubation with different [GSH]2/[GSSG] ratios. Lower, measured redox equilibrium constant of TMX4. The apparent equilibrium constant between TMX4 and glutathione was determined by the nonlinear least square fitting of the data in the upper panel.
Next, the redox potential of the TMX4-Trx-like domain was determined using maleimide alkylation. Recombinant TMX4-Trx was incubated in glutathione buffer that contained various ratios of GSH to GSSG, and reduced and oxidized forms of TMX4-Trx were separated electrophoretically using a SDS-polyacrylamide gel after modification of free cysteines with AMS (Fig. 4B). The redox state of TMX4-Trx was dependent on the GSH/GSSG ratio. The redox equilibrium constant for TMX4-Trx and glutathione was calculated to be Keq = 77 mm. Using the Nernst equation, the redox potential of TMX4-Trx was determined to be −171.5 mV (pH 7.0, 30 °C), which was much more reductive than the redox conditions in the ER, where the [GSSG] to [GSH] ratio is ∼1:3 (2). These observations indicate that TMX4-Trx potentially works as a reductase in the redox environment of the ER lumen. The oxidase and isomerase activities of TMX4-Trx were also examined under ER redox conditions. Neither oxidase activity using reduced RNaseA nor isomerase activity using scrambled RNaseA was detected under the ER redox conditions (supplemental Fig. 2, C and D). Taken together, these observations indicate that TMX4-Trx behaves as a reductase in the ER redox environment.
TMX4 Has No Effect on NHK Degradation
One PDI-like reductase, ERdj5, is known to accelerate ERAD by reducing intermolecular (and possibly intramolecular) disulfide bonds (18). ERdj5 reduces the intermolecular disulfide of the ERAD substrate NHK, resulting in conversion of the NHK dimer to a monomer, which accelerates the ERAD of NHK. Considering that TMX4 may work as a reductase in the ER, we examined the possibility that TMX4 accelerates ERAD of NHK by reducing its dimeric disulfide bond.
To address this question, we examined the effect of knockdown of TMX4 on the degradation kinetics of NHK (supplemental Fig. 3). The degradation kinetics of NHK in cells treated with siRNA for TMX4 was almost the same as in control cells (supplemental Fig. 3B). Additionally, no effects on NHK dimer formation were detected (supplemental Fig. 3A). These results suggest that TMX4 is not involved in the reduction of NHK, which is required for the acceleration of NHK ERAD (33).
TMX4 Interacts with Calnexin and ERp57
Because TMX4 has reductase activity but no effect on ERAD of NHK, we next investigated complex formation of TMX4 with other cellular proteins. Protein folding complexes (including calnexin) and ERAD components (including SEL1L) can be separated by density gradient analysis (34). Endogenous TMX4 was detected in the same fraction as calnexin and was not present in the fraction containing degradation complex components, including SEL1L (Fig. 5A). TMX1, which has been reported to be involved in oxidative folding of newly synthesized proteins (25), was also present in the same fraction with TMX4 and calnexin (Fig. 5A). These results suggest that TMX4 may be included in a protein folding complex, which is consistent with the observation that TMX4 did not accelerate ERAD (supplemental Fig. 3).
FIGURE 5.
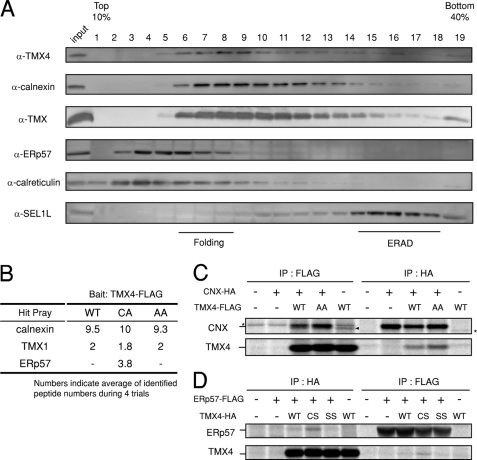
ER protein complexes containing TMX4. A, fractionation of endogenous TMX4 by sucrose density gradient centrifugation. HEK293 cells were lysed with 3% digitonin and fractionated using a 10–40% sucrose density gradient. The fractions were separated by SDS-PAGE and analyzed by Western blotting with the indicated antibodies. B, proteome analysis of TMX4-interacting proteins. TMX4-FLAG (bait)-overexpressed HEK293 cells were lysed mechanically. Cell lysate was subjected to immunoprecipitation with anti-FLAG antibody followed by liquid chromatography/mass spectrometry analysis. Average number of detected peptides derived from each protein in four experiments was shown. WT, wild type. C, co-immunoprecipitation (IP) of TMX4 with calnexin (CNX). HEK293 cells overexpressing TMX4-FLAG and calnexin-HA were labeled for 16 h with [35S]methionine/cysteine at 24 h after transfection. Cells were lysed with 3% digitonin and subjected to immunoprecipitation. Catalytic cysteine mutant of TMX4 (TMX4-AA) was used to check whether interaction between TMX4 and calnexin was independent of disulfide bond formation via CXXC motif or not. Arrowhead indicates endogenous calnexin. This band was confirmed to correspond to the endogenous calnexin. D, co-immunoprecipitation of TMX4 with ERp57. HEK293 cells overexpressing TMX4-HA and ERp57-FLAG were labeled for 1 h with [35S]methionine/cysteine. Cells were lysed with 1% Nonidet P-40 and subjected to immunoprecipitation. Catalytic cysteine mutants of TMX4 (TMX4-CS and TMX4-SS) were used to see whether this interaction was dependent of covalent binding via its catalytic cysteines or not. Asterisk shows nonspecific band.
A proteome analysis, in which TMX4-FLAG overexpressed in HEK293 cells was immunoprecipitated with anti-FLAG antibody and was followed by liquid chromatography/mass spectrometry analysis, showed that both calnexin and ERp57 interacted with TMX4 (Fig. 5B). Immunoprecipitation analysis was performed to confirm interactions of TMX4 with calnexin and ERp57 after overexpression of tagged proteins; calnexin and ERp57 were co-precipitated with TMX4 and vice versa (Fig. 5, C and D). Interestingly, endogenous calnexin was also co-immunoprecipitated with TMX4-FLAG (Fig. 5C).
Binding of TMX4 with calnexin was CXXC motif-independent because an AXXA mutant of TMX4, in which the CXXC motif was mutated to AXXA (TMX4-AA), could also bind to calnexin (Fig. 5C). On the contrary, interaction between TMX4 and ERp57 was interestingly CXXC-dependent; the CXXS mutant of TMX4 (TMX4-CS) bound to ERp57 more strongly compared with wild type TMX4 (Fig. 5D). The CXXA or CXXS mutant is known to interact with downstream substrates for much longer period through semistable intermediate state named mixed-disulfide bonds (35). These data suggest that TMX4 may be involved in a productive folding complex consisting of calnexin and ERp57 either through noncovalent bonds with calnexin or through mixed-disulfide bond with ERp57 and that TMX4 may serve as a donor of electrons to ERp57 in the electron transfer pathway during the folding process of nascent proteins in the ER.
DISCUSSION
We report here the identification of a novel protein, termed TMX4, that resides in the ER membrane. The protein is a type I transmembrane protein that functions as an ER membrane reductase and possesses a CPSC motif within a thioredoxin fold that faces the ER lumen. TMX1 and TMX3 are also TMX family proteins and ER membrane isomerases (21–24). TMX2 contains an SXXC motif in its Trx-like domain and is therefore thought to be inactive in terms of oxidoreductase activity (6). As shown here, TMX4 is the first and only reductase among TMX family proteins.
The Trx-like domain of TMX4 possesses the CPSC motif (supplemental Fig. 1), which faces the ER lumen, as determined by topological analysis and Endo H treatment (Fig. 2). An in vitro analysis using a recombinant Trx-like domain of TMX4 revealed that TMX4-Trx has reductase activity and can potentially work as a reductase in the redox environment of the ER lumen (Fig. 4B). Furthermore, redox state analysis using HeLa cells strongly suggests that the Trx-like domain of TMX4 should be involved in catalyzing reduction reactions because the recombinant TMX4-Trx domain has a reducing redox potential (Fig. 4B), and some portion of TMX4 exists as a reduced form in living cell (Fig. 3B). Oxidized and some reduced form of TMX4 was observed in living cells (Fig. 3), whereas many Trx superfamily proteins are known to exist primarily in the reduced state in the ER (36). These results suggest that TMX4 may have unique features compared with other oxidoreductases such as PDI, ERp57, or ERp72.
The precise cellular function of TMX4 as a reductase in the oxidative conditions of the ER will be a most interesting question to examine. We can hypothesize two possibilities: 1) TMX4 could be an ERAD-enhancing reductase like ERdj5 (18), or 2) TMX4 may work as a reducing regulator during productive folding, possibly in cooperation with ERp57 (17). The former possibility can be ruled out because sucrose density gradient analysis clearly showed that endogenous TMX4 was not found in the ERAD complex that includes SEL1L (Fig. 5A) (34). In addition, TMX4 knockdown had no effect on the degradation of the α1-antitrypsin NHK variant, a well known ERAD substrate (supplemental Fig. 3). However, TMX4 can interact with calnexin and ERp57, as determined by co-immunoprecipitation (Fig. 5, B–D). Sucrose density gradient analysis also revealed that TMX4 was present in the fraction containing calnexin and TMX1, a known productive folding complex (Fig. 5A). Taken together, these data suggest that TMX4 is involved in productive folding together with calnexin and ERp57.
There are two obvious possibilities for TMX4 function in the productive folding complex: 1) TMX4 directly reduces folding substrates for correct disulfide formation, similar to ERp57, or 2) TMX4 works as a regulator in the productive folding complex by reducing some components, such as ERp57, TMX1, or some other component in the complex. Calnexin cooperates with ERp57 (13, 37), which can isomerize or reduce incorrect disulfides. This complex has a central role in oxidative folding of glycoproteins. Reduction of β1 integrin, a substrate of ERp57, was reported to be required for proper folding (17). TMX4 (Keq = 77 mm) is potentially a stronger reducing enzyme compared with ERp57 (a domain, Keq = 3.3 mm; a′ domain, Keq = 1.5 mm (38) and binds to ERp57 via its catalytic cysteines (Fig. 5D); thus, TMX4 could work as a reductase that either reduces the key oxidoreductase ERp57 or reduces other substrates.
Recently, TMX1 was reported as an unconventional inhibitor of major histocompatibility complex class I heavy chain ERAD. TMX1 was postulated to cooperate with calnexin in protein retention in the ER, followed by refolding of misfolded proteins (25). Although TMX1 and TMX4 are similar, especially in the luminal domain (43.9% similarity), we identified a striking difference between TMX1 and TMX4 in oxidoreductase activities. TMX1 was thought to be an isomerase in the ER (24), whereas the in vitro redox equilibrium constant of TMX4 suggests that it would be a reductase in the ER. The redox state of endogenous TMX4 is more oxidative than that of TMX1 (25). These results indicate that, once the CXXC motif of TMX4 is reduced, it would more powerfully reduce other proteins compared with TMX1.
During preparation of this manuscript, Roth et al. (39) published the first report characterizing TMX4. They have reported that TMX4 is a ubiquitously expressed, type I transmembrane protein in the ER, which is consistent with our present data shown in Fig. 2. Furthermore, an RQR motif in the TMX4 cytosolic domain was reported to be essential for its ER localization because the deletion of RQR motif caused the transport of TMX4 to the cell surface. In melanoma cells, redox state of endogenous TMX4 is more oxidizing than TMX1 and TMX3, which is consistent with our in vitro results (TMX4 redox potential, Fig. 4B). However, the oxidoreductase activity of TMX4 was not determined in their study; and our study, including the in vitro oxidoreductase analysis, sheds light on the biological functions of TMX4. Identification of client substrates of TMX4 will be very important for determining the cellular function of TMX4.
Supplementary Material
Acknowledgments
We thank Drs. K. Maegawa and K. Inaba for help with the in vitro redox assay.
This work was supported by Grants-in-aid for Creative Scientific Research 19G50314 and Scientific Research 19058008 (to K. N.), by the Uehara Memorial Foundation and the Takeda Foundation (to J. H.), by the New Energy and Industrial Technology Development Organization (to T. N.), and by a fellowship from the Japan Society for the Promotion of Science (to K. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- ER
- endoplasmic reticulum
- Trx
- thioredoxin
- PDI
- protein disulfide isomerase
- ERAD
- ER-associated degradation
- TMX
- transmembrane Trx-like protein
- DTT
- dithiothreitol
- HA
- hemagglutinin
- TF
- trigger factor
- PNS
- postnuclear supernatant
- Endo H
- endoglycosidase H
- si
- small interfering
- AMS
- 4-acetamido-4′-maleidylstilbene-2,2′-disulfonic acid
- mPEG2K-mal
- methoxypolyethylene glycol (average molecular weight 2000)-maleimide
- NHK
- α1-antitrypsin null Hong Kong variant.
REFERENCES
- 1.Sevier C. S., Kaiser C. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 836–847 [DOI] [PubMed] [Google Scholar]
- 2.Hwang C., Sinskey A. J., Lodish H. F. (1992) Science 257, 1496–1502 [DOI] [PubMed] [Google Scholar]
- 3.Sevier C. S., Kaiser C. A. (2006) Antioxid. Redox Signal. 8, 797–811 [DOI] [PubMed] [Google Scholar]
- 4.Hebert D. N., Molinari M. (2007) Physiol. Rev. 87, 1377–1408 [DOI] [PubMed] [Google Scholar]
- 5.Appenzeller-Herzog C., Ellgaard L. (2008) Biochim. Biophys. Acta 1783, 535–548 [DOI] [PubMed] [Google Scholar]
- 6.Ellgaard L., Ruddock L. W. (2005) EMBO Rep. 6, 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkinson B., Gilbert H. F. (2004) Biochim. Biophys. Acta 1699, 35–44 [DOI] [PubMed] [Google Scholar]
- 8.Klappa P., Ruddock L. W., Darby N. J., Freedman R. B. (1998) EMBO J. 17, 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pirneskoski A., Klappa P., Lobell M., Williamson R. A., Byrne L., Alanen H. I., Salo K. E., Kivirikko K. I., Freedman R. B., Ruddock L. W. (2004) J. Biol. Chem. 279, 10374–10381 [DOI] [PubMed] [Google Scholar]
- 10.Mezghrani A., Fassio A., Benham A., Simmen T., Braakman I., Sitia R. (2001) EMBO J. 20, 6288–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L., Li S. J., Sidhu A., Zhu L., Liang Y., Freedman R. B., Wang C. C. (2009) J. Biol. Chem. 284, 199–206 [DOI] [PubMed] [Google Scholar]
- 12.Sevier C. S., Kaiser C. A. (2008) Biochim. Biophys. Acta 1783, 549–556 [DOI] [PubMed] [Google Scholar]
- 13.Molinari M., Helenius A. (1999) Nature 402, 90–93 [DOI] [PubMed] [Google Scholar]
- 14.Oliver J. D., van der Wal F. J., Bulleid N. J., High S. (1997) Science 275, 86–88 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Baig E., Williams D. B. (2006) J. Biol. Chem. 281, 14622–14631 [DOI] [PubMed] [Google Scholar]
- 16.Kang S. J., Cresswell P. (2002) J. Biol. Chem. 277, 44838–44844 [DOI] [PubMed] [Google Scholar]
- 17.Jessop C. E., Chakravarthi S., Garbi N., Hämmerling G. J., Lovell S., Bulleid N. J. (2007) EMBO J. 26, 28–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D. Y., Nagata K. (2008) Science 321, 569–572 [DOI] [PubMed] [Google Scholar]
- 19.Anelli T., Sitia R. (2008) EMBO J. 27, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng X., Zhang C., Chen J., Peng S., Cao Y., Ying K., Xie Y., Mao Y. (2003) Biochem. Genet. 41, 99–106 [DOI] [PubMed] [Google Scholar]
- 21.Haugstetter J., Maurer M. A., Blicher T., Pagac M., Wider G., Ellgaard L. (2007) J. Biol. Chem. 282, 33859–33867 [DOI] [PubMed] [Google Scholar]
- 22.Haugstetter J., Blicher T., Ellgaard L. (2005) J. Biol. Chem. 280, 8371–8380 [DOI] [PubMed] [Google Scholar]
- 23.Matsuo Y., Akiyama N., Nakamura H., Yodoi J., Noda M., Kizaka-Kondoh S. (2001) J. Biol. Chem. 276, 10032–10038 [DOI] [PubMed] [Google Scholar]
- 24.Matsuo Y., Nishinaka Y., Suzuki S., Kojima M., Kizaka-Kondoh S., Kondo N., Son A., Sakakura-Nishiyama J., Yamaguchi Y., Masutani H., Ishii Y., Yodoi J. (2004) Arch. Biochem. Biophys. 423, 81–87 [DOI] [PubMed] [Google Scholar]
- 25.Matsuo Y., Masutani H., Son A., Kizaka-Kondoh S., Yodoi J. (2009) Mol. Biol. Cell 20, 4552–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert N., Freedman R. B. (1985) Biochem. J. 228, 635–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyles M. M., Gilbert H. F. (1991) Biochemistry 30, 619–625 [DOI] [PubMed] [Google Scholar]
- 28.Yoshida H., Haze K., Yanagi H., Yura T., Mori K. (1998) J. Biol. Chem. 273, 33741–33749 [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto K., Yoshida H., Kokame K., Kaufman R. J., Mori K. (2004) J. Biochem. 136, 343–350 [DOI] [PubMed] [Google Scholar]
- 30.Kishigami S., Akiyama Y., Ito K. (1995) FEBS Lett. 364, 55–58 [DOI] [PubMed] [Google Scholar]
- 31.Wu H. H., Thomas J. A., Momand J. (2000) Biochem. J. 351, 87–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Appenzeller-Herzog C., Ellgaard L. (2008) Antioxid. Redox Signal. 10, 55–64 [DOI] [PubMed] [Google Scholar]
- 33.Hosokawa N., Wada I., Natsuka Y., Nagata K. (2006) Genes Cells 11, 465–476 [DOI] [PubMed] [Google Scholar]
- 34.Hosokawa N., Wada I., Nagasawa K., Moriyama T., Okawa K., Nagata K. (2008) J. Biol. Chem. 283, 20914–20924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motohashi K., Kondoh A., Stumpp M. T., Hisabori T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jessop C. E., Bulleid N. J. (2004) J. Biol. Chem. 279, 55341–55347 [DOI] [PubMed] [Google Scholar]
- 37.Jessop C. E., Tavender T. J., Watkins R. H., Chambers J. E., Bulleid N. J. (2009) J. Biol. Chem. 284, 2194–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frickel E. M., Frei P., Bouvier M., Stafford W. F., Helenius A., Glockshuber R., Ellgaard L. (2004) J. Biol. Chem. 279, 18277–18287 [DOI] [PubMed] [Google Scholar]
- 39.Roth D., Lynes E., Riemer J., Hansen H. G., Althaus N., Simmen T., Ellgaard L. (2010) Biochem. J. 425, 195–205 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.