Abstract
HDAC10 belongs to the class II histone deacetylase family; however, its functions remain enigmatic. We report here that the HDAC10 protein complex contained deacetylated chaperone protein hsc70, and HDAC10 relieved repression of melanogenesis by decreasing the repressional activity of two transcriptional regulators, paired box protein 3 (Pax3) and KRAB-associated protein 1 (KAP1). HDAC10 physically interacted with Pax3 and KAP1 in a ternary complex and maintained Pax3 and KAP1 in a deacetylated state. Deacetylated Pax3 and KAP1 derepressed promoters of microphthalmia-associated transcription factor (MITF) and melanocyte-specific tyrosinase-related protein 1 and 2 (TRP-1 and TRP-2), three genes of the melanogenesis cascade, in a trichostatin A-sensitive manner. Co-occupancy of melanogenic promoters by HDAC10, Pax3, and KAP1 only happened in cells of the melanocyte lineage, and KAP1 facilitated nuclear enrichment of HDAC10. Finally, cellular melanin content correlated directly with the expression level and activity of HDAC10. Our results not only show that HDAC10 regulates melanogenesis but also demonstrate that the transcriptional activities of Pax3 and KAP1 are intimately linked to their acetylation status.
Keywords: Histones/Deacetylase, Transcription/Regulation, Chromatin Immunoprecipitation (ChiP), Differentiation, Protein-Protein Interactions, HDAC10, Hsc70, KAP1, Pax3, Melanogenesis
Introduction
It is well established that histone acetyltransferases and histone deacetylases (HDACs)3 dynamically modify the N termini of core histones, change the chromatin structure, and modulate transcription. Histone acetyltransferases transfer acetyl groups from acetyl-CoA to the ϵ-amino group of lysine residues, thereby neutralizing the positive charge of the histone tails and decreasing their affinity for DNA. HDACs, in contrast, catalyze the hydrolysis of N-acetyllysines and restore the positive charge to the histone tails (for review, see Ref. 1). Several coactivators have histone acetyltransferase activity, whereas HDACs are found in corepressor complexes (2, 3). Therefore, histone acetyltransferases and HDACs are important gene regulators by changing local chromatin structures and the accessibility of DNA to other transcriptional regulators.
Previous reports demonstrated that histone acetyltransferases/HDACs modify not only core histones but also nonhistone proteins, most of which are transcription factors (for review, see 4). Acetylation/deacetylation of transcription factors may affect their DNA-binding activities, protein stability, or protein-protein interactions, thereby contributing to proper transcriptional regulation.
HDAC10 is a member of the class II HDAC family (5–8). The expression level of HDAC10 decreases in patients with lung cancer (9), suggesting that HDAC10 is important for maintaining the normal behavior of cells. Although HDAC10 represses transcription when tethered to a target promoter, a fundamental understanding of the function of HDAC10 remains elusive. In this report, we show that HDAC10 interacted with nonhistone proteins and kept them in a deacetylated form. Deacetylated heat shock protein 70 was a component of the HDAC10 stable protein complex. More important, in cells of melanogenic lineage, deacetylated paired box protein 3 (Pax3) and Krüppel-associated box (KRAB)-associated protein 1 (KAP1) derepressed promoters of microphthalmia-associated transcription factor (MITF) and melanocyte-specific tyrosinase-related protein 1 and 2 (TRP-1 and TRP-2), three genes that control the melanogenesis program. HDAC10 not only formed a ternary complex with Pax3 and KAP1 on these melanogenic promoters but also directly affected cellular melanin content. This is the first report demonstrating that HDAC10 regulates melanogenesis by decreasing the repressional activity of transcriptional regulators Pax3 and KAP1, which demonstrates that the repressional activity of Pax3 and KAP1 is affected by their acetylation status.
EXPERIMENTAL PROCEDURES
Plasmids
To express hemagglutinin (HA)-tagged, FLAG-tagged, glutathione S-transferase (GST)-tagged, and GFP-tagged HDAC10, the coding region of human HDAC10 (a gift from H. Y. Kao) (7) was subcloned into pcDNA3.1-HA, pM, pGSTag, and pEGFP (Clontech). Deletional mutations of GST-HDAC10 were made using restriction enzymes. 6×PRS-9Luc was made by subcloning the Pax3-binding sites from 6×PRSCAT (10) into pGL3 (Promega). Gal4-KAP1 was made by inserting the coding region of human KAP1 into a pM expression vector, which expresses a chimeric Gal4-KAP1 protein in mammalian cells. Plasmid constructs expressing FLAG-HDAC4, FLAG-HDAC5, and FLAG-HDAC6 have been described (11). HA-KAP1 was constructed by inserting the coding region of KAP1 into pcDNA3.1-HA (3). The TRP-1 promoter construct, which contains a segment of the TRP-1 promoter corresponding to nucleotide positions −336 to +114, was made by PCR-amplifying a segment of the genomic DNA from HEK293 and then subcloning into pGL3 (Promega). The MITF promoter construct was made similarly and contains a segment of the MITF promoter corresponding to nucleotide positions −382 to +95. TRP-2 promoter-Luc was a gift from D. Lang (12). All plasmid constructs were confirmed by sequencing.
FLAG-Pax3, FLAG-Pax3N, FLAG-Pax3C, GST-Pax3, HA-Pax3, FLAG-HP1β, and FLAG-KAP1 expression vectors have been described (13). TKLuc and G5TKLuc have been described previously (14).
HDAC10 Protein Complex Purification
An anti-FLAG immunoaffinity column was prepared using anti-FLAG M2 affinity gel (Sigma) following the manufacturer's suggestions. Approximately for every 6 × 107 HEK293 cells, 5 μg of the plasmid expressing FLAG-HDAC10 fusion protein was transfected using the calcium phosphate coprecipitation method (15). 48 h after transfection, cells were harvested by scraping. Cells were subsequently lysed by adding PBS plus 0.1% Nonidet P-40 and then briefly sonicating. Cell lysate obtained from approximately 4.8 × 109 cells was applied to an equilibrated FLAG column of 1-ml bed volume to allow for adsorption of the protein complex to the column resin. After binding, the column was washed with cold PBS plus 0.1% Nonidet P-40. FLAG peptide (Sigma) was applied to the column as described by the manufacturer to elute the FLAG-HDAC10 protein complex. Fractions of 1 bed volume were collected. A mock complex was purified simultaneously from HEK293 cells transfected with the empty FLAG vector as control.
In Vitro Protein-Protein Interaction Assays
GST fusion proteins were expressed in Escherichia coli strain DH5α and captured onto glutathione-agarose beads (Sigma). Plasmid constructs expressing either HA-tagged or FLAG-tagged proteins were transfected into HEK293 cells. Cells were then lysed in PBS plus 0.2% Nonidet P-40 for 0.5 h, and the resulting cell extracts containing HA-tagged or FLAG-tagged proteins were mixed with the beads in the presence of PBS plus 0.2% Nonidet P-40 at 4 °C for 1 h. Beads were then washed extensively in PBS plus 0.2% Nonidet P-40. Bound proteins were eluted by boiling in Laemmli sample buffer, separated by SDS-PAGE, and detected by Coomassie Blue staining and Western blot analysis.
Glycerol Gradient Sedimentation
2 × 107 cells were lysed in 1 ml of PBS plus 0.2% Nonidet P-40. 100 μl of cleared lysate was applied onto a 4-ml glycerol gradient (5–45%) and spun at 368,000× g in a Beckman SW55Ti rotor for 24 h. Fractions of 100 μl were collected, and 15 μl of each fraction was analyzed by Western blotting (16).
Immunoprecipitation, Ternary Complex Detection, and Western Blot Analysis
Immunoprecipitation of FLAG-tagged proteins were carried out as described previously (3). Western blot analyses were performed using standard protocols. To detect the HDAC10·Pax3·KAP1 ternary complex, tagged proteins were expressed in HEK293 cells. Cells were harvested and lysed in a buffer containing 50 mm Tris (pH 8.0), 50 mm NaCl, 5 mm MgCl2, 10% glycerol, and 0.5% Nonidet P-40. The lysates were immunoprecipitated with an anti-FLAG antibody and then eluted using a FLAG peptide. The eluates were used in a second immunoprecipitation against the HA tag. The final immunoprecipitates were analyzed by Western blotting using an anti-Myc antibody as described previously (17).
Cell Culture, Transfection, and Luciferase Assay
HEK293 cells and B16F10 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and penicillin-streptomycin. 106 cells were seeded onto 60-mm-diameter tissue culture dishes. 16 h later, 0.5 μg of pRL-TK, 5 μg of the reporter plasmid, and 10 μg of the overexpression plasmid(s) were transfected into cells using the calcium phosphate coprecipitation method (15) or Lipofectamine (Invitrogen). 48 h after transfection, cells were harvested, and a luciferase assay was performed using the Dual Luciferase Assay system (Promega).
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP experiments were done essentially as described previously (13, 18) with the 6×PRS-9 reporter plasmid (6×PRS-9Luc) or the TK-luciferase reporter containing Gal4-binding sites (G5TKLuc) and appropriate expression constructs. Immunoprecipitated DNA fragments were amplified by PCR using specific primer pairs (5′-gagctctccggatccaagcttgc-3′ and 5′-cttccagcggatagaatggcgccg-3′).
Fluorescence Microscopy
HEK293 cells were seeded on chamber slides and grown for 18 h. 5 μg of expression plasmids for various proteins were transfected into cells using the calcium phosphate coprecipitation method (15). 48 h later, cells were washed with PBS, fixed with 4% paraformaldehyde, rinsed with PBS again, and dried. One drop of antifade mounting medium with 4′,6′-diamidino-2-phenylindole (Vector) was added to the cells before coverslips were applied. Images were observed and analyzed using a fluorescence microscope (14).
HDAC10 Short Hairpin RNA (shRNA)
The shRNA expression vector specifically for HDAC10 was obtained commercially with a target sequence 5′-aaacccagggctgccttggaaaag-3′ (Gendiscovery).
Melanin Content Assay
Melanin content of cultured B16F10 murine melanoma cells was measured according to Oka et al. (19). B16F10 cells were seeded at a density of 5 × 103 cells/6-cm culture plate. 24 h later, the cells were transfected with the plasmids indicated. 48 h after transfection, cells were washed with PBS once, lysed with 200 μl of 1 m NaOH, and solubilized by boiling for 10 min. Melanin content was gauged by spectrophotometric analysis at 400 nm absorbance. 500 nm trichostatin A (TSA) was added to cells at 24 h before harvesting. Readings of spectrophotometric analysis were normalized against cell number. Cell numbers were monitored by trypan blue dye exclusion assay.
RESULTS
HDAC10 Protein Complex
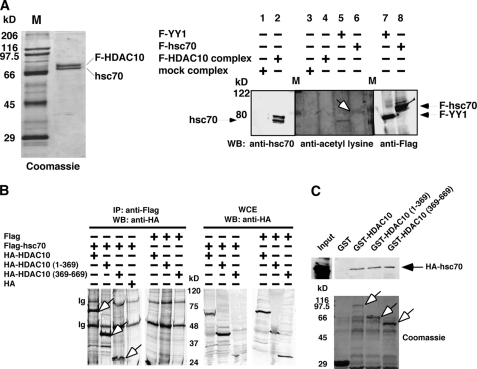
To understand the function of HDAC10, we obtained an HDAC10 protein complex using affinity purification (Fig. 1A, left). One major protein in the HDAC10 complex was heat shock cognate protein 70 (hsc70), whose specific interaction with HDAC10 was confirmed by immunoprecipitation assay and GST pulldown assay (Fig. 1, B and C). Interestingly, hsc70 was acetylated when purified by itself from cells (Fig. 1A, right, lane 6) but lost the acetylation mark when purified as a component of the HDAC10 complex (lane 4). Although direct evidence that HDAC10 deacetylates hsc70 was inconclusive, this result suggests that the targets of HDAC10 include nonhistone proteins.
FIGURE 1.
HDAC10-associated hsc70 is unacetylated. A, left, composition of the FLAG-HDAC10 complex from HEK293 cells. Right, hsc70 in the HDAC10 complex is unacetylated. Samples from the FLAG-HDAC10 complex or mock complex were probed with an anti-acetyllysine antibody. Immunoprecipitated FLAG-YY1 from HEK293 cells served as a positive control for acetylation. An open arrow indicates that immunoprecipitated FLAG-hsc70 was acetylated in cells. WB, Western blotting. B, HDAC10 interacts with hsc70. Hsc70 was expressed in HEK293 cells as a FLAG fusion protein and immunoprecipitated (IP) by anti-FLAG antibodies. Coprecipitated HA-HDAC10 and HDAC10 deletion mutants were detected by Western blotting. HDAC10 (1–369) contains amino acids 1–369 of HDAC10. HDAC10 (369–669) contains amino acids 369–669 of HDAC10. Open arrows indicate the positions of HA-HDAC10 deletion constructs that interacted with FLAG-hsc70. Ig, immunoglobulins; WCE, whole cell extract. C, HEK293-expressed HA-hsc70 was captured by immobilized GST-HDAC10 and GST-HDAC10 deletion mutants. Open arrows indicate the positions of various GST-HDAC10 deletion mutants.
HDAC10 Specifically Interacts with Unacetylated Pax3
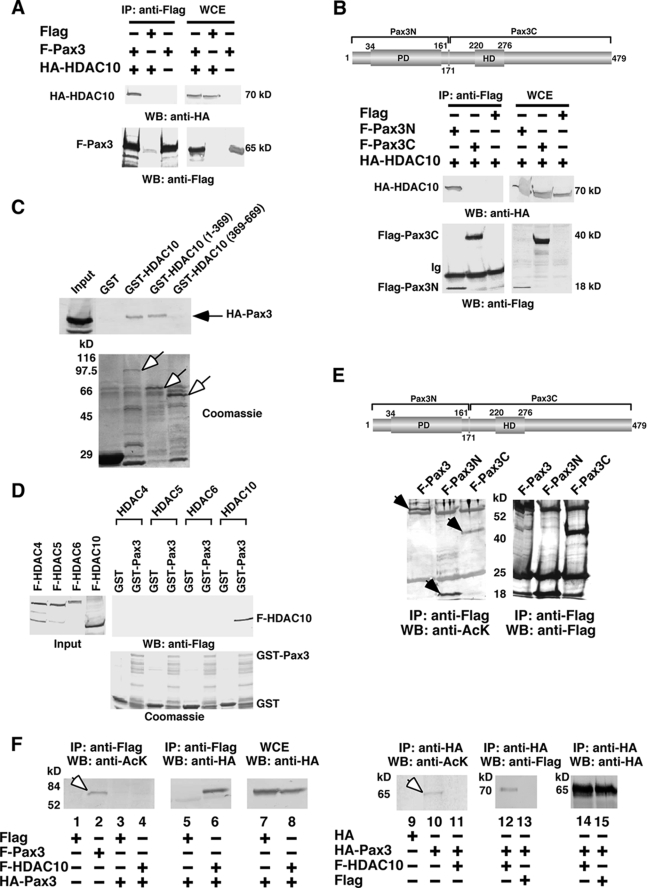
Meanwhile, we speculated that, without obvious DNA-binding domains, HDAC10 is recruited to target promoters through protein-protein interactions with other DNA-binding factors. After screening a panel of transcription factors in coimmunoprecipitation assays, we found that HDAC10 specifically interacted with Pax3, which is an important transcription factor for the normal development of neural crest, skeletal muscles, and melanocyte stem cells (for review, see Ref. 20). HDAC10 and Pax3 showed specific interactions at the paired domain of Pax3 and the putative HDAC catalytic domain of HDAC10 (Fig. 2, A–C). Pax3 failed to interact with HDAC4, HDAC5, and HDAC6, three other class II HDACs (Fig. 2D) or any of the class I HDACs (data not shown).
FIGURE 2.
HDAC10-associated Pax3 is unacetylated. A, HDAC10 interacts with Pax3. Anti-FLAG immunoprecipitates (IP) from HEK293 cell extracts expressing the indicated combinations of proteins were probed with anti-HA antibodies by Western blot (WB) analysis. The whole cell extract (WCE) for each lane was loaded with 1/10 the amount of the extract used in the immunoprecipitation reaction to confirm the expression of proteins. B, HDAC10 interacts with the paired domain-containing N terminus of Pax3 in coimmunoprecipitation assay. C, Pax3 interacts with the catalytic domain of HDAC10 in GST pulldown assay. HEK293-expressed HA-Pax3 was captured by immobilized GST-HDAC10 and GST-HDAC10 (1–369), but not by GST-HDAC10 (369–669). HDAC10 (1–369) contains mainly the putative catalytic domain of HDAC10. Input represents 1/10 the amount of the expressed proteins used in each binding reaction. Open arrows indicate the positions of various GST-HDAC10 deletion mutants. D, Pax3 interacts specifically with HDAC10. HEK293-expressed FLAG-HDAC10, but not FLAG-HDAC4, FLAG-HDAC5, or FLAG-HDAC6, was captured by immobilized GST-Pax3. E, Pax3 is an acetylated protein in cells. A schematic diagram of Pax3 is provided. Arabic numerals indicate amino acid numbers. HEK293 cell extracts expressing FLAG-Pax3, FLAG-Pax3N, or FLAG-Pax3C were immunoprecipitated with anti-FLAG antibodies. The immunoprecipitates were divided in two for Western blotting using either anti-acetyllysine antibodies or anti-FLAG antibodies. Arrows indicate the positions of acetylated proteins. F, HDAC10 interacts with unacetylated Pax3. Immunoprecipitates from HEK293 cell extracts expressing various combinations of Pax3 and HDAC10 proteins were probed by anti-acetyllysine antibodies. Lanes 2 and 10 serve as control for the detection of acetylated Pax3, with arrows indicating their positions.
In Western blot analysis using anti-acetyllysine antibodies, we found that Pax3 in cells is an acetylated protein (Fig. 2E). We also found that, in the HDAC10 immunoprecipitate, Pax3 was unacetylated (Fig. 2F, lane 4), indicating that the fraction of cellular Pax3 bound to HDAC10 as detected by coimmunoprecipitation is specifically enriched for an unacetylated form. This result reinforces our previous finding that the targets of HDAC10 include nonhistone proteins.
HDAC10 Interacts with Unacetylated KAP1
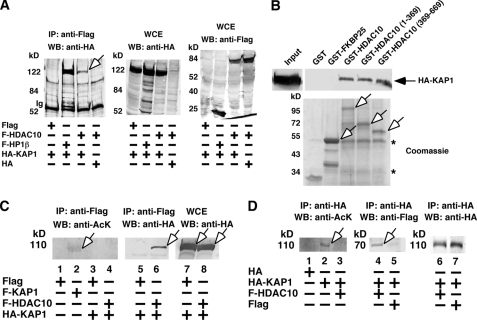
It has been demonstrated that Pax3 recruits a KRAB domain-containing corepressor called KAP1 to repress transcription (13), and we found that indeed HDAC10 interacted with KAP1 (Fig. 3, A and B). Interestingly, KAP1 was also detected as an acetylated protein (Fig. 3C, lane 2). Although KAP1 readily coimmunoprecipitated with FLAG-HDAC10 (lane 5), HDAC10-bound KAP1 was unacetylated (lane 3). To eliminate the possibility that acetylation of KAP1 was contributed by the lysines on the FLAG tag, we tested HA-tagged KAP1 and found that it was acetylated (Fig. 3D, lane 2). Importantly, we did not detect any acetylated KAP1 when HDAC10 was overexpressed (lane 3), and this was not due to failure of expression or interaction (lane 4), indicating that KAP1 is deacetylated in the presence of bound HDAC10.
FIGURE 3.
HDAC10-associated KAP1 is unacetylated. A, FLAG-HDAC10 interacts with HA-KAP1 in HEK293 cells. FLAG-HP1β was used as a positive control for interaction with KAP1. An open arrow indicates the position of HA-KAP1 coimmunoprecipitated with FLAG-HDAC10. IP, immunoprecipitation; WB, Western blotting; WCE, whole cell extract. B, HEK293-expressed HA-KAP1 was captured by immobilized GST-HDAC10, GST-HDAC10 (1–369), and GST-HDAC10 (369–669). Input represents the total amount of the expressed proteins used in each binding reaction. Open arrows indicate the positions of various GST-HDAC10 deletion mutants and GST-FKBP25, which is GST fused to immunophilin FKBP25 serving as a negative control. Stars indicate nonspecific protein bands. C, KAP1 is an acetylated protein, and HDAC10 interacts with unacetylated KAP1 in HEK293 cells. Arrows indicate the positions of FLAG-KAP1 (acetylated) (lane 2) and HA-KAP1 (lanes 6-8). D, KAP1 is unacetylated in the presence of overexpressed HDAC10. Arrows indicate the positions of HA-KAP1 (acetylated) (lane 2) and FLAG-HDAC10 (lane 4).
HDAC10, Pax3, and KAP1 Form a Ternary Complex
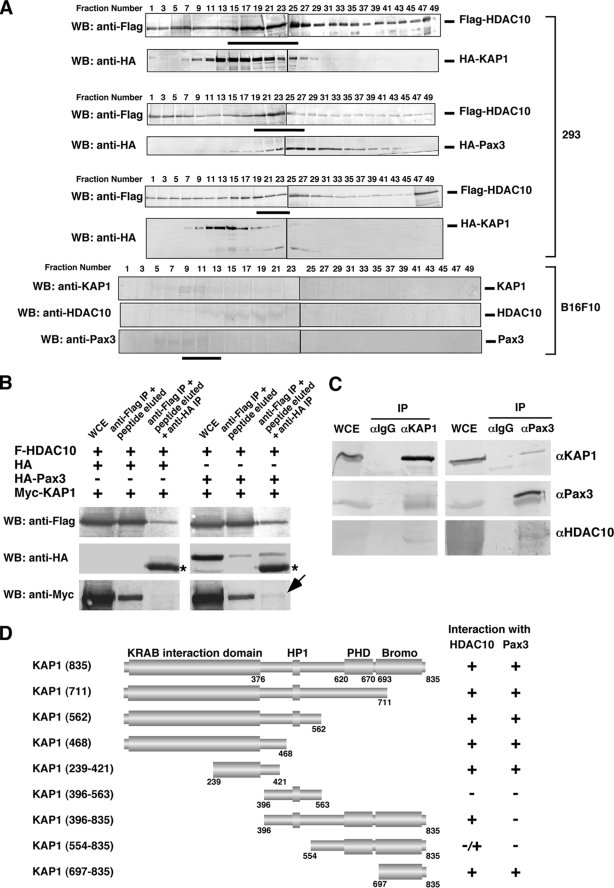
To understand further how HDAC10, Pax3, and KAP1 interact in the cell, we first performed glycerol gradient analysis to analyze protein distribution in cell extracts. Using overexpressed proteins in HEK293 cells, we found that the distribution of HDAC10 and KAP1 overlapped in fractions 15–25. For HDAC10 and Pax3, the overlap was from fraction 19 to fraction 27. When HDAC10, KAP1, and Pax3 coexpressed, they coexisted in fractions 19–23, consistent with the individual pairwise analyses (Fig. 4A, top panels). In lysates from B16F10 cells, the endogenously expressed KAP1, HDAC10, and Pax3 also showed significant coexistence, suggesting that these three proteins form a stable complex in the cell (Fig. 4A, bottom panels). Next, using serial coimmunoprecipitation assays, we found that HDAC10 formed a ternary complex with Pax3 and KAP1 (Fig. 4B). Endogenous interactions among HDAC10, Pax3, and KAP1 were detected in Pax3-expressing B16F10 melanoma cells (Fig. 4C). Coimmunoprecipitation experiments revealed that both the KAP1-HDAC10 interaction domain and the KAP1-Pax3 interaction domain mapped to the central region of KAP1 (amino acids 239–421) and the C-terminal Bromo domain of KAP1, consisting with the idea that HDAC10, KAP1, and Pax3 coexist in a ternary complex (Fig. 4D and supplemental Fig. S1).
FIGURE 4.
HDAC10, KAP1, and Pax3 form a ternary complex. A, glycerol gradient analysis of HDAC10, KAP1, and Pax3. Top panels, lysates from HEK293 cells expressing combinations of proteins as indicated were sedimented through a 5–45% glycerol gradient and analyzed by Western blotting (WB). Bottom panels, lysates from B16F10 cells expressing endogenous Pax3, KAP1, and HDAC10 were analyzed. Thick lines indicate overlaps of the distribution of the proteins. B, HDAC10, Pax3, and KAP1 exist in the same complex. HEK293 cell lysates expressing FLAG-HDAC10, HA-Pax3, and Myc-KAP1 were serially immunoprecipitated (IP) and eluted. An arrow indicates the presence of KAP1 in the final immunoprecipitate. Asterisks indicate the positions of IgG shown as a result of sequential anti-HA immunoprecipitation and anti-HA Western blotting. WCE, whole cell extract. C, endogenous interaction among HDAC10, Pax3, and KAP1. B16F10 cells were immunoprecipitated then probed with the antibodies indicated on the right. D, summary of coimmunoprecipitation results shows that the KAP1-HDAC10 interaction domain and the KAP1-Pax3 interaction domain overlap.
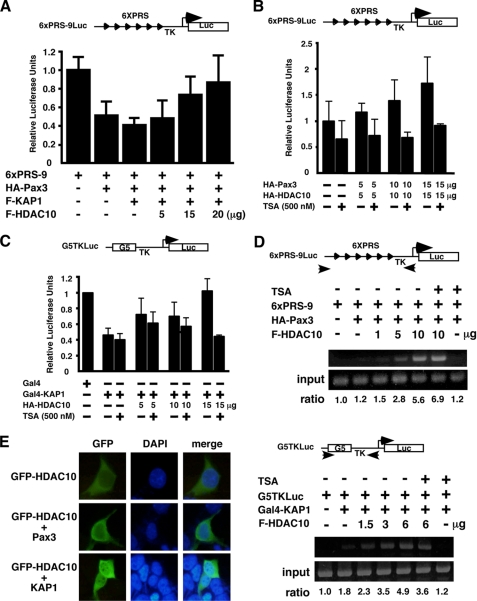
HDAC10 Attenuates the Repressional Activity of Pax3 and KAP1
Increasing amounts of HDAC10 decreased the transcriptional repression activity of Pax3 and KAP1 on 6×PRS-9, a heterologous promoter with six Pax3-binding sites (Fig. 5A). To test whether derepression was associated with the histone deacetylase activity of HDAC10, we used TSA, a histone deacetylase inhibitor (21), in reporter assays. TSA alone did not derepress the 6×PRS-9 promoter (supplemental Fig. S2). However, the derepressional effect of HDAC10 on Pax3 was erased in the presence of TSA (Fig. 5B). TSA also diminished the derepressional effect of HDAC10 on KAP1 when KAP1 was expressed as a Gal4 fusion protein recruited to a promoter containing Gal4-binding sites (Fig. 5C). However, ChIP assay results showed that increasing amounts of HDAC10 increased the amount of Pax3 or KAP1 on target promoters by an undefined mechanism, and TSA had little effect on the promoter occupancy by Pax3 or KAP1 in the absence of HDAC10 (Fig. 5D), precluding the possibility that HDAC10 abolishes the repressional activity of Pax3 and KAP1 by preventing their association with the promoters. Interestingly, although HDAC10 located in both the nucleus and the cytoplasm with a cytoplasmic enrichment (Fig. 5E) (6–8), KAP1, but not Pax3 alone, significantly increased the nuclear fraction of HDAC10 (Fig. 5E), suggesting that HDAC10 participates in KAP1-mediated transcriptional activity. Together, these results show that HDAC10 decreases the activities of repressor Pax3 and corepressor KAP1 on Pax3-target promoters, and this phenomenon is diminished by histone deacetylase inhibitor TSA.
FIGURE 5.
HDAC10 regulates the transcriptional activities of Pax3 and KAP1. A, HDAC10 relieves the transcriptional repression activity of Pax3 and KAP1 on a Pax3 target promoter. HEK293 cells were transfected with the expression constructs indicated and a reporter plasmid 6×PRS-9Luc with six tandem repeats of paired domain recognition sequences upstream of the thymidine kinase (TK) promoter. The total amounts of plasmids used were kept uniform using parental expression plasmids if needed. B, HDAC10 decreases Pax3-mediated repression in a TSA-sensitive manner. C, HDAC10 decreases KAP1-mediated repression in a TSA-sensitive manner. G5TKLuc is a luciferase reporter plasmid containing five tandem Gal4-binding sites. D, HDAC10 increases promoter occupancy by Pax3 or KAP1. DNA fragments bound by HA-Pax3 or Gal4-KAP1 were obtained by immunoprecipitation using anti-FLAG antibodies to bring down FLAG-HDAC10 and associated Pax3 or KAP1 with their target promoter fragments. These promoter fragments were subsequently PCR-amplified using specific primers whose positions are indicated. E, KAP1 promotes nuclear enrichment of HDAC10. GFP-HDAC10, either alone or together with an expression construct of Pax3 or KAP1, was transfected into HEK293 cells. Cells were fixed, and the subcellular distribution of GFP-HDAC10 was analyzed by fluorescence microscopy.
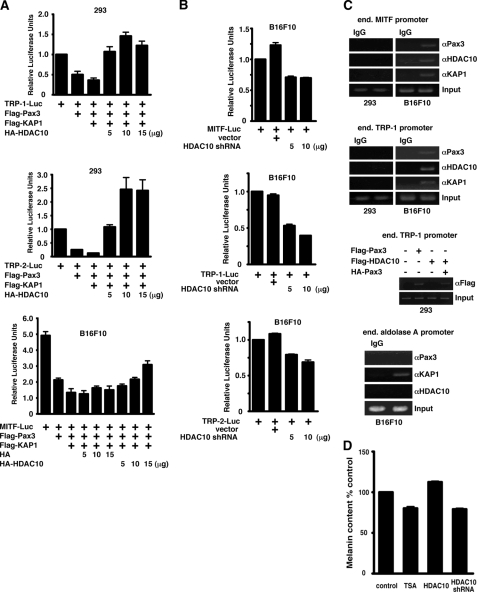
HDAC10 Relieves Repression of the Melanogenesis Program
Pax3 regulates expression of MITF (22, 23), TRP-1, and TRP-2 (12, 24). TRP-1 and TRP-2 encode two tyrosinase-related proteins with tyrosinase and DOPAchrome tautomerase activities and control the synthesis of eumelanins, which are melanin pigments that absorb UV light and scavenge free radicals (25). We found that in HEK293 cells or in B16F10 melanoma cells, Pax3 repressed expression from TRP-1, TRP-2, and MITF promoters, KAP1 further repressed these three promoters, and HDAC10 relieved repression by Pax3 and KAP1 in a dose-dependent manner (Fig. 6A). In B16F10 cells, knocking down the expression of endogenous HDAC10 with shRNA effectively caused repression of the MITF, TRP-1, and TRP-2 promoters (Fig. 6B). ChIP assays showed that physiological levels of HDAC10, Pax3, and KAP1 occupied the endogenous MITF and TRP-1 promoters in B16F10 cells, but not in HEK293 cells (Fig. 6C, top two panels) where very little Pax3 or HDAC10 was expressed, demonstrating that the melanogenesis program is derepressed in cells of the melanocyte lineage. The same assay condition was applied to HEK293 cells expressing exogenous Pax3, KAP1, and HDAC10, and the result confirmed that the failure to detect the HDAC10·Pax3·KAP1 complex in HEK293 cells was not an artifact because HA-Pax3 recruited FLAG-HDAC10 to the endogenous TRP-1 promoter (Fig. 6C, middle panel). In contrast, endogenous aldolase A promoter, which is regulated by KAP1 but not Pax3 (26), showed no detectable binding of Pax3 or HDAC10 (Fig. 6C, bottom panel). Furthermore, in B16F10 cells that were actively producing melanin, overexpression of HDAC10 resulted in a higher melanin content as shown by melanin content assay, whereas decreasing the level of HDAC10 by shRNA and administration of HDAC inhibitor TSA both led to a lower amount of cellular melanin (Fig. 6D). These results demonstrate that in cells of the melanocyte lineage, HDAC10 negatively regulates the repressional activities of Pax3 and KAP1, essentially activating the melanogenesis program.
FIGURE 6.
HDAC10 activates the melanogenesis program by decreasing the repressional activities of Pax3 and KAP1. A, KAP1 potentiates the ability of Pax3 to repress expression of TRP-1, TRP-2, and MITF, and HDAC10 reverts the repressional activity of Pax3 and KAP1 in HEK293 cells and B16F10 melanoma cells. TRP-1, TRP-2, and MITF promoter constructs were linked to a luciferase reporter. Total amounts of plasmids used were kept uniform using parental expression plasmids if needed. B, knocking-down HDAC10 decreases expression of MITF, TRP-1, and TRP-2. MITF, TRP-1, and TRP-2 promoter constructs and an shRNA construct specifically for HDAC10 were transfected into B16F10 cells. Vector, empty shRNA vector. C, ChIP assays show that HDAC10, Pax3, and KAP1 occupy MITF and TRP-1 promoters in B16F10 melanoma cells, but not in HEK293 cells. The endogenous (end.) aldolase A promoter, which is regulated by KAP1 but not by Pax3, was bound by KAP1 and not by Pax3 or HDAC10. IgG, isotype-matched IgGs as controls for immunoprecipitation. D, cellular melanin content directly correlates with HDAC10 activity. Melanin content was normalized against cell number and is expressed as percent control.
DISCUSSION
Biological Functions of HDAC10
HDAC10 was cloned in 2002 based on sequence homology to HDACs 1–8 (5) or to selected class II HDACs (6–8) and was shown to posses deacetylase activities toward acetylated histones or H4 peptides (5–8). However, biochemical analyses have failed to provide a link between the catalytic activity of HDAC10 and its cellular function. By analyzing the HDAC10 protein complex and proteins interacting with HDAC10, we found that HDAC10 decreased the transcriptional repression activity of Pax3 and KAP1 on melanogenic promoters, in essence activating the melanogenesis program, by keeping Pax3 and KAP1 in deacetylated forms. Therefore, we propose that association with HDAC10 constitutes a novel mechanism of active preservation of derepression by maintaining the deacetylation status of transcriptional regulators that function in delicate developmental switches.
Pax3, KAP1, and Melanogenesis
Mutations of Pax3 result in subtypes 1 and 3 of Waardenburg syndrome as they cause auditory-pigmentary impairment directly associated with neural crest-derived melanocyte deficiency (27). During melanogenesis, the final steps of eumelanin production are controlled by protein products from TRP-1 and TRP-2 genes (25). The expression of TRP-2 is directly activated by MITF (28, 29), whose gene expression is activated by Pax3 and Sox10 (22, 23, 30, 31), thus constituting the melanogenesis cascade. Interestingly, Pax3 also controls gene expression of TRP-1 and TRP-2 (12, 24), and KAP1 acts as a corepressor for Pax3 (13). A recent finding shows that although Pax3 activates MITF gene expression by cooperating with Sox10, it also at the same time inhibits downstream gene expression such as TRP-2 by competing with MITF for promoter occupancy (12). In other words, Pax3 simultaneously acts as a primer for lineage determination and an agent of prevention of terminal differentiation. Our findings that HDAC10 relieves the repressional program of Pax3 by keeping Pax3 and its corepressor KAP1 deacetylated could represent a crucial switch in the transcriptional regulatory activity of Pax3 in differentiation programs such as the melanogenesis cascade.
Upstream Deacetylase(s)
Our experimental data strongly support the hypothesis that HDAC10 maintains the deacetylation status of hsc70, Pax3, and KAP1; however, we are uncertain whether HDAC10 actively deacetylates these proteins. Therefore, which enzyme functions as an upstream deacetylase remains to be a critical question. It has been shown that HDAC10 interacts with HDAC2 and HDAC3 (5, 8), and one recent report showed that HDAC1 coimmunoprecipitates with acetylated Pax3 in a medulloblastoma cell line with stably transfected Pax3 (32). However, in our laboratory we observed that Pax3 failed to interact with HDAC1, HDAC2, HDAC3, or HDAC8 in coimmunoprecipitation experiments.4 We believe that class I HDACs are not involved in deacetylation of Pax3 and HDAC10-mediated derepression of Pax3 target genes. Nevertheless, we do not exclude the possibility that other HDACs also participate in the modulation of the activities of proteins targeted by HDAC10. In fact, Mayanil et al. (32) showed that SirT1 binds to transforming growth factor-β promoter fragments where Pax3 binds, suggesting that SirT1 or other NAD+-dependent HDACs might participate in HDAC10-mediated regulatory events as upstream deacetylases of target proteins.
Regulation of the HDAC10, Pax3, and KAP1 Ternary Complex
Our result that the KAP1-HDAC10 interaction domains largely overlapped with the KAP1-Pax3 interaction domains (Fig. 4D) suggests that any disruption of the interaction between any two of the three proteins would result in rapid disassembly of the ternary complex, consistent with the idea that the HDAC10·Pax3·KAP1 complex plays a decisive role in melanogenesis. However, questions remain such as upon which developmental signal is the ternary complex formed and disassembled. Among the three proteins, HDAC10 is expressed ubiquitously (5, 7, 8), KAP1 is a general corepressor, and only Pax3 shows distinct developmentally regulated expression pattern. We suspect that not only the expression pattern of Pax3 determines where the HDAC10·Pax3·KAP1 complex forms, the acetylation and subsequent deacetylation events of Pax3 might also prove to be crucial to the timing of the formation of the ternary complex. Therefore, finding the regulatory signals that cue the acetylation and deacetylation of Pax3 (and KAP1) will likely to be key to answer the question of how the formation of the HDAC10·Pax3·KAP1 ternary complex is regulated.
General Roles of the Class IIb HDACs
HDAC10, like HDAC6, mainly exists in the cytoplasm, suggesting that HDAC10 may target nonhistone proteins other than transcription factors, similar to the case of HDAC6. By deacetylating α-tubulin, HDAC6 regulates cell migration, chemotaxis, and immune synapse formation (for review, see 33). By changing the acetylation level of cortactin, HDAC6 regulates the F-actin-binding ability of cortactin and cell motility (34). Furthermore, by deacetylating chaperone hsp90 and affecting binding and subsequent degradation of client proteins with hsp90, HDAC6 participates in gene transcription activated by glucocorticoid receptor (35) and in the modulation of the activity of pro-growth and pro-survival proteins in leukemia cells (36). Our finding that HDAC10 forms a protein complex with another chaperone hsc70 (Fig. 1) could be more than just an exciting coincidence. Moreover, HDAC10 is recruited to the nucleus in the presence of KAP1 (Fig. 5E), suggesting that the subcellular localization of HDAC10 is regulated to complete its function on different targets in different subcellular locales. Therefore, in addition to specific functions described in this report and other published work, it is possible that HDAC6 and HDAC10, members of the class IIb HDACs (37), affect the acetylation status of molecular chaperones and influence their downstream activities such as modulation of the action of nuclear hormone receptors and other regulatory proteins.
Supplementary Material
Acknowledgments
We thank Ronald Evans and Hung-Ying Kao for the HDAC10 cDNA, Eishou Matsuda for the KAP1 cDNA, and Deborah Lang for the TRP-2 promoter construct.
This work was supported by National Health Research Institutes Grant NHRI-9212SI (to W.-M. Y.) and by National Science Council Grants NSC96-2311-B-005-006-MY3 (to W.-M. Y.) and NSC95-2311-B-468-001-MY3 (to Y.-L. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
I-L. Lai, T.-P. Lin, Y.-L. Yao, C.-Y. Lin, M.-J. Hsieh, and W.-M. Yang, unpublished results.
- HDAC
- histone deacetylase
- Pax3
- paired box protein 3
- KAP1
- KRAB-associated protein 1
- MITF
- microphthalmia-associated transcription factor
- TRP
- tyrosinase-related protein
- HA
- hemagglutinin
- GST
- glutathione S-transferase
- PBS
- phosphate-buffered saline
- ChIP
- chromatin immunoprecipitation
- shRNA
- short hairpin RNA
- TSA
- trichostatin A.
REFERENCES
- 1.Kingston R. E., Narlikar G. J. (1999) Genes Dev. 13, 2339–2352 [DOI] [PubMed] [Google Scholar]
- 2.Jepsen K., Rosenfeld M. G. (2002) J. Cell Sci. 115, 689–698 [DOI] [PubMed] [Google Scholar]
- 3.Yao Y. L., Yang W. M. (2003) J. Biol. Chem. 278, 42560–42568 [DOI] [PubMed] [Google Scholar]
- 4.Glozak M. A., Sengupta N., Zhang X., Seto E. (2005) Gene 363, 15–23 [DOI] [PubMed] [Google Scholar]
- 5.Fischer D. D., Cai R., Bhatia U., Asselbergs F. A., Song C., Terry R., Trogani N., Widmer R., Atadja P., Cohen D. (2002) J. Biol. Chem. 277, 6656–6666 [DOI] [PubMed] [Google Scholar]
- 6.Guardiola A. R., Yao T. P. (2002) J. Biol. Chem. 277, 3350–3356 [DOI] [PubMed] [Google Scholar]
- 7.Kao H. Y., Lee C. H., Komarov A., Han C. C., Evans R. M. (2002) J. Biol. Chem. 277, 187–193 [DOI] [PubMed] [Google Scholar]
- 8.Tong J. J., Liu J., Bertos N. R., Yang X. J. (2002) Nucleic Acids Res. 30, 1114–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osada H., Tatematsu Y., Saito H., Yatabe Y., Mitsudomi T., Takahashi T. (2004) Int. J. Cancer 112, 26–32 [DOI] [PubMed] [Google Scholar]
- 10.Chalepakis G., Jones F. S., Edelman G. M., Gruss P. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 12745–12749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grozinger C. M., Hassig C. A., Schreiber S. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lang D., Lu M. M., Huang L., Engleka K. A., Zhang M., Chu E. Y., Lipner S., Skoultchi A., Millar S. E., Epstein J. A. (2005) Nature 433, 884–887 [DOI] [PubMed] [Google Scholar]
- 13.Hsieh M. J., Yao Y. L., Lai I. L., Yang W. M. (2006) Biochem. Biophys. Res. Commun. 349, 573–581 [DOI] [PubMed] [Google Scholar]
- 14.Yang W. M., Yao Y. L., Seto E. (2001) EMBO J. 20, 4814–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham F., van der Eb A. J. (1973) Virology 52, 456–467 [DOI] [PubMed] [Google Scholar]
- 16.Ogawa H., Ishiguro K., Gaubatz S., Livingston D. M., Nakatani Y. (2002) Science 296, 1132–1136 [DOI] [PubMed] [Google Scholar]
- 17.Liu F., Pouponnot C., Massagué J. (1997) Genes Dev. 11, 3157–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orlando V., Strutt H., Paro R. (1997) Methods 11, 205–214 [DOI] [PubMed] [Google Scholar]
- 19.Oka M., Ichihashi M., Chakraborty A. K. (1996) J. Invest. Dermatol. 106, 377–378 [DOI] [PubMed] [Google Scholar]
- 20.Lang D., Powell S. K., Plummer R. S., Young K. P., Ruggeri B. A. (2007) Biochem. Pharmacol. 73, 1–14 [DOI] [PubMed] [Google Scholar]
- 21.Yoshida M., Kijima M., Akita M., Beppu T. (1990) J. Biol. Chem. 265, 17174–17179 [PubMed] [Google Scholar]
- 22.Potterf S. B., Furumura M., Dunn K. J., Arnheiter H., Pavan W. J. (2000) Hum. Genet. 107, 1–6 [DOI] [PubMed] [Google Scholar]
- 23.Watanabe A., Takeda K., Ploplis B., Tachibana M. (1998) Nat. Genet. 18, 283–286 [DOI] [PubMed] [Google Scholar]
- 24.Galibert M. D., Yavuzer U., Dexter T. J., Goding C. R. (1999) J. Biol. Chem. 274, 26894–26900 [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi T., Vieira W. D., Potterf B., Sakai C., Imokawa G., Hearing V. J. (1995) J. Cell Sci. 108, 2301–2309 [DOI] [PubMed] [Google Scholar]
- 26.Medugno L., Florio F., De Cegli R., Grosso M., Lupo A., Costanzo P., Izzo P. (2005) Gene 359, 35–43 [DOI] [PubMed] [Google Scholar]
- 27.Tachibana M. (1999) J. Investig. Dermatol. Symp. Proc. 4, 126–129 [DOI] [PubMed] [Google Scholar]
- 28.Aksan I., Goding C. R. (1998) Mol. Cell. Biol. 18, 6930–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertolotto C., Buscà R., Abbe P., Bille K., Aberdam E., Ortonne J. P., Ballotti R. (1998) Mol. Cell. Biol. 18, 694–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bondurand N., Pingault V., Goerich D. E., Lemort N., Sock E., Le Caignec C., Wegner M., Goossens M. (2000) Hum. Mol. Genet. 9, 1907–1917 [DOI] [PubMed] [Google Scholar]
- 31.Verastegui C., Bille K., Ortonne J. P., Ballotti R. (2000) J. Biol. Chem. 275, 30757–30760 [DOI] [PubMed] [Google Scholar]
- 32.Mayanil C. S., Pool A., Nakazaki H., Reddy A. C., Mania-Farnell B., Yun B., George D., McLone D. G., Bremer E. G. (2006) J. Biol. Chem. 281, 24544–24552 [DOI] [PubMed] [Google Scholar]
- 33.Valenzuela-Fernández A., Cabrero J. R., Serrador J. M., Sánchez-Madrid F. (2008) Trends Cell Biol. 18, 291–297 [DOI] [PubMed] [Google Scholar]
- 34.Zhang X., Yuan Z., Zhang Y., Yong S., Salas-Burgos A., Koomen J., Olashaw N., Parsons J. T., Yang X. J., Dent S. R., Yao T. P., Lane W. S., Seto E. (2007) Mol. Cell 27, 197–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovacs J. J., Murphy P. J., Gaillard S., Zhao X., Wu J. T., Nicchitta C. V., Yoshida M., Toft D. O., Pratt W. B., Yao T. P. (2005) Mol. Cell 18, 601–607 [DOI] [PubMed] [Google Scholar]
- 36.Bali P., Pranpat M., Bradner J., Balasis M., Fiskus W., Guo F., Rocha K., Kumaraswamy S., Boyapalle S., Atadja P., Seto E., Bhalla K. (2005) J. Biol. Chem. 280, 26729–26734 [DOI] [PubMed] [Google Scholar]
- 37.Verdin E., Dequiedt F., Kasler H. G. (2003) Trends Genet. 19, 286–293 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.