Abstract
Host defense against infection is induced by Toll-like and interleukin (IL)-1 receptors, and controlled by the transcription factor NF-κB. Our earlier studies have shown that IL-1 activation impacts cytoskeletal structure and that IL-1 receptor (IL-1RI) function is substrate-dependent. Here we identify a novel regulatory component, TILRR, which amplifies activation of IL-1RI and coordinates IL-1-induced control with mechanotransduction. We show that TILRR is a highly conserved and widely expressed enhancer of IL-1-regulated inflammatory responses and, further, that it is a membrane-bound glycosylated protein with sequence homology to members of the FRAS-1 family. We demonstrate that TILRR is recruited to the IL-1 receptor complex and magnifies signal amplification by increasing receptor expression and ligand binding. In addition, we show that the consequent potentiation of NF-κB is controlled through IL-1RI-associated signaling components in coordination with activation of the Ras GTPase. Using mutagenesis, we demonstrate that TILRR function is dependent on association with its signaling partner and, further, that formation of the TILRR-containing IL-1RI complex imparts enhanced association of the MyD88 adapter during ligand-induced activation of NF-κB. We conclude that TILRR is an IL-1RI co-receptor, which associates with the signaling receptor complex to enhance recruitment of MyD88 and control Ras-dependent amplification of NF-κB and inflammatory responses.
Keywords: Cytoskeleton/Actin, Proteoglycans Structure, Receptors/Cytokine, Receptors/Structure-Function, Signal Transduction/Adapter Proteins, Interleukin-1 Receptor Type 1, NF-kappaB Signaling, Receptor Complex Formation and Stoichiometry
Introduction
Immune and inflammatory response are regulated by Toll-like and IL-12 receptors (1–4). Their activation is initiated by ligand binding and co-receptor association to the receptor complex, followed by recruitment of distinct sets of adapter proteins to the cytoplasmic Toll/IL-1 receptor domain, common to all members of the family.
The IL-1 receptor subfamily is a continuously growing group of regulatory proteins, named after the initially identified type I signaling receptor (IL-1RI) (5, 6). Its activation is regulated by the proinflammatory cytokines IL-1α and IL-1β and the receptor antagonist, IL-1ra, and by association of the IL-1R accessory protein (AcP) with the receptor complex (6). IL-1 responses are, in addition, linked to events related to cell attachment and the cytoskeleton and controlled by the small GTPases (7, 8). Receptor activation, followed by recruitment of the MyD88 adapter and by association of the receptor-associated kinases (IRAKs) and TRAF6, leads to induction of the mitogen-activated protein kinase cascade and the NF-κB pathway (1–3, 9, 10).
The importance of receptor complex composition and stoichiometry in adapter protein recruitment and TIR signaling is well recognized (4, 11–13). Our earlier studies have identified a cell surface heparan sulfate proteoglycan, which is recruited to the IL-1RI complex in a substrate-dependent manner (14, 15). Here we report on the cloning and characterization of this novel co-receptor, TILRR (Toll-like/IL-1 receptor regulator). We show that TILRR associates with the extracellular domain of IL-1RI and magnifies NF-κB-regulated inflammatory responses by potentiating recruitment of MyD88 and linking TIR activation with mechanotransduction by controlling induction of the Ras GTPase.
EXPERIMENTAL PROCEDURES
Tissue Culture
Murine and human cell lines and primary cells were cultured under conventional conditions. HeLa cells, endothelial cells (HMEC-1, CDC, Ades, Lawley, and Candal), fibroblasts (NIH 3T3 and primary human gingival fibroblasts, HGF), monocytes (THP1, J774, BL413, ML-1, HL-60, U937, MOLT4, and K562), macrophages (Raw 264.7), embryonic kidney (HEK293), mouse epithelial cells (IL-1RI-transformed C127 cells), and primary human peripheral blood mononuclear cells (PBMCs) were cultured at 37 °C and 5% CO2 in DMEM (Invitrogen) with 10% fetal calf serum (Invitrogen or Cambrex), penicillin (50 units/ml), and streptomycin (50 μg/ml) (Invitrogen) or in MCDB 131 (Invitrogen) with 10% fetal calf serum, epidermal growth factor (10 ng/ml), hydrocortisone (1 μg/ml), and l-glutamine (2 mm) (all from Sigma) and the C127, in addition, with G418 (500 μg/ml). Cells were detached in EDTA (5 mm, Sigma) and plated on bare plastic or on fibronectin (10 μg/ml; Sigma) to enhance levels of endogenous TILRR (luciferase assays, 96-well plates (6.0 × 103 cells/well); confocal microscopy, 8-well chamber slides (12.0 × 103 cells/well); Western analysis, 24-well plates (35 × 103 cells/well); enzyme-linked immunosorbent assays, 6-well plates (2 × 105 cells/well); radioreceptor assays and immunoprecipitation, 10-cm dishes (1.0 × 106 cells)) and stimulated with IL-1β (1 nm) (a kind gift of Dr. Steve Pool (National Institute for Biological Standards and Control)), TNFα (10 ng/ml) (Promega), PDGFBB (25 ng/ml) (R&D Systems), or TGF-β2 (3 ng/ml) (R&D Systems) 24–48 h after transfection.
Protein Identification
In-gel digestion and MALDI-TOF was carried out as previously (16), and data were analyzed by Profound, MASCOT, MOWSE, and PeptIdent. Sequence analysis and alignments used PROSITE, ClustalW, and Lasergene (DNAStar).
cDNA Constructs
cDNA clones of human TILRR (GenBankTM number BC031064) were generated in pBluescript (Geneservice MGC Clone 33379), pcDNA3.1 (Invitrogen), and pMONO (Invivogen) and expressed as EGFP fusion proteins (N- or C-terminal) using Gateway® cloning vectors (Invitrogen). Constructs containing IκBαEGFP, IL-1RI chimera, DNMyD88, N17Ras, DDIRAK-1, DNTRAF6, the IL-8 promoter (IL-8luc and IL-8EGFP), TNF receptor (Invivogen), platelet-derived growth factor receptor β, or TGFβ R2 (Invitrogen) were used as described (8, 17–20).
siRNA Constructs
Multiple siRNAs were made against the mouse and human TILRR sequence. siRNA constructs were made against the TILRR mouse sequence, targeting regions separated by 230 base pairs (5′-CUCUCUCCACAGGUCUUUGTT-3′ (sense)/5′-CAAAGACCUGUGGAGAGAGTT-3′ (antisense) and 5′-CCAGACUGUAACAUCAUCCTT-3′ (sense)/5′-GGAUGAUGUUACAGUCUGGTT-3′ (antisense)) (Eurogentec) and verified by RT-PCR and Western analysis. Similarly, constructs against human TILRR, specifically designed to target a unique TILRR sequence at the 5′-untranslated region, were synthesized (Dharmacon) (5′-CUGGCACUCACUCAAGCUUUU-3′ (sense)/5′-PAAGCUUGAGUGAGUGCCAGUU-3′ (antisense) and 5′-AUUAGAUAUUCAAGAUGGCUU-3′ (sense)/5′-PGCCAUCUUGAAUAUCUAAUUU-3′ (antisense)) and were verified by RT-PCR and Western analysis. Scrambled siRNA (5′-GUUAACGAUGAUCGAUAAUTT-3′ (sense)/5′-AUUAUCGAUCAUCGUUAACTT-3′ (antisense)) (Qiagen) or random siRNA with no genomic equivalent (Eurogentec), which demonstrated no more than a 10% reduction in IL-1-induced activity, were used as controls.
Transfections
Cells were transfected by conventional methods, including using calcium phosphate (19) or Jetsi (Eurogentec) or Nucleofect (Amaxa). Constant levels of cDNA were obtained by adding relevant amounts of empty vector. Controls included cultures transfected with appropriate amounts of empty vector alone. Similarly, experiments using TILRR siRNA included controls with relevant levels of random or scrambled siRNA. Transfection levels were assessed in parallel cultures by using an EGFP-containing control vector and microscopy.
Luciferase Assay
Activity was determined using the Dual-Luciferase reporter assay system (Promega).
RNA Extraction
Total RNA was extracted from various cells using the RNEasy kit (Qiagen).
First Strand cDNA Synthesis
First strand cDNA synthesis was performed using a high capacity cDNA archive kit (Applied Biosystems).
RT-PCR
RT-PCR used first strand cDNA as template with TILRR-specific primers (5′-agagccctgcctgtggtaac-3′ (forward)/5′-gaaggggaatgcaagagtgtgata-3′ (reverse)) or primers for glyceraldehyde-3-phosphate dehydrogenase (5′-aactttggtatcgtggaaggac-3′ (forward)/5′-tggtcgttgagggcaatg-3′ (reverse)).
Quantitative RT-PCR
Quantitative RT-PCR used first strand cDNA as template with TILRR-specific primers/probe and glyceraldehyde-3-phosphate dehydrogenase primers/probe or actin primers/probe (Sigma) and 2× Mastermix (Eurogentec). Experiments included non-RT and water-only controls, neither of which produced a detectable signal.
Mutagenesis
Mutations were introduced in the TILRR core protein and in the extracellular domain of IL-1RI through alanine substitutions at sites selected based on secondary structure predictions (Lasergene Software; Garnier-Robson prediction) and using the QuikChange II mutagenesis kit (Stratagene). Similarly, alanine substitutions of serine residues were introduced at predicted GAG attachment sites (112 and 417) within the TILRR core protein. Mutations of the IL-1RI-TIR domain were introduced as previously (17). All sequences were confirmed by PCR and Western analysis, functionality was tested by luciferase assays, and cell surface expression of wild type receptors and of IL-1RI and TILRR mutants was confirmed by fluorescence-activated cell sorting analysis.
Protein Extraction
Cells were harvested as described (14), using EDTA (5 mm), and extracted by resuspending (50 μl/106 cells) in radioimmune precipitation buffer (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or in extraction buffer (1% Triton X-100), containing 2 mm phenylmethylsulfonyl fluoride, 1 μm pepstatin, 1 μm leupeptin, and incubated on ice (15 min), and DNA was sheared by passages through a hypodermic needle. Following centrifugation (13,000 rpm, 4 °C), supernatant was mixed with an equal volume of 2× sample buffer (0.1 m Tris-HCl) and denatured (95 °C, 5 min).
Immunoprecipitation
Cross-linked samples were extracted using radioimmune precipitation buffer or Triton X-100-containing extraction buffer, as above, total protein was determined, and equal amounts were immunoprecipitated, using an anti-IL-1RI antibody (2 μg/ml; Santa Cruz Biotechnology, Inc.) and protein G-Sepharose beads (50 μl/ml, 4 °C, overnight) (Sigma) or Protein A/G-agarose beads (20 μl/ml, 4 °C, overnight; Santa Cruz Biotechnology, Inc.), as described (14). Controls included precipitations in the absence of the primary antibody.
Western Analysis
Samples were resolved on SDS-PAGE (4, 6, 8, or 4–12%) and transferred to polyvinylidene difluoride, using conventional procedures. TILRR was detected by rabbit polyclonal anti-TILRR (1:1000, custom Eurogentec). Incubation with the antibody and treatment as outlined below identified the functional endogenous TILRR at 70 kDa, demonstrated its reduction by the TILRR-specific siRNA, and recognized the exogenous TILRR, induced by the TILRR cDNA. The only other band observed was significantly weaker, corresponding to the non-functional variant at 40 kDa (data not shown). Similarly, IL-1RI, MyD88, and β-actin were detected using anti-IL-1RI (1:1000), anti-MyD88 (1:1000), and anti-β-actin (1:1000 or 1:2000, loading control), respectively (Santa Cruz Biotechnology). Incubation with a relevant horseradish peroxidase-conjugated secondary antibody (1:5000; Santa Cruz Biotechnology) was followed by ECL visualization. Controls included incubation with secondary antibody alone. Analysis of immunoprecipitated samples, in addition, used IgG as a loading control.
Phalloidin Staining
Cells transfected with a mock construct, TILRR cDNA, or N17Ras or TILRR siRNA were incubated with IL-1β (1 nm, 1 h) or medium alone (control), fixed (paraformaldehyde, 3.7%, room temperature), permeabilized (0.1% Triton X-100, −20 °C), stained with rhodamine-tagged phalloidin (20 min, room temperature; Invitrogen), and analyzed by confocal microscopy.
FACS Analysis
Cells were transfected with a mock construct (10 μg/1 × 106 cells) or wild type or mutant IL-1RI construct (12 μg/1 × 106 cells) or wild type or mutant TILRR construct (10 μg/1 × 106 cells). Twenty-four hours after transfection, cells were fixed (2% paraformaldehyde, 15 min, 4 °C) and incubated with primary anti-IL-1RI (4 μg/ml, 40 min, 4 °C) or anti-TILRR (4.5 μg/ml, 40 min, 4 °C) or with normal rabbit IgG (control) and with fluorescein isothiocyanate-conjugated anti-rabbit antibody (10 μg/ml, 40 min, 4 °C) and analyzed by FACS.
Protein Quantitation
Total protein extracts were quantitated using the BCA assay (Pierce).
Enzyme-linked Immunosorbent Assay
IL-6 protein levels were measured by ELISA Ready-SET-Go! (eBioscience).
Single Cell Analysis
Cells transfected with IκBα-EGFP were stimulated with IL-1β (1 nm, 60 min), and levels of the fusion protein were determined by continuous monitoring by confocal microscopy, as described (19).
IL-1 Binding
IL-1α was radiolabeled using Chloramine-T, and cells were incubated with 125I-IL-1, as described (14), and used in radioreceptor assays, and nonspecific binding was determined by incubation in the presence of a 100-fold excess cold ligand, subtracted from the data. In addition, incubated cells were extracted and separated by SDS-PAGE, and dried gels were allowed to expose films, as described (14).
Cross-linking
Cells were incubated with amine-directed bis(sulfosuccinimidyl)suberate (membrane-impermeable, 5 mm, 2 h, 4 °C) or DSS (membrane-permeable, 5 mm, 30 min, room temperature) (Pierce), as described (14).
Ras Activation
Ras activation was determined using the EZ-DetectTM Ras activation kit (Pierce). HeLa cells were transfected, as above, and after 24 h were stimulated with IL-1β (1 nm). Active Ras was immunoprecipitated, using the Ras-binding domain of Raf 1 fused to GST, and detected by Western blot analysis using an anti-Ras antibody.
Statistical Analysis
p values were calculated using GraphPad Prism.
RESULTS
TILRR Is a Unique, Widely Expressed Protein, Which Controls IL-1RI-induced Inflammatory Responses
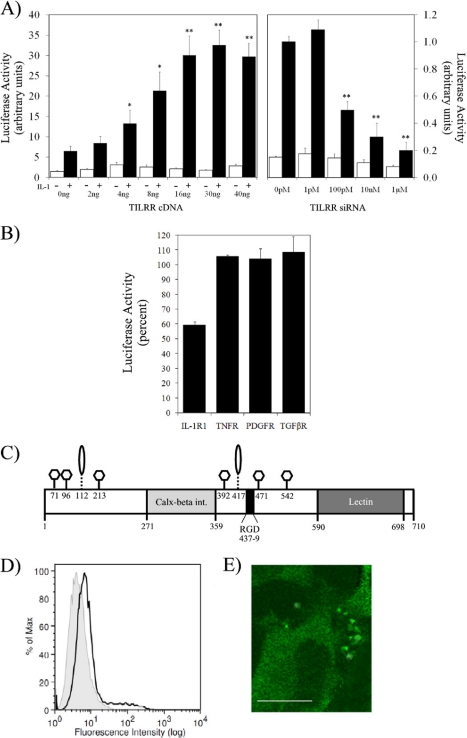
TILRR was detected because of its substrate-dependent association with IL-1RI (14), and identified through peptide mapping based on MALDI-TOF (Fig. 1A). Data base searching, using the obtained sequences DVDSGGVAYQHSGGGAR, VAELPLADKVESTTDLHFLR, and TGHWEWIGGEPVAFTNWR identified a gene present in humans (GenBankTM number BC031064) and mice (GenBankTM number AK052415.1), which mapped to human chromosome 9, location 9p22.3. Quantitative RT-PCR demonstrated expression of TILRR in a wide range of primary cells and cell lines of lymphocytic and mesenchymal origin from humans and mice (Fig. 1B). Further, Western analysis identified a TILRR core protein of 70–80 kDa in the same cell types (Fig. 1C). Inconsistencies between mRNA and protein levels, together with slight variations in size, are consistent with cell type-specific translational or post-translational processing.
FIGURE 1.
TILRR is a widely expressed protein of 70–80 kDa. A, the TILRR core protein sequence. Shown is the amino acid sequence of mouse and human TILRR protein identified from partial trypsin digests of mouse epithelial cells (C127). Samples were purified by immunoprecipitation and SDS-PAGE and analyzed by MALDI-TOF and using Profound, MASCOT, MOWSE, and PeptIdent. *, methionine at residue 7. B, TILRR mRNA is expressed in a variety of cell lines. RNA extracted from various cell lines and primary human periferal mononuclear cells (PBMC 1 and 2), was used to synthesize a first strand cDNA. TILRR and glyceraldehyde-3-phosphate dehydrogenase (human) or actin (mouse) mRNAs were quantitated using quantitative RT-PCR. Data are expressed relative to glyceraldehyde-3-phosphate dehydrogenase and actin, respectively, and represent mean ± S.E. of two experiments. C, the TILRR core is a 70–80-kDa protein. Total protein was extracted from a range of cell lines, as indicated, followed by separation using SDS-PAGE (8%). Levels of TILRR were determined by Western analysis using a TILRR-specific polyclonal antibody (1:1000) or secondary antibody alone (control), followed by horseradish peroxidase-conjugated secondary antibody (1:5000), and visualized by ECL. Levels of β-actin were similarly determined by incubating with an anti-actin antibody (1:1000) and used as a loading control. A representative gel from one of four experiments is shown.
Multiple alignments of the TILRR core protein using ClustalW, revealed significant homology with members of the FRAS1 family of proteoglycans (21). Specifically, TILRR is a protein encoded by an alternatively spliced mRNA species from the highly conserved FREM1 gene (supplemental Fig. S1). GeneScan predictions and expressed sequence tag analysis suggested that the gene gives rise to multiple alternatively spliced mRNAs. Subsequent bioinformatics analysis of both the human and murine FREM1 locus revealed that other potential variants would encode for proteins with molecular masses of around 40 kDa, significantly lower than 70–80 kDa determined for TILRR. In addition, further analysis revealed that these smaller species would lack the functionally relevant N-terminal GAG attachment site at residue 112 (supplemental Fig. S2).
Sequence analysis identified two potential start sites upstream of the open reading frame, which were both tested in functional assays. These demonstrated that the most potent form of TILRR, with a start site at the methionine at residue 7 (marked in Fig. 1A), caused a nearly 7-fold concentration-dependent potentiation of IL-1-induced IL-8 activation (Fig. 2A). Conversely, increasing concentrations of a TILRR siRNA, demonstrated to inhibit TILRR expression by up to 60–80% (supplemental Fig. S3), resulted in a successive reduction in promoter activation of up to 80%, which, as the cDNA-induced amplification, affected ligand-controlled levels only. In contrast, similar screening of a series of receptor systems, including PFGFRβ, TNF receptor, and TGFβR, demonstrated no effects of TILRR expression on induction of IL-8 activity (Fig. 2B).
FIGURE 2.
TILRR is a cell surface glycosylated protein, which potentiates IL-1RI activities. A, TILRR expression correlates with TIR-induced gene activity. HeLa cells were co-transfected with pIL-8-luc (0.2 μg/well (34 μg/106)) and TK-RL (0.075 μg/well (13 μg/106)) together with TILRR cDNA (0–40 ng/well (0–7 μg/106 cells)) or empty vector (control) (left panel). Other cultures were transfected with pIL-8-luc and TK-RL, as above, together with increasing levels of TILRR siRNA, or random siRNA (control), as indicated (right panel). Cells were incubated in the absence (□) or presence (■) of IL-1 (1 nm, 6 h), and luciferase activity was measured. Data are expressed relative to levels in unstimulated cells transfected with the relevant concentrations of empty vector or random siRNA and show mean ± S.E. of two experiments. *, p < 0.05; **, p < 0.01. B, TILRR uniquely regulates IL-1RI function. HeLa cells were used to screen a series of receptor systems (IL-1RI, TNFR, PDGFR, TGFβR) for effects of TILRR on IL-8 promoter activity. Cells were transfected with random or TILRR-specific siRNA (100 pm) and stimulated with respective ligand at saturated levels (IL-1β (1 nm), TNFα (10 ng/ml), PDGF (25 ng/ml), TGFβ (3 ng/ml)). Data are expressed as percentage of activation in the presence of the relevant control (containing Random siRNA) and demonstrate effects on IL-1RI function only. C, schematic outline of the TILRR core protein. The protein contains a GPI anchor in its C-terminal lectin domain, GAG attachment sites (ovals), multiple N-linked glycosylation sites (hexagons), and an integrin-binding site (RGD). D and E, TILRR is localized at the cell surface. D, HeLa cells (106 cells/10-cm dish) were plated for 48 h and then fixed (2% paraformaldehyde on ice) and stained using normal rabbit IgG (control, gray) or a TILRR specific polyclonal antibody (white), followed by a fluorescein isothiocyanate-tagged secondary antibody, and analyzed by flow cytometry. Data show one experiment of four. E, confocal micrograph of HeLa cells transfected with EGFP-TILRR (0.6 μg/well (12 μg/106 cells)). Shown is a representative micrograph demonstrating cell surface localization of the TILRR fusion protein. Bar, 3 μm.
Analysis of the 710-amino acid TILRR sequence revealed GAG attachment sites at residues 112 and 417 and a series of putative N-linked glycosylation sites at residues 71, 96, 213, 392, 471, and 542. In addition, the protein contains an integrin calcium-binding domain and a prominent C-terminal lectin domain, consistent with membrane association (Fig. 2C). The LOCtree prediction (available on the Columbia University Web site) gave a confidence level of 100% for extracellular localization and identified a putative signal peptide sequence at residues 5–14. Cell surface localization of TILRR was confirmed by fluorescence-activated cell sorting analysis and by confocal microscopy using an EGFP-tagged construct, which revealed a punctuate pattern at extended processes (Fig. 2, D and E).
TILRR Controls Activation of Inflammatory Genes through TIR Regulatory Components in a Variety of Cell Types
TILRR's impact at the level of IL-1RI function was confirmed using a series of TIR domain alanine-scanning mutants. We have demonstrated previously that substitutions at sites Asp369 and Arg411 cause a 50–60% reduction in IL-1 induced NF-κB activity, whereas mutations of Tyr374 and Lys428 have no significant effects (17). Here we show that blocking TILRR expression caused about a 40% reduction in IL-1-induced activation through the wild type receptor and through both of the Box 2 receptor mutants (R411A and K428A) (Fig. 3A). In contrast, activation levels in the Box 1 receptor mutants (D369A and Y374F) were unaffected by the TILRR siRNA, demonstrating that substitutions in this membrane-proximal part of the TIR domain render the receptor insensitive to TILRR expression. TILRR signaling through the IL-1RI complex was further supported by experiments demonstrating abrogation of the induced amplification in the presence of DDIRAKI and DNTRAF6 (Fig. 3B).
FIGURE 3.
TILRR controls TIR-induced inflammatory gene activation by IL-1 in various cell types. A, TILRR signals through Box 1 of the TIR domain. HeLa cells were co-transfected with pIL-8-luc (34 μg/106) and TK-RL (13 μg/106) and wild type (WT) or TIR domain mutants of IL-1RI (22 μg/106 cells), in the presence of TILRR-specific siRNA (+) or random siRNA (−) at 100 pm, stimulated (IL-1β1 nm, 6 h), and assayed for luciferase activity. Data show effects of mutants, as described previously (17), and reduced activities through the wild-type receptor and the Box 2 mutants in the presence of TILRR siRNA and represent mean ± S.E. of two experiments. *, p < 0.05. B, TILRR activation is controlled through IL-1RI regulatory components IRAK1 and TRAF6. Cells transfected with IL-8-luc, as above, and with DDIRAK1 (6 μg/106 cells) or DNTRAF6 (6 μg/106 cells), in the presence or absence of TILRR cDNA (3 μg/106 cells), as indicated, were stimulated with IL-1β (1 nm, 6 h). Data show mean ± S.E. of two experiments. **, p < 0.01. C–E, TILRR controls IL-1RI-induced gene regulation in a variety of cells. C, cells (black, Raw 264.7; gray, C127; white, 3T3) were co-transfected with pIL-8-luc (34 μg/106) and TK-RL (13 μg/106) and random- or TILRR-specific siRNA (100 pm, 1 nm), as indicated, and stimulated with IL-1 (1 nm, 6 h), and luciferase activity was measured. Random siRNA had no impact over a range of concentrations. Data are expressed relative to levels in mock-transfected, unstimulated cells and show mean ± S.E. of two experiments. *, p < 0.05; **, p < 0.01. D, endothelial cells were transfected with IL-8-EGFP (40 μg/106 cells) and random siRNA (100 pm) (♦) or TILRR siRNA (100 pm) (■). Single cell images were obtained by continuous recording during stimulation (IL-1, 1 nm), and cytoplasmic fluorescence was determined at the time points indicated, using NIH Image. Data, averaging readings from two experiments, including 30 cells, are expressed relative to initial cytoplasmic levels, S.E. ± 7%, p < 0.01 at 6–8 h. E, human endothelial cells (HMEC-1; left) and primary gingival fibroblasts (right) were transfected with TILRR-specific siRNA (0 pm, 100 pm, and 1 nm), as indicated, and stimulated with IL-1 (1 nm, 6 h), and levels of IL-6 determined by an enzyme-linked immunosorbent assay. Random siRNA had no impact over a range of concentrations. Data show levels of protein synthesized by 106 cells transfected with siRNA and represent mean ± S.E. of two experiments per graph. *, p < 0.05; **, p < 0.005.
Assessing the effect of TILRR on a variety of cell types demonstrated that blocking its expression resulted in a 50–60% reduction in IL-1-induced activation of IL-8 in mouse macrophage, epithelial, and fibroblast cell lines (Fig. 3C). Similarly, TILRR siRNA caused a nearly 70% reduction in IL-1-induced IL-8 activity in human endothelial cells (Fig. 3D) and a 50–75% decrease in IL-6 production by human endothelial cells and human primary fibroblasts (Fig. 3E).
TILRR Controls Cell Structure and Potentiates TIR-induced NF-κB through Activation of the Ras GTPase
Effects on the regulation of NF-κB were confirmed by demonstrating that blocking TILRR expression caused a 60% reduction in IL-1-induced IκBα-phosphorylation at peak levels after 5 min of stimulation (Fig. 4A). In addition, the presence of the TILRR siRNA caused a decrease in inhibitor degradation from 50–60% in control cultures, to 10–20%, during 60 min of stimulation (Fig. 4B).
FIGURE 4.
TILRR controls cell structure and activates the Ras GTPase to potentiate IL-1RI-induced NF-κB. A, TILRR controls IL-1-induced IκBα phosphorylation. HeLa cells were transfected with random (■) or TILRR siRNA (▴) (100 pm) and stimulated (IL-1, 1 nm) for the times indicated and analyzed by Western blotting, using a phosphospecific anti-IκBα antibody or anti-β-actin (loading control). Data are expressed relative to levels in unstimulated samples for each condition and represent mean ± S.E. of two experiments, p < 0.05 at the 5 min peak. B, TILRR controls IL-1-induced IκBα degradation. Confocal micrographs of HeLa cells were transfected with IκBα-EGFP (17 μg/106 cells) alone or in the presence of random or TILRR-specific siRNA (100 pm) and stimulated with IL-1 (1 nm). Quantitation, using NIH Image or Image J, shows levels of IκBα-EGFP cytoplasmic fluorescence in the presence of TILRR siRNA (■) or in the absence of siRNA (♦) indistinguishable from those measured in the presence of random siRNA. Data are from three experiments, including 55 cells, S.E. ± 10–25%, p < 0.05 at 60 min. Bar, 10 μm. C, IL-1-induced changes in cell structure are controlled by TILRR. Cells were transfected with TILRR cDNA, siRNA, or the dominant negative N17Ras and incubated with medium alone or IL-1 (1 nm, 1 h), as indicated. Following fixation, actin filaments were stained using rhodamine-tagged phalloidin, and samples were analyzed by confocal microscopy. The experiments demonstrate that in unstimulated cells (panels 1–3) TILRR cDNA causes pronounced cell rounding with reductions in cell size and extended processes (arrows, compare panels 1 and 2), whereas cells transfected with the dominant negative N17Ras have an appearance similar to that of mock-transfected cells (arrows, compare panels 1 and 3). Further, the data show that these changes (arrows) are induced by IL-1 (compare panels 1 and 4) but do not occur during stimulation in the presence of TILRR siRNA or N17Ras, where cells demonstrate an appearance similar to that observed in unstimulated mock-transfected cultures (compare panels 5 and 6 with panel 1). Bar, 2 μm. D, TILRR amplification is regulated through the Ras GTPase. Cells were co-transfected with pIL-8-luc (34 μg/106 cells) and TK-RL (13 μg/106 cells) alone (−) or together with TILRR cDNA (+) (4 μg/106 cells) and increasing levels of N17Ras, as indicated, and incubated in medium alone or stimulated with IL-1 (1 nm, 6 h). Data are expressed relative to activity in mock-transfected, unstimulated samples and represent mean ± S.E. of two experiments. *, p < 0.05; **, p < 0.01. E, TILRR controls IL-1RI-induced Ras activation. Cells were plated on fibronectin, transfected with random (♦) or TILRR-specific (■) siRNA (100 pm), and stimulated with IL-1 (1 nm) for various times, as indicated. Immunoprecipitated active Ras was detected by Western blot and quantitated by NIH Image, and data were expressed relative to levels in unstimulated cultures, using actin as the loading control, and represent Mean ± S.E. of two experiments, p < 0.05 at 2.5 min.
Our earlier studies showed that IL-1 stimulation causes substrate-dependent alterations in cell shape and cytoskeletal organization and activation of the Ras GTPase (7, 8). In addition, they revealed that these changes are induced under conditions demonstrated to potentiate TILRR/IL-1RI association and prompted analysis of TILRR involvement in mechanotransduction (14). The experiments showed that increasing TILRR expression causes structural alterations similar to those induced by IL-1 stimulation, characterized by loss of extended processes and cell rounding (Fig. 4C), and, further, that blocking endogenous TILRR eliminated the IL-1-induced changes in cell morphology, demonstrating the same effects as observed in the presence of the dominant negative N17Ras (Fig. 4C). Subsequent experiments revealed a successive reduction in TILRR-induced activation in the presence of increasing levels of N17Ras (Fig. 4D). Further, they showed that blocking TILRR expression caused a 75% reduction in IL-1-induced activation in the Ras GTPase (Fig. 4E).
TILRR Function Requires Association with the Signaling Receptor
Radioreceptor experiments demonstrated a 41 ± 7% reduction in IL-1 binding in the presence of TILRR siRNA (Fig. 5A). Gel electrophoresis confirmed that this corresponded to a decrease of 58.5 ± 4.5% in the high molecular weight IL-1RI complex, containing up to 70% of the ligand, which was used to identify TILRR (14) (Fig. 5B). The presence of IL-1RI and TILRR within the complex and its regulation by TILRR expression were confirmed by immunoprecipitation and Western analysis, which demonstrated reductions in the high molecular weight complex of 57 ± 5% in the presence of TILRR siRNA (Fig. 5C). Additional experiments using a series of high molecular weight markers confirmed the size of the complex to be 300–350 kDa, the same size as that determined previously (14) (data not shown).
FIGURE 5.
TILRR controls IL-1RI function by increasing ligand binding and receptor complex levels. A, TILRR expression controls ligand binding. HeLa cells were plated on fibronectin and transfected with random (▿) or TILRR siRNA (100 pm) (▴). Specific cell surface receptor binding was determined by incubating with radiolabeled 125I-IL-1 (2 h, 4 °C) at the concentrations indicated (S.E. ± 7%). B, TILRR expression controls formation of a 300–350-kDa IL-1RI complex. HeLa cells, plated on fibronectin and transfected with TILRR siRNA or random siRNA (100 pm) as in (A), were incubated with 125I-IL-1 (1 nm, 2 h, 4 °C) and cross-linked using bis(sulfosuccinimidyl)suberate, and extracts were analyzed by SDS-PAGE (8%). One experiment of three is shown (S.E. ± 7%). C, TILRR and IL-1RI associate to form a 300–350-kDa complex. HeLa cells transfected with empty vector (Mock) or TILRR cDNA (10 μg/106 cells) or TILRR siRNA (100 pm) and incubated (IL-1, 1 nm) as in A and B, were cross-linked and immunoprecipitated with an anti-IL-1RI antibody (2 μg/ml) and analyzed by SDS-PAGE (4–12%). Levels of TILRR and IL-1RI were determined by Western blotting, using IgG as the loading control. One representative gel is shown. D, TILRR control of IL-1RI function is dependent on the structure of the signaling receptor extracellular domain. Cells were co-transfected with pIL-8-luc (34 μg/106 cells), TK-RL (13 μg/106 cells), wild type TILRR (3.6 μg/106 cells), and wild type or mutant IL-1RI (22 μg/106 cells), as indicated, and stimulated with IL-1 (1 nm, 6 h), and extracts were assayed for luciferase activity. Data are expressed relative to activation in mock-transfected, IL-1-stimulated samples and represent mean ± S.E. of two experiments. *, p < 0.01; **, p < 0.001. E, TILRR function is dependent on association with IL-1RI. Cells were transfected with a mock construct or wild type TILRR (10 μg/106 cells) together with wild type IL-1RI (WT) or the N319A IL-1RI mutant (10 μg/106 cells), as indicated, and incubated with 125I-IL-1 (1 nm, 2 h, 4 °C), and cross-linked extracts were analyzed by SDS-PAGE. Quantitation of levels of the high molecular weight complex was carried out using scanned autoradiograms and NIH Image, and data are expressed relative to levels of the 97 kDa band (IL-1RI + ligand) and represent mean ± S.E. of two experiments. **, p < 0.01.
To assess the significance of IL-1RI association in TILRR function, residues in the extracellular domain of the signaling receptor, selected based on their predicted effect on secondary structure, were subjected to alanine substitution. Mutations I41A, I69A, and N319A, while demonstrating levels of cell surface expression similar to the wild type (supplemental Fig. S4A), caused reductions of about 50–70% in the TILRR-amplified response (Fig. 5D). In contrast, substitutions V148A, L170A, and L295A, used as controls, had no impact (Fig. 5D). Alanine substitution of the membrane proximal residue Asn319, predicted to affect α-helical, turn, and coil regions of the protein, had the most pronounced effects on TILRR amplification and was used in a series of subsequent experiments. These showed that the substitution had no impact on IL-1-induced promoter activation in the absence of TILRR (data not shown). Additional analysis using cross-linking to assess the effect of the 319 mutant on IL-1RI association demonstrated a pronounced reduction in formation of the high molecular weight TILRR/IL-1RI-containing complex, consistent with a link between TILRR function and IL-1RI association (Fig. 5E).
TILRR Association with IL-1RI Enhances Inflammatory Gene Regulation by Potentiating MyD88 Recruitment and MyD88-dependent Activation of NF-κB
Further analyses were carried out using a series of TILRR mutants. These demonstrated that alanine substitution of the predicted glycosaminoglycan binding site at residue 112 caused a 50% reduction in IL-1-induced promoter activation in the presence of TILRR, consistent with a role for the fibronectin heparin-binding region in TILRR function (supplemental Fig. S2) (14). Further, introducing alanine substitutions of residues within the TILRR core protein, selected based on their predicted impact on the secondary structure, identified three mutations (D58A, D448A, and Q558A), which, while demonstrating the same level of cell surface expression as the wild type receptor (supplemental Fig. S4B), resulted in a 30–60% reduction in IL-1-induced activation (Fig. 6A). In comparison, substitutions of residues His82, Asp119, His480, and Asp534, used as controls, caused no more than a 10% decrease in activity. Parallel experiments demonstrated that substitution of the membrane-proximal residue Asp448, with predicted effects on α-helical, turn, and coil regions of the protein, significantly reduced formation of the TILRR-containing IL-1RI complex, in agreement with a relevance of signaling receptor association to TILRR function (Fig. 6B).
FIGURE 6.
TILRR controls MyD88-dependent signaling and MyD88 association. A, TILRR function is dependent on the structure of its core protein. HeLa cells were co-transfected with wild type (WT) or mutant TILRR (3.6 μg/106 cells), together with pIL-8-luc (34 μg/106 cells) and TK-RL (13 μg/106 cells), and stimulated with IL-1 (1 nm, 6 h), and luciferase activity was measured. Data are expressed relative to activity in mock-transfected cells and represent mean ± S.E. of two experiments. *, p < 0.05; **, p < 0.001. B, functionally impaired TILRR mutants do not associate with IL-1RI. HeLa cells were transfected with empty vector (Mock) or wild type TILRR or D448 TILRR (10 μg/106 cells), as indicated, and incubated with 125I-IL-1 (1 nm, 2 h, 4 °C) and cross-linked bis(sulfosuccinimidyl)suberate, and extracts were analyzed by SDS-PAGE. Quantitation of scanned autoradiograms by NIH Image shows levels of the high molecular weight, TILRR-containing complex, expressed relative to levels of the 97 kDa band (IL-1 RI + ligand) and represent mean ± S.E. of two experiments. *, p < 0.05. C, TILRR amplification of IL-1RI responses is MyD88-dependent. HeLa cells were co-transfected with pIL-8-luc (34 μg/106 cells) and TK-RL (13 μg/106 cells) in the presence of TILRR cDNA (11 μg/106 cells) or the relevant mock construct and increasing concentrations of DN MyD88, as indicated, stimulated with IL-1 (1 nm, 6 h), and assayed for luciferase activity. Data are expressed relative to activity in mock-transfected, unstimulated cells and represent mean ± S.E. of two experiments. *, p < 0.01; **, p < 0.001. D, TILRR increases association of MyD88 with IL-1RI. HeLa cells transfected with empty vector (Mock) or TILRR cDNA (TILRR, 10 μg/106cells) were stimulated with IL-1 (1 nm) for 0 (−) or 20 (+) min, as indicated, subjected to membrane-permeable cross-linking (DSS), immunoprecipitated using an IL-1RI antibody (2 μg/ml), and analyzed by SDS-PAGE (4–12%). Levels of MyD88 and IL-1RI were determined by Western blotting, using IgG as the loading control. One representative gel is shown. Quantitation was done using NIH Image, and data are expressed as levels of MyD88 relative to IL-1RI for each condition and represent mean ± S.E. of two experiments. *, p < 0.05; **, p < 0.001.
Subsequent experiments to assess the impact of TILRR association on adapter protein usage showed that a dominant negative MyD88 caused a concentration-dependent reduction in IL-1-induced activation in the presence of the TILRR cDNA, with total abrogation at high enough levels (Fig. 6C). Impacts on adapter protein recruitment were assessed by Western analysis following membrane permeable cross-linking and immunoprecipitation (Fig. 6D). This showed that enhancing TILRR expression to levels demonstrated to induce formation of the high molecular weight IL-1RI complex potentiated TIR association of MyD88 by 4-fold, resulting from a 2-fold enhancement of IL-1 receptor levels (see Fig. 5, A–C) and a 2-fold increase in the MyD88/IL-1RI-ratio (Fig. 6D).
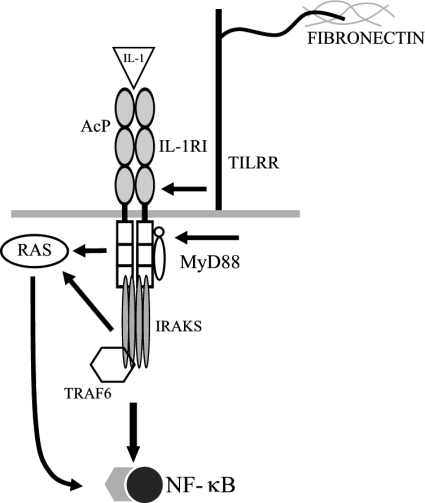
The data show that TILRR, a cell surface proteoglycan, associates with IL-1RI to potentiate NF-κB activation and inflammatory responses by increasing ligand binding and receptor expression and, further, that signal amplification is due to enhanced recruitment of the MyD88 adapter to the signaling receptor complex and to coordination of activities through the IL-1RI TIR domain and associated regulatory components with induction of the Ras GTPase (Fig. 7).
FIGURE 7.
TILRR enhances MyD88 recruitment to IL-1RI to coordinate TIR- and Ras-dependent activation. TILRR association with IL-1RI increases ligand binding and receptor complex formation and potentiates recruitment of the MyD88 adapter to the TIR domain to increase signal amplification at the level of the receptor complex and associated regulatory components and to direct activation of the Ras GTPase and magnify induction of NF-κB and inflammatory genes.
DISCUSSION
We report on a novel IL-1RI co-receptor, TILRR, which associates with the signaling receptor complex to potentiate recruitment of the MyD88 adapter, control induction of the Ras GTPase, and amplify activation of NF-κB and inflammatory genes.
Earlier studies identified TILRR as a heparan sulfate proteoglycan, recruited to the IL-1 receptor complex at focal adhesions (14). Involvement of TILRR in structural control is consistent with its role in induction of substrate-dependent alterations in cell shape and in activation of the Ras GTPase through IL-1RI (7, 8). The results thus demonstrate a link between TILRR expression and induction of Ras activity and amplification of NF-κB and suggest that TILRR functions as a coordinator of IL-1-induced responses and mechanotransduction (22). This is further supported by its homology with members of the FRAS1 family and indicates a role for TILRR in regulation of inflammatory control in the context of tissue structure and development (21). Its recruitment to the receptor complex may thus regulate IL-1RI function and cellular responses in a topographical and time-dependent manner (23).
A role for TILRR in ligand binding is consistent with its lack of effect on unstimulated levels. Further, it is supported by the reduced effect of TILRR on IL-1 responses, following substitutions at residues 39 and 66, within the IL-1R ligand-binding domain (24). It is also consistent with the reduction in binding affinity (Ka) and increase in downstream activation caused by TILRR/IL-1RI association, characteristic of cell surface glycoprotein and proteoglycan co-receptors (14, 25, 26). TILRR may thus have a role similar to that of β-glycan and gp130, which present ligand to their respective system signaling receptors (27, 28). In addition, the increase in cell surface binding may be a consequence of the enhanced receptor expression and caused by a reduced rate of internalization, due to retention of IL-1RI at focal adhesions, following TILRR association (14).
Effects on receptor function are expected to derive from alterations in receptor complex composition and conformation following TILRR association. With up to 70% of the receptor-bound ligand present within the high molecular weight, TILRR-containing complex, such changes are likely to have significant consequences for receptor dimerization, signal amplification, and downstream control (4, 14). Detailed analysis of effects of TILRR association on complex composition and on association of regulatory components will be carried out in future studies using crystallography, in combination with surface plasmon resonance and saturation transfer difference NMR.
Sequential changes in signaling receptor conformation following TILRR association are likely to underlie the enhanced recruitment of the MyD88 adapter and the increased signal amplification (4). In addition, such changes may facilitate downstream signaling cross-talk and link TIR-induced activation with substrate-dependent control and structural regulation through the GTPases (29, 30). This is consistent with the requirement of TILRR in IL-1-induced activation of the Ras GTPase and suggests that the TILRR·IL-1RI complex may constitute a platform for controlling structurally sensitive inflammatory responses by coordinating IL-1-regulated activities with mechanotransduction (31, 32).
A link between structural regulation at the level of signaling cross-talk and the increase in MyD88-dependent activation is consistent with the observed changes in the actin cytoskeleton. Our earlier studies have demonstrated that such Ras-controlled changes in cytoskeletal organization, induced by IL-1, involve phosphorylation of the transmembrane linkage protein talin and alterations at focal adhesions (7, 8). This type of effect on substrate interactions and actin reorganization by cytokines such as TNF has been demonstrated to be adapter-controlled and fundamental in the regulation of biological responses and cell behavior during development and wound healing (33–35). In addition, alterations in cell shape and the actin cytoskeleton will provide an efficient system for controlling NF-κB at the level of the IκBα·NF-κB complex, by allowing rapid changes in the available pool of IκBα during activation (36). Future studies will determine the significance of TILRR in the kinetics of complex dissociation, specifically in relation to overall network control and gene induction profiles.
We here identify a substrate-dependent and structurally regulated IL-1RI co-receptor, TILRR, which associates with the signaling receptor to potentiate activation of NF-κB. We demonstrate that TILRR association and ensuing changes in receptor complex composition potentiate ligand binding and recruitment of MyD88 and identify a role for TILRR in control of the Ras-GTPase and cytoskeletal organization.
We conclude that TILRR controls amplification of inflammatory responses by linking TIR-induced activities with mechanotransduction and expect it to be relevant to a range of diseases characterized by dysregulation of inflammatory responses. Future studies will focus on the role of TILRR in signaling cross-talk and in regulation of gene activation profiles under physiological and pathological conditions.
Supplementary Material
Acknowledgments
We thank Dr. Marta Muzio (Instituto Medico Nigri, Milan, Italy) for the kind gift of the DN MyD88, Prof. Allan Hall (Memorial Sloan-Kettering Cancer Center, New York) for the kind gift of the dominant negative N17Ras, Prof. Rob Read (University of Sheffield) for kindly providing peripheral blood mononuclear cells, and Dr. Steven Pool for the kind gift of IL-1.
This work was supported by Biotechnology and Biological Sciences Research Council Grants BB/C515798/1 and BBS/B/04056 and British Heart Foundation Grants PG/2000094 and PG/07/094/23742 (to E. E. Q.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- IL
- interleukin
- TNF
- tumor necrosis factor
- TGF
- transforming growth factor
- MALDI-TOF
- matrix-assisted laser desorption/ionization time-of-flight
- siRNA
- small interfering RNA
- RT
- reverse transcription
- EGFP
- enhanced green fluorescent protein
- PBMC
- peripheral blood mononuclear cell.
REFERENCES
- 1.Akira S., Takeda K. (2004) Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 2.Beutler B. (2004) Nature 430, 257–263 [DOI] [PubMed] [Google Scholar]
- 3.O'Neill L. A. (2006) Curr. Opin. Immunol. 18, 3–9 [DOI] [PubMed] [Google Scholar]
- 4.Gay N. J., Gangloff M. (2007) Annu. Rev. Biochem. 76, 141–165 [DOI] [PubMed] [Google Scholar]
- 5.Dinarello C. A. (2005) J. Exp. Med. 201, 1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sims J. E. (2002) Curr. Opin. Immunol. 14, 117–122 [DOI] [PubMed] [Google Scholar]
- 7.Qwarnström E. E., MacFarlane S. A., Page R. C., Dower S. K. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 1232–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caunt C. J., Kiss-Toth E., Carlotti F., Chapman R., Qwarnstrom E. E. (2001) J. Biol. Chem. 276, 6280–6288 [DOI] [PubMed] [Google Scholar]
- 9.Muzio M., Ni J., Feng P., Dixit V. M. (1997) Science 278, 1612–1615 [DOI] [PubMed] [Google Scholar]
- 10.Wesche H., Henzel W. J., Shillinglaw W., Li S., Cao Z. (1997) Immunity 7, 837–847 [DOI] [PubMed] [Google Scholar]
- 11.Gay N. J., Gangloff M., Weber A. N. (2006) Nat. Rev. Immunol. 6, 693–698 [DOI] [PubMed] [Google Scholar]
- 12.Akira S., Yamamoto M., Takeda K. (2003) Biochem. Soc. Trans. 31, 637–642 [DOI] [PubMed] [Google Scholar]
- 13.O'Neill L. A., Bowie A. G. (2007) Nat. Rev. Immunol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 14.Vallés S., Tsoi C., Huang W. Y., Wyllie D., Carlotti F., Askari J. A., Humphries M. J., Dower S. K., Qwarnström E. E. (1999) J. Biol. Chem. 274, 20103–20109 [DOI] [PubMed] [Google Scholar]
- 15.Valles S., Caunt C. J., Walker M. H., Qwarnstrom E. E. (2002) Lab. Invest. 82, 855–862 [DOI] [PubMed] [Google Scholar]
- 16.Ashcroft A. (2003) Nat. Prod. Rep. 2, 202–215 [DOI] [PubMed] [Google Scholar]
- 17.Slack J. L., Schooley K., Bonnert T. P., Mitcham J. L., Qwarnstrom E. E., Sims J. E., Dower S. K. (2000) J. Biol. Chem. 275, 4670–4678 [DOI] [PubMed] [Google Scholar]
- 18.Wyllie D. H., Kiss-Toth E., Visintin A., Smith S. C., Boussouf S., Segal D. M., Duff G. W., Dower S. K. (2000) J. Immunol. 165, 7125–7132 [DOI] [PubMed] [Google Scholar]
- 19.Yang L., Ross K., Qwarnstrom E. E. (2003) J. Biol. Chem. 278, 30881–30888 [DOI] [PubMed] [Google Scholar]
- 20.Kiss-Toth E., Wyllie D. H., Holland K., Marsden L., Jozsa V., Oxley K. M., Polgar T., Qwarnstrom E. E., Dower S. K. (2006) Cell. Signal. 18, 202–214 [DOI] [PubMed] [Google Scholar]
- 21.McGregor L., Makela V., Darling. S. M., Vrontou S., Chalepakis G., Roberts C., Smart N., Rutland P., Prescott N., Hopkins J., Bentley E., Shaw A., Roberts E., Mueller R., Jadeja S., Philip N., Nelson J., Francannet C., Perez-Aytes A., Megarbane A., Kerr B., Wainwright B., Woolf A. S., Winter R. M., Scambler P. J. (2003) Nat. Genet. 34, 203–208 [DOI] [PubMed] [Google Scholar]
- 22.Geiger B., Spatz J. P., Bershadsky A. D. (2009) Nat. Rev. Mol. Cell Biol. 10, 21–33 [DOI] [PubMed] [Google Scholar]
- 23.Kreuger J., Spillmann D., Li J. P., Lindahl U. (2006) J. Cell Biol. 174, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vigers G. P., Anderson L. J., Caffes P., Brandhuber B. J. (1997) Nature 386, 190–194 [DOI] [PubMed] [Google Scholar]
- 25.Schlessinger J., Lax I., Lemmon M. (1995) Cell 83, 357–360 [DOI] [PubMed] [Google Scholar]
- 26.Fukushima K., Ikehara Y., Yamashita K. (2005) J. Biol. Chem. 280, 18056–18062 [DOI] [PubMed] [Google Scholar]
- 27.López-Casillas F., Wrana J. L., Massagué J. (1993) Cell 73, 1435–1444 [DOI] [PubMed] [Google Scholar]
- 28.Tanaka M., Miyajima A. (2003) Rev. Physiol. Biochem. Pharmacol. 149, 39–52 [DOI] [PubMed] [Google Scholar]
- 29.Zeisel M. B., Druet V. A., Sibilia J., Klein J. P., Quesniaux V., Wachsmann D. (2005) J. Immunol. 174, 7393–7397 [DOI] [PubMed] [Google Scholar]
- 30.Kamon H., Kawabe T., Kitamura H., Lee J., Kamimura D., Kaisho T., Akira S., Iwamatsu A., Koga H., Murakami M., Hirano T. (2006) EMBO J. 25, 4108–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hahn C., Schwartz M. A. (2009) Nat. Rev. Mol. Cell Biol. 10, 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang N., Tytell J. D., Ingber D. E. (2009) Nat. Rev. Mol. Cell Biol. 10, 75–82 [DOI] [PubMed] [Google Scholar]
- 33.Cumberbatch M., Dearman R. J., Kimber I. (1997) Immunology 92, 388–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banno T., Gazel A., Blumenberg M. (2004) J. Biol. Chem. 279, 32633–32642 [DOI] [PubMed] [Google Scholar]
- 35.Haubert D., Gharib N., Rivero F., Wiegmann K., Hösel M., Krönke M., Kashkar H. (2007) EMBO J. 26, 3308–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pogson M., Holcombe M., Smallwood R., Qwarnstrom E. E. (2008) PLoSOne 3, e2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.