Abstract
Bacillus anthracis genome harbors a toxin-antitoxin (TA) module encoding pemI (antitoxin) and pemK (toxin). This study describes the rPemK as a potent ribonuclease with a preference for pyrimidines (C/U), which is consistent with our previous study that demonstrated it as a translational attenuator. The in silico structural modeling of the PemK in conjunction with the site-directed mutagenesis confirmed the role of His-59 and Glu-78 as an acid-base couple in mediating the ribonuclease activity. The rPemK is shown to form a complex with the rPemI, which is in line with its function as a TA module. This rPemI-rPemK complex becomes catalytically inactive when both the proteins interact in a molar stoichiometry of 1. The rPemI displays vulnerability to proteolysis but attains conformational stability only upon rPemK interaction. The pemI-pemK transcript is shown to be up-regulated upon stress induction with a concomitant increase in the amount of PemK and a decline in the PemI levels, establishing the role of these modules in stress. The artificial perturbation of TA interaction could unleash the toxin, executing bacterial cell death. Toward this end, synthetic peptides are designed to disrupt the TA interaction. The peptides are shown to be effective in abrogating TA interaction in micromolar range in vitro. This approach can be harnessed as a potential antibacterial strategy against anthrax in the future.
Keywords: Antimicrobial Peptides, Apoptosis, Drug Resistance/Prokaryotic, Enzymes/Catalysis, Gene, Peptides/Interactions, Toxins, Toxins/Drugs
Introduction
Toxin-antitoxin (TA)3 modules in prokaryotes consist of two adjacent open reading frames encoding a stable toxin and a cognate labile antitoxin (1, 2). The vulnerability of antitoxin to proteolysis demands a constant de novo synthesis of antitoxin to maintain a steady-state level that can sequester the toxin and circumvent its deleterious activity (3). A negative feedback homeostatic loop also exists, wherein antitoxin acts as a transcriptional repressor in conjunction with the toxin as a co-repressor (4). A multitude of functions have been ascribed to the chromosome-encoded TA loci, primarily as stress managers (5–7). It has been envisaged that these modules respond to intracellular stress facilitating phenotypic switching of the bacterial cell to a quasidormant stage amenable to survival under extreme conditions (8–12). However, a recent study demonstrates that TA loci do not impart selective advantage to the cells grown in stress, indicating that these genes can innocuously be deleted from the genome (13).
Several families of two component TA loci have been identified on bacterial chromosomes, namely relBE, higBA, mazEF, ccdAB, vapBC, parDE, phd-doc, and yoeB-yefM. The toxins of these families share sequence and structural similarity to some extent, but the corresponding antidotes do not, thus indicating their distinct evolutionary origin (2). However, despite their structural similarity and common ancestral origin, their downstream targets are quite different. CcdB and ParE function as gyrase poison, whereas MazF, Kid, YdcE, and YoeB are potent ribonucleases (14–22). VapC toxins represent a structurally distinct family of ribonucleases having an α-helical pilT N-terminal domain (PIN domain) (23).
Our previous study had identified and characterized a pemIK TA module from Bacillus anthracis (4). This study was conducted to further delineate its mechanism of action. PemK is identified as a ribonuclease, and the probable catalytic residues essential for its activity are also proposed. Our study illustrates the cardinal significance of this TA module in stress. We also present evidence that peptide inhibitors can disrupt TA interaction, which can be exploited as a potential antibacterial strategy. The ubiquity of TA systems in other prokaryotes and their absolute absence from eukaryotes make them attractive targets for developing novel antibacterial therapeutics.
EXPERIMENTAL PROCEDURES
Plasmid pET-28a(+) from Novagen (Madison, WI) was used for heterogeneous gene expression. Escherichia coli strains DH5α and BL21 (λDE3) (Novagen) were used as hosts for cloning and expression, respectively. Avirulent strain of B. anthracis (Sterne 34F2, pXO1+, pXO2−) was used in this study. Oligonucleotides were synthesized by Sigma.
In Vitro Ribonuclease Activity of the rPemK
Gel-based Assay
The function of PemK of B. anthracis was inferred by performing a BLAST search against the nonredundant data base at NCBI with the proteins possessing high similarity with PemK of B. anthracis.
Total RNA was isolated from E. coli and B. anthracis using an RNeasy mini kit (Qiagen, Germany). Ten μg of cellular RNA was incubated with 2 μg of the rPemK at 25 °C for 15 min. The ability of the rPemI to neutralize the toxicity of rPemK was examined. A fixed concentration of rPemI (20 μm) was co-incubated with rPemK in varying molar stoichiometric ratios at 4 °C for 16–18 h prior to the addition of RNA. Samples were subjected to agarose gel electrophoresis, and RNA was visualized by ethidium bromide staining. The heat-denatured rPemK (boiled for 30 min) was also checked for its ability to cleave RNA.
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) analysis of the rPemK was carried out using a Bruker Daltonic Ultraflex TOF/TOF system, which was calibrated using calibration standard II (Bruker Daltonik GmbH, Germany) at the Centre for Genomic Application, New Delhi, India. Protein identification was carried out using the MASCOT search engine (Matrix Science, London, UK), and the NCBI nonredundant data base was used for peptide search.
Fluorimetric Assay
Chimeric oligonucleotides with a specific ribonucleotide (rA, rG, rU, and rC) dually labeled with 6-carboxyfluorescein and black hole quencher 1 on the 5′- and 3′-ends, respectively, were purchased from Integrated DNA Technologies (Coralville, IA) (24). Fluorescence was measured using a spectrofluorimeter (Cary Eclipse, Varian, Australia) at an excitation of 485 nm and emission at 530 nm in a 0.5-min time interval for 3 h. The slit width was set as 10 nm for both the excitation and emission filters. Each fluorescent-tagged oligonucleotide substrate (20 μm) was added in a 96-well microtiter plate in the absence or presence of 500 ng of the rPemK, and an increase in the fluorescence, if any, was monitored after every 0.5-min interval. Autodegradation of the substrates was subtracted from the experimental readings. RNase A (20 units) and bovine serum albumin (500 ng), included as positive and negative controls, respectively, were incubated with 20 μm of the rC.
The exact molar stoichiometric ratio of the rPemI-rPemK interaction was assessed. For this, the rPemK (10 μm) was incubated with varying molar concentrations of the rPemI (0–50 μm) at 4 °C for 16–18 h prior to the addition of 30 μm rC, and the fluorescence quenching was recorded. Initial slopes were monitored for each case (RFI/min). The percent fluorescence quenching was calculated from the control containing 30 μm rC and 10 μm rPemK, whose fluorescence was considered as 100%. Similarly, the rPemI (50 μm) was titrated with increasing rPemK (0–500 μm) followed by the addition of 30 μm rC. The fluorescence obtained with the rPemI (50 μm)-rPemK (500 μm) complex with 30 μm rC was taken as 100%.
Effect of rPemK on Double-stranded DNA
Plasmids (1 μg) representing circular double-stranded DNA of different sizes (ranging from 3 to 10 kb) and a linear double-stranded DNA (600 bp) were incubated with 2 μg of the rPemK for 15 min at 25 °C. The samples were analyzed on a 1.5% agarose gel.
Ribosome-binding Assay
Ribosomes were prepared essentially as described previously (25). Prior to use, the protein samples (rPemK and rAlr) were centrifuged at 200,000 × g for 3 h to remove any insoluble material. The ribosome-binding assay mixture containing ribosomes (500 μg) and rPemK was incubated at 4 °C for 60 min and centrifuged at 200,000 × g for 3 h to pellet the ribosomes and any associated proteins. The rPemI and rPemK were preincubated at 4 °C overnight to facilitate the reconstitution of the rPemI-rPemK complex. The pellets were resuspended, and an equal amount of protein was fractionated on SDS-PAG. The electrophoresed proteins were analyzed by Western blotting using specific antibodies directed against the protein and His6 monoclonal antibody. The rAlr (alanine racemase, ∼45-kDa purified recombinant protein of B. anthracis) was taken as a negative control.
Molecular Modeling and Structural Validation of the Model of PemK
Multiple sequence alignment of the toxins (B. subtilis, YdcE; E. coli, Kid) was constructed using ClustalW (26). The final multiple sequence alignment was viewed using BioEdit (27), and the similarity with the well characterized Kid toxin was used to predict the catalytic base of B. anthracis PemK. Initial fold recognition was done using the SVM-Fold server (28) followed by structural modeling using the Swiss-Model Workspace (29). The structure validation of the designed PemK model was done using Procheck (30, 31). The Verify3D program was also used for quality assessment of the model (32).
Prediction of Catalytic Site Residues of PemK
By exploiting the designed model of PemK, the favorably positioned residue in the structure was proposed as the probable catalytic acid. Additional conserved stabilizing residues were also proposed from this analysis.
Site-directed Mutagenesis
Point mutations were generated using the QuikChange site-directed mutagenesis kit (Stratagene). Briefly, the pETSAT plasmid harboring the wild type pemK (4) was used as a template for DpnI-based mutagenesis. The transformants obtained were subjected to dideoxy sequencing to confirm the incorporation of the specific mutations only at the desired positions and absence of any other random mutation(s) elsewhere. The oligonucleotides used for specific Q21A, H59A, H59K, E78A, E78D, and Q79A mutations are shown in the Table 1. All the mutant proteins were purified under native conditions using Ni2+-nitrilotriacetic acid affinity chromatography as described previously (4). Equal proteins (2 μg) were tested for their potential to function as ribonuclease as described. The catalytic coefficients, Km and Vmax, of the mutants were calculated using double-reciprocal Lineweaver-Burk plot and were compared with the wild type rPemK employing increasing concentrations of the fluorigenic substrates (rC and rU).
TABLE 1.
The sequences of the oligonucleotides employed in this study
The oligonucleotides obtained in the lyophilized form were dissolved in 1× Tris-EDTA buffer, pH 8.0, before use. The restriction enzyme sites in the primers used for the cloning purposes are underlined and marked in boldface. F is forward, and R is reverse.
| Primers used | Sequence (5′–3′) | Study purpose |
|---|---|---|
| Q21A (F) | TGGTTCTGAGGCCGGAGGTGTTC | Site-directed mutagenesis |
| Q21A (R) | GAACACCTCCGGCCTCAGAACCA | Site-directed mutagenesis |
| H59A (F) | GAAATTACCCACTGCCGTGGAAATTG | Site-directed mutagenesis |
| H59A (R) | CAATTTCCACGGCAGTGGGTAATTTC | Site-directed mutagenesis |
| E78A (F) | TTATTTTACTTGCCCAGATTCGAAC | Site-directed mutagenesis |
| E78A (R) | GTTCGAATCTGGGCAAGTAAAATAA | Site-directed mutagenesis |
| Q79A (F) | ATTTTACTTGAGGCCATTCGAACAATC | Site-directed mutagenesis |
| Q79A (R) | GATTGTTCGAATGGCCTCAAGTAAAAT | Site-directed mutagenesis |
| H59K (F) | AAATTACCCACTAAGGTGGAAATT | Site-directed mutagenesis |
| H59K (R) | AATTTCCACCTTAGTGGGTAATTT | Site-directed mutagenesis |
| E78D (F) | GTTATTTTACTTGATCAGATTCGA | Site-directed mutagenesis |
| E78D (R) | TCGAATCTGATCAAGTAAAATAAC | Site-directed mutagenesis |
| Δ1–17 (F) | GCCGGATCCATGGTAACGGAATTGGACGGAATTGGA | Coning of PemI deletion, ELISA |
| 17/30/50 (R) | GCCGTCGACTTACCCCCCGCTAACTAAGCGTTC | Cloning of PemI deletion, ELISA |
| Δ1–30 (F) | GCCGGATCCAAGAATCGCCATGAACTAATTTGC | Cloning of PemI deletion, ELISA |
| Δ1–50 (F) | GCCGGATCCAAGAAACGCTACCAACATGAATCAATG | Cloning of PemI deletion, ELISA |
| 1–70 (F) | GCCGGATCCGTGTCCGAATCAAGTGTAACTACTG | Cloning of PemI deletion, ELISA |
| 1–70 (R) | GCCGTCGACATTAATTTTTCCCATTTCAATGTACC | Cloning of PemI deletion, ELISA |
| pemK (F) | TGCAGACCTTTCCCCAGTT | Real time PCR |
| pemK (R) | TGACAAGAACCGGACGAACA | Real time PCR |
| pemI (F) | TGCCAGGCAACACAACTGTT | Real time PCR |
| pemI (R) | TGATTCATGTTGGTAGCGTTTCTT | Real time PCR |
| dna gyr (F) | GATGAGCAAGAGGTAGCGACAGT | Real time PCR |
| dna gyr (R) | TCGCTCGTTTCTTCCTCATCA | Real time PCR |
Ability of the rPemK Mutants to Interact with the rPemI
ELISA was performed by coating 500 ng of the rPemK (wild type) and its mutants in 0.1 m sodium carbonate coating buffer, pH 9.6, and incubating for 16 h at 4 °C in a 96-well plate as described. 500 ng of the rPemI was then added and incubated for 1 h at 37 °C. The bound protein was captured by anti-rPemI antiserum raised in mouse (1:8000). Subsequent incubation with anti-mouse IgG-horseradish peroxidase (1:10,000) was carried out for 1 h at 37 °C. The color was developed by the addition of 3,3′,5,5′-tetramethylbenzidine substrate, and the absorbance was read at 630 nm in a microplate reader (Bio-Rad). The absorbance of the rPemI-rPemK complex was taken as 100%, and the extent of binding of the rPemK mutants to the rPemI was calculated.
Spectral Characterization of the rPemK Mutants
The influence of alanine substitution on the tertiary structure of the mutant proteins in comparison with the wild type rPemK was assessed. For this, the rPemK and mutants were analyzed for their fluorescence emission maxima at 25 °C in 10 mm potassium phosphate buffer, pH 8.0, at a concentration of 50 μg/ml. Emission spectrum was obtained by exciting the samples at 280 nm and monitoring emissions from 295 to 350 nm. The slit widths were kept as 20 nm for both the filters.
To study the effect of mutations on the secondary structure of the rPemK mutants in comparison with the wild type protein, CD spectroscopy was employed as described using 100 μg/ml of the proteins (4). The CD signals were converted to molar ellipticity by the following relationship: θm = θo × 100/lc, where θm is molar ellipticity (in degrees per cm2 per dmol); θo is the observed ellipticity (in degrees); l is the path length (in centimeters), and c is the molar concentration.
Proteinase K-sensitive Assay
For proteinase K-sensitive assay, 1 mg/ml of the rPemK and rPemI were mixed with proteinase K (10 μg/ml) in 20 mm Tris-HCl, pH 7.4, 300 mm NaCl with 10% glycerol. Also, the rPemI-rPemK complex mixed in an equimolar ratio was also subjected to the proteinase K digestion. The samples were incubated at 37 °C for different time intervals. Reaction was terminated with 1 mm phenylmethylsulfonyl fluoride before subjecting the samples to SDS-PAGE followed by Coomassie staining.
The effect of proteinase K digestion on the functionality of the rPemK and rPemI individually and in a complex was assessed both qualitatively and quantitatively. For this, 10 μg of the total bacilli RNA was incubated at 25 °C for 15 min with the proteinase K-treated (10 μg/ml for 45 min) rPemK, rPemI, and rPemI-rPemK complex. Also, the proteinase K (10 μg/ml for 45 min)-treated rPemI was allowed to form a complex with the rPemK to generate rPemI(proteinase K)-rPemK complex, which was further incubated with the RNA as described above. The samples were electrophoresed on agarose gel and visualized by ethidium bromide staining. The samples were also incubated with 20 μm of the rC fluorigenic substrate, and the extent of cleavage was monitored and plotted in each case. Appropriate experimental controls were taken in both the cases as indicated under the “Results.”
Gel Filtration Chromatography
A Sephacryl-200 HiPrep16/60 FPLC column (GE Healthcare) equilibrated with 10 mm Tris-HCl, pH 8.0, was calibrated with standard proteins (1 mg/ml), bovine serum albumin (66.2 kDa), ovalbumin (45.0 kDa), carbonic anhydrase (31.0 kDa), and lysozyme (14.4 kDa). 1 mg of the purified proteins (rPemK and rPemI) individually and as a complex were loaded onto the column. The flow rate was maintained at 0.5 ml/min, and the elution volumes were recorded.
Native PAGE
The rPemK (25 and 50 μm) was mixed with the rPemI (50 μm) in binding buffer (50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 1 mm dithiothreitol, and 5% glycerol) at 4 °C for 1 h and subjected to native PAGE. The bands were further sliced from the gel, processed, and subjected to second dimension electrophoresis on a 16% SDS-PAG. The protein bands were visualized by staining with Coomassie Brilliant Blue.
Far-Western Blotting/Blot Overlay Assay
The rPemI (14 kDa) was separated on 15% SDS-PAG and transferred to a nitrocellulose membrane followed by renaturation by incubating the blot at 4 °C for 12 h in renaturation buffer (50 mm Tris-HCl, pH 7.5, 100 mm KCl, 5 mm MgCl2, 0.5% Triton X-100, 10% glycerol). The blot was overlaid with an equimolar amount of the rPemK in renaturation buffer for 1 h. The rPemI was detected with the anti-rPemK antiserum (1:10,000) followed by the addition of a 1:10,000 dilution of sheep anti-mouse IgG-alkaline phosphatase-conjugated secondary antibody. Immunoreactive bands were visualized by 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium dye substrate.
Fluorescence Spectroscopy, ANS Binding
Fresh ANS stock (1 mm, Sigma) was prepared in analytical grade ethanol. The rPemI and rPemK (25 μm) were titrated by increasing the ANS (0–200 μm) until the fluorescence intensity no longer changed. The fluorescence of bound ANS was determined by exciting the sample at 340 nm, and emission was recorded in the range of 400–600 nm. The saturating ANS concentration (140 μm) was used for further experiments. ANS (140 μm) was added to an equimolar complex of the rPemI-rPemK, and the change in the ANS fluorescence was compared with the ANS fluorescence observed with either of the proteins.
Fluorescence Polarization
Fluorescence polarization was carried out using a spectrofluorimeter equipped externally with a polarizing accessory. Excitation wavelength was 280 nm, and the emission spectrum ranged from 310 to 450 nm. The change in the polarization of the rPemK (10 μm) and the rPemI-rPemK complex (10 μm each) was recorded. The G factor (grating correction factor) for each sample was also analyzed, and the polarization values were normalized by using the following equations: G factor = IHV/IHH·polarization = IVV − GIVH/IVV + G IVH. To determine the stoichiometry of the rPemI-rPemK interaction, 20 μm of the rPemI was titrated with increasing concentrations of the rPemK (0–50 μm), and the changes in the polarization signal were calculated.
Fluorescence Quenching Studies
Acrylamide quenching experiment was performed at 25 °C. Fresh stock of 6 m acrylamide was prepared, and the rPemK was titrated by adding 2 μl of acrylamide each time with continuous stirring. Emission of the fluorophore was monitored at 315 nm by exciting the samples at 280 nm. Intensity data after quencher addition were averaged and corrected for the background emission. The Stern-Volmer (KSV) constant for measuring the accessibility of the fluorophore was calculated using the Stern-Volmer equation, F0/F = 1 + KSV [Q], where F and F0 are the emission intensities of the protein in the presence and absence of quencher, respectively; [Q] is the concentration of quencher (acrylamide), and KSV is the slope when data are plotted as F0/F versus [Q]. Similarly, for assessing the interaction of the rPemK with the rPemI, the recombinant proteins were preincubated for 16–18 h at 4 °C prior to the titration with 6 m acrylamide. The slopes obtained for the rPemK and the rPemI-rPemK complex were compared.
Circular Dichroism Spectroscopy
CD spectra of the rPemK, rPemI, and their complex (rPemI-rPemK) (0.1 mg in 10 mm phosphate-buffered saline, pH 7.4) were obtained using Jasco J-815 spectropolarimeter at 25 °C using a 1-mm cell and wavelengths between 200 and 250 nm at a scanning speed of 20 nm/min. Ten successive spectra were accumulated in each case and averaged, followed by the base-line correction using the buffer. The fraction of the secondary structure in each protein was deconvoluted using the K2d software.
CD spectrum was acquired in the near-UV range (250–300 nm) to assess the stoichiometry of protein interaction. Titration of the rPemK (20 μm) was performed with increasing concentrations of the rPemI (5–35 μm). The antitoxin spectrum was taken as the control, and base-line corrections were performed with buffer and the rPemK. Three successive spectra were recorded in a 1-cm cell and wavelengths between 250 and 330 nm at a scan speed of 20 nm/min. The CD values (millidegrees) were plotted on the y axis versus the calculated stoichiometry ([rPemK]/[rPemI]) on the x axis.
Red Edge Excitation Spectroscopy (REES)
To determine the stoichiometry of the rPemI-rPemK interaction, change in the REES upon interaction of the rPemK with rPemI was calculated by exciting the samples at 270 nm and recording the emission maximum ranging from 310 to 400 nm with an excitation increment of 5 nm and excitation and emission slit widths being 2.5 nm. The data were accumulated in a three-dimensional mode, and the excitation stop was set at 305 nm. The rPemI (20 μm) with buffer was considered as control and was subtracted from the REES obtained with experimental sample containing the rPemK. Similarly, change in the REES was analyzed upon interaction of the rPemI (20 μm) with the rPemK (0–50 μm) to determine the stoichiometry.
Real Time Reverse Transcription-PCR (Quantitative PCR)
Overnight-grown B. anthracis cultures were diluted to an absorbance (A600) of 0.1 in LB medium and grown at 37 °C until exponential phase (A600 of 0.6). An aliquot of the untreated cells was taken as control before stress exposure. Nutritional stress was induced by the addition of serine hydroxamate, a competitive inhibitor of seryl-tRNA synthetase (1 mg/ml). For heat shock and UV stress, cultures were either transferred to a water bath preset at 40 °C with gentle shaking for 1 h or kept under UV radiation for 1 h. Antibiotic stress was induced by adding chloramphenicol to a final concentration of 30 μg/ml. Mitomycin C (1 μg/ml), a DNA-damaging agent, was added in the culture medium. Cells harvested from each condition were analyzed for changes in the levels of specific pemI and pemK transcript/protein(s).
The first cDNA strand was reverse-transcribed from 5 μg of DNase-treated RNA isolated from B. anthracis under normal and stress conditions using high capacity cDNA Protein Data Bank code kit (Applied Biosystems) employing random hexamers. Gene-specific primers were designed using the Primer Express software, version 2.0. The reaction was performed in a 20-μl volume containing 10 μl of 2× SYBR Green Mastermix (Applied Biosystems), 300 nm of primers, and 4 μl of 10-fold diluted reverse transcription products. The PCRs were run in ABI 7500 using the following program: 50 °C for 2 min, 95 °C for 10 min, and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Following PCR amplification, the reactions were subjected to a temperature ramp to create the dissociation curve, which measures the changes in fluorescence as a function of temperature, by which the nonspecific products can be detected. The dissociation program was 95 °C for 15 s, 60 °C for 15 s followed by 20 min of slow ramp from 60 to 95 °C. For each gene, cDNA dilution curves were generated to calculate the individual real time PCR efficiencies that were determined by measuring the values of the slopes obtained. Three technical replicates of each reaction were run and dna gyrase, as the constitutively expressed gene, was used as internal control for data normalization (33). Quantification of the relative changes in gene expression was performed using the 2−ΔΔCT method. The data were tabulated as fold change in gene expression normalized to a housekeeping gene and relative to the control.
Immunoblotting
To assess the effect of stress on the expression profile of PemK and PemI, the cells were harvested post stress induction. Equal amount of protein was loaded on 12% SDS-PAG, and the proteins were transferred onto nitrocellulose membrane. The blots were probed with anti-rPemK (1:10,000), anti-rPemI (1:8000), and anti-DNA gyrase antisera (1:10,000). The captured antibody was detected by incubation of a 1:10,000 dilution of sheep anti-mouse IgG-alkaline phosphate-conjugated antibody. The blot was developed with 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium dye substrate.
Construction of Deletion Variants of PemI
Several deletions within the pemI gene were constructed to generate truncated fragments of PemI protein. The primers used for the study are shown in Table 1. The DNA fragments were amplified using B. anthracis genomic DNA, and the amplified DNA fragments were digested with BamHI and SalI. The fragments were subsequently ligated to pET-28(a) expression vector digested with the same enzymes to obtain pATΔ16(n), pATΔ29(n), and pATΔ49(n) (where (n) denotes truncations from the N terminus) and pATΔ25(c) (where (c) denotes deletions from the C terminus). The recombinant constructs were transformed into E. coli BL21 (λDE3), and the protein expression was analyzed after induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 4 h. The PemI fragments were purified using Ni2+-nitrilotriacetic acid affinity chromatography, quantitated using Bradford reagent, and tested for their ability to bind to the rPemK using ELISA as described previously (4). The absorbance obtained for the rPemK with full-length rPemI was taken as reference (100%), and the percentage binding for the deletions was calculated.
Design of Peptides and ELISA to Study Disruption of the rPemI-rPemK Interaction
By examining the Phyre-modeled partial antitoxin structure, secondary structure prediction, and similarity with the available structure of the MazEF complex (34), the critical residues at the C terminus involved in interaction with the toxin were predicted. To evaluate the efficacy of the peptides to disrupt the rPemI-rPemK interaction, in vitro, ELISA was performed by coating the wells with 200 ng of the rPemI followed by addition of pre-formed rPemK (200 ng)-peptide(s) (10 nm to 10 μm) complexes. The decrease in its binding to the bait protein (rPemI) was studied by employing anti-rPemK antiserum (1:10,000). The captured antibody was further probed with horseradish peroxidase-conjugated secondary antibody (1:10,000). The color was developed by addition of 3,3′,5,5′-tetramethylbenzidine substrate, and absorbance was read at 630 nm. The absorbance obtained for the interaction of both the proteins in the absence of peptide was taken as 100%. NSP-15, a 15-amino acid-long unrelated random nonspecific peptide, was used in this study as a control. Also, a related N-terminal octapeptide, NTP-8, derived from the sequence of PemI was included. The physicochemical parameters for the peptides were deduced using the ProtParam tool from ExPASy.
Impact of Peptides on the Ribonuclease Activity
The effect of peptides on the ribonuclease activity of the rPemK was also assessed. For this, RNA (5 μg)/rC (20 μm) was added to a preformed complex (which was kept for 16–18 h at 4 °C) of peptides (125 μm) and rPemK (20 μm). This was incubated for 2 h at 37 °C. The RNA remaining after 2 h was analyzed on a 1.5% formaldehyde gel and was quantitated using Quantity One one-dimensional analysis densitometric software (Bio-Rad). Untreated RNA and RNA incubated with 20 μm rPemK were taken as controls for visualizing the uncleaved and cleaved RNA, respectively. The percent of RNA left was calculated in each case by considering the intensity of the untreated RNA as 100%. The ability of each sample to cleave the fluorigenic substrate was also monitored as described above, and the RFI(AU) obtained with the rC (20 μm) and 20 μm rPemK was taken as 100%. The statistical significance (p value) between the indicated groups was calculated by the Student's t test.
RESULTS
Molecular Modeling and Structure Validation of the PemK of B. anthracis
The homology model generated for the PemK of B. anthracis using the Swiss-Model Workspace was validated using Procheck. The Ramachandran plot of the predicted model revealed that 92.8% of the residues lie in the most favored regions, and 6.2% of the residues lie in the additionally allowed regions, whereas 1.0% of the residues lie in the generously allowed regions (supplemental Fig. S1). Absence of any residues in the disallowed regions indicated an overall good quality of the predicted model. All Ramachandrans showed only 2 labeled residues out of 232 residues, whereas chi1 and chi2 plots showed no labeled residues. The main chain bond lengths were 100% within the limits, and main chain bond angles were 99.7% within limits. The main chain and side chain parameters were concentrated in the “better” region. No bad contacts were detected in the modeled PemK dimer. The reliability of the obtained model can be assessed by the score of G factor, which should be above −0.5. The G factor value for the dihedrals was found to be −0.07 and for the covalent bonds was found to be 0.39, and the overall value was 0.11. In the Verify3D scan, the designed model showed that 96.5% of the residues had an average three-dimensional ID score of >0.2, which is indicative of a good quality model (32).
PemK Is a Ribonuclease
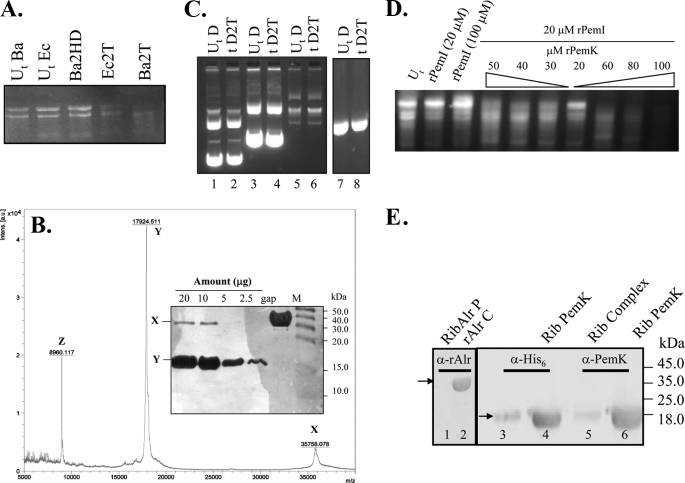
A high degree of structural similarity (96%) of PemK of B. anthracis to the YdcE protein of B. subtilis, which is an endoribonuclease (20), indicated a similar function for PemK. Also, our previous study demonstrated that overexpression of the PemK in vivo was detrimental to the protein synthesis, which was depicted by a reduced [35S]methionine incorporation (4). To evaluate the exact molecular mechanism underlying the observed translation arrest, the ability of the rPemK to cleave cellular RNA was assessed. The rPemK was shown to degrade a cellular preparation of RNA, including the 23 S and 16 S RNA from both E. coli and B. anthracis without any host specificity (Fig. 1A, 4th and 5th lanes). The heat-inactivated rPemK was unable to degrade the RNA (Fig. 1A, 3rd lane). MALDI-TOF mass spectrometry analysis confirmed the purity of the rPemK preparation. The MALDI profile showed three detectable peaks, corresponding to m/z of 35,758.078, 17,924.511, and 8960.117 (depicted as “X,” “Y,” and “Z,” respectively) (Fig. 1B). The identification of both the peaks, X and Y revealed them as PemK of B. anthracis (corresponding to the dimeric and monomeric forms, respectively). The third peak, Z, obtained at m/z of 8960.117 in fact depicts the doubly charged species (Z, M2H+) in contrast to the singly charged monomer obtained at 17,924.511 (Y, MH+), and singly charged dimer at 35,758.078 (X, 2MH+). Fig. 1B, inset, represents the silver staining of the rPemK in increasing concentrations, indicating the presence of only two bands at ∼36 and ∼18 kDa without any contaminating proteins, corroborating with the MALDI results. Collectively, the results confirmed the absence of any extraneous contaminant(s) in the rPemK that could confer the ribonuclease activity. The rPemK had no effect on E. coli DNA, demonstrating its specific action on RNA (Fig. 1C).
FIGURE 1.
A, PemK is a ribonuclease. Ten μg of RNA isolated from both E. coli and B. anthracis (abbreviated as Ec and Ba, respectively) were added to the rPemK at 25 °C for 15 min. 1st and 2nd lanes, untreated RNA (UtBa and UtEc) without the rPemK; 3rd lane, 2 μg of the heat-denatured (Ba2HD) rPemK with B. anthracis cellular RNA; 4th and 5th lanes, 2 μg of the rPemK with E. coli (Ec2T) and B. anthracis RNA (Ba2T), respectively. B, MALDI-TOF mass spectrometry profile of the rPemK of B. anthracis. X depicts a peak (m/z) at 35,758.078 indicating dimeric species (2MH+), and Y depicts a major peak at 17,924.511 indicating predominance of monomeric species (MH+). Z depicts a peak at m/z of 8960.117 corresponding to the doubly charged species, M2H+. The inset shows silver-stained SDS-PAG loaded with the indicated amounts of protein (μg); M depicts the molecular mass standards, and gap is glyceraldehyde-3-phosphate dehydrogenase (∼38.0 kDa) of B. anthracis loaded as a control protein. C, PemK cannot cleave any form of DNA. D depicts DNA; 2T depicts 2 μg of the rPemK; Ut and t represent untreated and treated DNA, respectively. Lanes 2, 4, and 6 represent circular double-stranded DNA (4, 6, and 9 kb) with 2 μg of the rPemK. Lanes 1, 3, and 5 represent untreated plasmids (UtD). Lanes 7 and 8 represent 600-bp linear double-stranded DNA (UtD) and with 2 μg of the rPemK (tD2T), respectively. D, inhibition of the RNA cleavage upon addition of the rPemI. 1st lane, B. anthracis total RNA untreated (Ut); 2nd and 3rd lanes, B. anthracis total RNA with 20 and 100 μm of rPemI alone; 4th to 7th lanes, B. anthracis RNA and 20 μm rPemI with decreasing concentrations of the rPemK (50 to 20 μm); 8th to 10th lanes, B. anthracis RNA and 20 μm rPemI with increasing concentrations of rPemK (60–100 μm). E, ribosome (Rib) binding activity of the rPemK in an in vitro assay. Antibodies directed against either the His6 tag (1:5000) or anti-PemK antibody (1:10,000) were used. Pellet fraction of alanine racemase (rAlr) with ribosomes is shown in lane 1; alanine racemase as a loading control (rAlr C) (lane 2) was probed with a 1:5000 dilution of anti-alanine racemase polyclonal antiserum; rPemI-rPemK complex in an equimolar ratio is shown in lane 3, and rPemK is shown in lane 4. Lanes 5 and 6 were similar to lanes 3 and 4, respectively, but probed with anti-rPemK polyclonal antibody.
rPemI Inhibits the rPemK-mediated Ribonuclease Activity
To evaluate whether the rPemI in vitro can neutralize the toxicity appended with the toxin, we incubated the rPemK in various ratios with rPemI prior to the addition of RNA. It was demonstrated that the rPemI by itself has no intrinsic ribonuclease activity (Fig. 1D, 2nd and 3rd lanes) at either of the concentrations (20 or 100 μm). The rPemI-rPemK complex in a 1:1 ratio was competent enough to bring about a maximum of ∼97% inhibition in the RNase activity (Fig. 1D, 7th lane). However, the rPemI-rPemK complexes in 1:2 or 1:1.5 molar ratios could only inhibit the ribonuclease activity to some extent, i.e. 40 and 20% of the ribonuclease activity still remained in each case (Fig. 1D, 5th and 6th lanes). Upon further increasing the amount of the rPemK corresponding to the molar stoichiometries of 2.5:1 to 3:1, 4:1, and 5:1 (rPemK:rPemI), a significant increase in the RNase activity was observed (Fig. 1D, 4th and 8th to 10th lanes, respectively).
The ability of the rPemK to bind to ribosomes was also ascertained, and it was observed that the rPemK was able to bind to ribosomes suggesting its possible role in translation attenuation (Fig. 1E, lanes 4 and 6). The absence of an intense band for the rPemK (Fig. 1E, lanes 3 and 5) indicates that rPemI inhibited the ribosome binding activity of the rPemK because the complex showed drastic inhibition in ribosomal binding.
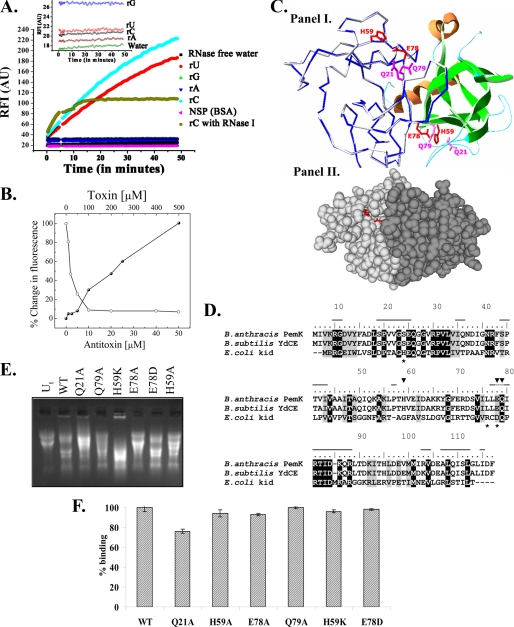
The chimeric fluorigenic substrates consist of a single RNA base flanked by short (∼5 bases) sequences of DNA labeled with a fluorophore on one end and a quencher on the other end. The close proximity of fluorophore and quencher in an intact probe obliterates any fluorescence, and the cleaved probe results in an enhanced fluorescence. The ability of the rPemK to cleave the RNA substrates was evaluated. The results obtained pointed toward preferential cleavage of “C” and “U” residues by the rPemK (Fig. 2A) demonstrating that the toxin is a pyrimidine-specific ribonuclease. Fig. 2A, inset, depicts negligible autofluorescence of the chimeric substrates.
FIGURE 2.
A, specificity of the rPemK ribonuclease. 20 μm oligonucleotide substrates were used. rA (filled blue inverted triangles), rG (filled green triangles), rC (filled blue diamonds), and rU (filled red circles) were incubated with the rPemK (500 ng), and an increase in 6-carboxyfluorescein fluorescence was monitored by exciting the samples at 485 nm and recording the emission at 530 nm as a function of time. Intrinsic autofluorescence of the substrates is shown as an inset. Brown circles and pink triangles depict ribonuclease A from bovine pancreas, and 1 μg of bovine serum albumin (BSA) was added to a solution of 20 μm rC substrate as a positive and a negative control, respectively. NSP, nonspecific protein. B, inhibition of ribonuclease activity of the rPemK by rPemI at 1:1 stoichiometry. 30 μm of the oligonucleotide substrate rC was incubated either with the rPemK (10 μm) alone or with the rPemI (0–50 μm), and an increase in 6-carboxyfluorescein fluorescence was monitored by exciting the samples at 485 nm and recording the emission at 530 nm as a function of time. The fluorescence obtained with 10 μm rPemK was taken as 100%, and the percentage decrease in the fluorescence intensities was plotted in each case and depicted as open circles. Also, the rPemI (50 μm) was titrated against increasing rPemK concentrations (0–500 μm), and the percent increase in fluorescence intensity in each case was calculated as described and depicted as closed circles. C, panel I, structural modeling of the PemK dimer. The monomer on the left shows the Cα trace of the PemK model (blue) superimposed upon the YdcE (gray). The monomer on the right is shown as a schematic. The helices are shown in orange, sheets in green, and loop regions in cyan. The catalytic base, Glu-78, the catalytic acid, His-59 (both shown as red sticks), and the stabilizing residues, Gln-21 and Gln-79 (both as magenta sticks), are shown. Panel II, two PemK monomers are shown in a space fill mode in different shades of gray. The side chains of the catalytic residues, His-59 and Glu-78, are shown in red sticks. The figure rendering was accomplished using PyMOL (Delano Scientific). D, multiple sequence alignment of PemK, YdcE, and Kid. The similar residues are shaded gray, and the identical residues are shaded black. The dashes on top of the sequence of B. anthracis PemK show the residues involved in forming the dimer interface. The asterisks below the kid sequence denote the catalytic residues in Kid, and the inverted triangles indicate the catalytic residues of the PemK as inferred from the sequence comparison and examination of the modeled PemK structure. E, ribonuclease activity of the rPemK mutants. 10 μg of B. anthracis RNA was incubated with 2 μg of the wild type rPemK (2nd lane), Q21A.rPemK (3rd lane), Q79A.rPemK (4th lane), H59K.rPemK (5th lane), E78A.rPemK (6th lane), E78D.rPemK (7th lane), and H59A.rPemK (8th lane). 1st lane (Ut, untreated RNA) contains 10 μg of intact RNA isolated from B. anthracis. F, wild type (WT) rPemK and its mutants are able to interact with the rPemI proficiently. 500 ng of the rPemK and its mutants were coated followed by the addition of 500 ng of the rPemI. The bound protein was captured by mouse anti-rPemI polyclonal antiserum. The absorbance of the captured anti-mouse IgG-horseradish peroxidase was measured after substrate addition at 630 nm. The binding of the wild type rPemK to the rPemI was taken as 100%. The bars depict mean ± S.D. of three independent experiments.
The enzymatic assay was further extended to delineate the exact stoichiometry required for the proteins to form a catalytically inactive complex. As is evident, the maximum fluorescence quenching upon increasing rPemI was observed when the proteins were reconstituted in a 1:1 ratio. There is no further decrease in the RFI upon a further increase in the rPemI, which indicates that the rPemI was able to chelate all the free rPemK molecules when present in an equimolar ratio (Fig. 2B, open circles). The graph in Fig. 2B also represents the experiment carried out in a different manner, where the rPemI was titrated with increasing concentrations of the rPemK. It could be seen clearly that the fluorescence intensity started to increase only when the rPemK was in excess and remained uncomplexed in solution to mediate the ribonuclease activity. This appeared when the proteins crossed 1:1 molar stoichiometry (Fig. 2B, closed circles).
The Michaelis-Menten constants (Km) of the rPemK for both the substrates, rU and rC, were calculated to be 5.16 ± 0.76 and 7.11 ± 0.36 mm, respectively, from the linear double-reciprocal plot. The Vmax values obtained for both the substrates were 200 ± 5 and 212 ± 10 μmol/min/mg (Table 2).
TABLE 2.
Comparison of kinetic coefficients of wild type and catalytic mutants of the PemK of B. anthracis using rU and rC fluorigenic substrates
| Mutants of PemK | KmrU | KmrC | VmaxrU | VmaxrC | kcatrU | kcatrC | (kcat/Km)rU | (kcat/Km)rC |
|---|---|---|---|---|---|---|---|---|
| mm | mm | μmol/min/mg | μmol/min/mg | s−1 | s−1 | (mmol/liter−1) s−1 | (mmol/liter−1) s−1 | |
| rPemK | 5.16 ± 0.76 | 7.11 ± 0.36 | 200 ± 5 | 212 ± 10 | 3 × 105 | 4 × 105 | 6 × 104 | 6 × 104 |
| Q21A | 7.21 ± 0.56 | 10.03 ± 0.45 | 150 ± 6 | 160 ± 8 | 2 × 105 | 3 × 105 | 2.8 × 104 | 3 × 104 |
| H59A | 10.0 ± 0.2 | 13.2 ± 0.5 | 50 ± 3 | 55 ± 4 | 6 × 104 | 8 × 104 | 6 × 103 | 6 × 103 |
| E78A | >50 mm | >50 mm | Cannot be determined | Cannot be determined | ||||
| Q79A | 5.43 ± 0.32 | 8.09 ± 0.17 | 180 ± 8 | 175 ± 6 | 3 × 105 | 2 × 105 | 6 × 104 | 3 × 104 |
| E78D | 5.22 ± 0.22 | 6.2 ± 0.3 | 220 ± 12 | 219 ± 15 | 4 × 105 | 4 × 105 | 8 × 104 | 6 × 104 |
| H59K | 6.06 ± 0.15 | 6.3 ± 0.2 | 218 ± 9 | 198 ± 6 | 4 × 105 | 3 × 105 | 6 × 104 | 5 × 104 |
Prediction and Determination of the Catalytic Residues of PemK
Sequence analysis and fold recognition for the PemK revealed a high degree of structural similarity to the YdcE protein (Protein Data Bank code 1NE8) of B. subtilis and thus classifies the protein in the Kid/PemK family of proteins. A ribbon view of the modeled PemK dimer superposed on the experimentally determined structure of the YdcE is shown in Fig. 2C, panel I. 232 Cα atoms of the modeled PemK dimer were superimposed on the dimer of code 1NE8, with a root mean square deviation of 0.1 Å.
For prediction of the catalytic residues, the PemK of B. anthracis was aligned with YdcE and Kid. The alignment obtained is shown in Fig. 2D. PemK shows 43% sequence similarity to the Kid for which the binding site has been well characterized. It has been shown that the residues Asp-75, Arg-73, and His-17 form the active site of the Kid toxin, wherein Asp-75 acts as the catalytic base and Arg-73 is the catalytic acid (18). Examination of the multiple sequence alignment showed that only the residue corresponding to catalytic base, Asp-75, was conserved in B. anthracis PemK and YdcE. The corresponding residue in PemK was Glu-78. From the three-dimensional structure of PemK, the only positively charged residue located in the close proximity of Glu-78 was found to be His-59. The position of conserved Glu-78 side chains and the probable catalytic acid (His-59) are shown in the modeled toxin dimer (Fig. 2C, panel I). Also, as shown in Fig. 2C, panel II, these proposed catalytic residues lie in a solvent accessible cavity on the surface of the modeled toxin dimer. Additional conserved stabilizing residues were predicted, as shown in Fig. 2C, panel I. Gln-79, the probable stabilization residue located near Glu-78 and His-59 proposed by analogy with the active site of Kid, is also shown. The Gln-21 residue was predicted to be present in a loop lying close to this triad, which was speculated to function as an additional stabilizing residue. Therefore, these four residues predicted to be involved in the catalysis were substituted with alanine. Also, Glu-78 and His-59 were mutated to a similarly charged negative and positive residue, aspartate (E78D) and lysine (H59K), respectively. The effect of these substitutions on the ribonuclease activity was investigated.
The mutations were introduced in the pemK, and the mutant proteins were expressed and purified to near-homogeneity (supplemental Fig. S2); the yields of the recombinant proteins were comparable with the wild type alleviating the possibility of any toxicity associated with them. RNA was resistant to degradation by the addition of E78A.rPemK (Fig. 2E, 6th lane), whereas Q21A.rPemK and Q79A.rPemK were able to degrade the RNA comparable with the wild type protein (Fig. 2E, 3rd and 4th lanes, respectively). The H59A.rPemK was able to degrade the RNA albeit to a lesser extent than the wild type rPemK (Fig. 2E, 8th lane). Interestingly, the mutants H59K.rPemK and E78D.rPemK, although harboring a different side chain and imparting a similar charge, were able to mediate the RNA cleavage comparable with the wild type rPemK (Fig. 2E, 5th and 7th lanes, respectively).
The Km value was increased drastically for the rPemK harboring E78A substitution (E78A.rPemK) with a subsequent reduction in the Vmax values. The mutant did not show any activity with either of the substrates (rU/rC) even at 50 mm concentration. The H59A mutation in the rPemK was also deleterious to the ribonuclease activity. The apparent Km value was twice (10 ± 0.2 mm for rU and 13.2 ± 0.5 mm for rC) as compared with the wild type rPemK (5.16 ± 0.76 mm for rU and 7.11 ± 0.36 mm for rC). Also, Vmax value was significantly reduced with a decrease in the catalytic efficiency (kcat/Km) by 10-fold (Table 2). Consistent with the gel-based qualitative assay, H59K.rPemK and E78D.rPemK were found to be as effective as the wild type rPemK (Table 2). The Q21.rPemK and Q79.rPemK mutants were also analyzed for their potential to cleave the RNA substrates. It was observed that the activity of these mutants was not significantly affected in comparison with the wild type protein, suggesting that these residues are not involved directly in the reaction mechanism.
The residues His-59 and Glu-78 of the PemK neither constitute the hydrophobic core of the monomer nor are involved in the dimer formation (Fig. 2C, panel II). Thus, the mutagenesis of these residues into a small neutral residue like alanine is not expected to alter the tertiary structure. In concordance with this, similar fluorescence emission maxima and CD spectra were obtained for the wild type rPemK and its mutants (supplemental Fig. S3 and Fig. S4, respectively). Therefore, it could be inferred that the loss of enzymatic activity in the mutants is solely due to the amino acid substitutions and is not due to any other gross significant structural perturbations or alterations.
rPemK Catalytic Mutants Bind to rPemI Efficiently
The efficiency of the catalytic mutants of the rPemK to bind to the rPemI was investigated. It was found that the H59A.rPemK and E78A.rPemK were competent enough to bind to the rPemI with equal efficiency as that observed for the wild type rPemK (Fig. 2F). The same was true for the Q79A.rPemK mutant. Contrarily, Q21A.rPemK mutant showed a slightly compromised binding to the rPemI (76%) when compared with the wild type rPemK.
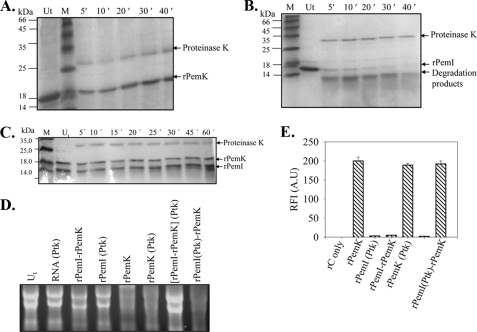
Differential Proteolytic Susceptibilities of the rPemK and rPemI
Proteinase K sensitivity assay was carried out to ascertain differences in the stability of the rPemK and rPemI. The two proteins showed considerable differences in their susceptibilities to proteolysis, when treated with proteinase K. The rPemK resisted proteolysis for over a period of 45 min (Fig. 3A). Unlike rPemK, the rPemI was degraded within 5–10 min, and degradation products could be seen that were absent in the untreated rPemI (Fig. 3B). Interestingly, the rPemI in a complex with the rPemK was resistant to proteinase K digestion, which indicated that the rPemI attained stability upon interaction with the rPemK (Fig. 3C). However, a mild degradation was observed only after 60 min of treatment. An additional band at 27 kDa in the protease-treated samples corresponds to the proteinase K.
FIGURE 3.
PemI is sensitive to proteinase K treatment unlike PemK. Coomassie Blue-stained SDS-PAG depicting the proteolysis of the rPemK and the rPemI with proteinase K (10 μg/ml) was analyzed at the indicated time intervals. Lane Ut represents untreated proteins, rPemK (A), rPemI (B), and the rPemI-rPemK complex (C). M indicates the molecular mass standards. The indicated proteins were incubated either with 10 μg of total bacterial RNA (D) or with 20 μm rC fluorigenic substrate, and the RFI(AU) are plotted on the y axis that reflects the extent of substrate cleavage (E). The rPemK (Ptk) and rPemI (Ptk) denotes proteinase K-treated rPemK and rPemI, respectively; and (rPemI-rPemK) (Ptk) depicts preformed rPemI-rPemK complex treated with proteinase K, and rPemI(Ptk)-rPemK depicts rPemI treated with proteinase K prior to the formation of complex with the rPemK.
The effect of proteinase K digestion on the catalytic activity of the rPemK and rPemI was investigated. The proteinase K-treated rPemK retained its ribonuclease activity suggesting that it remains functionally unperturbed (Fig. 3D, 6th lane). It was observed that the pretreatment of the rPemI with proteinase K followed by the addition of the rPemK (rPemI (proteinase K)-rPemK) failed to protect the rPemK-mediated ribonuclease activity indicating that the rPemI underwent proteolysis (Fig. 3D, 8th lane). On the contrary, when the preformed rPemI-rPemK complex was subjected to proteolysis, the rPemK-associated ribonuclease activity was abrogated (Fig. 3D, 7th lane). This suggested that the rPemI in conjunction with the rPemK formed a proteinase K-resistant complex, wherein the rPemI was able to inhibit the rPemK-mediated ribonuclease activity. The controls, including the effect of proteinase K on RNA (Fig. 3D, 2nd lane), proteinase K-treated rPemI (4th lane), and the untreated rPemI-rPemK complex (3rd lane), were found comparable with the untreated RNA (1st lane).
The qualitative findings were further substantiated with the quantitative assay; wherein the cleavage of the rC substrate was reduced to minimum when an equimolar rPemI-rPemK preformed complex was treated with proteinase K (Fig. 3E). Although the ribonuclease activity of the proteinase K-digested rPemK was unaffected, the rPemI was substantially degraded and was unable to confer any protection to the rPemK-mediated ribonuclease activity.
Interaction of the rPemI-rPemK in Vitro
In our earlier studies, we had shown that the toxin-antitoxin formed a complex in vivo using the glutathione S-transferase pulldown assay (4). To ascertain whether they retain the ability to form a complex in vitro, gel filtration was employed. The interaction resulted in the formation of a complex having a larger Stokes radius with lesser retention time as compared with the individual entities. The rPemK (∼18 kDa) and the rPemI (∼14 kDa) alone were eluted at volumes corresponding to 115 and 131 ml, respectively. But when they were loaded as a preformed complex, it eluted at a much lower elution volume, Ve = 80 ml. The peak fractions containing both the proteins were electrophoresed on SDS-PAG to confirm the presence of the proteins. The bands at ∼18 and ∼14 kDa were observed when individual proteins were loaded; however, the complex contained both the proteins (data not shown).
Native PAGE was also performed to confirm the in vitro TA interaction. As is evident, the rPemK displayed migration in the gel (Fig. 4A, panel I, lane 4) unlike the rPemI (panel I, lane 1). This was attributed to the positive charge on the rPemI due to its pI value equivalent to 9.16, in contrast to the rPemK, which has a theoretical pI of 4.2. The rPemI-rPemK complex with a slightly reduced mobility could be seen (Fig. 4A, panel I, lanes 2 and 3). Also, the intensity of the complex was enhanced upon increasing the rPemK concentration. A second dimension SDS-PAG analysis for the complex formed in the native PAG (Fig. 4A, panel I, lane 2) revealed the presence of two bands at 18 and 14 kDa, corresponding to the rPemK and the rPemI, respectively (Fig. 4A, panel II, lane 1). Also, the far-Western analysis with the rPemI as a bait protein further confirmed its interaction with the rPemK (Fig. 4B, lane 1).
FIGURE 4.
rPemK and the rPemI interact in vitro. A, native PAG. Panel I, lane 1, rPemI alone; lane 2, rPemI (50 μm), rPemK (50 μm) complex; and lane 3, rPemI (50 μm), rPemK (25 μm) complex. The slower migrating complex (c) is shown by an arrowhead and the free rPemK (f) by an arrow. Panel II, gel band corresponding to the complex (c) in lane 2 of panel I was sliced and electrophoresed on 16% SDS-PAG. Lane M represents molecular mass standards as follows: lane 1, arrows indicating ∼18- and ∼14-kDa bands corresponding to the rPemK and rPemI, respectively, separated from the complex on SDS-PAG. B, blot overlay assay. Lane 1, rPemI was run on a 12% SDS-PAG and was transblotted onto nitrocellulose membrane followed by incubation with an equimolar rPemK. The blot was probed with anti-rPemK antibody followed by addition of anti-mouse alkaline phosphatase-conjugated secondary antibody. The bands were revealed by addition of 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium dye substrate solution. Lane M, molecular mass standards. C, CD spectroscopy. Conformational change following complex formation was assessed by far-UV CD spectra of 20 μm of both the proteins alone and in complex. D, ANS binding. ANS (140 μm) was added to an equimolar complex of the rPemI and rPemK, and the change in the ANS fluorescence was measured and compared with the ANS fluorescence observed with either of the proteins. Inset shows ANS fluorescence for the proteins rPemK and rPemI (25 μm) at 450 nm obtained by continuous ANS titration with 0–200 μm as a function of protein folding. E, acrylamide quenching. The rPemK alone and the rPemI-rPemK complex was titrated using 6 m acrylamide, and the emission of the fluorophore was monitored at 315 nm by exciting the samples at 280 nm. The ratio of emission intensity (F0/F) in the absence of quencher (F0) and F at a given quencher concentration was plotted in each case, and the slopes obtained for the rPemK (open circles) and the rPemI-rPemK complex (closed circles) were calculated.
Conformational Changes during the rPemI-rPemK Interaction
To determine whether the rPemK and rPemI undergo a conformational change following their in vitro interaction, the rPemI-rPemK complex was analyzed by far-UV CD and compared with respect to the individual proteins. As is evident, the CD spectrum for the complex did not overlap with the spectra obtained for either of the proteins, indicating that a structural change has occurred following their interaction (Fig. 4C). The two negative peaks were observed for the rPemI at 217 and 208 nm, which correspond to the β-sheets and random coils, respectively. The rPemK exhibits two negative bands at 208 and 222 nm, ascribed to the α-helices. The secondary structure content deconvolution for the rPemI-rPemK complex revealed a modest transition of random coils (58 to 55%) to β-sheets (18–23%) post-interaction (Fig. 4C).
The anilinonaphthalene group of ANS fluoresces by interacting with the surface-exposed nonpolar (hydrophobic) residues in the protein. The ability of the ANS to bind to the rPemK and rPemI was assessed by titrating both the proteins with increasing concentrations of ANS. It was found that the rPemI was able to bind to ANS efficiently in comparison with the rPemK (Fig. 4D, inset). A substantial decrease in the ANS fluorescence intensity for the rPemI-rPemK complex suggested that the rPemI attained some tertiary structure upon rPemK binding (Fig. 4D).
Fluorescence polarization of a molecule depends on its molecular geometry and rigidity. Therefore, a change in the fluorescence polarization was anticipated upon the rPemI-rPemK interaction. A modest increase in the fluorescence polarization (FP) was observed for the rPemI-rPemK complex (FP, 0.13) in comparison with the PemK alone (FP, 0.12), suggesting the formation of a high molecular weight complex that did not depolarize the incident plane polarized light.
Acrylamide-induced fluorescence quenching was employed to examine whether the rPemK undergoes any change upon rPemI binding. The Stern-Volmer constant (which is indicative of the accessibility of fluorophore residues to the quencher in the solvent environment) for the rPemK was calculated from the slope of the plot and was found to be 6 × 10−4 m, compared with 4 × 10−4 m obtained for the rPemI-rPemK complex. This indicates that binding of the rPemK to rPemI introduces a conformational change in the rPemK, which partly protects the fluorophore from the quencher (Fig. 4E). Thus, binding of the rPemK to the rPemI changes the disposition of fluorophore from partially exposed to a much more buried state.
Stoichiometry of rPemI-rPemK Interaction
To establish the molar ratio of the rPemI-rPemK interaction, CD spectroscopy (Fig. 5A), fluorescence polarization (Fig. 5B), and red edge excitation spectroscopy (Fig. 5C) were employed. When a fluorophore is present in a polar and nonviscous environment, fluorescence emission maximum of the fluorophore is independent of the wavelength of excitation, but if it is present in a viscous (motionally restricted) environment, the emission maximum of the fluorophore shifts toward the higher wavelength, which is observed as a red shift (increase) in the excitation wavelengths. It was assumed that the rPemI-rPemK complex will be less dynamic and more rigid in nature compared with the individual proteins alone. Hence, the trapped solvent molecules (water) present in the near vicinity of fluorophore will also get motionally restricted; therefore, an increase in the REES was expected. Thus, the changes in the REES were used as a probe to determine the protein-protein interaction and stoichiometric ratio as well.
FIGURE 5.
Determination of stoichiometry of TA interaction. A, fluorescence polarization. B, CD spectroscopy. C, red edge excitation spectroscopy. The point of intersection of both the lines in a Job Plot is a direct measure of stoichiometry of interaction that is plotted on the x axis.
It was found that the rPemI-rPemK interacts in a molar stoichiometry of 1:1, i.e. one molecule of the rPemK interacts with one molecule of the rPemI. This was in agreement with the stoichiometric data obtained from the gel and fluorigenic substrate-based ribonuclease assay, which also demonstrated that the rPemI-rPemK forms a functional entity at a molar stoichiometry of 1:1. However, the possibility of formation of hetero-oligomers in a 1:1 stoichiometry as X2Y2, X3Y3, and XnYn cannot be ruled out.
Role of TA Module in Stress
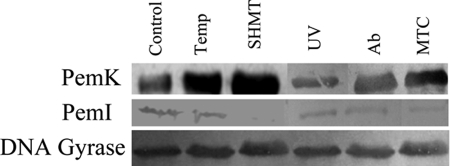
Real time PCR was employed to specifically quantitate the levels of the pemI/pemK transcript in B. anthracis cells grown under stress conditions. The comparative CT method of relative quantitation was validated by determining the efficiencies of the target and reference (endogenous control) amplifications, which were found to be approximately equal; the slopes were close to −3.32 (data not shown). The fold change in the pemK and pemI transcripts upon stress induction is depicted in Table 3. It was observed that the stress had a similar effect on both the transcripts, which was attributed to the fact that both the genes are located adjacent to each other and are co-transcribed as bicistronic mRNA.4 Nutrient deprivation in the cells was able to bring about a maximal increase in the pemI-pemK transcript followed by antibiotic addition. The other stress inducers, viz. high temperature, mitomycin C, and UV irradiation, generated an almost comparable increase in the pemIK transcript levels.
TABLE 3.
Real time PCR to quantitate the differences in the mRNA levels of pemK and pemI upon stress induction
Quantification of the relative changes in gene expression was performed using the 2−ΔΔCT method, and the data were plotted as fold change in gene expression normalized to a housekeeping gene, dnagyrase, and relative to the control (untreated cells). RQ is relative quantitation; MTC is mitomycin C; SHMT is serine hydroxamate.
| Stress | Fold change (RQ) |
|
|---|---|---|
| pemK | pemI | |
| Temperature | 2.53 ± 0.56 | 2.13 ± 0.13 |
| Antibiotic | 3.67 ± 0.23 | 3.02 ± 0.19 |
| MTC | 2.42 ± 0.75 | 2.02 ± 0.41 |
| SHMT | 5.83 ± 1.24 | 4.38 ± 0.91 |
| UV | 2.01 ± 0.22 | 2.20 ± 0.31 |
The effect of stress on the expression level of the TA proteins was also established. It is evident that the PemK was up-regulated with a concomitant decline in the levels of PemI (Fig. 6). DNA gyrase as an endogenous control was used as a loading control in each case.
FIGURE 6.
Levels of PemK and PemI proteins under stress conditions. Immunoblotting was performed by electrophoresing the crude cell lysates obtained under all the conditions and probed with anti-rPemK and anti-rPemI. Anti-DNA gyrase was used followed by addition of anti-mouse alkaline phosphatase-conjugated secondary antibody. The blot was revealed by addition of 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium dye substrate solution. SHMT, serine hydroxamate; Temp, temperature; MTC, mitomycin C.
Peptides to Disrupt the rPemI-rPemK Interaction
In case of the modeled PemK toxin, it could be seen that the monomer folding as well as the dimer organization is largely similar to that of the experimentally determined MazEF structure. Indeed, it was found that the modeled toxin dimer superimposed well with the MazF dimer (root mean square deviation of 1.6 Å between 165 equivalent Cα atoms) (Fig. 7A). It is thought that although the overall antitoxin structure may be very different, its mode of interaction with the toxin would remain similar to that established for the MazEF (34). Thus, a series of peptides based on the probable interacting regions in the antitoxin were designed, and their efficacy in disrupting TA interaction was evaluated.
FIGURE 7.
A, superposition of PemK of B. anthracis and MazF of E. coli and critical sites of interaction between the MazE and MazF. The model generated for the PemK was superimposed on the MazF dimer. The two monomers of the MazF are shown in cyan and green, and MazE is shown in orange. The ribbon of the PemK dimer is shown in black. The two most critical sites of interaction (labeled as Site 1 and Site 2) between MazE and MazF in the crystal structure are shown. Trp-73 of MazE in Site 1 is shown. Residues 66–73 (blue) of MazE bind to Site 1, and residues 55–62 (yellow) bind to Site 2. These were used to propose peptide II and VI, respectively. The figure rendering was done with PyMOL. B, C terminus of PemI is implicated in interaction with the PemK. The binding ability of the rPemK to the deletion variants of PemI was quantitated in a standard ELISA. The binding of the full-length rPemI to the rPemK was taken as 100%. The results reflect mean ± S.D. of three independent estimations. The lengths of the deletion fragments in base pairs and amino acids (a.a.) are shown at the right. C, sequence of the PemI of B. anthracis showing secondary structure prediction and rationale for design of peptides to disrupt the PemI-PemK interaction. The residues with predicted extended structure are underlined, and the helical residues are shaded. The deletion mutants Δ29 amino acids (shown as solid arrow) and Δ49 amino acids (shown as dotted arrow) were used to determine the C-terminal part of the PemI involved in the PemK binding. Their ability to bind to the rPemK is shown as percentage in parentheses. The PemI regions on which the peptides I, III, IV, and V are based is also shown. The N-terminal octapeptide (NTP-8) is shown in a box as a nonspecific but related peptide used as a control. Site 2-specific peptides showed slight protection in RNA degradation. RNA (5 μg) (D)/rC (20 μm) (E) was added to a preincubated complex of 125 μm of the indicated peptides (x axis) and 20 μm PemK (overnight at 4 °C) for 2 h at 37 °C. The percentage of remaining RNA was quantitated by a densitometer and plotted as % RNA left with the untreated RNA being 100% (D). The RFI(AU) was monitored at 630 nm for each sample, and units obtained with the rC (20 μm) and 20 μm rPemK were taken as 100% (E). For NSP-15 and NTP-8, 15- and 8-amino acid-long nonspecific peptides were used in this study. The bars depict mean ± S.D. of three independent experiments. The Student's t test was employed to calculate the statistical significance between the indicated groups, denoted with an asterisk, which represents a p value of <0.005.
The MazEF crystal structure showed that the interactions between the MazE and MazF are primarily mediated by the C terminus of MazE, which wraps around the MazF homodimer, crossing over the edge of dimer interface to interact with one of the MazF monomers (34). Four major interaction sites have been determined for the MazE and MazF of E. coli, of which two are the most critical sites. Site 1, consisting of C-terminal residues of MazE, includes one invariant tryptophan (Trp-73) and forms a β-strand when bound to MazF. Site 2 from the MazEF complex represents the most extensive contact interface between MazE and MazF with the largest amount of solvent-accessible area buried upon interaction. This site consists of residues of MazE in a helical conformation, overlying a crevice formed by both the MazF monomers. The locations of these critical sites in the MazF and the binding residues of MazE are shown in Fig. 7A.
Our results with the deletion variants of the rPemI demonstrate that the deletion of the first 29 amino acids from the N terminus of the PemI did not alter its ability to interact with the rPemK, whereas deletion of residues up to 49 amino acids lowered the toxin binding ability (Fig. 7B). Also, PemI devoid of its 25 amino acids from the C terminus was incompetent to interact with the rPemK, indicating that the C terminus of PemI is implicated in toxin binding. Therefore, 7–8-residue-long peptides (a) corresponding to the predicted helical region and (b) determined to be in the region necessary for the toxin binding were predicted systematically. Thus, peptide III (Leu-Leu-Phe-Gln-His-Leu-Thr-Glu) similar to residues 44–51, peptide IV (Lys-Arg-Tyr-Gln-His-Glu-Ser-Met) similar to residues 52–59, and peptide V (Arg-Arg-Gly-Tyr-Ile-Glu-Met-Gly) similar to residues 60–67 of PemI were proposed. To bind to Site 1, peptide I (Val-Glu-Arg-Leu-Val-Ser-Gly-Gly) corresponding to the residues 88–95 of PemI was proposed. The location of the determined deletion mutants, the predicted helical regions, and the regions used for design of peptides are shown in Fig. 7C.
Two peptides, II and VI based on the sequence of MazE that binds to Site 1 and Site 2, respectively, were also proposed. Peptide II (Asn-Leu-His-Arg-Asn-Ile-Trp) was proposed based on the C-terminal residues 66–73 of the MazE (Fig. 7A). Peptide VI (Lys-Ala-Glu-Leu-Val-Asn-Asp-Ile) similar to the helical part of MazF (chain C, residues 55–62, Protein Data Bank code 1UB4) binds to the interface of the MazE toxin dimer as shown by the crystal structure (34).
All the designed peptides were capable of inhibiting the toxin-antitoxin interaction to some extent (Table 4). Our results show that the maximum disruption is achieved by peptides III, V, IV, and VI (in decreasing order). Strikingly, all of them have been predicted to possess helical conformation and may bind to PemK region corresponding to Site 2 of MazE. A lower disruption of toxin-antitoxin binding was achieved with peptide II and I (in decreasing order). Because the N-terminal region of the PemI is not involved in interaction with PemK and does not constitute the binding interface, NTP-8, an octapeptide corresponding to the 1–8 residues of PemI, was tested as a nonspecific but related peptide for its ability to disrupt the rPemI-rPemK interaction. It did not inhibit the TA interaction, which further confirmed the specificity of the peptides in disrupting the TA interaction. The inability of an unrelated peptide (NSP-15) to inhibit the rPemI-rPemK interaction further demonstrated the specificity of the peptides used in this study. The physicochemical parameters evaluated for both the control peptides (NSP-15 and NTP-8) fell within the range of the experimental peptides in all the aspects including length, pI, and hydrophobicity (supplemental Fig. S5).
TABLE 4.
Sequence, characteristics, and potential of designer peptides to inhibit the rPemI-rPemK interaction in vitro as assessed by competitive ELISA using 2 μm peptides
NSP-15 denotes a 15-amino acid-long nonspecific random unrelated peptide used as a negative control. NTP-8, a related octapeptide derived from the N terminus of PemI (1–8 amino acids), was also included as a control.
| Site no. | Peptide sequence | Solubility in water | % inhibition |
|---|---|---|---|
| I (Site 1) | VERLVSGG | Partially insoluble | 11 ± 1 |
| II (Site 1) | NLHRNIW | Soluble | 20 ± 2 |
| III (Site 2) | LLFQHLTE | Soluble | 35 ± 2 |
| IV (Site 2) | KRYQHESM | Soluble | 25 ± 2 |
| V (Site 2) | RRGYIEMG | Partially insoluble | 30 ± 1 |
| VI (Site 2) | KAELVNDI | Soluble | 22 ± 1 |
| NSP-15 | LLKKIPKDVLEIYSE | Soluble | 0 |
| NTP-8 | MSESSVTT | Soluble | 0 |
The effect of Site 1 and Site 2 peptides on the ribonuclease activity of the rPemK was also assessed. It was observed that only peptide III and peptide V were able to inhibit the ribonuclease activity of the rPemK to a significant extent (p < 0.005), albeit at a higher concentration (125 μm) (Fig. 7D). On the contrary, the Site 1-specific peptides (I and II) and nonspecific peptides (NSP-15 and NTP-8) when incubated with the rPemK had nil protective efficacies and degraded RNA like rPemK (Fig. 7D). This was further substantiated by investigating the ability of the peptides to inhibit the cleavage of the rC fluorigenic substrate. As is evident from Fig. 7E, peptides III and V were able to bind to the Site 2 and inhibit the ribonuclease activity to a significant extent (p < 0.005). It is also noteworthy here that these two peptides offered maximal TA disruption in this study (Table 4, peptide III, 35 ± 2%, and peptide V, 30 ± 1%). The other two Site 2 peptides (IV and VI) showed only a minimal protection, which is again in line with the magnitude of inhibition they conferred to the rPemI-rPemK interaction, which is 25 ± 2 and 22 ± 1%, respectively (Table 4). As a control, the peptides alone were also tested for their possible ribonuclease activity, and it was found that none of the peptides had a deleterious effect on the RNA (supplemental Fig. S6).
DISCUSSION
We had previously demonstrated that the overexpression of B. anthracis PemK was toxic in both homologous/heterologous hosts and displayed a drastic inhibition of protein synthesis with no effect on DNA replication (4). In view of its high similarity to the YdcE, an endoribonuclease of B. subtilis, this study demonstrates PemK as a catalytically active ribonuclease with a base specificity for pyrimidines (C/U). Thus, the reduced translational rate observed upon PemK induction in vivo could be attributed to its ribonuclease activity and a possible “bystander” cleavage of cellular mRNAs encoding essential genes (4). Analogous to the Kid and YdcE, B. anthracis PemK is also expected to form a symmetrical dimer leading to the formation of two RNA-binding sites. However, in the case of Kid, it was established that only one of the two RNA-binding sites can be occupied at a time (18, 20). It still remains to be elucidated that whether PemK can accommodate one or two RNA molecules.
The cleavage mechanism of Kid has been studied in considerable detail, and the active site residues responsible for catalysis have been proposed (18). The catalytic acid, Arg-73 and catalytic base, Asp-75, of Kid have been proposed to deprotonate the 2′-OH group of uracil, which then performs a nucleophilic attack on the electrophilic phosphorous. The transphosphorylation reaction terminates once the catalytic acid Arg-73 donates a hydrogen atom to the adenosine 5′-O. However, it has been reported that the catalytic acid (Arg-73) of Kid is not even conserved within the Kid homologues (18). Parallel to this, our results indicate the absence of a positively charged residue in B. anthracis PemK (Leu-76) at the position corresponding to Arg-73 of Kid. Therefore, His-59, the only positively charged residue found to be present in the catalytic cleft, was speculated to act as the catalytic acid, which became evident from our mutagenesis studies. Also, it was found to be conserved among the sequence homologues of PemK (data not shown). The sequence alignment has shown that most related toxins possess an acidic residue (Asp or Glu), which can function as a catalytic base (18). The substitution of Asp or Glu with Gln (glutamine) in ChpBK resulted in reduced ribonuclease activity in comparison with MazF, which possesses an Asp-76 (35, 36). This study shows that B. anthracis PemK possesses Glu-78 as its catalytic base. Apart from the Asp-75 and Arg-73 in the Kid toxin, an additional residue His-17 has been proposed to stabilize the Kid-RNA complex but is not directly involved in the Kid-mediated toxicity (18). The presence of a functionally similar residue, serine, in B. anthracis PemK (Fig. 2D) and in other Kid homologues at the position corresponding to His-17 is speculated to stabilize the reaction intermediate (18).
Thus, our study reveals that the PemK of B. anthracis is a ribonuclease that exploits His-59 and Glu-78 as its catalytic acid-base couple. Interestingly, our mutagenesis studies indicate that the Lys-59 to Glu-78 (H59K.rPemK) and His-59 to Asp-78 (E78D.rPemK) mutants retain their ribonuclease activity comparable with the wild type rPemK (His-59 to Glu-78). The in silico substitution of histidine and glutamate to lysine and aspartate at the 59th and 78th position, respectively, also revealed that the mutated side chains can be accommodated well in the catalytic pocket without any steric hindrance. This is in further agreement with the occurrence of lysine (Lys-41) as the catalytic acid in RNase A and aspartate (Asp-75) as the catalytic base in Kid (18), unlike His-59 and Glu-78 in B. anthracis PemK. Thus, the substitution of His-59 of PemK by Lys or Glu-78 by Asp conferred efficient ribonuclease activity pointing toward the significance of the acid-base couple in governing the overall catalysis. It can also be inferred that the substrate (RNA)-binding site of the PemK overlaps with the antitoxin-binding site because the presence of antitoxin could bring about a significant reduction in the ribonuclease activity of the PemK. Similarly, it has been reported for the C terminus of MazE that it is extended and bears a negative charge, and thereby it can mimic the RNA polyanion and could compete with the MazF to occupy its active site (36). The active site of PemK can thus be visualized as a pocket that can accommodate an extended strand of RNA as its catalytic substrate. The active site is present as a cleft that is formed at the interface of the two PemK dimers and it overlaps with the Site 2 of the MazEF complex (Fig. 7A).
By analogy with the crystal structure of MazEF, Glu-78 and His-59 of PemK lie within Site 2 and make contact with the helical region of the antitoxin (supplemental Fig. S7). However, the modeling studies demonstrate that Glu-78 is likely to make contact with only one residue of MazE (His-68), whereas His-59 is able to interact with both His-68 and Asn-66 of MazE (supplemental Fig. S8). This constitutes only a small fraction of the large contact interface between toxin and antitoxin that encompasses both Site 1 and Site 2. Therefore, a single amino acid substitution in the PemK mutants (His-59 to Ala and Glu-78 to Ala) was not expected to alter its binding characteristics to the rPemI. Our results for the H59A.rPemK and E78A.rPemK also demonstrate the same. However, a loss of binding was observed for the Q21A.rPemK. The molecular modeling of the PemK of B. anthracis indicated that the Gln-21 lies in a loop region. By analogy to the MazF, this loop differs considerably in the free and antitoxin-bound form. In the latter form, the loop adopts an open conformation, thus allowing the C terminus of the antitoxin to bind to the toxin. In view of the reduced rPemI binding ability of the Q21A.rPemK mutant, it can be speculated that this residue is involved in stabilization of the PemI-PemK complex. However, the exact role of this residue requires detailed knowledge about the structure of these proteins in a complex state.
It has been postulated for the relBE TA family that RelB is a ribonuclease, but it requires presentation of its substrate on the ribosomes (2). The RelE interacts with RelB making it bulky enough to block its entry into the ribosomal A site. Contrarily, MazF of MazEF family binds and cleaves free RNA independent of ribosomes but with a marked codon specificity (9, 19, 37). The Doc of Phd-Doc system mediates translation inhibition by interacting with 30 S ribosomal subunit (38). The mechanism of action of the PemK of B. anthracis is slightly different because it is able to interact with free RNA and also the ribosomes individually. Previously, our results only reveal that ribosomes are sequestered by the toxin, which might account for the cytotoxicity associated with the PemK; however, it still remains elusive whether PemK binds to ribosomes via the 30 S or 50 S subunit. A detailed structural characterization of PemK in complex with PemI or with its target (RNA or ribosomes) will further help to understand its precise mechanism of action.
We envisage that the different decay rates of toxin and antitoxin, the latter being highly vulnerable to the action of specific proteases, form the basis of physiological functioning of the TA module. We had previously established that the proteins possess differential susceptibility to heat (4). The antitoxin was found to be relatively thermolabile when compared with toxin with a faster unfolding rate, which intrigued us to study their protease sensitivities. It has been well documented for the MazE of E. coli that in the absence of MazF, the C terminus adopts an unstructured conformation that makes it vulnerable to several proteases. ProtParam analysis of the PemI revealed it to be quite unstable when compared with the PemK. The Phyre disorder prediction analysis revealed that the PemI possesses a poor hydrophobic core with no definite structure (supplemental Fig. S9). Here, we have shown that the proteinase K efficiently degraded the rPemI when compared with the rPemK. The elongated conformation of PemI at the C terminus and lack of tertiary structure might account for its high degree of protease susceptibility. The two proteins, rPemK and rPemI, showed substantial difference in their ANS binding potential. An enhanced binding observed for the rPemI further substantiates its disordered conformation (Fig. 4D, inset).
Until now, the role of TA modules is debatable and seems paradoxical. Induction of mazEF operon in the presence of ppGpp, a stress alarmone, has been correlated with programmed cell death (3). However, the restoration of the toxicity by the induction of transcription of the corresponding antidote, MazE (39), conflicts with the former hypothesis and points toward its bacteriostatic role rather than a bactericidal role. The other groups have shown that the cells devoid of the mazEF module are more significantly resistant to stress, although the stresses include relatively short exposure to antibiotics, high temperature, DNA damage agents, and oxidative stress (40, 41). A contradiction was put forth against this hypothesis stating that the stress response system reduces the translation rate to benefit the cells by preventing energy consumption, but initiation of a cell death program in response to such short stress conditions was assumed to be less evident and questionable (42). However, a plethora of evidence has accumulated that indicates the role of TA modules in stress. The sigB operon of Staphylococcus aureus is co-transcribed with the mazF gene in response to heat shock (8, 9). Similarly, genes encoding PemK (NE0974 and NE0975) were up-regulated ≥2-fold both under zinc and chloroform stress in Nitrosomonas europa (10). Transcriptomic profiling of Sulfolobus solfataricus on temperature shift (80–90 °C) revealed the up-regulation of vapBC genes, and knocking out a specific vapBC locus rendered the host susceptible to heat shock (11). An interesting study by Tsilibaris et al. (13) posed a challenge to the most widely accepted notion about the role of TA loci in stress physiology; this prompted us to investigate the role of pemIK locus of B. anthracis in stress conditions. Our data indicate that the pemIK module of B. anthracis functions as a stress regulator. Induction of the pemI-pemK transcript in stress leads to an increased level of PemK. The parallel degradation of PemI, which is shown to repress the operon,4 thereby marks the prolific activation of the operon. Therefore, we speculate that stress acts as an auto-inducer of the operon that probably leads to activation of an X molecule that either could be a protease for which the antitoxins are a lucrative target or a key factor/regulator that could sense stress within the cells and arrest the pemIK operon in an “on” state. Also, we have worked only on a single module from B. anthracis; however, the possibility of existence of several others is not without precedent. It is plausible that the effect observed could be due to a concerted interplay of several unexplored TA modules from B. anthracis, because the regulation of the apoptotic machinery is governed by an intricately interwoven network culminating in cell death.
The ability of microbes to develop resistance determinants in a short span of time necessitates development of effective drugs against B. anthracis, a potential bioterror weapon. The toxins of TA systems have been given an analogy of “intracellular molecular time bombs” whose release from their cognate antitoxins could trigger bacterial apoptosis (14). Thus, it was envisioned that the disruption of TA interaction by peptides/chemical molecule holds an enormous reservoir for the development of potential antimicrobials. However, there is a need to develop high affinity compounds that can efficiently displace the toxin from the antitoxin-generating “antitoxin-compound” adduct, unveiling the toxin to act on its intracellular target. Because of the large region buried by the complex formed between proteins, it is usually difficult to disrupt their interaction with the help of small peptides or organic molecules. However, the TA complexes are uniquely suited for this approach because the antitoxin lacks a hydrophobic core. Especially at the C terminus, it assumes an elongated state that attains an ordered state upon binding with toxin. Toward our goal of disrupting the TA interaction, we tested the efficacy of a few peptides in abrogating TA interaction. The peptides were designed to mimic the two most probable sites of TA interaction. Our studies demonstrate 10–35% inhibition of the rPemI-rPemK interaction with 2 μm of the peptides. Thus, we demonstrate for the first time that it is feasible to disrupt the TA interaction with the help of small octapeptides.
A higher inhibition was achieved with all the helical peptides thought to mimic the Site 2 of rPemI-rPemK interaction. In contrast, the peptides thought to disrupt interaction at Site 1 showed lower activity. This could be attributed to either the absence of Site 1 in PemK or due to the unsuitability of the peptides designed. In this context, Trp-73 at Site 1 is the most intimate site of interaction between MazE and MazF, but no such hydrophobic residues are present at the C terminus of PemI. Trp-73 occupies a hydrophobic pocket on the surface that is lined by the following hydrophobic residues: Val-15, Arg-29, Ala-31, Cys-48, Pro-50, Lys-79, Ile-81, and Arg-86 in MazF. The corresponding residues are Ala-11, Arg-25, Val-27, Val-44, Ala-46, Arg-81, Ile-83, and Arg-87 and are highly conserved in sequences similar to B. anthracis PemK. Thus, the presence of a hydrophobic pocket similar to that in MazF is probable. Therefore, more specific and high affinity peptides designed to bind to Site 1 are required. This is also important because Site 2 coincides with the active site of the B. anthracis PemK. Peptide inhibitors that work by binding at this site may also interfere with the ribonuclease/bactericidal activity of PemK. It was found that the most active TA interaction disruption peptides (III and V), having a predicted helical conformation and ability to bind to Site 2, showed a slight inhibition of the ribonuclease activity of the PemK. This was expected because the antitoxin-binding site of the structurally similar Kid partly overlaps with the RNA-binding site at Site 2 (43). However, the inhibition of ribonuclease activity with these peptides could only be attained at a 60-fold higher concentration as required for the disruption of TA interaction. This shows the specificity of the designed peptides. However, peptides predicted to bind to Site 1, expected to be exclusive for antitoxin binding alone, did not show any inhibition of the ribonuclease activity. Thus, small molecule inhibitors that bind to the Site 1 are more suitable as therapeutic agents for anthrax. Our preliminary results, although not shown here, also demonstrate the bactericidal activity associated with these peptides in the micromolar range.
The determination of the crystal structure of the PemI-PemK complex and PemK in conjunction with its substrate/target (RNA) will shed light on the exact molecular mechanisms underlying the physiological functioning of this locus. This will further facilitate the development of novel and efficacious antimicrobials that can be targeted to enter the bacterial cells to block TA interaction specifically and elicit a robust antibacterial response.
Supplementary Material
Acknowledgments
We sincerely thank Shivani Agarwal (Gene Regulation Laboratory, School of Biotechnology, Jawaharlal Nehru University, Delhi, India) for help in few experiments. Nikita Chopra is duly acknowledged for valuable input in the modeling studies. The animal experiment(s) carried out for raising polyclonal antiserum were with the approval of the Institutional Animal Ethics Committee, Jawaharlal Nehru University, India (IAEC-JNU Project Code 05/2008).
This work was supported by the Department of Biotechnology, Delhi, India.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
S. Agarwal, N. K. Mishra, S. Bhatnagar, and R. Bhatnagar, unpublished results.
- TA
- toxin-antitoxin
- rPemK
- recombinant His6-tagged toxin
- rPemI
- recombinant His6-tagged antitoxin
- RFI (A.U.)
- relative fluorescence intensity in arbitrary units
- ANS
- 1-amino-2-naphthol-4-sulfonic acid
- ELISA
- enzyme-linked immunosorbent assay
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- REES
- red edge excitation spectroscopy
- PAG
- polyacrylamide gel.
REFERENCES
- 1.Engelberg-Kulka H., Amitai S., Kolodkin-Gal I., Hazan R. (2006) PLoS Gen. 2, e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buts L., Lah J., Dao-Thi M. H., Wyns L., Loris R. (2005) Trends Biochem. Sci. 30, 672–679 [DOI] [PubMed] [Google Scholar]
- 3.Gerdes K., Christensen S. K., Løbner-Olesen A. (2005) Nat. Rev. Microbiol. 3, 371–382 [DOI] [PubMed] [Google Scholar]
- 4.Agarwal S., Agarwal S., Bhatnagar R. (2007) FEBS Lett. 581, 1727–1734 [DOI] [PubMed] [Google Scholar]
- 5.Christensen S. K., Mikkelsen M., Pedersen K., Gerdes K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14328–14333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolodkin-Gal I., Engelberg-Kulka H. (2006) J. Bacteriol. 188, 3420–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandey D. P., Gerdes K. (2005) Nucleic Acids Res. 33, 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mittenhuber G. (1999) J. Mol. Microbiol. Biotechnol. 1, 295–302 [PubMed] [Google Scholar]
- 9.Fu Z., Donegan N. P., Memmi G., Cheung A. L. (2007) J. Bacteriol. 189, 8871–8879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park S., Ely R. L. (2008) Arch. Microbiol. 189, 541–548 [DOI] [PubMed] [Google Scholar]
- 11.Tachdjian S., Kelly R. M. (2006) J. Bacteriol. 188, 4553–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemos J. A., Brown T. A., Jr., Abranches J., Burne R. A. (2005) FEMS Microbiol. Lett. 253, 251–257 [DOI] [PubMed] [Google Scholar]
- 13.Tsilibaris V., Maenhaut-Michel G., Mine N., Van Melderen L. (2007) J. Bacteriol. 189, 6101–6108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kedzierska B., Lian L. Y., Hayes F. (2007) Nucleic Acids Res. 35, 325–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J., Zhang Y., Zhu L., Suzuki M., Inouye M. (2004) J. Biol. Chem. 279, 20678–20684 [DOI] [PubMed] [Google Scholar]
- 16.Muñoz-Gómez A. J., Santos-Sierra S., Berzal-Herranz A., Lemonnier M., Díaz-Orejas R. (2004) FEBS Lett. 567, 316–320 [DOI] [PubMed] [Google Scholar]
- 17.Christensen S. K., Pedersen K., Hansen F. G., Gerdes K. (2003) J. Mol. Biol. 332, 809–819 [DOI] [PubMed] [Google Scholar]
- 18.Kamphuis M. B., Bonvin A. M., Monti M. C., Lemonnier M., Muñoz-Gómez A., van den Heuvel R. H., Díaz-Orejas R., Boelens R. (2006) J. Mol. Biol. 357, 115–126 [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Zhang J., Hoeflich K. P., Ikura M., Qing G., Inouye M. (2003) Mol. Cell 12, 913–923 [DOI] [PubMed] [Google Scholar]
- 20.Pellegrini O., Mathy N., Gogos A., Shapiro L., Condon C. (2005) Mol. Microbiol. 56, 1139–1148 [DOI] [PubMed] [Google Scholar]
- 21.Tsuchimoto S., Nishimura Y., Ohtsubo E. (1992) J. Bacteriol. 174, 4205–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamada K., Hanaoka F. (2005) Mol. Cell. 19, 497–509 [DOI] [PubMed] [Google Scholar]
- 23.Daines D. A., Wu M. H., Yuan S. Y. (2007) J. Bacteriol. 189, 5041–5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang N. R., Hergenrother P. J. (2007) Anal. Biochem. 371, 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galvani C., Terry J., Ishiguro E. E. (2001) J. Bacteriol. 183, 2700–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tippmann H. F. (2004) Brief. Bioinform. 5, 82–87 [DOI] [PubMed] [Google Scholar]
- 28.Melvin I., Ie E., Kuang R., Weston J., Stafford W. N., Leslie C. (2007) BMC Bioinformatics 8, Suppl. 4, S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnold K., Bordoli L., Koop J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 30.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 31.Morris A. L., MacArthur M. W., Hutchinson E. G., Thornton J. M. (1992) Proteins 12, 345–364 [DOI] [PubMed] [Google Scholar]
- 32.Luthy R., Bowie J. W., Eisenberg D. (1992) Nature 356, 83–85 [DOI] [PubMed] [Google Scholar]
- 33.Vetter S. M., Schlievert P. M. (2005) Antimicrob. Agents Chemother. 49, 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamada K., Hanaoka F., Burley S. K. (2003) Mol. Cell 11, 875–884 [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y., Zhu L., Zhang J., Inouye M. (2005) J. Biol. Chem. 280, 26080–26088 [DOI] [PubMed] [Google Scholar]
- 36.Li G. Y., Zhang Y., Chan M. C., Mal T. K., Hoeflich K. P., Inouye M., Ikura M. (2006) J. Mol. Biol. 357, 139–150 [DOI] [PubMed] [Google Scholar]
- 37.Pimentel B., Madine M. A., de la Cueva-Méndez G. (2005) EMBO J. 24, 3459–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu M., Zhang Y., Inouye M., Woychik N. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5885–5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pedersen K., Christensen S. K., Gerdes K. (2002) Mol. Microbiol. 45, 501–510 [DOI] [PubMed] [Google Scholar]
- 40.Sat B., Hazan R., Fisher T., Khaner H., Glaser G., Engelberg-Kulka H. (2001) J. Bacteriol. 183, 2041–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hazan R., Sat B., Engelberg-Kulka H. (2004) J. Bacteriol. 186, 3663–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Condon C. (2006) Mol. Microbiol. 61, 573–583 [DOI] [PubMed] [Google Scholar]
- 43.Kamphuis M. B., Monti M. C., van den Heuvel R. H., López-Villarejo J., Díaz-Orejas R., Boelens R. (2007) Protein Pept. Lett. 14, 113–124 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.