Abstract
The synthesis of melanin pigments, or melanogenesis, is regulated by the balance of a variety of signal transduction pathways. Among these pathways, p38 MAPK signaling was found to be involved in stress-induced melanogenesis and to be activated by α-melanocyte-stimulating hormone (α-MSH) and ultraviolet irradiation. Previous studies have shown that α-MSH-stimulated melanogenesis can be inhibited by blocking p38 MAPK activity with SB203580, a pyridinyl imidazole compound. Consistent with this, we observed that pyridinyl imidazoles (SB203580 and SB202190) inhibited both basal and α-MSH-induced melanogenesis in B16 melanoma cells. However, SB202474, which has no ability to inhibit p38 MAPK activity and is usually used as a negative control compound in p38 MAPK studies, also suppressed melanin synthesis induction. Furthermore, the independence of the p38 kinase pathway from the repression of melanogenesis by pyridinyl imidazole compounds was also confirmed by small interfering RNA experiments. Interfering with p38 MAPK expression surprisingly stimulated melanogenesis and tyrosinase family protein expression. Although the molecular mechanism(s) by which p38 promotes the degradation of melanogenic enzymes remain to be determined, the involvement of the ubiquitin-proteasome pathway was demonstrated by co-treatment with the proteasome-specific inhibitor MG132 and the relative decrease in the ubiquitination of tyrosinase in cells transfected with p38-specific small interfering RNA.
Keywords: Cell, Signal Transduction, Signal Transduction/Protein Kinases/Cyclic Nucleotide, Signal Transduction/Protein Kinases/MAP, Cell Differentiation, Cyclic AMP (cAMP), Melanogenesis, Pigmentation
Introduction
Skin pigmentation, which results from the production and distribution of melanin in the epidermis, is the major physiological defense against solar irradiation. In mammalian melanocytes, melanins are synthesized within melanosomes that contain three major pigment enzymes: tyrosinase and tyrosinase-related protein-1 (TRP-1)2 and dopachrome tautomerase (DCT), also known as tyrosinase related protein-2 (TRP-2) (1–5). Melanin synthesis is stimulated by a large number of effectors, including cAMP-elevating agents (forskolin, isobutylmethylxanthine, α-MSH) (6–8), cholera toxin (9), UV light (10–12), placental total lipid fraction (13), lupeol (14), lipopolysaccharide (15), GSK3β signaling pathway blocker (16), rosmarinic acid (17), the phosphatidylinositol 3-kinase inhibitor LY294002 (18), and the MEK inhibitor PD98059 (19). The balance of a variety of signal transduction pathways regulates melanogenesis. Thus far, one of the most important signaling pathways found to induce melanogenesis is the cAMP/protein kinase A (PKA) pathway. Cyclic AMP, through the activation of PKA and cAMP-responsive element binding protein 1 transcription factors, up-regulates the expression of microphthalmia-associated transcription factor (Mitf), the master regulator of melanogenesis that controls the production of the melanogenic enzymes (tyrosinase, TRP-1, and DCT) (20–23) at the mRNA level. In addition, cAMP activates the ERK pathway, which also plays a key role in melanin synthesis (24), at least in part through the regulation of Mitf activation and stability (25, 26). The role of Mitf in determining the extent of melanocyte pigmentation has also been shown by studies utilizing a variety of depigmenting agents that have demonstrated preceding reductions in Mitf expression (18, 27, 28). Although Mitf-mediated transcriptional activation of pigmentation genes is essential for the control of melanocyte cellular differentiation (24), the up-regulation of tyrosinase, TRP-1, and DCT gene transcription in mature melanocytes does not completely explain melanogenesis stimulation by cAMP. For example, cAMP signaling can increase the stability of tyrosinase mRNA and also the enzyme activity of preexisting tyrosinase protein, suggesting regulation via post-transcriptional events (27).
The cGMP pathway can also increase melanin production. This pathway is activated by NO, which is released by keratinocytes irradiated by UVB (29–31). In human melanocytes, the protein kinase C-dependent pathway has emerged as an intracellular signaling pathway regulating melanogenesis (32, 33). Park et al. (34) reported that protein kinase C can phosphorylate tyrosinase at two serine residues in the cytoplasmic domain and that this phosphorylation can enhance tyrosinase activity. Among the pathways involved in the synthesis of melanin pigments, p38 MAP kinase signaling was recently found to be involved in stress-induced melanogenesis (35). A series of melanogenic stimuli such as α-MSH, UV irradiation, lipopolysaccharide, and placental total lipid fraction promotes a sustained increase of phospho-p38 MAPK active form (13, 15, 35, 36). However, the effective contribution of p38 MAPK in melanogenesis is not completely understood. Corre et al. (35) demonstrated that the p38-activated USF-1 transcription factor is responsible for UV-induced expression of two genes upstream in the pigmentation cascade: pro-opionmelanocortin and melanocortin 1 receptor (MC1R), which encode a precursor of α-MSH and its receptor, respectively. Because it has been reported that α-MSH regulates MC1R expression and function in mouse melanoma cells (37), it is possible that the activation of p38 may also contribute to the melanogenic rate under conditions of hormonal stimulation by increasing receptor density after its activation and internalization. Even if these data strengthen the idea that p38 MAPK is implicated in the regulation of important steps in melanocyte differentiation, the involvement of p38 mitogen-activated protein kinase in α-MSH-induced melanogenesis in B16 melanoma cells is, at the moment, exclusively demonstrated using a small cell-permeant inhibitor of p38 MAP kinase (SB203580) (38, 39), which is a member of the pyridinyl imidazole class of compounds. On the other hand, several studies have demonstrated that inhibition of the MAP kinase pathway leads to stimulation of melanogenesis. Treatment of B16 melanoma cells with MEK inhibitors blocks ERK activation and increases steady-state mRNA levels of mRNAs and the corresponding protein expression levels of the melanogenic antigens melan-A/Mart-1, gp100, and tyrosinase (40). In addition, the specific MEK inhibitor PD98059 stimulates tyrosinase activity and potentiates the effect of cAMP on melanogenesis (2). Moreover, ERK activation by sphingosine-1-phosphate, ceramide, or sphingosylphorylcholine is responsible for reducing melanogenesis by down-regulating Mitf (39–42), indicating that activation of the ERK pathway can lead to attenuation of melanogenesis. Altogether, these studies suggest that there are not conclusive data regarding the MAPK contribution to melanogenesis.
In this study, we present evidence that down-regulation of p38 expression leads to an increase in the levels of differentiation-associated markers such as melanin synthesis and the expression of tyrosinase and related proteins. Our experiments demonstrate that the mechanism involved in p38-mediated regulation of melanogenesis is the ubiquitin-proteasome pathway, where melanogenic enzymes are degraded.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
B16-F0 murine melanoma cells were purchased from the American Type Culture Collection and were maintained in Dulbecco's modified Eagle's medium with 4 mm l-glutamine + 7% heat-inactivated fetal bovine serum and antibiotics (Invitrogen) at 37 °C in a 5% CO2 atmosphere. For the induction studies, the cells were plated, and 24 h later, the medium was removed, and the cells were then cultured in Dulbecco's modified Eagle's medium with 2% heat-inactivated fetal bovine serum and antibiotics with or without pharmacological treatment in the absence of phenol red. Normal human melanocytes (NHM) isolated from several independent Caucasians foreskins were maintained in M-254 medium with human melanocyte growth supplements (Cascade Biologics, Inc.) and used for passages 2–6. NHM were treated with compounds for 5 days before analysis because melanogenesis is a much longer process in these cells than in B16 melanoma cells. SB202190 (4-(4-fluorophenyl)-2-(4-hydroxyphenyl)-5-(4-pyridyl)-1H-imidazole), SB203580 (4-(4-fluorophenyl)-2-(4-methylsulfinylphenyl)-5-(4-pyridyl)-1H-imidazole), actinomycin D, cycloheximide, α-MSH, dimethyl sulfoxide, l-3,4-dihydroxyphenylalanine (l-DOPA), and MG132 (benzyloxycarbonyl-Leu-Leu-leucinal) were purchased from Sigma-Aldrich. SB202474 (4-ethyl-2-(p-methoxyphenyl)-5-(4′-pyridyl)-1H-imidazole) was from Calbiochem.
Melanin Content Determination
Extracellular melanin release was measured as previously described (16). Briefly, B16-F0 cells were seeded at a density of 1 × 104/cm2 and incubated overnight, prior to α-MSH and pharmacological administration. The plates were then incubated for additional 72 h, after which 200 μl of the medium were removed, and the absorbance was measured spectrophotometrically at 405 nm using a plate reader. The cells were then washed twice with ice-cold phosphate-buffered saline, lysed with radioimmune precipitation assay buffer or with phosphate-buffered saline containing 1% Triton X-100, and centrifuged at 10,000 × g for 10 min. The supernatants were analyzed for protein concentration, and the pellets were solubilized in 200 μl of 1 m NaOH. Following an incubation period of 2 h at 60 °C, the absorbance was measured spectrophotometrically at 405 nm using a plate reader. Standard curves using synthetic melanin (0–250 μg/ml) were prepared in duplicate for each experiment. Melanin production was calculated by normalizing the total melanin values with protein content (μg of melanin/mg of protein) and reported as a percentage of control. For this purpose protein content was determined using Bradford dye reagent (Bio-Rad) that has been demonstrated as the best procedure in the presence of synthetic and natural melanin (45).
siRNA Transfection
p38 siRNA (m2) duplex (Santa Cruz Biotechnology Inc., Santa Cruz, CA), SignalSilencing® pool p38 MAP kinase siRNA (Cell Signaling) and siRNA-Mitf SiGENOME SMARTpool (Dhamacon) were used to interfere with p38 and Mitf expression, respectively. For dose-dependent experiments, the siRNA transfection protocol suggested by the manufacturer was optimized as follows: 5.0 × 104 cells were plated on 12-well dishes and left to grow overnight. The following day, the cells were transfected with 10, 30, or 60 pmol of siRNA dimers (p38) and 1, 3, or 6 μl of transfection reagent (Santa Cruz Biotechnology Inc.) mixed in Opti-MEM (Invitrogen). An equivalent amount of nonspecific siRNA was used as a negative control. Twenty-four hours following transfection, α-MSH was added to some samples in agreement with the experimental design. For all of the following experiments in which the B16 cells were incubated with multiple treatments after transfection, the cells were transfected with Amaxa Nucleofector System to ensure identical siRNA efficiency among the plates, because in this case the cells were transfected altogether in a single cuvette and plated immediately after nucleofection. For this purpose, we transferred 200 pmol of p38-specific siRNA into 1.6 × 106 cells using the Amaxa nucleofector cell line kit R (Program P-031). Preliminary experiments demonstrated that this optimized protocol produces transfection efficiency similar to the maximum obtained by lipofection (60 pmol/5.0 × 105). For double interfering experiments, 200 pmol of p38 siRNA dimers were mixed with a dose of 200 pmol of Mitf siRNA to transfect 1.6 × 106 cells. To obtain the same interfering efficiency in all samples, 200 pmol of nonspecific duplex were added in single transfected cells. The interference efficiencies were evaluated by Western blot and/or quantitative PCR in all experiments.
Tyrosinase Assay
Tyrosinase enzyme activity was estimated by measuring the rate of l-DOPA oxidation as previously described (16) with slight modifications. Briefly, the cells were treated with p38-specific siRNA or control siRNA for 72 h in Dulbecco's modified Eagle's medium containing 2% (v/v) fetal bovine serum ± α-MSH. At the end point, the cells were solubilized with phosphate buffer (pH 6.8) containing 1% Triton X-100. The cells were then disrupted by freezing and thawing, and the lysates were clarified by centrifugation at 10,000 × g for 10 min. After protein quantification and adjustment of protein concentrations with lysis buffer, 80 μl of each lysate (each containing the same amount of protein) were aliquoted into the wells of a 96-well plate, and 20 μl of 5 mm l-DOPA were then added to each well. The absorbance was measured spectrophotometrically at 475 nm following a 20-min incubation period at 37 °C. The measurement was repeated five times.
Western Blot Analysis
Whole cell extracts were prepared with radioimmune precipitation assay buffer (Tris-buffered saline, 0.5% deoxycholate, 0.1% SDS, 1% Triton X-100) containing Complete Mini protease inhibitor mixture (Roche Applied Science). The protein concentration was determined using the Bradford dye reagent (Bio-Rad) that has been demonstrated as the best procedure in pigment cells. Aliquots of cell lysates were separated by electrophoresis on SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and then treated with the appropriate antibodies. Anti-Mitf, anti-p38, anti-Ub(P4D1) (Santa Cruz Biotechnology Inc.), and anti-P-p38 (Cell Signaling) were used at 1:500; anti-tyrosinase, anti-TRP-1, and anti-TRP-2/DCT antibodies were used at 1:3,000 (Santa Cruz Biotechnology Inc.); and anti-β-tubulin (Sigma-Aldrich) was used at 1:15,000. Horseradish peroxide-conjugated goat anti-mouse, goat anti-rabbit, and bovine anti-goat immunoglobulin (Santa Cruz Biotechnology Inc.) were used at 1:5,000, 1:10,000, and 1:3,000, respectively. Antibody complexes were detected by chemiluminescence (ECL; Amersham Biosciences). Western blot assays were representative of at least three experiments. The protein levels were quantified by measuring the optical densities of specific bands using a GS-800 calibrated image densitometer (Bio-Rad).
Immunoprecipitation
Cell extracts (400 μg of total protein in 500 μl of lysis buffer) were incubated with 40 μl of protein A/G-agarose (Santa Cruz Biotechnology Inc.) under continuous mixing for 2 h at 4 °C. After centrifugation (2,000 × g for 3 min at 4 °C), the supernatants were used as precleared cell extracts. The precleared cell extracts were incubated with 1.6 μg of the anti-tyrosinase antibody. After overnight continuous mixing at 4 °C, 40 μl of protein A/G-agarose were added for 2 h at 4 °C. The immunocomplexes were precipitated by brief centrifugation, and the pellets were washed three times with 500 μl of lysis buffer. Finally, the absorbed proteins were eluted with 30 μl of Tris-glycine SDS sample buffer containing 2-mercaptoethanol at 95 °C for 5 min. Each supernatant was separated on 10% Tris-glycine SDS gels and then transferred to nitrocellulose membranes. Ub(P4D1) was used to detect ubiquitinated tyrosinase precipitated with anti-tyrosinase antibody. The level of ubiquitinated tyrosinase was quantified by measuring the optical densities of specific bands as reported in the protocol for Western blot analysis.
Semi-quantitative Reverse Transcription-PCR
Total RNA was extracted using an RNeasy mini kit (Qiagen). cDNA was synthesized from 1 μg of total RNA in the presence of 0.5 μg/reaction of oligo(dT)15 primer using the Improm IITM reverse transcription system (Promega). First strand cDNA (1 μl) was amplified using a iQ5 light cycler (Bio-Rad). Aliquots (1 μl) of the reverse transcription products were amplified in a reaction mixture (15 μl) containing Maxima SYBR Green qPCR Master mix and 25 pmol of forward and reverse primers. Bio-Rad optical system software was used to determine the mRNA levels. Data analysis was done by normalization to β-actin mRNA levels. PCRs were done in triplicate. Forward and reverse primers were as follows: p38, AATGGAGTCCTGAGCACCTG-3′ and 5′-GACAGACGAACAGACAGACAC-3′; Mitf, 5′-GTATGAACACGCACTCTCGAG-3′ and 5′-CAGGAGTTGCTGATGGTAAG-3′; tyrosinase, 5′-GGCCAGCTTTCAGGCAGAGGT-3′ and 5′-TGGTGCTTCATGGGCAAAATC-3′; TRP-1, 5′-AAGCAGACATCCAACAACACTAG-3′ and 5′-GCAAGAGTTCAGAACACAGGTC-3′; DCT, 5′-GCAAGAGATACACGGAGGAAG-3′ and 5′-CTAAGGCATCATCATCATCACTAC-3′; and β-actin, 5′-GACAGGATGCAGAAGGAGATTACT-3′ and 5′-TGATCCACATCTGCTGGAAGGT-3′.
Statistical Analysis
Statistical significance was determined by Student's t tests. The minimal level of significance was p ≤ 0.05 (*) or p ≤ 0.01 (**).
RESULTS
Effect of MAPK Inhibitors on α-MSH-stimulated Melanogenesis
In in vitro culture, melanin synthesized intracellularly is secreted into the extracellular culture fluid and represents at the specific times of incubation used in this study a considerable amount of total melanin produced (13, 16, 46, 47). For this reason, the melanin released into the medium of each well was determined spectrophotometrically in addition to intracellular melanin quantification (see “Experimental Procedures” for details). Previous studies have shown that α-MSH-stimulated melanogenesis can be inhibited by blocking p38 MAPK activity with SB203580, a pyrimidinyl imidazole compound (13, 15, 23, 39, 48). In this study, stimulation of B16 cells with α-MSH (1 μm) caused a 5.6-fold increase in total melanin production (Fig. 1A). In accordance with the literature, co-treatment with SB203580 (1–20 μm) caused a significant reduction in α-MSH-induced melanogenesis (p < 0.05). Similarly, the pyrimidinyl imidazole SB202190 decreased melanogenesis at the same concentrations. However, co-treatment with the structural analog SB202474, which has no effect on p38 MAPK activity and is commonly included as a negative control when SB203580 or its analog SB202190 are used to investigate the involvement of p38 MAPK (49), caused a similar inhibition of α-MSH-stimulated pigmentation (p < 0.05) (Fig. 1A). Similar results were also obtained using the cAMP elevating agent forskolin (supplemental Fig. S1). In addition, the effect of each compound on the basal level of melanogenesis in B16 cells was tested (Fig. 1A). In this case the cells were treated with compounds for 96 h before analysis because spontaneous melanogenesis is a much longer process. The results demonstrated that all pyridinyl imidazole compounds caused a dose-dependent reduction of basal melanogenesis also in unstimulated B16 cells. Accordingly, with the concept that pyridinylimidazoles may block the biological activity of p38 kinase by binding to the ATP pockets of active and inactive forms of the enzyme with equal affinity without affecting its rate of activation (50), we did not observe alteration of the level of p38 expression or activation (Fig. 1B). In unstimulated and α-MSH-stimulated cells, the presence of compounds reduced tyrosinase enzymatic activity as measured by l-DOPA oxidation (Fig. 1C). Interestingly, the strong reduction of melanin produced in the presence of compounds corresponds to a moderate decrease of tyrosinase activity. We then verified the results obtained in B16 melanoma cells in NHM. The results obtained confirmed that p38 inhibitors and their negative control SB202474 significantly repress melanin synthesis even in normal human melanocytes (supplemental Fig. S2).
FIGURE 1.
Effect of pyridinyl imidazoles compounds on melanin synthesis in B16 melanoma cells. A, following incubation with increasing concentrations (1, 2.5, 5, 10, and 20 μm) of pyridinyl imidazoles for 72 h, the extracellular and intracellular levels of melanin were determined separately by measuring the absorbance at 405 nm. Standard curves of synthetic melanin were used to extrapolate the absolute values of melanin content. The total amount of melanin was calculated for each experimental point by adding the extracellular and intracellular melanin values after normalization for protein content. B16 cells were also treated with pyridinyl imidazoles compounds in absence of α-MSH. B, the effect of compounds on p38 activation and expression was estimated performing Western blot analysis of controls and cells treated with pyridinyl imidazoles (20 μm) for 24 h. As a positive control of immunoblot, B16 cells were treated with anisomycin, a specific activator of p38. C, samples treated as in B were also evaluated for tyrosinase activity by measuring l-DOPA oxidation at several different time points. In addition, the end point (80 min) absorbance measured spectrophotometrically at 475 nm is reported to compare control and hormone-stimulated cells. The data show the means ± S.D. of three experiments performed in duplicate. *, p ≤ 0.05; **, p ≤ 0.01 versus control.
Effects of p38 Silencing on Melanogenesis
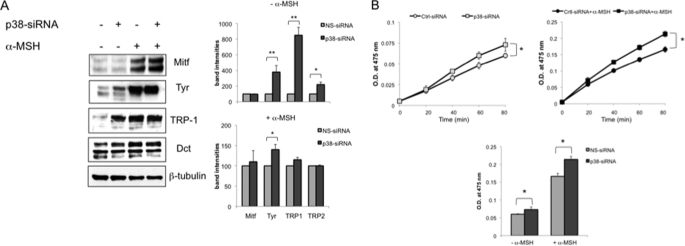
Treatment with p38 inhibitors SB202190 and SB203580, as well as with SB202474, the inactive compound in the presence or absence of α-MSH, was effective in reduce melanogenesis, suggesting that this effect is due to an activity of these molecules that is unrelated to their inhibition of p38 MAPK activity. To confirm that the p38 MAPK pathway is not involved in imidazole compound-induced reduction of melanogenesis, we used siRNA instead of drugs to inhibit p38 MAPK activity. B16 cells were transiently transfected with siRNA for p38 or control nonspecific siRNA (NS-siRNA) at three different doses of siRNA duplex (10, 30, or 60 pmol). The experiments were performed using two different p38-specific duplexes obtaining similar results (see “Experimental Procedures”). Twenty-four hours after transfection, the cells were treated with α-MSH or not. As a consequence of reduced activity or expression of p38 MAPK, we observed a slight but significant (p < 0.05) stimulation of melanin synthesis (Fig. 2A). Western blot analysis confirmed that p38 protein levels were significantly reduced in cells transfected with p38 siRNA (Fig. 2B). Interestingly, α-MSH stimulation seems to improve p38 siRNA efficiency, suggesting that α-MSH could also participate in the regulation of p38 protein expression.
FIGURE 2.
The effect of siRNA on melanogenesis. A, B16 cells were transfected with 10, 30, or 60 pmol of p38 siRNA dimers. p38 siRNA and nonspecific control siRNA-transfected cells (NS-siRNA) were treated or not by adding α-MSH 1 × 10−7 24 h after transfection. The total amount of melanin was calculated after 72 h for each experimental point by adding the extracellular and intracellular melanin values after normalization for protein content. The data show the means ± S.D. of five independent experiments. B, immunoblots of anti-p38 of cells treated as in A were used to quantify RNA interference efficiency. Variation in protein loading was determined by blotting with anti-β-tubulin antibody.
Effect of p38 Silencing on Melanogenic Protein Expression
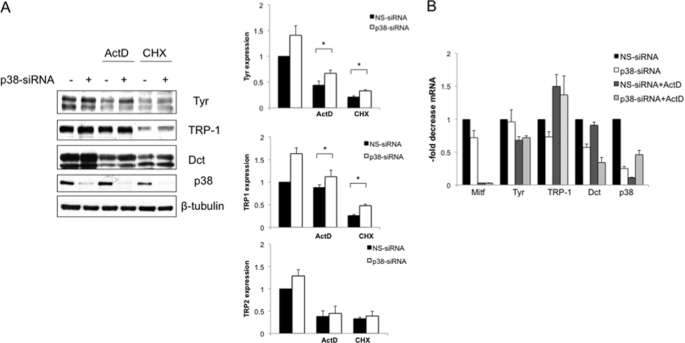
Next, we examined the effect of p38 silencing on the protein levels of Mitf and the three melanocyte-specific enzymes: tyrosinase, TRP-1, and DCT, after 96 h of siRNA. The observed increase in pigmentation was associated with a significant stimulation of tyrosinase expression (Fig. 3A). In addition, we also observed a strong induction of TRP-1 expression and a small increase of DCT protein level. However, we did not observe a significant change in Mitf protein expression. In accordance with melanin production, the effect of imidazole compounds on melanogenic enzymes expression was greater in unstimulated cells compared with α-MSH-treated cells. Western blotting revealed that tyrosinase appeared mostly as an ∼80-kDa species, the size of the mature, fully Golgi-processed form (51, 52). To confirm the functional activation of the rate-limiting enzyme in melanin synthesis, we assayed tyrosinase activity by quantifying l-DOPA oxidation. Consistent with the Western blot assay, the results obtained showed that in B16 cells, tyrosinase activity was significantly increased (p < 0.05) by p38 down-regulation (Fig. 3B). We next examined whether p38-mediated regulation of melanogenic proteins was due to accelerated transcription of their genes. First, we performed a time course experiment to establish the exact kinetics of Mitf and melanogenic enzyme up-regulation following α-MSH stimulation in B16 cells. As shown in Fig. 4A, Mitf mRNA increased in a time-dependent manner, reaching maximum induction (9.6-fold) after 6 h of treatment, followed by a gradual decline to the basal level by 24 h. The mRNA levels of Mitf downstream molecules tyrosinase and TRP-1 were found to be up-regulated during the entire time of observation, whereas DCT mRNA transcription was slightly modified by α-MSH. Next, B16 cells treated with control siRNA or with siRNA specific for p38 alone or in association with α-MSH for 6 or 24 h were lysed for quantitative PCR. As shown in Fig. 4B, analysis of mRNA extracted from cells treated with p38 siRNA showed no significant change of melanogenic gene mRNA compared with control or α-MSH-treated cells, suggesting that p38 acts at the protein level. In this case, the amount of p38 mRNA was used as a control for siRNA efficiency. To investigate whether the effects of p38 down-regulation on melanogenesis might be related to changes in the rate of degradation of melanogenic proteins, we exposed cells transfected with NS-siRNA or p38 siRNA to the transcriptional inhibitor actinomycin D (Act D) or to the translational inhibitor cycloheximide for 8 h, and the extracted proteins were analyzed by Western blotting. As expected, these antibiotics inhibited the de novo synthesis of tyrosinase, TRP-1, and DCT proteins, although it is important to note that TRP-1 expression was slightly modified by Act D treatment. Decelerated depression of melanogenic enzymes expression was observed in cells pretreated with p38 siRNA (Fig. 5A). In addition, we use cycloheximide to block protein synthesis and to demonstrate that at several different time points the level of residual tyrosinase expressed is higher in cells transfected with p38 siRNA (supplemental Fig. S3). Western blot analysis demonstrated that at all of the time points considered. Therefore, p38 repression induces melanogenesis in a manner that is independent of de novo mRNA and protein synthesis. By contrast, the levels of tyrosinase and related protein mRNAs in cells treated with Act D or not and transfected p38 siRNA were found to be similar to those of cells transfected with NS-siRNA (Fig. 5B). Interestingly, the dramatic decrease of Mitf mRNA observed after 8 h of transcriptional inhibition did not rapidly correspond to a reduction of the mRNA levels of tyrosinase and related proteins. Consistent with the Western blot results, quantitative analysis of mRNA demonstrated that, at the time point examined, TRP-1 gene expression was not affected by Act D treatment, suggesting that the corresponding mRNA is quite stable. These results are consistent with the result for Mitf transcription factor expression described in Fig. 3A and confirm that p38 regulates melanogenic enzymes at the protein level.
FIGURE 3.
The effect of p38 silencing on melanogenic protein expression and activity. A, expression of tyrosinase, TRP-1, and DCT proteins in B16 cells after 72 h of treatment with p38 siRNA (60 pmol). Total cellular proteins (20 μg/lane) were subject to 10% SDS-PAGE. Variation of loading was determined by blotting with anti-β-tubulin antibody. The Western blot assays are representative of at least three experiments. B, tyrosinase activity by l-DOPA oxidation was measured using cell lysates obtained from cells treated with 60 pmol of duplex as described in A. The values represent the means ± S.D. of three representative experiments performed in duplicate. *, p ≤ 0.05.
FIGURE 4.
The effect of p38 silencing on Mitf and melanogenic enzymes mRNA transcription. A, semi-quantitative real time PCR to measure the kinetics of Mitf, tyrosinase, TRP-1, and DCT mRNA increase following α-MSH treatment was performed by using the SyBr Green detection system. The graphs show fold differences in transcript abundance in α-MSH-stimulated cells compared with untreated cells. B, total RNA was purified from p38 siRNA and NS-siRNA-transfected cells after 6 and 24 h of α-MSH treatment (or not), and the levels of mRNA were analyzed by semi-quantitative real time PCR. The results shown in A and B were normalized by the β-actin mRNA levels. The values shown represent the means ± S.D. of three independent experiments.
FIGURE 5.
Increase of tyrosine family proteins is independent from de novo protein synthesis. A, increase of tyrosine family proteins is independent from de novo protein synthesis. B16 cells transfected with NS-siRNA or p38 siRNA were exposed to the Act D (1 μg/ml) to inhibit RNA synthesis or to cycloheximide (CHX, 1 μg/ml) to inhibit protein synthesis for 8 h. Total cellular proteins were extracted after 8 h. Western blotting analysis was performed using 20 μg of each sample. Densitometric scanning of band intensities obtained from three separate experiments were used to quantify change of proteins expression (control value taken as 1-fold in each case). Q p ≤ 0.05 versus control. B, total RNA extracted from samples treated as in A were analyzed for p38, Mitf, tyrosinase, TRP-1, and Dct mRNA expression. The results shown were normalized by the β-actin mRNA levels. The values shown represent the means ± S.D. of three independent experiments.
p38 Down-regulation-dependent Induction of Melanogenesis Is Mitf-independent
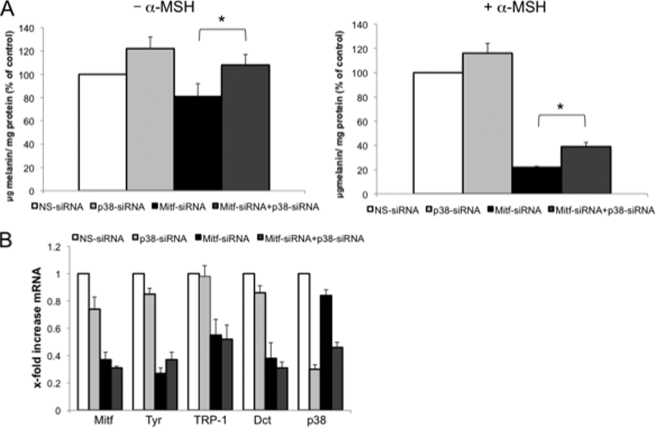
Considering that a crucial involvement of Mitf in the regulation of pigmentation without any accompanying change of mRNA levels of the melanogenic enzymes has been proposed (36–52), we investigated the possibility that Mitf, directly or indirectly, regulates the melanogenic enzyme expression observed with p38 siRNA at a step other than mRNA. To rule out any participation of Mitf in p38 down-regulation-induced melanogenesis, we double-transfected B16 cells with Mitf-siRNA and p38 siRNA in the presence or absence of α-MSH stimulation. In the absence of hormonal stimulation in B16 cells, Mitf siRNA only slightly affected melanogenesis (0.8-fold decrease) (Fig. 6A), probably because of the stability of melanogenic enzymes until the experimental end point (96 h). As expected, Mitf-specific siRNA strongly decreased melanin synthesis in α-MSH-stimulated cells (0.2-fold decrease). However, at the same time, melanogenesis inhibition observed in the absence of Mitf protein was partially but significantly rescued by p38 down-regulation in the basal (1.3-fold increase; Mitf-siRNA+p38 siRNA versus Mitf-siRNA) or hormone-stimulated condition (1.4-fold increase; Mitf-siRNA+p38 siRNA versus Mitf-siRNA) (Fig. 6A). Quantitative PCR analysis confirmed that the decrease in melanogenic enzyme mRNA occurring in the absence of Mitf expression was not rescued by p38 siRNA, excluding again the increased production and/or stabilization of mRNA (Fig. 6B). Analysis of mRNA was also used to verify siRNA efficiency and to exclude reduction of Mitf down-regulation in double-transfected cells. This experiment clearly demonstrates that Mitf does not directly influence stimulation of melanogenesis by p38 down-regulation, even though Mitf plays an important role in determining tyrosinase and trp mRNA and protein levels. In this sense, it is important to emphasize that Mitf exerts an indirect but fundamental influence on melanogenesis regulation even during p38 inhibition by determining the amount of mature tyrosinase protein that can be stabilized by p38 kinase activity inhibition.
FIGURE 6.
The melanogenic effect of p38 silencing is Mitf-independent. A, B16 cells were double-transfected with Mitf-siRNA and p38 siRNA or single transfected, and melanin content was measured as described above in cells were treated (or not) with α-MSH after 72 h of treatment. The values shown represent the means ± S.D. of three independent experiments. B, total RNA was extracted after 24 h of interfering to perform semi-quantitative real time PCR of tyrosinase, TRP-1, and DCT. Analysis of Mitf and p38 mRNA was included to ensure that double and single transfection produced the same level of interference. The values shown represent the means ± S.D. of three independent experiments.
Involvement of the Ubiquitin-Proteasome Pathway in the p38-induced Regulation of Tyrosinase Expression
Because the Western blot analysis of the relative abundance of melanogenic enzymes indicated potential effects of p38 on protein turnover, we next examined whether the p38-dependent control of tyrosinase expression was due to regulation of its proteolytic degradation by the proteasome. We used anisomycin, a well characterized and relatively specific activator of p38, alone or in association with the membrane-permeable proteasome inhibitor MG132, to demonstrate that activation of p38 induces melanogenic enzyme degradation. To confirm that anisomycin activates p38, the phosphorylation of p38 on Thr180/Tyr182 was measured. As shown in Fig. 7A, exposure of B16 cells to anisomycin caused tyrosinase degradation. In addition, that degradation was inhibited when cells were pretreated with 150 nm of MG132, confirming proteasome involvement. B16 cells were also preincubated with the p38 MAPK-specific inhibitors SB202190 and SB203580 or the inactive analog SB202474 10 min before stimulation with anisomycin to verify the role of p38. Preincubation with SB202190 and SB203580 strongly inhibited anisomycin-induced tyrosinase degradation. In contrast, SB202474, which has no ability to inhibit p38 MAPK activity, did not influence melanogenic protein stability. Similar analyses applied to tyrosinase-related proteins confirmed that p38 regulates TRP-1 stability (Fig. 7A) and in some extends DCT degradation (data not shown). Another major effect of anisomycin is that it is a translational inhibitor of protein synthesis; however, this effect does not appear to be a problem in the present context, because of the slight tyrosinase protein decrease observed after the same time of treatment (3 h) with the translational inhibitor cycloheximide. (data not shown). Consistent with Western blot analysis, the decrease of tyrosinase activity induced by anisomycin was prevented by MG132 or SB202190 and SB203580 pretreatment (Fig. 7B). We then verified the results obtained in B16 melanoma cells in normal human melanocytes. The results shown in Fig. 7 (C and D) demonstrated that also in NHM stimulation of p38 increased proteasome-mediated tyrosinase and TRP-1 degradation and that inhibition of p38 restored tyrosinase stability. Interestingly, for unknown reasons, the same experiment did not show a similar reproducible regulation of DCT (data not shown). To corroborate that p38 is required for anisomycin-induced melanogenic enzyme degradation, we depleted p38 with duplex siRNA before anisomycin treatment. As shown in Fig. 7E, abrogation of expression of p38 reduced the efficiency of proteasome degradation observed after incubation with anisomycin. Activating phosphorylation occurred in the residual p38 protein in samples treated with p38 siRNA, explaining the incomplete rescue of anisomycin-dependent degradation observed in those samples. Altogether, these results indicate that p38 might control tyrosinase turnover by regulating its proteasome-dependent degradation.
FIGURE 7.
Involvement of proteasomes in the p38 MAPK regulation of tyrosinase stability. A, Western blot analysis of cell lysates (20 μg of protein/lane) of cells treated with anisomycin (5 μm) to induce p38 MAPK activity alone or in association with the membrane-permeable proteasome inhibitor MG132 (150 nm) for 3 h. The cells were also pretreated with SB202190, with SB203580 (20 μm) to inhibit p38 kinase activity, or with SB202474 (20 μm) as a negative control. The data are expressed as percentages of control and are the mean values ±S.D. of triplicate determinations. B, tyrosinase activity by l-DOPA oxidation was measured using cell lysates obtained from cells treated as described in A. The end point (80 min) absorbance was measured spectrophotometrically at 475 nm. C and D, results described in A and B were confirmed with NHM. E, Western blot analysis of cell lysates (20 μg of protein/lane) of NS-siRNA and p38 siRNA-transfected cells treated or not with anisomycin in association with proteasome inhibitor MG132. Densitometric analysis was performed as described above.
Regulatory Effect of p38 on Ubiquitinated Tyrosinase
Because the selectivity of protein degradation is determined mainly at the stage of ubiquitin ligation, we evaluated the effect of p38 silencing on tyrosinase ubiquitination. To do that, we first stabilized ubiquitinated proteins, which would normally be immediately degraded by proteasome, and we used an incubation time of 3 h with MG132 at 150 nm to accomplish this. First, we compared the relative amounts of on cellular ubiquitination in general following MG132 treatment. As shown in Fig. 8A, no significant differences were observed in p38 siRNA-transfected cells compared with mock transfected cells. We then performed immunoprecipitation analysis to clarify whether p38 MAPK regulates the ubiquitination of tyrosinase. Cell extracts were immunoprecipitated with the tyrosinase antibody, and Western blotting of ubiquitin probed proteins was performed with Ub(P4D1). Quantification of bands corresponding to tyrosinase revealed that down-regulation of p38 decreased the level of ubiquitinated tyrosinase to 0.7-fold compared with the control (Fig. 8B). Then the membrane was stripped to quantify the amount of tyrosinase immunoprecipitated in each sample. Because the degradation of tyrosinase was prevented by MG132, similar band intensities of tyrosinase were obtained (Fig. 8C). These results demonstrate that p38 MAPK activity directly or indirectly regulates tyrosinase protein levels by enhancing its ubiquitination.
FIGURE 8.
Regulatory effects of p38 on the ubiquitination of tyrosinase. A, whole cell lysates were loaded with 10 μg/lane of total cell extracts to evaluate the effect of p38 silencing on cellular general ubiquitination. The cells were transfected with NS-siRNA or p38 siRNA and then treated with MG132 (150 nm) for 3 h. B, cell extracts were immunoprecipitated (IP) with anti-tyrosinase antibody (IP), and Western blotting (WB) of ubiquitinated probed Ub(P4D1) was performed. Molecular masses, indicated on the left of the Western blots, are expressed in kilodaltons. B and C, after stripping the same membrane was reprobed with anti-tyrosine antibody. The experiment was performed two times with similar results.
DISCUSSION
MAPKs are evolutionarily conserved enzymes connecting cell surface receptors to critical regulatory targets within cells. p38 MAPK is a stress-regulated protein kinase belonging to the MAPK superfamily in addition to c-Jun N-terminal kinase and extracellular signal-regulated kinases. p38 MAPK has been shown to play a key role in the regulation of cellular responses to external stress signals. It is activated by diverse stimuli that include ultraviolet light, irradiation, heat shock, osmotic stress, and proinflammatory cytokines. Pharmacological inhibitors have accelerated studies concerning the role of p38 MAPK in many different cellular processes, including proliferation, differentiation, apoptosis, cellular senescence, transcriptional regulation, and cytoskeletal reorganization (53–55). The two pyridinyl imidazole compounds, SB202190 and SB203580, have been shown to inhibit p38 MAPK, but they have no effect on related kinases such as extracellular signal-regulated protein kinase end c-Jun N-terminal kinase (51). In contrast, SB202474, which has no effect on p38, is commonly included as a negative control when SB203580 and SB202190 are used to study the involvement of p38 MAPK. From these studies emerged some sporadic evidence that pyridynil imidazole compounds have unexpected targets in addition to p38, and consequently they were found to be too nonspecific to assess the physiological roles of p38 MAPK (56, 57).
A number of studies have reported the involvement of p38 MAPK in melanogenic differentiation (13, 15, 24, 38, 39, 48). However, the role of p38 MAPK in controlling melanogenesis is not fully understood. We observed here that pyridinyl imidazoles inhibited both basal and α-MSH-induced melanogenesis. At first, we suspected that the p38 MAPK pathway might be involved in melanogenesis suppression. However, SB202474, which has no ability to inhibit p38 MAPK activity and is widely used as a negative control compound in p38 MAPK studies, also suppressed melanin synthesis induction. Furthermore, the independence of the p38 kinase pathway from melanogenesis repression by pyridinyl imidazole compounds was also confirmed by siRNA experiments. In fact, p38 MAPK siRNA surprisingly stimulated melanogenesis. From these results, we conclude that the p38 MAPK pathway is not involved in inhibition by SB202190 and SB203580 and that pyridinyl imidazole compounds have a target molecule(s) besides p38 MAPK that might be important for pigmentation. Previous studies described repression of melanogenesis by p38 activity inhibition using the p38 inhibitor SB203580 (38, 39, 48). It is our concern that no one study regarding melanogenesis included SB202474 as a negative control or siRNA as a positive control.
While investigating the involvement of p38 MAPK in the regulation of melanogenesis, we observed a very interesting function of p38 MAPK in the regulation of melanogenic protein expression. The levels of intracellular proteins are regulated by a balance between their synthesis and degradation, and this is also true for tyrosinase family proteins. Tyrosinase and TRP-1 are degraded endogenously at least in part by proteasomes (52, 58–60), which are multicatalytic protease complexes that selectively degrade intracellular proteins. Previously, Ando et al. (58) reported that fatty acids, the major components of cell membranes, are able to regulate the proteasomal degradation of tyrosinase by relative increases or decreases in ubiquitinated tyrosinase (59, 60). Consistently, we demonstrated that, in B16 cells and in NHM, p38 modulates melanogenic enzyme abundance, reinforcing the concept that post-transcriptional regulation might play a crucial role in the melanogenic pathway. In fact, stimulation of p38 MAPK activity by anisomycin and p38 siRNA affects the steady-state level of tyrosinase and trp proteins oppositely. Although the precise mechanism(s) by which p38 promotes melanogenic enzyme degradation still remain to be determined, the involvement of the ubiquitin-proteasome pathway is supported by co-treatment with the proteasome-specific inhibitor MG132. Recent reports indicate that p38 activation targets D-type cyclins for ubiquitin-proteasome degradation (61, 62). Interestingly, the magnitude of p38-dependent regulation of melanogenic enzyme expression is variable among these proteins. Altogether, our experiments indicate that TRP-1 is more sensitive to p38-dependent control. Dct protein stability is modestly influenced by p38 MAPK activity in B16 melanoma cells, whereas this regulation does not seem to have an important role in homologous human protein TRP-2 expression. In agreement with previous reports, our experiments indicate that tyrosinase family proteins may be regulated independently of each other as well as independently of Mitf (23, 63, 64). Furthermore, another recent study revealed that, in human neuroblastoma cells, prolonged induction of tyrosinase led to the activation of c-Jun N-terminal kinase, p38 mitogen-activated protein kinase, and mitogen-activated protein kinase kinase, and that the ubiquitin ligase Parkin could protect cells from the neurotoxicity of reactive oxygen species overproduction caused by tyrosinase activity (65). Similarly, it is tempting to speculate that α-MSH-induced stimulation of p38 activation might be a feedback system that controls tyrosinase family protein turnover, thereby balancing the strong induction via the cAMP/PKA pathway. Consequently, melanogenic proteins ubiquitination triggered by α-MSH stimulation could reset the pathway to prestimulatory status and modulate the strength and duration of α-MSH signaling. Our study demonstrated that in addition to extensively studied post-translational modification mechanisms such as glycosylation, melanogenic enzyme ubiquitination plays a central role in melanogenesis regulation. In agreement with the data presented in this study, Englaro et al. (2) demonstrated that in B16 cells, MAPK pathway activation is not required for cAMP-induced melanogenesis and that inhibition of the mitogen-activated protein ERK1/2 pathway increases tyrosinase activity. Moreover, cAMP-elevating agents stimulate the activation of the mitogen-activated proteins ERK1/2 (2, 25), which in turn promote phosphorylation of Mitf on serine 73 and serine 409 (27). Dually phosphorylated Mitf displays increased transcriptional activity and enhanced proteasome-dependent degradation at the same time (24). Also in this case, elevation of intracellular cAMP primes a retrocontrol mechanism that prevents excessive melanin synthesis. Hence, cAMP-elevating agents seem to trigger at least two antagonistic molecular events, one ending in the stimulation of melanin synthesis and a second one decreasing melanogenesis through the activation of the MAP kinase pathway. Pretreatment of cells with inhibitors of MEK1/2 that regulate extracellular signal-regulated kinases was found to potentiate the α-MSH-induced stimulation of p38 MAP kinase, suggesting that there may be some feedback between ERK1/2 MAP kinase inhibition and p38 MAP kinase activation (66). A seesaw-like balance between ERK and p38 phosphorylation is also demonstrated by the administration of SB203580, which increases phosphorylation of ERK (66–68). We also observed in this study a significant and persistent activation of ERK1/2 MAP kinase when B16 cells were treated with high doses of pyridinil imidazole compounds (data not shown). In conclusion, the present study has firmly demonstrated both the involvement of p38 in proteasome-dependent tyrosinase, TRP-1, and DCT degradation (Fig. 9) and p38-independent inhibition of melanogenesis by pyridinyl imidazole compounds. Consequently, a prerequisite for unambiguous interpretation of melanogenesis experiments involving these inhibitors is the knowledge of possible additional effects on the melanogenic pathway.
FIGURE 9.
Model of contribution of p38 MAPK signal transduction in melanogenesis. The activation of p38 by UV irradiation could directly stimulate melanogenesis increasing Usf-1-dependent transcription of tyrosinase gene (11). Elevation of cAMP content leads to PKA activation and stimulation of Mitf transcription resulting in stimulation of tyrosinase, TRP-1, and DCT gene expression. Stimulation of MC1R activates ERK1/2 cascade promoting Mitf transcriptional activity and its degradation at the same time. Stimulation of MC1R by α-MSH also phosphorylates p38 MAPK. We propose that similar to ERK1/2, p38 activation could repress melanin synthesis, preventing excessive production of melanin synthesis through the accelerate degradation of melanogenic enzymes (dotted lines/arrows). p38, as well as ERK1/2, might represent a feedback system that controls tyrosinase family protein turnover, balancing the strong induction via the cAMP/PKA pathway.
Supplementary Material
This work was supported in part by Grant onc-ord/32/07 from the Ministero della Salute (Italy).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- TRP
- tyrosinase-related protein
- α-MSH
- α-melanocyte-stimulating hormone
- siRNA
- small interfering RNA
- DCT
- dopachrome tautomerase
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- MEK
- MAPK/ERK kinase
- PKA
- protein kinase A
- Mitf
- microphthalmia-associated transcription factor
- MC1R
- melanocortin 1 receptor
- NHM
- normal human melanocyte(s)
- l-DOPA
- l-3,4-dihydroxyphenylalanine
- NS-siRNA
- nonspecific siRNA
- Act D
- actinomycin D.
REFERENCES
- 1.Benedito E., Jiménez-Cervantes C., Perez D., Cubilana J. D., Solano F., Jiménez-Cervantes J., Meyer zum Gottesberge A. M., Lonzano J. A., Garcia Borron J. C. (1997) Biochim. Biophys. Acta 1336, 59–72 [DOI] [PubMed] [Google Scholar]
- 2.Englaro W., Bertolotto C., Buscà R., Brunet A., Pagès G., Ortonne J. P., Ballotti R. (1998) J. Biol. Chem. 273, 9966–9970 [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi T., Urabe K., Winder A., Jiménez-Cervantes C., Imokawa G., Brewington T., Solano F., García-Borrón J. C., Hearing V. J. (1994) EMBO J. 13, 5818–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winder A., Kobayashi T., Tsukamoto K., Urabe K., Aroca P., Kameyama K., Hearing V. J. (1994) Cell Mol. Biol. Res. 40, 613–626 [PubMed] [Google Scholar]
- 5.Yokoyama K., Suzuki H., Yasumoto K., Tomita Y., Shibahara S. (1994) Biochim. Biophys. Acta 1217, 317–321 [DOI] [PubMed] [Google Scholar]
- 6.Abdel-Malek Z., Swope V. B., Suzuki I., Akcali C., Harriger M. D., Boyce S. T., Urabe K., Hearing V. J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1789–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuller B. B., Lunsford J. B., Iman D. S. (1987) J. Biol. Chem. 262, 4024–4033 [PubMed] [Google Scholar]
- 8.Halaban R., Lerner A. B. (1977) Exp. Cell Res. 108, 111–117 [DOI] [PubMed] [Google Scholar]
- 9.Hornyak T. J., Hayes D. J., Ziff E. B. (2000) J. Invest. Dermatol. 115, 106–112 [DOI] [PubMed] [Google Scholar]
- 10.Abdel-Malek Z., Swope V., Smalara D., Babcock G., Dawes S., Nordlund J. (1994) Pigment Cell Res. 7, 326–332 [DOI] [PubMed] [Google Scholar]
- 11.Galibert M. D., Carreira S., Goding C. R. (2001) EMBO J. 20, 5022–5031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Im S., Moro O., Peng F., Medrano E. E., Cornelius J., Babcock G., Nordlund J. J., Abdel-Malek Z. A. (1998) Cancer Res. 58, 47–54 [PubMed] [Google Scholar]
- 13.Smalley K, Eisen T. (2000) FEBS Lett. 476, 198–202 [DOI] [PubMed] [Google Scholar]
- 14.Hata K., Hori K., Takahashi S. (2003) J. Biochem. 134, 441–445 [DOI] [PubMed] [Google Scholar]
- 15.Ahn J. H., Jin S. H., Kang H. Y. (2008) Arch. Dermatol. Res. 300, 325–329 [DOI] [PubMed] [Google Scholar]
- 16.Bellei B., Flori E., Izzo E., Maresca V., Picardo M. (2008) Cell Signal. 20, 1750–1761 [DOI] [PubMed] [Google Scholar]
- 17.Lee J., Kim Y. S., Park D. (2007) Biochem. Pharmacol. 74, 960–968 [DOI] [PubMed] [Google Scholar]
- 18.Jiménez-Cervantes C., Martínez-Esparza M., Pérez C., Daum N, Solano F, García-Borrón J. C. (2001) J. Cell Sci. 114, 2335–2344 [DOI] [PubMed] [Google Scholar]
- 19.Khaled M., Larribere L., Bille K., Ortonne J. P., Ballotti R., Bertolotto C. (2003) J. Invest. Dermatol. 121, 831–836 [DOI] [PubMed] [Google Scholar]
- 20.Aksan I., Goding C. R. (1998) Mol. Cell Biol. 18, 6930–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertolotto C., Abbe P., Hemesath T. J., Bille K., Fisher D. E., Ortonne J. P., Ballotti R. (1998) J. Cell Biol. 142, 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertolotto C., Buscà R., Abbe P., Bille K., Aberdam E., Ortonne J. P., Ballotti R. (1998) Mol. Cell Biol. 18, 694–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tachibana M., Takeda K., Nobukuni Y., Urabe K., Long J. E., Meyers K. A., Aaronson S. A., Miki T. (1996) Nat. Genet. 14, 50–54 [DOI] [PubMed] [Google Scholar]
- 24.Buscà R., Abbe P., Mantoux F., Aberdam E., Peyssonnaux C., Eychène A., Ortonne J. P., Ballotti R. (2000) EMBO J. 19, 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong G., Pawelek J. (1975) Nature 255, 644–646 [DOI] [PubMed] [Google Scholar]
- 26.Wu M., Hemesath T. J., Takemoto C. M., Horstmann M. A., Wells A. G., Price E. R., Fisher D. Z., Fisher D. E. (2000) Genes Dev. 14, 301–312 [PMC free article] [PubMed] [Google Scholar]
- 27.Bertolotto C., Bille K., Ortonne J. P., Ballotti R. (1998) Oncogene 16, 1665–1670 [DOI] [PubMed] [Google Scholar]
- 28.Chung S. Y., Seo Y. K., Park J. M., Seo M. J., Park J. K., Kim J. W., Park C. S. (2009) Biosci. Biotechnol. Biochem. 73, 1704–11710 [DOI] [PubMed] [Google Scholar]
- 29.Roméro-Graillet C., Aberdam E., Biagoli N., Massabni W., Ortonne J. P., Ballotti R. (1996) J. Biol. Chem. 271, 28052–28056 [DOI] [PubMed] [Google Scholar]
- 30.Roméro-Graillet C., Aberdam E., Clément M., Ortonne J. P., Ballotti R. (1997) J. Clin. Invest. 99, 635–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki M., Horikoshi T., Uchiwa H., Miyachi Y. (2000) Pigment Cell Res. 13, 248–252 [DOI] [PubMed] [Google Scholar]
- 32.Park H. Y., Russakovsky V., Ohno S., Gilchrest B. A. (1993) J. Biol. Chem. 268, 11742–11749 [PubMed] [Google Scholar]
- 33.Park H. Y., Russakovsky V., Ao Y., Fernandez E., Gilchrest B. A. (1996) Exp. Cell Res. 227, 70–79 [DOI] [PubMed] [Google Scholar]
- 34.Park H. Y., Perez J. M., Laursen R., Hara M., Gilchrest B. A. (1999) J. Biol. Chem. 274, 16470–16478 [DOI] [PubMed] [Google Scholar]
- 35.Corre S., Primot A., Sviderskaya E., Bennett D. C., Vaulont S., Goding C. R., Galibert M. D. (2004) J. Biol. Chem. 279, 51226–51233 [DOI] [PubMed] [Google Scholar]
- 36.Newton R. A., Cook A. L., Roberts D. W., Leonard J. H., Sturm R. A. (2007) J. Invest. Dermatol. 127, 2216–2227 [DOI] [PubMed] [Google Scholar]
- 37.Rouzaud F., Annereau J. P., Valencia J. C., Costin G. E., Hearing V. J. (2003) FASEB J. 17, 2154–2156 [DOI] [PubMed] [Google Scholar]
- 38.Singh S. K., Sarkar C., Mallick S., Saha B., Bera R., Bhadra R. (2005) Pigment Cell Res. 18, 113–121 [DOI] [PubMed] [Google Scholar]
- 39.Kim D. S., Park S. H., Kwon S. B., Na J. I., Huh C. H., Park K. C. (2007) Arch. Pharm. Res. 30, 581–586 [DOI] [PubMed] [Google Scholar]
- 40.Kono M., Dunn I. S., Durda P. J., Butera D., Rose L. B., Haggerty T. J., Benson E. M., Kurnick J. T. (2006) Mol. Cancer Res. 4, 779–792 [DOI] [PubMed] [Google Scholar]
- 41.Kim D. S., Kim S. Y., Chung J. H, Kim K. H., Eun H. C., Park K. C. (2002) Cell Signal. 14, 779–785 [DOI] [PubMed] [Google Scholar]
- 42.Kim D. S., Hwang E. S., Lee J. E., Kim S. Y., Kwon S. B., Park K. C. (2003) J. Cell Sci. 116, 1699–1706 [DOI] [PubMed] [Google Scholar]
- 43.Kim D. S., Park S. H., Kwon S. B., Li K., Youn S. W., Park K. C. (2004) Arch. Pharm. Res. 27, 334–339 [DOI] [PubMed] [Google Scholar]
- 44.Kim D. S., Park S. H., Kwon S. B., Park E. S., Huh C. H., Youn S. W., Park K. C. (2006) Pigment Cell Res. 19, 146–153 [DOI] [PubMed] [Google Scholar]
- 45.Borovansky J., Lipsova A., Kozakova M., Borovansky J. (2008) Pigment Cell Res. 21, 298–299 [Google Scholar]
- 46.Siegrist W., Eberle A. N. (1986) Anal. Biochem. 159, 191–197 [DOI] [PubMed] [Google Scholar]
- 47.Laskin J. D., Piccinini L., Engelhardt D. L., Weinstein I. B. (1982) J. Cell Physiol. 113, 481–486 [DOI] [PubMed] [Google Scholar]
- 48.Smalley K. S., Eisen T. G. (2002) Melanoma Res. 12, 187–192 [DOI] [PubMed] [Google Scholar]
- 49.Cuenda A., Rouse J., Doza Y. N., Meier R., Cohen P., Gallagher T. F., Young P. R., Lee J. C. (1995) FEBS Lett. 364, 229–233 [DOI] [PubMed] [Google Scholar]
- 50.Young P. R., McLaughlin M. M., Kumar S., Kassis S., Doyle M. L., McNulty D., Gallagher T. F., Fisher S., McDonnell P. C., Carr S. A., Huddleston M. J., Seibel G., Porter T. G., Livi G. P., Adams J. L., Lee J. C. (1997) J. Biol. Chem. 272, 12116–12121 [DOI] [PubMed] [Google Scholar]
- 51.Halaban R., Pomerantz S. H., Marshall S., Lambert D. T., Lerner A. B. (1983) J. Cell Biol. 97, 480–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halaban R., Cheng E., Zhang Y., Moellmann G., Hanlon D., Michalak M., Setaluri V., Hebert D. N. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6210–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang L., Karin M. (2001) Nature 410, 37–40 [DOI] [PubMed] [Google Scholar]
- 54.Lee J. C., Laydon J. T., McDonnell P. C., Gallagher T. F., Kumar S., Green D., McNulty D., Blumenthal M. J., Heys J. R., Landvatter S. W. (1994) Nature 372, 739–746 [DOI] [PubMed] [Google Scholar]
- 55.Godl K., Wissing J., Kurtenbach A., Habenberger P., Blencke S., Gutbrod H., Salassidis K., Stein-Gerlach M., Missio A., Cotten M., Daub H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15434–15439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang M., Wang Y., Collins M., Gu J. J., Mitchell B. S., Graves L. M. (2002) J. Biol. Chem. 277, 28364–28367 [DOI] [PubMed] [Google Scholar]
- 57.Shibazaki M., Takeuchi T., Ahmed S., Kikuchi H. (2004) J. Biol. Chem. 279, 3869–3876 [DOI] [PubMed] [Google Scholar]
- 58.Ando H., Watabe H., Valencia J. C., Yasumoto K., Furumura M., Funasaka Y., Oka M., Ichihashi M., Hearing V. J. (2004) J. Biol. Chem. 279, 15427–15433 [DOI] [PubMed] [Google Scholar]
- 59.Godbole D., Mojamdar M., Pal J. K. (2006) Cell Biol. Int. 30, 895–902 [DOI] [PubMed] [Google Scholar]
- 60.Watabe H., Valencia J. C., Yasumoto K., Kushimoto T., Ando H., Muller J., Vieira W. D., Mizoguchi M., Appella E., Hearing V. J. (2004) J. Biol. Chem. 279, 7971–7981 [DOI] [PubMed] [Google Scholar]
- 61.Casanovas O., Jaumot M., Paules A. B., Agell N., Bachs O. (2004) Oncogene 23, 7537–7744 [DOI] [PubMed] [Google Scholar]
- 62.Kida A., Kakihana K., Kotani S., Kurosu T., Miura O. (2007) Oncogene 26, 6630–6640 [DOI] [PubMed] [Google Scholar]
- 63.Fang D., Kute T., Setaluri V. (2001) Pigment Cell Res. 14, 132–139 [DOI] [PubMed] [Google Scholar]
- 64.Pavan W. J., Tilghman S. M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7159–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hasegawa T., Treis A., Patenge N., Fiesel F. C., Springer W., Kahle P. J. (2008) J. Neurochem. 105, 1700–1715 [DOI] [PubMed] [Google Scholar]
- 66.Hemesath T. J., Price E. R., Takemoto C., Badalian T., Fisher D. E. (1998) Nature 391, 298–301 [DOI] [PubMed] [Google Scholar]
- 67.Shimo T., Matsumura S., Ibaragi S., Isowa S., Kishimoto K., Mese H., Nishiyama A., Sasaki A. (2007) J. Cell Commun. Signal. 1, 103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Al-Shanti N., Stewart C. E. (2008) J. Endocrinol. 198, 243–252 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.