Abstract
The activity of many P-type ATPases is found to be regulated by interacting proteins or autoinhibitory elements located in N- or C-terminal extensions. An extended C terminus of fungal and plant P-type plasma membrane H+-ATPases has long been recognized to be part of a regulatory apparatus involving an autoinhibitory domain. Here we demonstrate that both the N and the C termini of the plant plasma membrane H+-ATPase are directly involved in controlling the pump activity state and that N-terminal displacements are coupled to secondary modifications taking place at the C-terminal end. This identifies the first group of P-type ATPases for which both ends of the polypeptide chain constitute regulatory domains, which together contribute to the autoinhibitory apparatus. This suggests an intricate mechanism of cis-regulation with both termini of the protein communicating to obtain the necessary control of the enzyme activity state.
Keywords: ATPases, Enzyme Mechanisms, H+-ATPase, Membrane Proteins, Proton Pumps, Autoinhibition
Introduction
Plasma membrane (PM)2 H+-ATPases are responsible for generating the essential electrochemical proton motive force across the plasma membrane of plant and fungal cells and are part of a large family of ion transport proteins termed P-type ATPases. These are all characterized by a transmembrane domain, a phosphorylation domain, a nucleotide-binding domain, and an actuator domain (1–3). Other members of the P-type ATPase family of proteins include the well studied animal Na+,K+-ATPase and Ca2+-ATPases. P-type ATPases seem to share a similar overall structure and a similar catalytic mechanism that involves a phosphorylated intermediate where the enzymes get phosphorylated from ATP on a conserved aspartate residue within the phosphorylation domain. Domain movements of the cytoplasmic domains are associated with the opening and closing of discrete ion-binding sites within the membranous part and ion translocation. The activities of many P-type ATPases are known to be regulated by secondary modifications and accessory proteins as well as cis-acting autoinhibitory domains. However, no high resolution structural information is at present available to aid in the molecular explanation of such autoinhibitory regulatory phenomena of P-type ATPases.
Autoinhibition of proteins is a widespread occurrence of enzymatic regulation identified among a variety of different protein structures and families (4). Autoinhibition involves discrete interactions between separable elements within a single polypeptide, where a cis-regulatory autoinhibitory domain negatively regulates the activity of other domains. The activity of fungal and plant PM H+-ATPases is regulated through such autoinhibitory domains located in the C terminus of the proteins. Removal of the C terminus by proteolytic digestion or truncation at the genetic level leads to release of the autoinhibitory restraints and thereby activation of the H+-ATPase and a marked (>50-fold) increase in its proton-transporting capacity (5–7). The only available three-dimensional crystal structure of an autoinhibited P-type ATPase, the PM H+-ATPase from plants, does not provide any information concerning the PM H+-ATPase regulatory mechanism as the terminal domains were not ordered in the crystals (3).
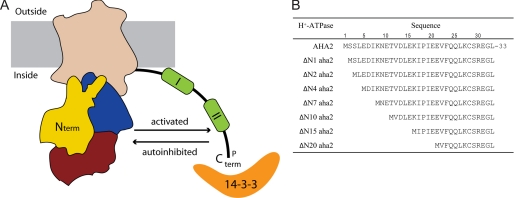
In the plant PM H+-ATPase, the C-terminal regulatory domain encompasses two autoinhibitory regions as well as several phosphorylation sites (Fig. 1A) (8–11). Phosphorylation of the penultimate threonine residue induces the binding of regulatory 14-3-3 proteins to the C terminus (12–15). This interaction is believed to cause a detachment of the autoinhibitory domain from an intramolecular binding site (reviewed in Ref. 16). This release of the regulatory domain goes along with a transformation of the enzyme from the low activity state to the high activity state (Fig. 1A). One 14-3-3 protein dimer has been demonstrated to bind two C-terminal peptides simultaneously, suggesting that the high activity state of the PM H+-ATPase could involve formation of dimers or multimeric complexes (17). Regulatory phosphorylatable residues have been identified in the fungal counterparts as well (18, 19). This could suggest that regulation of plant and fungal proton pumps shares mechanistic features. How initial phosphorylation events are regulated and which specific kinases that catalyze the reaction are largely unresolved questions, and structural information regarding the inhibited/activated form of PM H+-ATPases is lacking.
FIGURE 1.
The plant PM H+-ATPase and the truncated PM H+-ATPases constructed in this study. A, schematic representation of the PM H+-ATPase, which is composed of discrete domains, namely a transmembrane domain (wheat), a phosphorylation domain (blue), a nucleotide-binding domain (red), and an actuator domain (yellow). The N terminus (N term) is located at the actuator domain. The regulatory C-terminal extension contains two autoinhibitory regions (I and II), and activation of the PM H+-ATPase occurs by phosphorylation (P) of Thr-947 and subsequent binding of 14-3-3 protein. The figure is based on the three-dimensional crystal structure of the plant PM H+-ATPase, where structural information was obtained for residues (Val-12 to Ile-844) but not for the regulatory C terminus (C term) or the extreme N terminus. B, wild-type and N-terminal truncated PM H+-ATPases used in this study. The N-terminal truncated H+-ATPase mutants are named according to the number of deleted amino acid residues.
Much effort has been devoted to explain the molecular mechanism of C terminus-mediated pump regulation. In contrast, the N terminus of the PM H+-ATPase has not been investigated to any significant extent. Here we report that the extreme N terminus of the plant PM H+-ATPase directly participates in pump regulation. Our findings suggest a model in which both the N and the C termini constitute the cis-regulatory autoinhibitory domain and suggest that transformation of the PM H+-ATPase protein molecule from the low activity state to the high activity state involves a structural rearrangement of both domains.
EXPERIMENTAL PROCEDURES
Construction of Mutants
The multicopy vector Yep-351 (20) containing the full-length cDNA of the Arabidopsis thaliana AHA2 PM H+-ATPase isoform under the control of the PMA1 promoter (21) was used as in Ref. 22. All deletion and single point mutations were introduced by standard procedures using polymerase chain reaction, and all mutated sequences were verified by DNA sequencing.
Expression of PM H+-ATPases in the Yeast Saccharomyces cerevisiae
The yeast strain RS-72 was transformed and cultured essentially as described previously (23). In RS-72 (MATa ade1-100 his4-519 leu2-3,112), the natural constitutive promoter of the yeast endogenous PM H+-ATPase has been replaced by the galactose-dependent GAL1 promoter (24). Yeast complementation assays were performed on solid minimal media containing either galactose or glucose as described previously (8). The yeast cells were harvested, and plasma membranes were purified essentially as described (8), except that yeast cells were homogenized in 29% glycerol (v/v), 50 mm Mes-KOH, pH 6.5, 10 mm EDTA, 1.3 mm ATP, 50 mm KCl, 1 mm dithiothreitol, 0.4 mm phenylmethylsulfonyl fluoride, and 0.4 μg/ml pepstatin A.
The plasma membrane fraction was obtained by resuspension of the yeast total microsomal membrane fraction in 10% sucrose (w/w), 50 mm Mes-KOH, pH 6.5, 1 mm dithiothreitol, and 1 mm EDTA followed by centrifugation for 16 h at 30,000 rpm (SW 41 Ti rotor; Beckmann Instruments International S.A., Geneva, Switzerland) through a 10-ml sucrose step gradient (5 ml each of 43 and 53% (w/w) sucrose in 10 mm Mes-KOH, pH 6.5, 1 mm EDTA, 1 mm dithiothreitol). The plasma membrane fraction was hereafter collected from the (43%/53%) interface and resuspended in 20% glycerol (v/v), 50 mm Mes-KOH, pH 6.5, 1 mm EDTA, 1 mm dithiothreitol, and 0.4 mm phenylmethylsulfonyl fluoride before further biochemical testing.
SDS-PAGE, Western Blotting, and 14-3-3 Protein Overlay Assay
Immunological detection of the plant PM H+-ATPase was performed after SDS-PAGE and Western blotting with specific anti-H+-ATPase antibodies. For detection of phosphorylated threonine residues, an anti-phosphothreonine antibody (71-8200; Zymed Laboratories Inc.) was employed. Immunological detection of bound 14-3-3 protein was performed by a so-called 14-3-3 overlay assay as described (12). For immunological detection of expression, 10 μg of plasma membrane proteins were loaded in each well, whereas 25 μg of plasma membrane proteins were used for the detection of phosphothreonine residues and for the 14-3-3 overlay assay.
ATPase Assay
An ATPase assay was performed as described previously (8) on plasma-enriched fractions, and unless otherwise stated, the assay buffer contained 3 mm ATP.
RESULTS
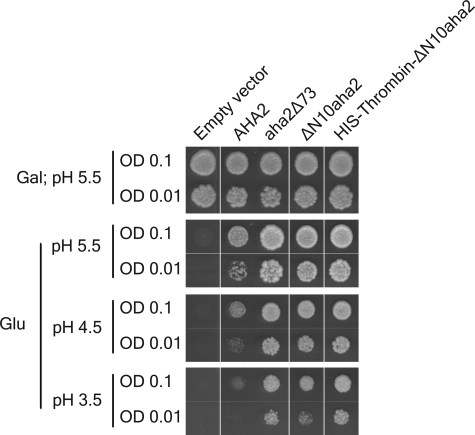
Previous efforts to purify the PM H+-ATPase in its autoinhibited state for biochemical and structural studies have not proven successful (3). Likewise, in our hands, placement of an affinity tag at the regulatory C terminus was not compatible with keeping the enzyme in its low activity state (data not shown). In an attempt to obtain three-dimensional structural information of the autoinhibited PM H+-ATPase, we constructed a mutant of the PM H+-ATPase with a cleavable affinity tag in the N terminus, inserted at position 11 (Fig. 2). We expected this mutant PM H+-ATPase to accumulate in yeast cells in its autoinhibited state just as the wild-type plant H+-ATPase. The autoinhibited plant PM H+-ATPase only supports slow yeast growth at pH 5.5 and is unable to support growth at acidic pH values such as pH 3.5 (25, 26). To our surprise, the N-terminally tagged pump protein was much more efficient in H+ transport when compared with the wild-type protein as determined from its capacity to complement the function of the endogenous yeast PM H+-ATPase, Pma1p, in the yeast strain RS-72 (Fig. 2). This result suggested that the placement of the affinity tag at the truncated N terminus somehow perturbed normal autoinhibition of the PM H+-ATPase.
FIGURE 2.
Modifications at the N terminus activate the plant PM H+-ATPase in a yeast complementation assay. A yeast complementation test was performed for no PM H+-ATPase, wild-type PM H+-ATPase (AHA2), C-terminal truncated PM H+-ATPase (aha2Δ73), N-terminal truncated PM H+-ATPase (ΔN10aha2), and a N-terminal truncated PM H+-ATPase with a His tag and a thrombin recognition sequence (His-Thrombin-ΔN10aha2). In the yeast strain RS-72, the endogenous yeast PM H+-ATPase, Pma1p, has been placed under the control of a galactose promoter, whereas the introduced, plasmid-borne plant PM H+-ATPases are under the control of the constitutive PMA1 promoter (24). Yeast growth on glucose is therefore dependent on a functional plasmid-borne PM H+-ATPase. Transformed yeast cells, RS-72, were spotted on either galactose-containing media (Gal) at pH 5.5 or glucose-containing media (Glu) at different pH values at two different concentrations (A600 = 0.1 and A600 = 0.01). Growth was recorded after 4 days at 30 °C. OD, optical density.
The Activity of the PM H+-ATPase Is Influenced by the Length of Its N Terminus
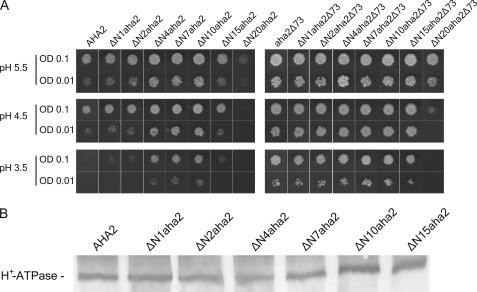
Next, we investigated the effect of introducing small deletions in the N terminus of the PM H+-ATPase as autoinhibitory domains are often characterized by their sensitivity to proteolytic attack or truncations at the genetic level (4). The resulting mutants (Fig. 1B) were expressed in yeast cells and tested for their ability to complement pma1 (Fig. 3A). The introduction of small deletions (1–10 amino acid residues in length) in the N terminus increased the ability of the plant PM H+-ATPase to support yeast growth, both at pH 5.5 and at more acidic pH values (Fig. 3A). Further extending the N-terminal deletions in length led to a reduction in the ability to complement the yeast pma1. Plasma membranes were purified from yeast expressing the wild-type AHA2 PM H+-ATPase and the N-terminal truncated PM H+-ATPase mutants. PM H+-ATPase expression level of the N-terminal deletion mutants was observed to be comparable with that of the wild-type PM H+-ATPase (Fig. 3B). Thus, the observed growth effects following removal of part of the N terminus of the PM H+-ATPase were not due to alterations in the amount of the proteins expressed.
FIGURE 3.
Activation level of the PM H+-ATPase by truncation of the N terminus increases with the length of the deletion up to 10 amino acid residues and does not give an additive effect when combined with a C-terminal truncation. A, the yeast strain RS-72 transformed with N-terminal truncated mutants of either the wild-type plant PM H+-ATPase or the C-terminal truncated form of the enzyme was spotted on galactose- or glucose-containing media as in Fig. 2. OD, optical density. B, plasma membrane-enriched fractions of yeast strains expressing various N-terminal truncated mutants of the PM H+-ATPase were tested for expression level by immunostaining.
To test whether the effect of deleting the N-terminal residues was additive to that of manipulating the C terminus, the same N-terminal deletions were introduced in a C-terminal truncated aha2Δ73 PM H+-ATPase background. This aha2Δ73 PM H+-ATPase is constitutively active due to the truncation of its regulatory C terminus (27). Deleting up to 10 amino acid residues in the N terminus of the aha2Δ73 PM H+-ATPase did not have any additional effect on the ability of the C-terminal truncated PM H+-ATPase to complement pma1. Further increasing the number of amino acid residues to be deleted reduced the ability of the C-terminal truncated AHA2 PM H+-ATPase to support yeast growth (Fig. 3A). Thus, the first 10 amino acid residues of the PM H+-ATPase appear not to be necessary for the basic proton-pumping apparatus but rather seem to be involved in the regulation of the PM H+-ATPase, whereas subsequent residues are required for catalytic function.
The N Terminus Constitutes a True Regulatory Domain
Activation of the PM H+-ATPase goes along with altered kinetic parameters for the PM H+-ATPase such as a shift in the pH optimum toward more neutral values and an increase in the apparent affinity for ATP, Km (23). To test whether the deletions in the N terminus of the PM H+-ATPase were reflected at the biochemical level, plasma membranes were purified from yeast expressing wild-type and mutant ΔN10aha2 PM H+-ATPase. The biochemical characterization revealed that the truncated ΔN10aha2 mutant had a higher apparent affinity for ATP than the wild type (Km ∼0.3 mm for the ΔN10 mutant when compared with Km ∼1.4 mm for the wild type) (Fig. 4A) and that its pH optimum was shifted toward more neutral value (pH ∼6.7 when compared with pH ∼6.2 for wild-type PM H+-ATPase) (Fig. 4B). This is consistent with the ΔN10 truncation leading to a biochemical activation of the membrane protein. Thus, the N terminus constitutes the kinetic hallmarks of a true regulatory domain of the PM H+-ATPase.
FIGURE 4.
The Δ N10aha2 PM H+-ATPase is activated biochemically. A, ATP dependence of ATP hydrolytic activity by wild-type PM H+-ATPase and the ΔN10aha2 PM H+-ATPase mutant at pH 7.0. B, pH dependence of ATP hydrolysis by wild-type PM H+-ATPase and the ΔN10aha2 PM H+-ATPase mutant. Purified plasma membrane-enriched fractions were analyzed in both A and B. □, wild-type AHA2; ■, ΔN10aha2. All experiments are represented as ± S.D.
Molecular Communication Between the N and the C Termini
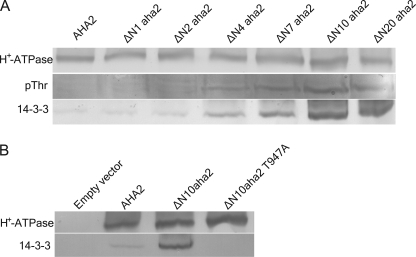
The activated plant PM H+-ATPase is known to be phosphorylated in vivo at the penultimate residue, Thr-947, which creates a binding site for activating 14-3-3 proteins. To investigate how the N terminus is linked to regulation of the PM H+-ATPase, purified yeast plasma membranes expressing AHA2 and N-terminal truncated mutants hereof were separated on SDS gels, transferred to nitrocellulose membranes, and probed with an antibody directed against phosphothreonine. Fig. 5 demonstrates that the successive removal of up to 10 amino acid residues from the N terminus of the plant PM H+-ATPase led to a concomitant increase in the threonine phosphorylation level of the PM H+-ATPases. As phosphorylation of Thr-947 is correlated with 14-3-3 protein binding, an in vitro 14-3-3 protein overlay assay was performed. Consistent with their increased phosphorylation level, the N-terminal truncated PM H+-ATPase mutants also displayed an increased ability to bind 14-3-3 proteins when compared with the wild type (Fig. 5A). Interestingly, both phosphorylation and 14-3-3 binding were proportional to the length of N-terminal truncation and pma1 complementation. That truncation at the N terminus of the H+-ATPase gives rise to specific 14-3-3 binding at the extreme C terminus was confirmed by constructing a PM H+-ATPase containing the ΔN10 truncation with a mutation of the penultimate threonine residue to an alanine (T947A), which should abolish phosphorylation of and 14-3-3 binding by the mutant PM H+-ATPase. Indeed, despite an expression level comparable with the wild type and the ΔN10aha2 mutant, the ΔN10aha2 T947A mutant did not bind 14-3-3 protein (Fig. 5B). Thus, truncations at the N terminus are directly linked to phosphorylation of Thr-947 and subsequent 14-3-3 protein binding at the extreme C terminus.
FIGURE 5.
Molecular communication between the N and the C termini. Plasma membrane-enriched fractions of yeast strains expressing the various mutants of the PM H+-ATPase were run on SDS-PAGE and transferred to nitrocellulose for immunostaining or overlay assay. A, N-terminal truncated mutants of the PM H+-ATPase show increased level of phosphorylation and bind more 14-3-3 proteins than the wild type. pThr, phosphothreonine. B, the high capacity for 14-3-3 binding of the ΔN10aha2 PM H+-ATPase is strictly dependent on the penultimate Thr-947 residue.
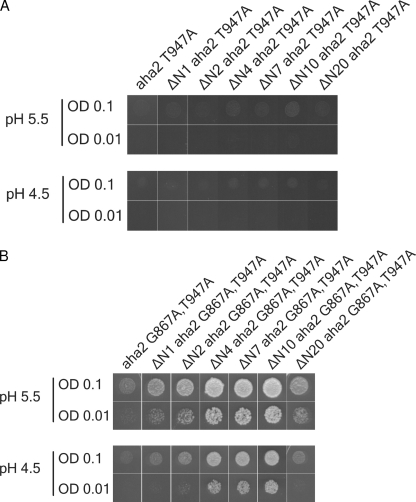
Mutagenesis of Thr-947 has previously been shown to abolish the activation by 14-3-3 proteins and results in a lack of complementation when the proteins are expressed in yeast. 14-3-3-independent activation can, however, occur if the T947A mutation is combined with mutations in both autoinhibitory regions of the C terminus, namely G867A (region I) together with an R913A mutation (region II) (22). Combining the N-terminal deletions with a PM H+-ATPase containing the T947A mutation was not sufficient to overcome the lack of activation by 14-3-3 proteins in the yeast complementation assay (Fig. 6A). Thus, no complementation was observed irrespective of the length of the N-terminal truncation. However, if N-terminal deletions were combined with both the T947A mutation and the G867A single point mutation in one of the autoinhibitory regions, an increased ability of the mutant PM H+-ATPases to support yeast growth was observed (Fig. 6B). In contrast, the corresponding PM H+-ATPase without an N-terminal deletion was not able to complement the function of Pma1p (Fig. 6B).
FIGURE 6.
Activation of the PM H+-ATPase by N-terminal truncations is dependent on Thr-947 phosphorylation but can be surpassed by a modification of autoinhibitory region I. A, a yeast complementation test of aha2 T947A and N-terminal truncated mutants hereof. B, a yeast complementation test of aha2 G867A,T947A and N-terminal truncated mutants hereof. All yeast cells were spotted in optical densities (OD) of 0.1 and 0.01 on solid glucose-containing medium with different pH (5.5 and 4.5).
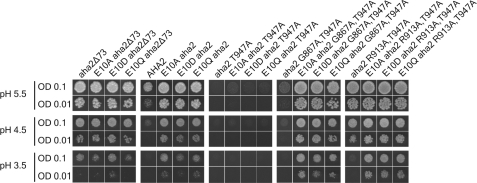
The N Terminus as Part of the Regulatory Domain Seems Conserved in the Plant Kingdom
In a large screen for regulatory mutants of the Nicotiana plumbaginifolia PMA2 PM H+-ATPase, it was observed that an N-terminal E14D mutation in PMA2 gives rise to an activation of the PM H+-ATPase (28). To test whether this residue, corresponding to Glu-10 in AHA2 and highly conserved among plant PM H+-ATPases (see the P-type ATPase Database), could mimic the effect of the N-terminal truncations in the AHA2 PM H+-ATPase, site-directed mutagenesis was employed to substitute this glutamic acid residue with aspartic acid (E10D), glutamine (E10Q), and alanine (E10A). Expression of these three mutants in the same yeast system as described above showed that the aha2E10 mutants indeed were able to support yeast growth to a greater extent than the wild type at pH 5.5 as well as at more acidic pH values (Fig. 7). Hence, the yeast growth phenotype of the Glu-10 mutants is the same as the phenotype observed upon deletion of the first 11 amino acid residues of the AHA2 PM H+-ATPase.
FIGURE 7.
Mutagenesis of the conserved Glu-10 residue in the N terminus of the PM H+-ATPase perturbs autoinhibition. RS-72 transformed with the indicated mutants was spotted on plates with different pH (5.5, 4.5, and 3.5) and in two different optical densities (OD), namely ODs of 0.1 and 0.01. Gly-867 resides in autoinhibitory region I, and Arg-913 resides in autoinhibitory region II (see Fig. 1A).
DISCUSSION
The plant plasma membrane H+-ATPase has long been known to be autoinhibited through a regulatory domain located at its C terminus. In this study, we have identified the N terminus of the PM H+-ATPase as a central component of the autoinhibitory apparatus of the proton pump.
The introduction of small deletions in the N terminus of the PM H+-ATPase improved the ability of the proton pump to support yeast growth. In this context, the N-terminal deletions could substitute for mutations in one of the autoinhibitory regions of the C terminus. The same yeast growth phenotype was observed if a point mutation was introduced at residue Glu-10 instead of a deletion of the first 10 N-terminal residues. Together these data indicate that the observed activation of the PM H+-ATPase occurs through a destabilization of the N-terminal region of the PM H+-ATPase.
The destabilization or complete removal of the N terminus of the PM H+-ATPase is coupled to secondary modifications at its extreme C terminus. Upon expression in yeast of the N-terminally truncated PM H+-ATPase mutants, we observed a marked increase in the phosphorylation level of the penultimate threonine residue and 14-3-3 protein binding, and both the phosphorylation level and the 14-3-3 binding were found to correlate with the length of the N-terminal deletion. Thus, modifications at the N terminus somehow result in an unmasking of the extreme C terminus, which makes the penultimate threonine residue accessible for protein kinase-mediated phosphorylation and subsequent activation of the PM H+-ATPase.
The Mechanism of Autoinhibition of PM H+-ATPases
With our finding that the N and the C termini of the PM H+-ATPase function together as cis-regulatory elements, the question arises as to how autoinhibition is achieved. Several residues within the central part of the PM H+-ATPase, which upon mutation give rise to an activated proton pump, have previously been identified (28, 29), and plotting these residues on the structure of the PM H+-ATPase reveals that many of these regulatory residues are found to cluster at the actuator domain (3). Based on this observation, it was proposed that the C terminus wraps around the proton pump at the actuator domain to inhibit domain movements and/or proton access to the membranous proton-binding site. Activation of the PM H+-ATPase in vivo may occur through a destabilization of the interaction between the N terminus and the rest of the PM H+-ATPase molecule, which ultimately leads to molecular modifications taking place at the C terminus. This destabilization could perhaps be achieved through post-translational modifications, small ligand binding, or possibly through the docking of a protein kinase. Several different stimuli have been found to affect the activity of the PM H+-ATPase in planta, such as light, hormones, toxins, and environmental stresses (reviewed in Ref. 16). In plants, multiple PM H+-ATPase isoforms are expressed, and the expression patterns of the isoforms have revealed that although some isoforms display tissue-specific localization, there is considerable overlap of the expression of isoforms in many cell types and tissues (30–36). One could speculate that structural variations in the N terminus permits various PM H+-ATPases to perceive activation stimuli differently. Thereby differential regulation of PM H+-ATPase isoforms within the same cell could be attained.
Is the Bilateral Autoinhibition Mechanism a General Feature of Autoinhibited P-type ATPases?
Several P-type ATPases are regulated by cis-acting elements located in either the N or the C terminus (5, 6, 37, 38). The plant PM H+-ATPase is, however, the first P-type ATPase found to possess autoinhibitory domains at both its termini. An open question is, therefore, whether such a bilateral autoinhibition mechanism could be a common characteristic for autoinhibited P-type ATPases. The yeast PM H+-ATPase, Pma1p, also contains a regulatory C terminus, albeit shorter than the corresponding C terminus in plant PM H+-ATPases. The yeast PM H+-ATPase has a much longer N-terminal segment than the plant PM H+-ATPase, proposed to serve as a voltage sensor based on a high propensity of negatively charged amino acid residues (39). Removal of the first 27 N-terminal residues of this yeast pump had no effect on the enzymatic activity or on the response toward changes in glucose metabolism (40). However, by combining this N-terminal truncation with the removal of the C-terminal regulatory domain, Mason et al. (40) found that this bilaterally truncated PM H+-ATPase was more activated than a pump devoid of only the C terminus. Longer truncations in the N terminus of the yeast PM H+-ATPase led to significant reductions in the expression level of the yeast PM H+-ATPase (6).
Plasma membrane Ca2+-ATPases are another example of autoinhibited P-type ATPases, where either the N terminus (plants) or the C terminus (animals) functions as a regulatory domain. Using fluorescence resonance energy transfer, it was recently observed that the N and the C termini of the PM Ca2+-ATPase, hPMCA4xb, are in close proximity in the autoinhibited conformation, and furthermore that the two termini appear to separate or reorient upon activation of the PM Ca2+-ATPase (41). This observation supports the idea that the involvement of both termini in the autoregulation might not be a unique feature of the plant PM H+-ATPase.
Many completely structurally diverse proteins have been found to possess bilateral autoinhibitory modules located at the N and the C termini that together constitute the autoinhibitory regulatory module, such as the eukaryotic transcription factor Ets1 and cyclin T1, a subunit of the human positive transcription elongation factor b (P-TEFb) (42, 43). In a working model of autoinhibition in P-type PM H+-ATPases, the N terminus and the C terminus together form the regulatory integration machinery. This model contains two key aspects. First, the PM H+-ATPase is inhibited in its catalytic cycle as the autoinhibitory parts of the C terminus prevent access of protons to the binding site and/or domain movements, and second, in the autoinhibited state, the regulatory threonine of the C terminus is kept buried within the protein structure by molecular interactions formed by the two termini.
Footnotes
- PM
- plasma membrane
- Mes
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Toyoshima C., Nakasako M., Nomura H., Ogawa H. (2000) Nature 405, 647–655 [DOI] [PubMed] [Google Scholar]
- 2.Morth J. P., Pedersen B. P., Toustrup-Jensen M. S., Sørensen T. L., Petersen J., Andersen J. P., Vilsen B., Nissen P. (2007) Nature 450, 1043–1049 [DOI] [PubMed] [Google Scholar]
- 3.Pedersen B. P., Buch-Pedersen M. J., Morth J. P., Palmgren M. G., Nissen P. (2007) Nature 450, 1111–1114 [DOI] [PubMed] [Google Scholar]
- 4.Pufall M. A., Graves B. J. (2002) Annu. Rev. Cell Dev. Biol. 18, 421–462 [DOI] [PubMed] [Google Scholar]
- 5.Palmgren M. G., Sommarin M., Serrano R., Larsson C. (1991) J. Biol. Chem. 266, 20470–20475 [PubMed] [Google Scholar]
- 6.Portillo F., de Larrinoa I. F., Serrano R. (1989) FEBS Lett. 247, 381–385 [DOI] [PubMed] [Google Scholar]
- 7.Venema K., Palmgren M. G. (1995) J. Biol. Chem. 270, 19659–19667 [DOI] [PubMed] [Google Scholar]
- 8.Axelsen K. B., Venema K., Jahn T., Baunsgaard L., Palmgren M. G. (1999) Biochemistry 38, 7227–7234 [DOI] [PubMed] [Google Scholar]
- 9.Nühse T. S., Stensballe A., Jensen O. N., Peck S. C. (2003) Mol. Cell Proteomics. 2, 1234–1243 [DOI] [PubMed] [Google Scholar]
- 10.Niittylä T., Fuglsang A. T., Palmgren M. G., Frommer W. B., Schulze W. X. (2007) Mol. Cell Proteomics. 6, 1711–1726 [DOI] [PubMed] [Google Scholar]
- 11.Duby G., Poreba W., Piotrowiak D., Bobik K., Derua R., Waelkens E., Boutry M. (2009) J. Biol. Chem. 284, 4213–4221 [DOI] [PubMed] [Google Scholar]
- 12.Fuglsang A. T., Visconti S., Drumm K., Jahn T., Stensballe A., Mattei B., Jensen O. N., Aducci P., Palmgren M. G. (1999) J. Biol. Chem. 274, 36774–36780 [DOI] [PubMed] [Google Scholar]
- 13.Svennelid F., Olsson A., Piotrowski M., Rosenquist M., Ottman C., Larsson C., Oecking C., Sommarin M. (1999) Plant Cell 11, 2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maudoux O., Batoko H., Oecking C., Gevaert K., Vandekerckhove J., Boutry M., Morsomme P. (2000) J. Biol. Chem. 275, 17762–17770 [DOI] [PubMed] [Google Scholar]
- 15.Olsson A., Svennelid F., Ek B., Sommarin M., Larsson C. (1998) Plant Physiol. 118, 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmgren M. G. (2001) Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 817–845 [DOI] [PubMed] [Google Scholar]
- 17.Ottmann C., Marco S., Jaspert N., Marcon C., Schauer N., Weyand M., Vandermeeren C., Duby G., Boutry M., Wittinghofer A., Rigaud J. L., Oecking C. (2007) Mol. Cell 25, 427–440 [DOI] [PubMed] [Google Scholar]
- 18.Lecchi S., Allen K. E., Pardo J. P., Mason A. B., Slayman C. W. (2005) Biochemistry 44, 16624–16632 [DOI] [PubMed] [Google Scholar]
- 19.Lecchi S., Nelson C. J., Allen K. E., Swaney D. L., Thompson K. L., Coon J. J., Sussman M. R., Slayman C. W. (2007) J. Biol. Chem. 282, 35471–35481 [DOI] [PubMed] [Google Scholar]
- 20.Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. (1986) Yeast 2, 163–167 [DOI] [PubMed] [Google Scholar]
- 21.Villalba J. M., Palmgren M. G., Berberián G. E., Ferguson C., Serrano R. (1992) J. Biol. Chem. 267, 12341–12349 [PubMed] [Google Scholar]
- 22.Jahn T. P., Schulz A., Taipalensuu J., Palmgren M. G. (2002) J. Biol. Chem. 277, 6353–6358 [DOI] [PubMed] [Google Scholar]
- 23.Regenberg B., Villalba J. M., Lanfermeijer F. C., Palmgren M. G. (1995) Plant Cell 7, 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cid A., Perona R., Serrano R. (1987) Curr. Genet. 12, 105–110 [DOI] [PubMed] [Google Scholar]
- 25.Palmgren M. G., Christensen G. (1993) FEBS Lett. 317, 216–222 [DOI] [PubMed] [Google Scholar]
- 26.de Kerchove d'Exaerde A., Supply P., Dufour J. P., Bogaerts P., Thinés D., Goffeau A., Boutry M. (1995) J. Biol. Chem. 270, 23828–23837 [DOI] [PubMed] [Google Scholar]
- 27.Buch-Pedersen M. J., Venema K., Serrano R., Palmgren M. G. (2000) J. Biol. Chem. 275, 39167–39173 [DOI] [PubMed] [Google Scholar]
- 28.Morsomme P., de Kerchove d'Exaerde A., De Meester S., Thinès D., Goffeau A., Boutry M. (1996) EMBO J. 15, 5513–5526 [PMC free article] [PubMed] [Google Scholar]
- 29.Morsomme P., Dambly S., Maudoux O., Boutry M. (1998) J. Biol. Chem. 273, 34837–34842 [DOI] [PubMed] [Google Scholar]
- 30.DeWitt N. D., Harper J. F., Sussman M. R. (1991) Plant J. 1, 121–128 [DOI] [PubMed] [Google Scholar]
- 31.DeWitt N. D., Sussman M. R. (1995) Plant Cell 7, 2053–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harper J. F., Manney L., DeWitt N. D., Yoo M. H., Sussman M. R. (1990) J. Biol. Chem. 265, 13601–13608 [PubMed] [Google Scholar]
- 33.Ewing N. N., Bennett A. B. (1994) Plant Physiol 106, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hentzen A. E., Smart L. B., Wimmers L. E., Fang H. H., Schroeder J. I., Bennett A. B. (1996) Plant Cell Physiol 37, 650–659 [DOI] [PubMed] [Google Scholar]
- 35.Moriau L., Michelet B., Bogaerts P., Lambert L., Michel A., Oufattole M., Boutry M. (1999) Plant J. 19, 31–41 [DOI] [PubMed] [Google Scholar]
- 36.Gévaudant F., Pétel G., Guilliot A. (2001) Planta 212, 619–626 [DOI] [PubMed] [Google Scholar]
- 37.Enyedi A., Verma A. K., Filoteo A. G., Penniston J. T. (1993) J. Biol. Chem. 268, 10621–10626 [PubMed] [Google Scholar]
- 38.Harper J. F., Hong B., Hwang I., Guo H. Q., Stoddard R., Huang J. F., Palmgren M. G., Sze H. (1998) J. Biol. Chem. 273, 1099–1106 [DOI] [PubMed] [Google Scholar]
- 39.Kühlbrandt W., Zeelen J., Dietrich J. (2002) Science 297, 1692–1696 [DOI] [PubMed] [Google Scholar]
- 40.Mason A. B., Kardos T. B., Monk B. C. (1998) Biochim. Biophys. Acta 1372, 261–271 [DOI] [PubMed] [Google Scholar]
- 41.Corradi G. R., Adamo H. P. (2007) J. Biol. Chem. 282, 35440–35448 [DOI] [PubMed] [Google Scholar]
- 42.Jonsen M. D., Petersen J. M., Xu Q. P., Graves B. J. (1996) Mol. Cell. Biol. 16, 2065–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fong Y. W., Zhou Q. (2000) Mol. Cell. Biol. 20, 5897–5907 [DOI] [PMC free article] [PubMed] [Google Scholar]