Abstract
Rhodopsins are photoreceptor proteins that have diverged from ligand-binding G protein-coupled receptors (GPCRs). Unlike other GPCRs, rhodopsins contain an intrinsic antagonist, 11-cis-retinal, which is converted to the agonist all-trans-retinal upon absorption of a photon. Through evolution, vertebrate rhodopsins have lost the ability of direct binding to the agonist, but some invertebrate and vertebrate non-visual rhodopsins have retained this ability. Here, we investigated the difference in the agonist-binding state between these rhodopsins to further our understanding of the structural and functional diversity of rhodopsins. Mutational analyses of agonist-binding rhodopsin showed that replacement of Ala-269, one of the residues constituting the antagonist-binding site, with bulky amino acids resulted in a large spectral shift in its active state and a great reduction in G protein activity, whereas these were rescued by subsequent replacement of Phe-208 with smaller amino acids. Although similar replacements in vertebrate rhodopsin did not cause a spectral shift in the active state, a similar reduction in and recovery of G protein activity was observed. Therefore, the agonist is located close to Ala-269 in the agonist-binding rhodopsin, but not in vertebrate rhodopsins, and Ala-269 with Phe-208 acts as a pivot for the formation of the G protein-activating state in both rhodopsins. The positions corresponding to Ala-269 and Phe-208 in other GPCRs have been reported to form part of an agonist-binding site. Therefore, an agonist-binding rhodopsin has the molecular architecture of the agonist-binding site similar to that of a general GPCR, whereas vertebrate rhodopsins changed the architecture, resulting in loss of agonist binding during molecular evolution.
Keywords: G Proteins/Coupled Receptors (GPCR), Methods/Site-directed Mutagenesis, Methods/Spectroscopy, Protein/Chemistry, Signal Transduction/G proteins, Vision/Rhodopsin
Introduction
Rhodopsin is a member of the family of G protein-coupled receptors (GPCRs)4 that have a seven-transmembrane α-helical structure. It contains the antagonist (inverse agonist) 11-cis-retinal covalently attached to its protein moiety, opsin. The agonist of rhodopsin is all-trans-retinal, but curiously, vertebrate rhodopsins have no ability to bind this agonist directly (1). Instead, the agonist all-trans-retinal is produced by photoisomerization of the antagonist 11-cis-retinal in the rhodopsin molecule upon photon absorption. Loss of the ability to directly bind exogenous agonist is probably the result of molecular evolution of vertebrate rhodopsin, which would have favored the decrease in background “noise” in the dark state (2). We have found that, in contrast to vertebrate rhodopsins, a non-vertebrate rhodopsin, amphioxus rhodopsin (3), and a vertebrate pineal UV light-sensitive pigment, lamprey parapinopsin5 (4), still have the ability to bind the agonist directly, and the active state formed by direct binding of agonist exhibited biochemical and spectroscopic properties indistinguishable from those of the photoproduct that was formed by irradiation of the antagonist-binding rhodopsin. In addition, the G protein-activating efficiencies of these rhodopsins were several dozen times lower than those of a vertebrate rhodopsin and similar to those of other Gi/Go-coupled GPCRs, although detailed quantitative comparison of the efficiencies between rhodopsins and other receptors was difficult (5). Therefore, in this study, we examined the difference in the molecular architecture between agonist-binding rhodopsins and vertebrate rhodopsins.
GPCRs stabilize their active and inactive states upon binding to agonist and antagonist (inverse agonist), respectively. Because the structures differ between the active and inactive states, it is reasonable to assume that some of the amino acid residues located close to the agonist are relatively far from the antagonist. Therefore, we hypothesized that identifying the amino acid residues that interact with the agonist but not with the antagonist would provide insight into the structural changes in the activation mechanism of GPCRs. In rhodopsins, the retinal agonist and antagonist in the active and resting (inactive) states exhibit unique and different absorption spectra due to their interactions with nearby amino acids residues (6). This property makes it possible to identify amino acid residues located near the agonist and/or antagonist in rhodopsins by site-directed mutagenesis combined with spectroscopic measurements.
In this study, using parapinopsin as an agonist-binding rhodopsin, we tried to identify amino acid residues whose mutations cause spectral changes in only the agonist-binding state. We examined whether or not these mutations affect G protein-activating efficiencies and whether the same phenomena are observed in vertebrate rhodopsins. We also investigated amphioxus rhodopsin (2, 3) as an invertebrate agonist-binding rhodopsin. Our findings clearly show that mutations of Ala-269 in helix VI cause spectral changes in the agonist-binding state. In addition, this residue forms a specific pair with Phe-208 in helix V and constitutes a pivot for the helical rearrangement in rhodopsin activation. Sequence analysis indicated that these amino acids correspond to the residues that interact with the agonists in other GPCRs. Although they are also essential for the helical rearrangement followed by G protein activation in bovine rhodopsin, introduction of the bulky amino acid into position 269 causes no spectral changes in the active state of bovine rhodopsin. We will discuss the significance of the amino acid pair in the formation of the active state and its displacement from the agonist-binding site in the course of molecular evolution.
EXPERIMENTAL PROCEDURES
Preparation of Rhodopsins
Parapinopsin and amphioxus rhodopsin tagged with the Rho 1D4 epitope and bovine rhodopsin were expressed in HEK293S cells as described (2–4). Site-directed mutants were prepared using the QuikChange kit (Stratagene) according to the manufacturer's instructions. Rhodopsins were constituted by mixing opsin-containing membranes with 11-cis-retinal. All procedures were carried out under dim red light at 4 °C. The pigment-containing membranes were prepared by sucrose floatation methods (5). The pigments were extracted using 1% n-dodecyl β-d-maltoside (DM) in buffer A (50 mm HEPES (pH 6.5) and 140 mm NaCl). The extract was incubated overnight with Rho 1D4 antibody-agarose. After washing with buffer B (0.02% DM in buffer A), rhodopsins were eluted with buffer B containing the C-terminal octadecapeptide of bovine rhodopsin. For Gi activation assay of parapinopsin, the pigments were extracted with 0.2% DM in buffer A from pigment-containing membranes.
Spectrophotometry
Absorption spectra were recorded with a Shimadzu MPS-2000 (for membrane samples) or UV-2400 (for purified samples) spectrophotometer. Parapinopsin, amphioxus rhodopsin, and bovine rhodopsin were irradiated with light from a 1-kilowatt tungsten-halogen lamp (Rikagaku Seiki), which had been passed through a UTVAF-50S-36V glass filter (Sigma Koki), a 501-nm interference filter (Optical Coatings Japan), and a Y-52 glass cutoff filter (Toshiba), respectively. The spectra of parapinopsin and amphioxus rhodopsin mutants were recorded at pH 6.5 and 0 °C. The spectra of the dark and active states of bovine rhodopsin were recorded using purified samples in the presence of DM at pH 6.5 and 20 °C. To measure the spectral shifts in metarhodopsin I (meta-I) of bovine rhodopsin, the spectra were recorded using membrane preparations at pH 8.9 and 0 °C.
G Protein Activation Assays
A radionucleotide filter binding assay, which measures GDP/GTPγS exchange by G protein (Gt (transducin) or Gi), was carried out at 0 °C (for parapinopsin) or 20 °C (for bovine rhodopsin) as described (2, 5). The assay mixture consisted of 50 mm HEPES (pH 6.5), 140 mm NaCl, 8 mm MgCl2, 1 mm dithiothreitol, 1 μm [35S]GTPγS, and 2 μm (Gt assay) or 4 μm (Gi assay) GDP. For the assay, crude extract of parapinopsin (final concentration of 10 nm) or purified bovine rhodopsin (final concentration of 2 nm) was used. The final concentration of DM was kept at 0.01%. The samples were irradiated or kept in the dark and then mixed immediately with Gi or Gt solution (final concentration of 600 nm). At selected times, an aliquot (20 μl) was removed and mixed with 200 μl of stop solution (20 mm Tris-Cl (pH 7.4), 100 mm NaCl, 25 mm MgCl2, 1 μm GTPγS, and 4 μm GDP). The mixture was filtered immediately through a nitrocellulose membrane filter. The membrane filter was washed three times with 200 μl of wash solution (20 mm Tris-Cl (pH 7.4), 100 mm NaCl, and 25 mm MgCl2) and air-dried. The amount of bound [35S]GTPγS was quantitated with a liquid scintillation counter. For comparison of G protein activation rates, incubation times were 1 and 2 min for bovine rhodopsin and parapinopsin, respectively. In the case of parapinopsin mutants A269E, A269Q, F208G/A269L, and F208S/A269L (see Figs. 2 and 4), the pigment concentration was 5 nm, and the incubation time was 4 min due to low expression levels. The difference in the activities before and after irradiation was normalized to that of the wild type (WT) under the same conditions. It should be noted that the G protein-activating abilities of amphioxus rhodopsin mutants could not be measured because the active states of the mutants were unstable under the conditions containing detergent DM.
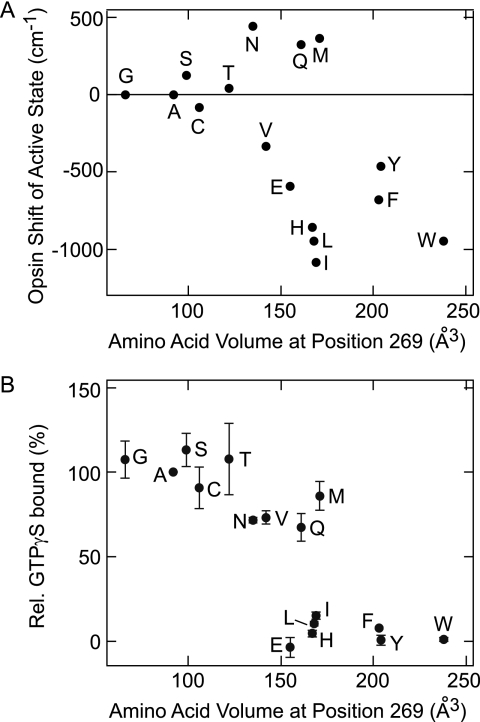
FIGURE 2.
Opsin shift and G protein-activating efficiency of the Ala-269 mutant active states of parapinopsin. A, relationship between the opsin shift in the photoproduct of the Ala-269 mutants and the volume of the amino acid introduced at position 269. B, correlation between the relative G protein activity of the Ala-269 mutants and the volume of the amino acid introduced at position 269. Single letters indicate amino acids introduced at position 269. A indicates WT. Plus and minus values represent red shift and blue shift values, respectively. Note that mutants A269D, A269K, A269P, and A269R were expressed too poorly to be analyzed. Each value of relative (Rel.) GTPγS bound is expressed as the mean ± S.E. of three separate experiments. The λmax values of the mutants are 493 nm (A269G), 496 nm (A269S), 491 nm (A269C), 494 nm (A269T), 485 nm (A269V), 504 nm (A269N), 479 nm (A269E), 501 nm (A269Q), 473 nm (A269H), 496 nm (A269S), 471 nm (A269L), 468 nm (A269I), 502 nm (A269M), 477 nm (A269F), 482 nm (A269Y), and 471 nm (A269W). The amino acid volumes are from Ref. 26.
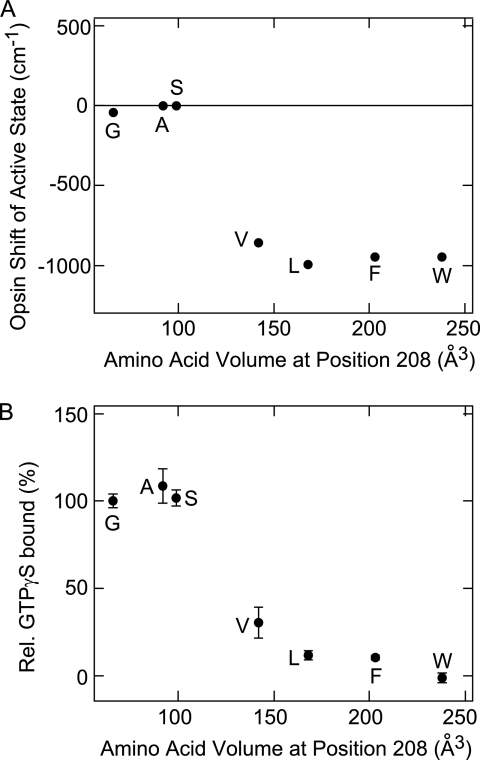
FIGURE 4.
Opsin shift and G protein-activating efficiency of the F208X/A269L double mutant active states of parapinopsin. A, correlation between the opsin shift in the active state of the F208X/A269L mutants and the volume of the amino acid introduced at position 208. B, correlation between the relative G protein activity of the F208X/A269L mutants and the volume of the amino acid introduced at position 269. Single letters indicate amino acids introduced at position 208. F indicates the A269L single mutant. Plus and minus values represent red shift and blue shift values, respectively. Each value of relative (Rel.) GTPγS bound is expressed as the mean ± S.E. of three separate experiments. The λmax values of the mutants are 492 nm (F208G/A269L), 493 nm (F208A/A269L), 493 nm (F208S/A269L), 473 nm (F208V/A269L), 470 nm (F208L/A269L), and 471 nm (F208W/A269L). The amino acid volumes are from Ref. 26.
RESULTS AND DISCUSSION
Identification of the Amino Acid Residue(s) Whose Mutation Causes Spectral Changes in the Active State
Structural analyses of rhodopsin intermediates by x-ray crystallography (7), NMR spectroscopy (8), and photoaffinity labeling (9) strongly suggested that the β-ionone ring of the chromophore changes its position during the activation pathway of rhodopsin. Thus, we focused on the amino acid residues situated near the β-ionone ring regions of antagonist and agonist. There are 10 amino acid residues whose side chains are located within 4.5 Å of the β-ionone ring of the antagonist 11-cis-retinal in the crystal structure of bovine rhodopsin (10). These residues are shown in Fig. 1A. Eight of the 10 residues (except positions 118 and 268) also have side chains located within 4.5 Å of the β-ionone ring in the crystal structure of an invertebrate squid rhodopsin (11). To explore the effect of changing the steric volume around this region, we constructed a series of parapinopsin mutants in which one of the amino acid residues at corresponding positions was replaced with another amino acid to change the volume and/or polarity, expecting that some of the mutations could induce spectral changes in only the agonist-binding states. We also prepared a similar mutant series of another agonist-binding rhodopsin, amphioxus rhodopsin.
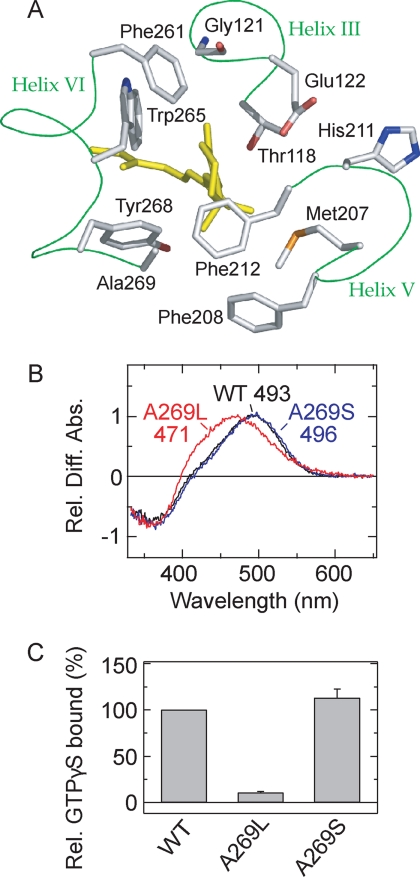
FIGURE 1.
Molecular properties of the active state of the parapinopsin Ala-269 mutant. A, 11-cis-retinal and amino acid residues around the β-ionone ring in the crystal structure of bovine rhodopsin (Protein Data Bank code 1U19) (10). 11-cis-Retinal (yellow) and amino acid residues within 4.5 Å of the β-ionone ring are shown. Phe-208 is also shown. Note that the backbone of the amino acid residues except Gly-121 is omitted. B, difference spectra of WT parapinopsin (black) and mutants A269L (red) and A269S (blue) in the membrane preparations. All difference absorbances at λmax of the active states were normalized to 1.0. The mutations introduced and the λmax values from the difference spectrum are also shown. Rel. Diff. Abs., relative difference absorbance. C, Gi activation rates of WT parapinopsin and mutants A269L and A269S. Each sample was purified with DM, and experiments were performed at 0 °C. Data are expressed as means ± S.E. of three separate experiments. Figs. 1A, 3A, 3B, and 6 were prepared using PyMOL.
The WT parapinopsin exhibits absorption maxima at 363 and 493 nm in the dark resting state and the active state, respectively (5). The difference spectrum calculated by subtracting the spectrum of the resting state from that of the active state shows positive and negative peaks that reflect the peaks of active and resting states, respectively (Fig. 1B). Among the 20 constructed mutants of parapinopsin, 16 mutants formed pigments when incubated with 11-cis-retinal. Thus, we measured spectral shifts in these mutants by irradiation with UV light, and the spectral data are summarized in supplemental Table S1. We also obtained spectral data for 20 amphioxus rhodopsin mutants by irradiation with 501-nm light (summarized in supplemental Table S1). It should be noted that the 20 constructed mutants of amphioxus rhodopsin all formed pigments.
Among the parapinopsin mutants, the A269L mutant showed an active-state spectrum considerably different from that of WT, whereas its resting-state spectrum was identical to that of WT (Fig. 1B). In contrast, the A269S mutant exhibited little change in the resting- and active-state spectra (Fig. 1B). These results suggest that introduction of a bulky (but not polar) amino acid residue at position 269 induces a specific interaction with the agonist all-trans-retinal. Similar changes in the spectroscopic properties were also observed in the Ala-269 mutants of amphioxus rhodopsin (supplemental Fig. S1). The A269L mutant of amphioxus rhodopsin exhibited an active-state spectrum largely perturbed from that of the original pigment, suggesting that introduction of the bulky group at this position caused a severe steric interaction with the chromophore. It is unclear why the A269L mutation caused a more prominent effect on the absorption spectrum of amphioxus rhodopsin compared with parapinopsin. Interestingly, the A269L mutant of parapinopsin exhibited almost no G protein activity, but the A269S mutant exhibited G protein activity similar to that of WT (Fig. 1C), indicating that introduction of the bulky amino acid at this position caused a great reduction in G protein activity.
To further investigate the mode of this interaction, we prepared a series of parapinopsin mutants in which Ala-269 was replaced with one of 19 amino acids, including leucine and serine. Replacements of Ala-269 with amino acids larger than 200 Å3 caused a large blue shift in the absorption spectra of the active states, whereas those with amino acids smaller than 130 Å3 did not alter the spectra significantly (Fig. 2A). Except for the replacements with glutamine, asparagine, and methionine, replacements with amino acids having volumes between 130 and 200 Å3 tended to the blue shift as the volume of the amino acid became large (Fig. 2A).6 Interestingly, replacement of Ala-269 with bulky amino acids, which caused a more than ∼500 cm−1 blue shift in the active-state spectra, also caused a severe reduction in G protein-activating efficiencies (Fig. 2B). Thus, introduction of bulky amino acid residue at position 269 tends to cause both a spectral shift in the active state and a severe reduction in G protein-activating ability.
Deletion of the methyl group by replacement of alanine with glycine at this position caused no change in the absorption spectrum and G protein-activating efficiency (Fig. 2, A and B). In addition, replacement with serine or cysteine, whose volume is similar to that of alanine, did not change the spectrum and G protein-activating efficiency (Fig. 2, A and B). Therefore, we propose that the above results can be accounted for as follows. As a result of replacement of Ala-269 with a bulky amino acid, a steric interaction between a newly introduced bulky amino acid and the residue that originally interacted with Ala-269 occurs, thereby resulting in a specific interaction of the bulky amino acid with the agonist, which causes an inadequate protein structure for G protein activation. This explanation is supported by the fact that spectral shifts caused by some mutations of amino acid residues proximal to Ala-269, such as Y268F and W265F, caused small but significant spectral shifts in the active states of parapinopsin and amphioxus rhodopsin (supplemental Table S1). These results support our idea that the agonist all-trans-retinal is located near Ala-269 and the proximal residues. We next set out to identify residue(s) that originally interacted with Ala-269 in the inactive “dark” state of the agonist-binding receptor parapinopsin.
Identification of the Amino Acid Residue That Pairs with Ala-269
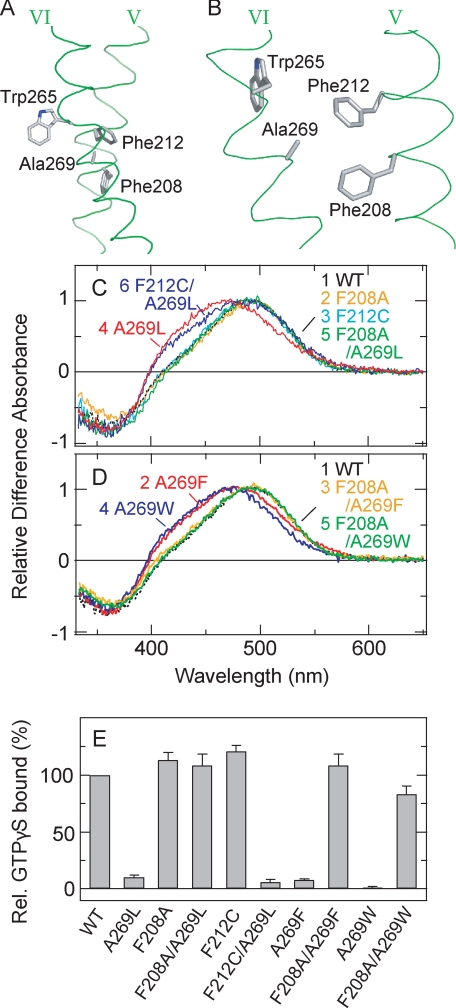
To obtain insight into the residue(s) that interacts with Ala-269 in parapinopsin, we searched for the amino acid residues located within 4.5 Å of Ala-269 in the crystal structure of bovine rhodopsin (10) and found that, in addition to the residues located near Ala-269 in helix VI, two residues, Phe-208 and Phe-212, present in helix V fit this criterion (Fig. 3, A and B). We also searched for the amino acid residues in the crystal structure of squid rhodopsin (11) and found that Phe-205, the corresponding residue of Phe-208 in bovine rhodopsin, also fits the criterion. Because both Phe-208 and Phe-212 are residues with bulky side chains, we expected that steric interaction should occur between one of these residues and a bulky amino acid introduced at position 269 instead of alanine and that this interaction might cause perturbation of the helical arrangements of helices V and VI that are essential for the formation of the G protein-activating state (12–14). Therefore, we tested whether the effect of replacing Ala-269 with a bulky amino acid would be compensated by replacing either of these phenylalanine residues with a smaller amino acid. Because mutant F212A showed a severe reduction in expression levels, we focused our studies instead on mutant F212C/A296L.
FIGURE 3.
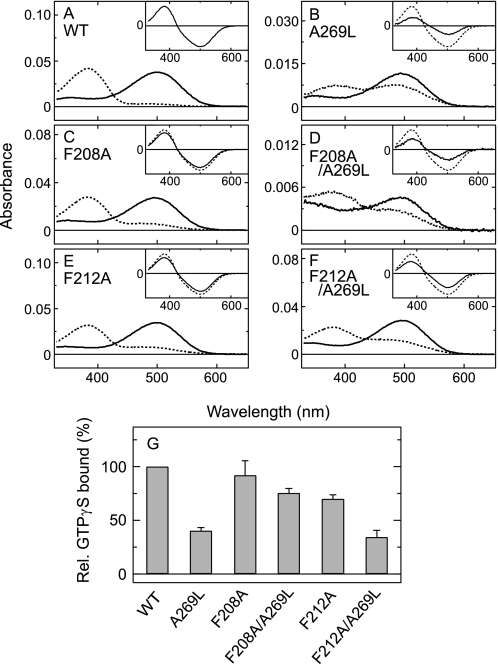
Effects of a second site replacement at positions 208 and 212 on the parapinopsin Ala-269 mutants. A and B, amino acid residues at positions 208, 212, and 269 in the crystal structure of the dark state of bovine rhodopsin (Protein Data Bank code 1U19) (10). Trp-265, which is highly conserved among rhodopsins and GPCRs, is also indicated. C, difference spectra of WT parapinopsin (curve 1) and mutants F208A (curve 2), F212C (curve 3), A269L (curve 4), F208A/A269L (curve 5), and F212C/A269L (curve 6) in the membrane preparations. D, difference spectra of WT parapinopsin (curve 1) and mutants A269F (curve 2), F208A/A269F (curve 3), A269W (curve 4), and F208A/A269W (curve 5) in the membrane preparations. In the C and D, all the difference absorbances at λmax of the active states were normalized to 1.0. Note that the WT and A269L mutant spectra in C and D are the same as those in Fig. 1B. E, comparison of the Gi activation rates of WT parapinopsin and mutants A269L, F208A, F208A/A269L, F212C, F212C/A269L, A269F, F208A/A269F, A269W, and F208A/A269W. Each sample was solubilized with DM, and experiments were performed at 0 °C. The difference in the activities before and after irradiation is normalized to that of WT. Data are expressed as means ± S.E. of three separate experiments. Rel., relative.
Replacement of Phe-208 with alanine in the A269L, A269F, and A269W mutants returned the absorption spectra and G protein-activating efficiencies back to those of WT (Fig. 3, C–E). A similar recovery of the absorption spectra was also observed in amphioxus rhodopsin (supplemental Fig. S2). In contrast, replacement of Phe-212 with cysteine did not cause such a recovery of the phenotype of the A269L mutant to that of WT (Fig. 3, C and E). These results strongly suggest that Ala-269 is located near Phe-208 in the active state and that replacement of Ala-269 with a bulky amino acid causes a steric interaction with Phe-208, resulting in inhibition of the formation of structure suitable for G protein activation. It should be noted that all of these replacements caused little spectral changes in the dark resting state (Fig. 3, C and D, and supplemental Fig. S3).
We then tried to establish whether the recovery of the active-state characteristics by the second F208A mutation was due to the decreasing volume of the amino acid residue at position 208. Our results show that introducing a small amino acid (Gly or Ser), but not a bulky amino acid (Val, Leu, or Trp), at position 208 recovered the spectral property of the active state of the A269L mutant and its G protein-activating efficiency (Fig. 4, A and B). These results further support the idea that the absence of steric interaction in the region near these residues is required for active-state formation.
Characterization of the Amino Acid Pair in Bovine Rhodopsin
We next examined whether or not similar mutational effects could be observed in bovine rhodopsin. Replacement of Ala-269 with leucine caused a great shift in equilibrium between meta-I and meta-II in favor of meta-I. However, the absorption spectra of meta-I and meta-II did not change significantly upon this replacement, i.e. meta-I showed only an ∼4-nm blue shift, and meta-II did not show the shift at all (Fig. 5B and supplemental Fig. S4B). Subsequent replacement of Phe-208 with alanine in the A269L mutant caused a slight shift in equilibrium in favor of meta-II, but the spectra of meta-I and meta-II did not change (Fig. 5D and supplemental Fig. S4B). Replacement of Phe-212 with alanine also caused a shift in equilibrium similar to that caused by replacement of Phe-208 (Fig. 5F and supplemental Fig. S4B). It should be noted that the single replacement of Phe-208 or Phe-212 did not change the equilibrium and spectra of meta-I and meta-II (Fig. 5, C and E, and supplemental Fig. S4A).
FIGURE 5.
Absorption spectra and G protein-activating abilities of the A269L mutant of bovine rhodopsin and the effects of a second site mutation of F208A or F212A. Shown are the absorption spectra before (solid curves) and after (dotted curves) irradiation of bovine WT rhodopsin (A) and mutants A269L (B), F208A (C), F208A/A269L (D), F212A (E), and F212A/A269L (F). Insets, difference absorption spectra of the mutants normalized by the amount of rhodopsin. Solid and dotted curves indicate mutant and WT, respectively. G, Gt activation rates of bovine WT rhodopsin and mutants A269L, F208A, F208A/A269L, F212A, and F212A/A269L. Each sample was purified with DM, and experiments were performed at 20 °C. Data are expressed as means ± S.E. of three separate experiments. Rel., relative.
In contrast to the little change in the absorption spectra, the G protein-activating efficiencies were considerably perturbed by the replacements, i.e. the A269L mutant of bovine rhodopsin showed G protein-activating efficiency that was ∼40% of WT (Fig. 5G). The subsequent F208A mutation recovered the efficiency to ∼80% of WT (Fig. 5G). On the other hand, the F212A mutation caused no change in the G protein-activating efficiency of the A269L mutant (Fig. 5G). These results indicate that adequate interaction between the amino acid pair at positions 208 and 269 is also important for G protein activation by bovine rhodopsin. It should be noted that the recovery of G protein-activating efficiency by the subsequent F208A mutation was not due to the recovery of the equilibrium between meta-I and meta-II because the F212A mutation of the A269L mutant also recovered the equilibrium but did not recover the G protein-activating efficiency (Fig. 5, F and G).
The above results clearly show that the introduction of the Ala-269 mutation into bovine rhodopsin caused no spectral change in the active state, whereas the same mutation in parapinopsin caused large spectral changes. However, G protein-activating efficiencies were disturbed by the mutations in both bovine rhodopsin and parapinopsin. In the basis of these results, we propose that, in the active state of bovine rhodopsin, the β-ionone ring of the chromophore is present in a position where it does not interact with the bulky amino acid introduced at position 269, whereas in the active state of parapinopsin, the β-ionone ring is in close proximity to position 269 so as to interact with the bulky amino acid introduced at this position. Separation of the β-ionone ring of the chromophore from position 269 in bovine rhodopsin upon absorption of a photon is consistent with the results of x-ray crystallography of lumirhodopsin (7) and NMR studies on metarhodopsin II (15). The difference in movement of the β-ionone ring of the chromophore from the pivot constituted by the specific pair of amino acids at positions 269 and 208 would cause a different amplitude of helical movement between parapinopsin and bovine rhodopsin (16). The differences in the conformational changes in chromophore and protein would be relevant to functional differences in G protein-activating efficiency and direct binding of the agonist all-trans-retinal.
Conservation of the Amino Acid Pair in Various Rhodopsins
In this study, we found that a “push-pull” relationship between amino acid residues at positions 269 and 208 (with the existence of a bulky amino acid at either of the positions) is important for the formation of the G protein-activating state. Thus, it is of interest to determine whether or not a similar relationship is conserved in various rhodopsins. Most rhodopsins possess bulky and small amino acids at positions 208 and 269, respectively (supplemental Fig. S5). On the other hand, the relationship is reversed in some Gq-coupled invertebrate rhodopsins, where small and bulky amino acid residues are present at positions 208 and 269, respectively (supplemental Fig. S5). Some rhodopsins, such as Xenopus melanopsin, Drosophila RH6, and Anopheles GPRop8, contain small amino acids at both positions (supplemental Fig. S5). We confirmed by use of parapinopsin that replacement of the bulky phenylalanine at position 208 with the small alanine caused no change in the spectrum and G protein-activating efficiency (Fig. 3, C and E). Therefore, we conclude that the important restriction is that bulky amino acids must not exist at both positions.
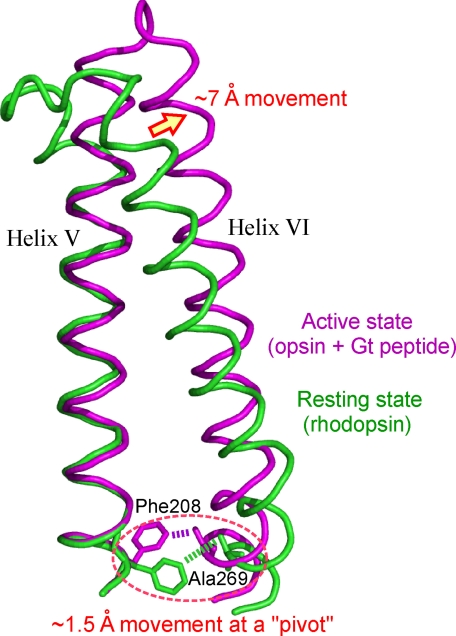
According to the crystal structures of bovine rhodopsin and its partial active state (opsin with a C-terminal peptide of the transducin α-subunit), the Cα–Cα distance between Ala-269 and Phe-208 changes ∼1.5 Å upon conversion to the partial active state (10, 17, 18). In contrast, the cytoplasmic region of helix VI would move away from the original position by ∼7 Å (10, 17, 18). Outward displacement of the cytoplasmic region of helix VI was also suggested from site-directed spin labeling studies (12, 13). These facts, coupled with our new results, enable us to speculate that the amino acid pair at positions 269 and 208 would work as a pivot to induce the outward shift in the cytoplasmic region of helix VI (Fig. 6), whereas its extracellular region does not move significantly (18, 19).
FIGURE 6.
Summary depicting how the interaction between residues at positions 208 and 269 serves as a pivot between helices V and VI based on the available crystal structures. The crystal structures of bovine rhodopsin (green; Protein Data Bank code 1GZM) and bovine opsin with a peptide derived from the C terminus of Gt (magenta; code 3DQB) are compared and superimposed. In the opsin structure, the cytoplasmic end of helix VI moved ∼7 Å compared with the rhodopsin structure. In contrast, the change in the Cα–Cα distance between Phe-208 and Ala-269 is only ∼1.5 Å.
Amino Acid Residues at Positions 208 and 269 in Other GPCRs
The amino acid residues that are located at the corresponding positions in rhodopsin also have important roles in other GPCRs. The β-adrenergic receptor contains important and highly conserved serine and phenylalanine residues at positions 208 and 269, respectively (Ser-204 and Phe-290 in the β-adrenergic receptor numbering system, respectively). Both residues were reported to interact directly with agonist, such as isoproterenol, and the interactions have been reported to lead to formation of the G protein-activating state (20, 21). It was also reported that, in the 5-HT2A (serotonin) receptor, the agonist 5-hydroxytryptamine interacts with amino acid residues at positions 208 and 269 (22, 23). Thus, it is reasonable to speculate that, in not only the agonist-binding rhodopsins but also various ligand-binding GPCRs, the amino acid residues at positions 208 and 269 are situated near the agonists and act as a pivot for the helical arrangements. The crystal structures of several GPCRs other than rhodopsin have recently been determined, but all the structures so far contain antagonists (inverse agonists) (24). Thus, further biochemical analyses in combination with structural determinations of the agonist-binding states will be necessary for the confirmation of the functional role of the specific pair found in this study.
In conclusion, our results provide novel insights into the amino acid residues responsible for regulation of rhodopsin activity. In addition, we speculate that the agonist-binding site of vertebrate rhodopsin changes its position from the pivot at positions 208 and 269. We previously proposed based on studies on the displacement of the counterion that vertebrate visual rhodopsins diverged from the agonist-binding rhodopsins (2, 5). Further analyses of the molecular mechanisms of the pivot will clarify the emergence of vertebrate visual rhodopsins in the course of molecular evolution of rhodopsins.
Supplementary Material
Acknowledgment
We thank Dr. David Farrens (Oregon Health & Science University) for critical reading of the manuscript.
This work was supported in part by grants-in-aid for scientific research from the Japanese Ministry of Education, Science, Sports, and Culture (to Y. S. and A. T.) and by the Yamada Science Foundation (to A. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Table S1.
Parapinopsins have been found as one of the non-visual opsins in the vertebrate opsin subfamily in the photosensitive pineal and parapineal organs of jawless fish, teleost fish, and amphibians (4, 25).
It is unclear how the replacements with glutamine, asparagine, and methionine caused a red shift in the active states of parapinopsin. In general, an opsin shift is the reflection of the electronic state of the chromophore, which would be affected by the electrostatic environment around the chromophore. Because the residues that induce the red shift from the WT have side chains containing atoms with electron-withdrawing properties, the presence of these residues would cause some effect on the electronic state of the chromophore, resulting in the red shift in the absorption maximum.
- GPCR
- G protein-coupled receptor
- DM
- n-dodecyl β-d-maltoside
- meta-I
- metarhodopsin I
- meta-II
- metarhodopsin II
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- WT
- wild type.
REFERENCES
- 1.Jäger S., Palczewski K., Hofmann K. P. (1996) Biochemistry 35, 2901–2908 [DOI] [PubMed] [Google Scholar]
- 2.Tsukamoto H., Terakita A., Shichida Y. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6303–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koyanagi M., Terakita A., Kubokawa K., Shichida Y. (2002) FEBS Lett. 531, 525–528 [DOI] [PubMed] [Google Scholar]
- 4.Koyanagi M., Kawano E., Kinugawa Y., Oishi T., Shichida Y., Tamotsu S., Terakita A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6687–6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terakita A., Koyanagi M., Tsukamoto H., Yamashita T., Miyata T., Shichida Y. (2004) Nat. Struct. Mol. Biol. 11, 284–289 [DOI] [PubMed] [Google Scholar]
- 6.Shichida Y., Imai H. (1998) Cell. Mol. Life Sci. 54, 1299–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamichi H., Okada T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12729–12734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel A. B., Crocker E., Eilers M., Hirshfeld A., Sheves M., Smith S. O. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10048–10053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borhan B., Souto M. L., Imai H., Shichida Y., Nakanishi K. (2000) Science 288, 2209–2212 [DOI] [PubMed] [Google Scholar]
- 10.Okada T., Sugihara M., Bondar A. N., Elstner M., Entel P., Buss V. (2004) J. Mol. Biol. 342, 571–583 [DOI] [PubMed] [Google Scholar]
- 11.Murakami M., Kouyama T. (2008) Nature 453, 363–367 [DOI] [PubMed] [Google Scholar]
- 12.Farrens D. L., Altenbach C., Yang K., Hubbell W. L., Khorana H. G. (1996) Science 274, 768–770 [DOI] [PubMed] [Google Scholar]
- 13.Altenbach C., Kusnetzow A. K., Ernst O. P., Hofmann K. P., Hubbell W. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7439–7444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobilka B. K. (2007) Biochim. Biophys. Acta 1768, 794–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahuja S., Crocker E., Eilers M., Hornak V., Hirshfeld A., Ziliox M., Syrett N., Reeves P. J., Khorana H. G., Sheves M., Smith S. O. (2009) J. Biol. Chem. 284, 10190–10201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukamoto H., Farrens D. L., Koyanagi M., Terakita A. (2009) J. Biol. Chem. 284, 20676–20683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J., Edwards P. C., Burghammer M., Villa C., Schertler G. F. (2004) J. Mol. Biol. 343, 1409–1438 [DOI] [PubMed] [Google Scholar]
- 18.Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., Hofmann K. P., Ernst O. P. (2008) Nature 455, 497–502 [DOI] [PubMed] [Google Scholar]
- 19.Struthers M., Yu H., Kono M., Oprian D. D. (1999) Biochemistry 38, 6597–6603 [DOI] [PubMed] [Google Scholar]
- 20.Strader C. D., Candelore M. R., Hill W. S., Sigal I. S., Dixon R. A. (1989) J. Biol. Chem. 264, 13572–13578 [PubMed] [Google Scholar]
- 21.Dixon R. A., Sigal I. S., Strader C. D. (1988) Cold Spring Harbor Symp. Quant. Biol. 53, 487–497 [DOI] [PubMed] [Google Scholar]
- 22.Roth B. L., Shoham M., Choudhary M. S., Khan N. (1997) Mol. Pharmacol. 52, 259–266 [DOI] [PubMed] [Google Scholar]
- 23.Johnson M. P., Wainscott D. B., Lucaites V. L., Baez M., Nelson D. L. (1997) Brain Res. Mol. Brain Res. 49, 1–6 [DOI] [PubMed] [Google Scholar]
- 24.Hanson M. A., Stevens R. C. (2009) Structure 17, 8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blackshaw S., Snyder S. H. (1997) J. Neurosci. 17, 8083–8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chothia C. (1984) Annu. Rev. Biochem. 53, 537–572 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.