Abstract
Steroidogenic acute regulatory protein-related lipid transfer (START) domains, found in 15 mammalian proteins termed StarD1–StarD15, are lipid-binding domains implicated in the intracellular lipid transport systems. In the present study, we analyzed the lipid ligand and function of StarD7. We found two variable forms of mammalian StarD7, termed StarD7-I and StarD7-II. Unlike StarD7-II, StarD7-I contained a mitochondrial-targeting sequence in its N terminus. Overexpressed StarD7-I tagged with V5/His in HEPA-1 cells was mainly observed in the mitochondria of cells prepared at low cellular density, but it was distributed in the cytoplasm of high density cells. StarD7-II was constantly distributed in the cytoplasm at any cellular density. Endogenous StarD7 in HEPA-1 cells and rat liver was also distributed in both the cytoplasm and the mitochondria. A protease K protection assay indicated that the mitochondrial StarD7 was associated with the outer mitochondrial membrane. The purified recombinant StarD7 specifically catalyzed the transfer of PC between lipid vesicles in vitro. Furthermore, the intracellular transport of fluorescent PC that was exogenously incorporated into the mitochondria was increased in cells that overexpressed StarD7-I. These results suggest that StarD7 facilitates the delivery of PC to mitochondria in non-vesicular system.
Keywords: Cell, Lipid/Phospholipid, Lipid/Phosphatidylcholine, Lipid/Transport, Membrane/Trafficking, Transport/Mitochondria
Introduction
The phospholipid components are variable, specialized, and important for the organization of lipid bilayers of plasma membranes and other cellular organelles. Phospholipid biosynthesis occurs in limited organelles such as the endoplasmic reticulum (ER),2 Golgi complex, and mitochondria (1). Therefore, the selective transport of newly synthesized lipids to their appropriate destinations is essential for the maintenance of functional membranes. The transport of budding vesicles from a donor compartment to an acceptor compartment is one of the key processes for lipid transport (2). Lipids can also be delivered and exchanged via several cytosolic proteins in a monomeric manner between the cytosolic membrane surfaces of different organelles. This exchange system performed by carrier proteins requires binding of the lipid from the donor membrane, passage through the cytoplasm, and subsequent insertion into the acceptor membrane. Cytosolic proteins containing specific lipid-binding domains that are capable of accelerating lipid exchange in vitro have been identified; these proteins include glycolipid transfer proteins (3), ceramide transport protein (CERT) (4), and members of the steroidogenic acute regulatory protein-related lipid transfer (START) domain family.
START domains, which contain ∼210 amino acid residues, bind to specific lipids, including phospholipids, sterols, and sphingolipids (5). In mammals, START domains are found in 15 distinct proteins, StarD1–StarD15, which can be classified into six families. As shown in the phylogenetic tree (Fig. 1A), StarD2/phosphatidylcholine (PC) transfer protein (PC-TP), StarD10, StarD11/ceramide transport protein, and StarD7/gestational trophoblastic tumor are recognized as similar (6). StarD2/PC-TP is a cytosolic protein that can specifically bind and transfer PC between membranes (7). StarD10, originally found as an overexpressed protein in breast cancer cells, is a PC and phosphatidylethanolamine (PE) transfer protein (8). StarD11/ceramide transport protein can transfer ceramide from ER to Golgi membranes (4). The binding of some phospholipids to StarD7 has been reported; however, the identification of the specific lipids that bind to StarD7 and the biological functions of StarD7 are not well understood.
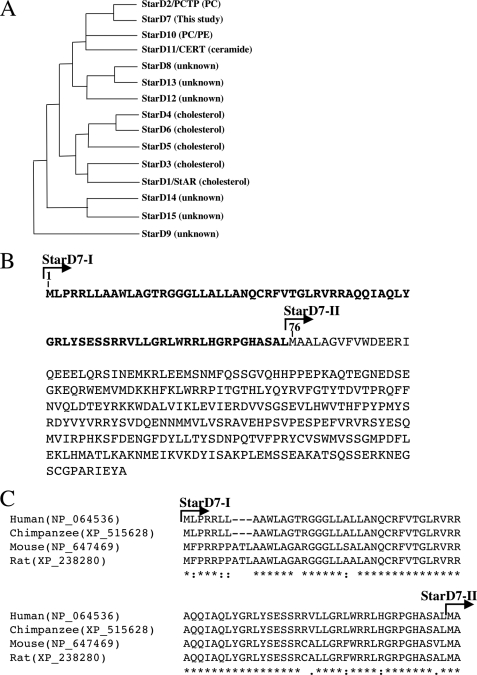
FIGURE 1.
Phylogenetic analysis of StarD families and mitochondrial-targeting sequences of StarD7-I. A, phylogenetic analysis and lipid ligands of StarD families. B, amino acid sequences of StarD7-I. The putative mitochondrial-targeting signal is indicated by boldface. The first Met of StarD7-II corresponds to Met76. C, mitochondria-targeting sequences at the N terminus of mammalian StarD7-I are aligned by using ClustalW (27). Identical and chemically similar amino acids are indicated by asterisks and dots, respectively. Gaps inserted into the sequences are indicated by dashed lines.
Mitochondria must import PC, the major constituent of both their inner and outer membranes, because they do not contain the sequential enzymes needed for PC production. Mitochondria do contain the enzymes needed to produce phosphatidylglycerol, cardiolipin, and PE (9). Soluble carrier proteins and direct contact zones between mitochondria and ER, called mitochondria-associated membranes (MAMs) (1), have been appear to be important for the efficient supply of PC from the ER or Golgi complex to mitochondria. However, the specific molecules responsible for the transport of PC to mitochondria have not been well characterized. In the present study, we show that StarD7 can exchange PC between vesicles in vitro. We also demonstrate that the intracellular transport of exogenously incorporated PC into the mitochondria is increased in cells that overexpressed StarD7. These findings suggest that StarD7 is involved in the intracellular transfer of PC to mitochondria in vivo.
EXPERIMENTAL PROCEDURES
Materials
A cDNA clone containing full-length human StarD7-I (Mammalian Gene Collection 16334), 1-palmitoyl-2-[12-{(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-amino}dodecanoyl]- sn-glycero-3-phosphocholine (C12-NBD-PC), 1-palmitoyl-2-[6- {(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-amino}hexanoyl]-sn-glycero-3-phosphocholine (C6-NBD-PC), lissamine rhodamine B-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt (rh-PE), anti-porin, anti-Complex Vα, and anti-Core 1 antibodies were purchased from Invitrogen. The Stealth siRNA, a 25-bp duplex oligoribonucleotide with a sense strand corresponding to nucleotides 1097–1121 of the mouse StarD7 mRNA (5′-GCCCUGCUCGGAUUGAGUAUGCUUA-3′) was also obtained from Invitrogen. We purchased 1-palmitoyl-2-[12-{(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino}dodecanoyl]-sn-glycero-3-phospho-l-serine (C12-NBD-PS), 1-myristoyl-2-[12-{(7-nitrobenz-2-oxa-1,3-diazol- 4-yl)amino}dodecanoyl]-sn-glycero-3-phosphoethanolamine (C12-NBD-PE), 1-stearoyl-2-oleoyl-sn-glycero-3-phosphocholine (C18:0–18:1 PC), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (C16:0–18:1 PC) from Avanti Polar Lipid. Egg yolk phosphatidic acid, N-[12-{(7-nitro-2–1,3-benzoxadiazol-4-yl)amino}dodecanoyl]sphingosine-1-phosphocholine (C12-NBD-SM), and anti-GM130 antibody were purchased from Sigma. Anti-GAPDH (glyceraldehydes-3-phosphate dehydrogenase) antibody was purchased from Abcam, and pre-coated silica gel 60 TLC plates were purchased from Merck.
Cell Culture, Expression, and Immunocytochemistry
The cDNA fragments containing human StarD7-I and -II were amplified by PCR (Platinum DNA polymerase, Invitrogen) with Mammalian Gene Collection 16334 as a template. The amplified fragments were cloned into pcDNA3.1/V5-His (Invitrogen). The expression vector for Green fluorescent protein (GFP) fused with the mitochondrial targeting sequence in the N terminus of StarD7-I was constructed by inserting the DNA sequence coding Met1–Leu75 into pEGFP-N3 (Novagen). HEPA-1 cells, a mouse hepatoma cell line, were in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum in a humidified incubator containing 5% CO2. Cells were transfected with the expression vectors by using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Twenty-four hours after transfection, cells were treated with 250 nm MitoTracker Red CMXRos and incubated at 37 °C for 30 min. Then, the cells were fixed with 4% paraformaldehyde in PBS for 15 min, washed with PBS, and permeabilized by 0.1% Triton X-100 in PBS for 5 min at room temperature. After treatment with blocking buffer (5% skim milk in PBS) for 15 min, the samples were incubated with anti-V5 antibody (Invitrogen) in blocking buffer at 4 °C for 16 h followed by anti-mouse IgG conjugated with Alexa 488 (Invitrogen) for 1 h. All confocal images were taken with a laser-scanning confocal microscope FV500 (Olympus).
Antibody Preparation and Western Blot Analysis
A rabbit (New Zealand White strain) was injected 5 times with 0.2 mg of the purified StarD7-I protein. HiTrap N-hydroxysuccinimide-activated HP Columns (Amersham Biosciences) covalently conjugated with the antigen were used to purify the specific IgGs after antiserum antibodies were precipitated with ammonium sulfate at 50%. To perform Western blot analysis, proteins separated by SDS-PAGE were transferred to nitrocellulose membranes by using a semi-dry blotter (Nihon Eido Co. Ltd.), and the membranes were incubated with 1% (w/v) skim milk in TBS for 1 h and washed three times with T-TBS (TBS containing 0.02% Tween 20). Then, the membranes were incubated with the purified antibodies for 12 h at 4 °C, washed three times with T-TBS, and incubated with horseradish peroxidase-conjugated anti-Rabbit IgGs for 1 h at room temperature. The membranes were washed three times with T-TBS and stained with a SuperSignal West Pico Substrate peroxidase staining kit (Pierce), according to the manufacturer's instructions.
Isolation of Mitochondria and Protease K Protection Assays
Mitochondria and cytosolic fractions were freshly prepared from HEPA-1 cells by using a Mitochondria Isolation kit (Pierce), according to the manufacturer's instructions. To isolate mitochondria from rat livers, tissues were homogenized with a motor-driven Potter homogenizer by 7 strokes in buffer A (20 mm Tris-HCl buffer, pH 8.0, 250 mm sucrose). After centrifugation at 500 × g for 10 min, crude mitochondria in the homogenates were pelleted at 8,000× g for 10 min. The pellets were resuspended in buffer A and applied to a discontinuous sucrose gradient with 1, 1.3, 1.5, and 1.6 m sucrose and centrifuged at 80,000× g for 1 h. The mitochondria-rich bands were collected and washed once with buffer A. Cytosolic fractions were prepared by ultracentrifugation of the post-mitochondrial supernatants for 1 h at 100,000× g. For protease K protection assays, mitochondria from HEPA-1 cells were incubated with 280 μg/ml protease K (Invitrogen) for 30 min on ice with or without 1% Triton X-100. The reactions were terminated by the addition of 1 mm phenylmethylsulfonyl fluoride.
Pulse-Chase Experiments
HEPA-1 cells transfected with StarD7-I cloned in the pCAG vectors (10) were incubated with 50 μm of carbonylcyanide-m-chlorophenylhydrazone (CCCP) for 30 min and then metabolically labeled with [35S]methionine and [35S]cysteine (Expre35S35S Protein Labeling Mix, PerkinElmer) at a concentration of 30 μCi/ml for 20 min in methionine and cysteine-free DMEM (Sigma) containing CCCP. Then, cells were incubated in isotope-free medium for another 3 h. Cell lysates solved in 20 mm Tris-HCl, pH 8.0, 0.5 m NaCl, 1% Nonidet P-40 were prepared and subjected to immunoprecipitation with anti-StarD7 antibody, and the resulting precipitates were separated by SDS-PAGE, subjected to autoradiography, and analyzed with a BAS-2000 bioimaging analyzer (Fujifilm).
Expression and Purification of Recombinant StarD7
StarD7-I (human) and -II (rat) were cloned into a bacterial expression vector, pET23a (Novagen), containing 6× His tags for the C terminus of the expressed proteins. In the expression vector for His-tagged StarD7-I, a 6× glycine linker sequence was inserted upstream of the His tag to increase the column-binding efficiency of the expressed protein. The constructs were transformed into Escherichia coli strain BL21(DE3)LysS (Novagen). The transformed cells were grown at 37 °C in 200 ml of Luria-Bertani medium with 100 μg/ml ampicillin to an optical density of 0.5 (A600), and then 0.1 mm isopropyl-β-d-thiogalactopyranoside was added to enhance the protein expression. After another 2 h of incubation at 37 °C, cells were harvested and then suspended and sonicated in 10 ml of 50 mm Tris-HCl buffer (pH 8.0) containing 0.5 mm phenylmethylsulfonyl fluoride. After centrifugation at 15,000 × g × 20 min, the supernatants were applied to a Ni-Sepharose 6 Fast Flow column (Amersham Biosciences), and the expressed proteins were eluted with 20 mm Tris-HCl buffer (pH 8.0) containing 500 mm imidazole. To purify the StarD7-I protein expressed in E. coli, the inclusion bodies were isolated and then dissolved in 8 m urea. Then, the protein solution was dialyzed against buffer consisting of 20 mm Tris-HCl (pH 8.0) and 150 mm NaCl to promote protein refolding prior to purification procedures. The open reading frame was also cloned into a wheat germ expression vector, pEU (CellFree Sciences), and the proteins were expressed with a Wheat Germ Expression H Kit (CellFree Sciences), according to the manufacturer's instructions, to obtain StarD7-I protein in a cell-free system.
Phospholipid Extraction Assay
HEPA-1 cells were grown in DMEM containing 0.5% fetal bovine serum and [14C]palmitic acid (0.5 μCi/ml, American Radiolabeled Chemicals, Inc.) for 16 h. The cells were washed with PBS, and then total cellular lipids were extracted by using the Bligh and Dyer method (11). The extracted lipids were used to prepare lipid vesicles by sonication in buffer B (20 mm Tris-HCl buffer, pH 8.0, 150 mm NaCl). The vesicles containing radiolabeled total cellular lipids were incubated with purified StarD7 (100 μg/300 nmol of phospholipid) for 1 h at 37 °C and then applied to Centricon 100-kDa cutoff filters (Millipore). After centrifugation at 3000 × g for 30 min, the protein-lipid complexes were passed through the filters and obtained in the filtrates. The radioactive lipids in the filtrates were separated by TLC with chloroform, methanol, and water (65:25:4, v/v) and analyzed with BAS-2000.
Phospholipid Transfer Assay
The lipid transfer activity was calculated with a fluorescence technique based on a resonance energy transfer mechanism with some modification (12). Briefly, donor phospholipid vesicles containing 1 mol % C12-NBD-PC, 5 mol % rh-PE, and 94 mol % C18:0–18:1 PC (500 μm of total phospholipids) were prepared by sonication in buffer B. Fluorescence (530 nm) of C12-NBD-PC in the donor vesicles excited by 464 nm was quenched by rh-PE. Acceptor vesicles were prepared by sonication of 95 mol % C18:0–18:1 PC and 5 mol % egg yolk phosphatidic acid (500 μm of total phospholipids) in buffer B. The reaction mixture containing 30 μl of acceptor vesicles, 15 μl of donor vesicles, and 15 μl of the purified StarD7-I or -II protein in 200 μl of buffer B was put into a cuvette and immediately monitored by a Hitachi F-4010 fluorescence spectrophotometer (excitation, 464 nm; emission, 530 nm) at room temperature for 5 min. The maximum intensity was obtained by the addition of Triton X-100 at 0.7% concentration.
Intracellular Trafficking of Fluorescent PC Analog in Living Cells
Trafficking of fluorescent PC analog in living cells was investigated as described previously with slight modifications (13). Lipid vesicles containing 40 mol % C6-NBD-PC and 60 mol % C16:0–18:1 PC (50 μm of total phospholipids) were prepared by sonication in serum-free DMEM. Cells were washed with cold DMEM without fetal bovine serum and incubated on ice in DMEM containing the lipid vesicles for 30 min. Then, cells were washed and incubated at 37 °C in serum-free DMEM containing 6 nm MitoTracker Red CMXRos for 30 min. For confocal microscopy, cells were fixed with 4% paraformaldehyde in PBS.
RESULTS
Identification of Mitochondrial-targeting Signal in StarD7
StarD7, also named GTT1 (GenBankTM accession number AF270647), was originally identified by Durand et al. as a gene highly expressed in gestational trophoblastic tumor (6). We performed a BLAST search of the human data base with reference to the GTT1 sequence and found a variant form of gestational trophoblastic tumor (NP_064536) containing 75 additional amino acids at the N terminus (Fig. 1B). For convenience, we refer to the larger protein as StarD7 type I (StarD7-I) and the originally reported protein as StarD7 type II (StarD7-II). The molecular masses of StarD7-I and StarD7-II were estimated to be 43.1 and 34.7 kDa, respectively.
The additional 75 amino acids in the N terminus of StarD7-I, namely Met1–Leu75, are enriched in Arg (18.7%), Leu (21.3%), and Ala (9%), and the predicted secondary structure is an amphipathic α helix. When the N terminus (Met1–Leu75) of StarD7-I was analyzed by a mitochondrial protein predicting program (MITOPRED program) (14), the confidence of the sequence as a mitochondrial localization signal was 99.0% (Fig. 1B). StarD7-II does not contain a mitochondrial localization signal. The second Met76 from the N terminus of StarD7-I is the first Met for the translation initiation site of StarD7-II, and a mitochondrial localization signal of StarD7-I is followed by this first Met. The alignment of the N-terminal sequences of mammalian StarD7-I are shown in Fig. 1C. All mammalian StarD7-I contain the putative mitochondrial-targeting signal at their N termini.
Proteolytic Processing of StarD7-I
Most mitochondrial proteins are commonly translated in cytosolic ribosomes as precursor proteins with an N-terminal targeting signal, which is proteolytically cleaved after localization into the mitochondria to yield a mature form (15). As shown in Fig. 2A, two protein bands at 48 and 39 kDa were observed in the lysates prepared from cells transfected with the expression vector for V5/His-tagged StarD7-I. The molecular mass of 48 kDa coincides with the molecular mass of StarD-I (43.1 kDa) fused with the V5/His tag (4.9 kDa). The molecular mass of 75 amino acids in the N terminus of StarD7-I is ∼8.4 kDa. Therefore, it appeared that the 48-kDa form was a precursor of StarD7-I that was processed into the 39-kDa mature form by the cleavage of the mitochondrial-targeting sequence. One band at ∼39 kDa, which coincides with the molecular mass of StarD-II (34.7 kDa) fused with the V5/His tag (4.9 kDa), was detected in the lysates of cells transfected with the expression vector for V5/His-tagged StarD7-II (Fig. 2A). Therefore, the molecular mass of StarD7-II was happened to be almost the same as the mature form of StarD7-I.
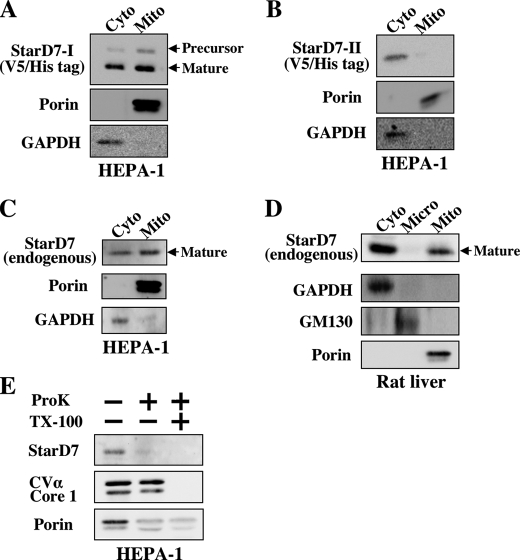
FIGURE 2.
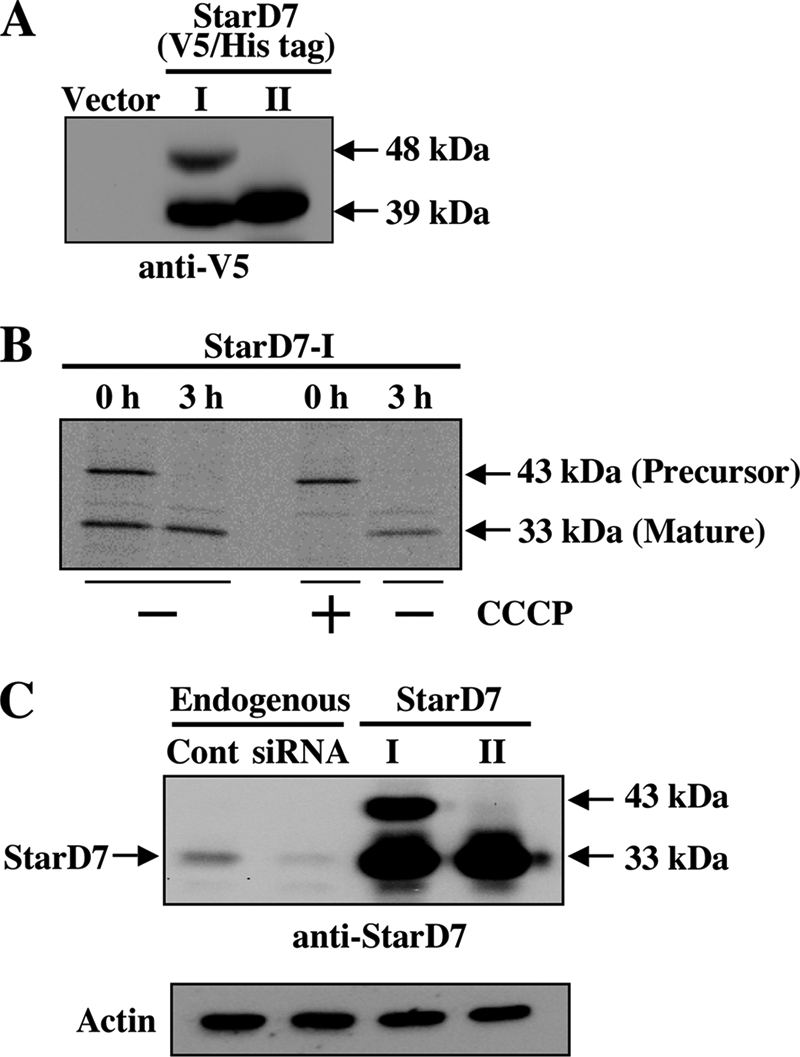
Proteolytic processing of StarD7-I. A, V5/His-tagged StarD7-I and -II were expressed in HEPA-1 cells (60–70% confluent), and cell lysates were analyzed by Western blotting with anti-V5 antibody. B, pulse-chase experiment. StarD7-I without tag was overexpressed in HEPA-1 cells, and proteins were pulse-labeled with 30 μCi/ml of [35S]Met and [35S]Cys for 20 min with or without CCCP. Then, cells were cultured in normal medium for 3 h. Proteins were immunoprecipitated with anti-StarD7 antibody, and precipitated proteins were separated by SDS-PAGE. C, molecular mass of endogenous StarD7. HEPA-1 cells were transfected with the StarD7-specific siRNA or the expression vector for StarD7-I or StarD7-II, and cell lysates were analyzed by Western blotting with anti-StarD7 antibody.
To verify the proteolytic processing of StarD7-I, we performed pulse-chase experiments. HEPA-1 cells overexpressing StarD7-I were pulse-labeled with [35S]methionine and [35S]cysteine for 20 min, and then chased for 3 h. Polyclonal antibodies to human StarD7 were prepared, and proteins were immunoprecipitated with the antibody and separated by SDS-PAGE. As shown in Fig. 2B (left), both the precursor (43 kDa) and the mature (33 kDa) forms were detected in StarD7-I-expressing cells just after the end of pulse labeling. Next, we assessed the effect of a protonophore, CCCP, to arrest the protein import into the mitochondria. As shown in Fig. 2B, only the precursor form (43 kDa) was detected just after pulse labeling in the presence of CCCP. The precursor form was converted to the mature form when CCCP was removed for 3 h. These results indicated that the precursor form of StarD7-I was imported into the mitochondria, and then the mature form was produced by the cleavage of a mitochondrial localization signal. The second Met76 of StarD7-I may not be an adequate translation initiation site, because a 33-kDa protein band was not detected in the cell lysates incubated with CCCP.
Then, the molecular mass of endogenous StarD7 in HEPA-1 cells was determined by Western blotting. As shown in Fig. 2C, the endogenous StarD7 was detected as a 33-kDa protein, which may be a mature form of StardD7-I. This band was diminished by the transfection of the StarD7-specific siRNA. In this experiment, the 43-kDa precursor form of StarD7-I was not detected, because the precursor may be processed rapidly to the mature form, and the amount of the precursor may be very low. It should be noted that the molecular mass of endogenous StarD7 was completely the same as the mature form of StarD7-I, and also StarD7-II. Thus, we could not distinguish the mature form of StarD7-I from StarD7-II by Western blot analyses.
Subcellular Fractionation and Protease K Protection Assay of Endogenous StarD7
To determine the localization of StarD7, subcellular fractionations were performed with HEPA-1 cells transfected with the expression vector for V5/His-tagged StarD7-I or -II. In this experiment, cells were cultured at 60–70% confluent. The purity of the mitochondrial or cytosolic fraction was determined with antibody against porin or GAPDH. Both the precursor (48 kDa) and the mature (39 kDa) forms were detected in cytoplasm and mitochondria (Fig. 3A) in cells overexpressing V5/His-tagged StarD7-I. In contrast, V5/His-tagged StarD7-II was distributed only in cytosol in cells overexpressing StarD7-II (Fig. 3B). These results indicate that StraD7-I could be moved into the mitochondria, but not StarD7-II. Therefore, a 33-kDa band reacted with anti-StarD7 antibody in mitochondria must be derived from the mature form of StarD7-I.
FIGURE 3.
Subcellular fractionation and protease K protection assay. Localization of the overexpressed V5/His-tagged StarD7-I (A) and StarD7-II (B) in HEPA-1 cells (60–70% confluent). Mitochondrial and cytoplasmic fractions were analyzed by Western blotting with anti-V5 antibody. The purity of the mitochondrial or cytosolic fraction was verified with anti-porin or anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody. C, localization of endogenous StarD7 in HEPA-1 cells (60–70% confluent). Mitochondrial and cytoplasmic fractions were analyzed by Western blotting with anti-StarD7 antibody. D, localization of endogenous StarD7 in rat liver. The purity of microsomal fraction was verified with anti-GM130 antibody. E, mitochondrial fractions from HEPA-1 cells were analyzed by protease K protection assay. StarD7 and porin were sensitive to protease K treatment, whereas complex Vα and core I were resistant. All proteins were digested by protease K in the presence of Triton X-100 (1% w/v).
The subcellular localization of endogenous StarD7 in HEPA-1 cells (60–70% confluent) was also determined. A 33-kDa protein band was detected in both the mitochondrial and cytosolic fractions of HEPA-1 cells (Fig. 3C). The same results were obtained with fractionated samples from rat liver (Fig. 3D). These results indicate that the mature form of StarD7-I derived from the precursor was surely expressed in HEPA-I cells and rat liver. The endogenous StarD7 was not detected in the microsomal fraction from rat liver by Western blot analysis (Fig. 3D). We think that the attachment of StarD7 to donor membranes may be done within a short period.
To determine the submitochondrial localization of the protein, isolated mitochondria from HEPA-1 cells were incubated with protease K. Proteins sensitive to the protease are suggested to be localized in the mitochondrial outer membrane. The mature form of StarD7-I and porin, a mitochondrial outer membrane protein, were sensitive to protease K (Fig. 3E). Complex Vα and Core 1, which are located in the inner mitochondrial membrane, were protected from the enzymes. These results demonstrate that the mature form of StarD7-I may be associated with the outer leaflet of mitochondria.
Phospholipid Extraction and Transfer Activity of StarD7
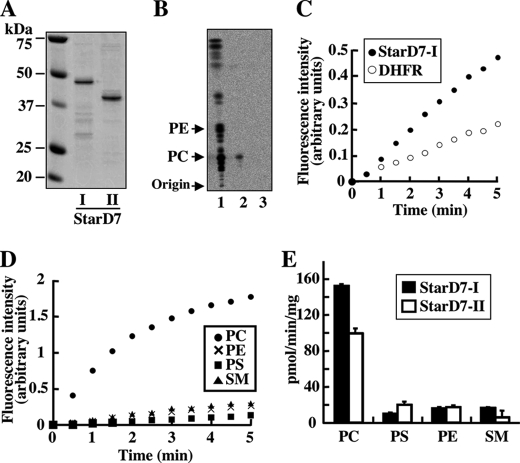
To identify the endogenous lipid ligands for StarD7, the lipid extraction activities of recombinant StarD7 (Fig. 4A) were examined in vitro. Bacterially expressed and purified recombinant StarD7-II was incubated with vesicles prepared from the [14C]palmitic acid-labeled total lipids extracted from HEPA-1 cells (Fig. 4B, lane 1). StarD7-lipid complexes were then separated from the remaining vesicles by using a 100-kDa cutoff filter. PC was specifically extracted from the vesicles by the purified proteins (Fig. 4B, lane 2). The radioactive phospholipid bands were not detected when boiled (inactivated) StarD7 was incubated with the vesicles (lane 3). This result indicates that StarD7 specifically binds and extracts PC from vesicle membranes. To confirm the intermembrane phospholipid-transfer activity of StarD7, fluorescence resonance energy transfer-based assays were performed. As shown in Fig. 4C, the PC-transfer activity of StarD7-I expressed in a wheat germ extract cell-free system was significantly greater than that of the dihydrofolate reductase control. The phospholipid-transfer activities of purified recombinant StarD7-I obtained from E. coli extracts were also investigated. As shown in Fig. 4D, StarD7-I had a much greater preference for PC as a lipid ligand as compared with PS, PE, and SM (∼5% of its PC-transfer activity). A comparison of the phospholipid ligand specificities of StarD7-I and StarD-II (Fig. 4E) revealed that both proteins preferred PC and that the specific activity of StarD7-I was slightly greater than that of StarD7-II. These results clearly indicate that StarD7 is a PC-specific lipid-transfer protein.
FIGURE 4.
Lipid extraction and transfer activities of StarD7. A, purified StarD7-I and -II from E. coli. B, lipid extraction activity of StarD7. Purified StarD7-II (100 μg) was incubated with vesicles prepared from total lipids extracted from HEPA-1 cells labeled with [14C]palmitic acid (300 nmol of total phospholipid). StarD7-lipid complexes were separated from the remaining lipid vesicles with 100-kDa cutoff filters. Radioactive lipids in the filtrates were extracted and analyzed by TLC with chloroform, methanol, and water (65:25:4, v/v). 1: total lipids; 2: non-boiled StarD7-II; 3: boiled StarD7-II. C, PC transfer activity of StarD7-I. The intermembrane transfer activities for fluorescent PC analog, C12-NBD-PC, from donor vesicles to acceptor vesicles were analyzed by a fluorescence resonance energy transfer-based assay. StarD7-I prepared by an in vitro translation system in wheat germ lysates was used. Dihydrofolate reductase (DHFR) is a control protein for the in vitro translation. D, specificity of the phospholipid transfer activities of StarD7-I. Phospholipid transfer activities from donor vesicles containing one of the fluorescent phospholipid analogs, C12-NBD-PC, C12-NBD-PE, C12-NBD-PS, or C12-NBD-SM, to acceptor vesicles were assessed with purified StarD7-I. The results are representative of several independent experiments. E, ligand specificities of StarD7-I and -II for several fluorescent phospholipids. Phospholipid transfer activities of both constructs were analyzed with a fluorescence resonance energy transfer-based assay. Values are the means ± S.D. of three independent experiments.
Intracellular Transfer of Fluorescence PC to Mitochondria in Living Cells
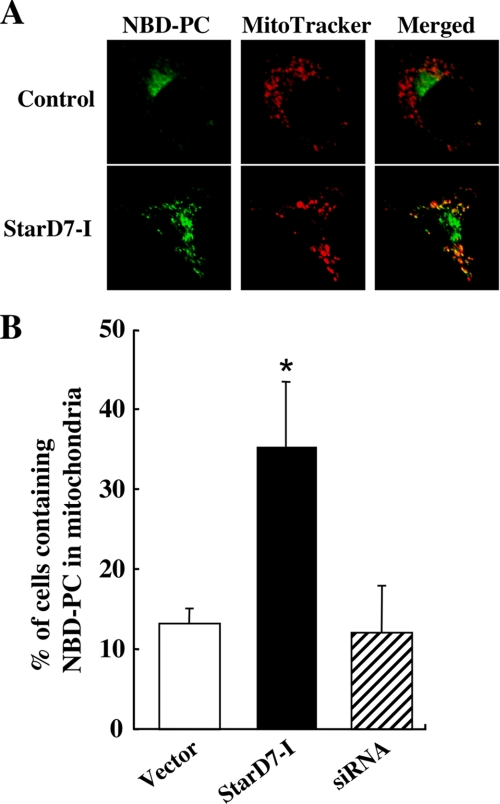
The distribution of the incorporated fluorescent PC in HEPA-1 cells that overexpress StarD7-I was investigated to determine if the protein can mediate PC transfer to mitochondria in living cells. Briefly, cells were analyzed by laser-scanning confocal microscopy after C6-NBD-PC-containing liposomes and MitoTracker Red were added to the medium (Fig. 5A). It was reported that the fluorescent PC in liposome was integrated into plasma membrane at 4 °C at first, and then internalized into intracellular membranes such as the ER and Golgi apparatus at 37 °C (13). As reported, the signals of fluorescent PC were predominantly observed in perinuclear regions that may contain the Golgi apparatus in control cells. In cells overexpressing StarD7-I, the fluorescence was observed in the perinuclear region/Golgi apparatus and intracellular vesicles that were co-localized with MitoTracker signals. Cells containing co-localized NBD-PC and MitoTracker were counted. As shown in Fig. 5B, a significantly greater percentage of cells showed co-localization of the two signals when StarD7-I was overexpressed. These results strongly suggest that StarD7-I can facilitate the transport of PC from the Golgi apparatus or some membranes to mitochondria in living cells.
FIGURE 5.
Intracellular transport of fluorescent PC analog in cells that overexpress StarD7-I. A, intracellular localization of fluorescent PC exogenously incorporated into HEPA-1 cells. Cells (60–70% confluent) transfected with empty vector or the expression vector for StarD7-I were incubated with lipid vesicles containing C6-NBD-PC (green) and then with MitoTracker Red (red). Cells were fixed and analyzed by confocal microscopy. Yellow indicates the co-localization of the green and red signals. B, quantification of cells showing the co-localization of NBD-PC and MitoTracker Red. Cells (60–70% confluent) transfected with empty vector, the expression vector for StarD7-I, or StarD7-specific siRNA were incubated with NBD-PC and MitoTracker. Values are means ± S.D. from four independent culture dishes. *, p < 0.01 as compared with the vector control.
To determine the importance of endogenous StarD7, HEPA-1 cells were transfected with StarD7-specific siRNA as shown in Fig. 2C, and the rate of PC transfer to mitochondria was compared with the vector control. However, as shown in Fig. 5B, the significant decrease in PC transfer to mitochondria compared with control was not determined in StarD7-knockdown cells. It could not be denied that the knockdown of endogenous StarD7 was not sufficient; however, there may be other important systems for PC transfer to mitochondria in living cells.
Intracellular Localization of StarD7-I and -II
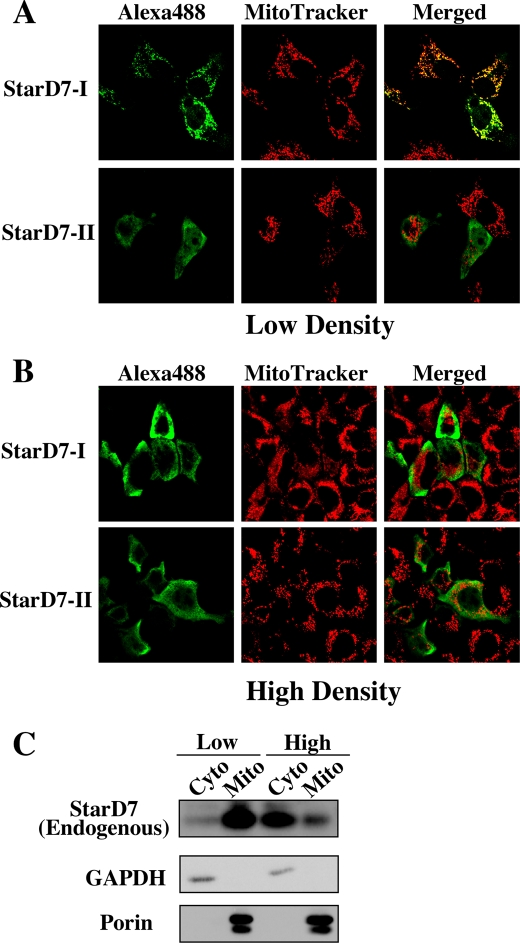
To investigate the precise intracellular localization of StarD7, StarD7-I and -II fused with a V5/His tag at the C terminus were expressed in HEPA-1 cells. As shown in Fig. 6A, the green signals of V5/His-tagged StarD7-I were co-localized with the red mitochondrial probe when cells were cultured at low density (20–30% confluent). However, StarD7-I was distributed in the cytoplasm when cells were plated at high density (100% confluent) (Fig. 6B). StarD7-II was distributed in the cytoplasm in both the conditions. To evaluate the dependence of localization of endogenous StarD7 on cell densities, fractionated samples of HEPA-1 cells cultured at low or high density were collected. As shown in Fig. 6C, endogenous StarD7 was mainly distributed in the mitochondria when cells were cultured at low density, and in cytoplasm when cultured at high density.
FIGURE 6.
Intracellular localization of StarD7-I and -II. HEPA-1 cells plated at low density (20–30% confluent) (A) or high density (100% confluent) (B) were transfected with the expression vector for V5/His-tagged StarD7-I or -II, and then immunostained with anti-V5 antibody followed by anti-mouse IgG Alexa488 (green) and MitoTracker Red (red). Yellow indicates the co-localization of their signals. C, subcellular fractionations of HEPA-1 cells cultured at different cellular densities. Mitochondrial and cytoplasmic fractions were prepared from cells cultured at low density (20–30% confluent) or high density (100% confluent) and analyzed by Western blotting with anti-StarD7 antibody. The purity of the mitochondrial or cytosolic fraction was verified by anti-porin or anti-GAPDH antibody.
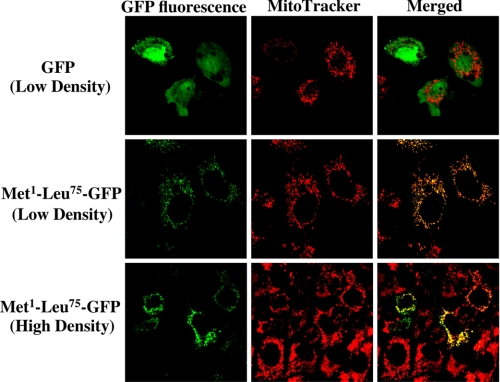
To confirm the N-terminal Met1–Leu75 of StarD7-I as a mitochondria targeting signal, we constructed an expression vector for the N-terminal Met1–Leu75 of StarD7-I fused to the N terminus of GFP. As shown in Fig. 7, GFP without the sequence was distributed in the cytoplasm and did not co-localize with the mitochondria. However, the GFP with the N terminus of StarD7 was co-localized with mitochondria when cells were cultured at both low and high densities. These results demonstrated that the N-terminal Met1–Leu75 of StarD7-I is indeed a mitochondrial-targeting signal.
FIGURE 7.
Intracellular localization of GFP fused with Met1–Leu75 at the N terminus. GFP (green) and chimeric GFP fused with Met1–Leu75 of StarD7-I at the N terminus were expressed in HEPA-1 cells cultured at low density (20–30% confluent) or high density (100% confluent) followed by treatment with MitoTracker Red (red) and analysis by confocal microscopy. Yellow indicates the co-localization of their signals.
DISCUSSION
PC, an essential compound in the mitochondrial membrane structure, must be transported from its sites of synthesis, such as the ER or Golgi apparatus, because mitochondria cannot synthesize PC. The precise mechanism by which PC is trafficked to mitochondria has not been elucidated. We performed a phylogenetic analysis (Fig. 1A) and found that StarD7 is part of a family that contains phospholipid-binding proteins; thus, we speculated that StarD7 may bind to phospholipids. Indeed, we found that StarD7 can transport PC from vesicles to other vesicles in vitro (Fig. 4). We also demonstrated that StarD7-I contains a mitochondrial-targeting signal at its N terminus (Fig. 1, B and C) and that the transfer of fluorescent PC to mitochondria was enhanced in cells that overexpress StarD7-I (Fig. 5). These results suggest that StarD7-I mediates the intracellular trafficking of PC to mitochondria.
Based on our results, we propose the following mechanism for the transfer of PC to mitochondria by StarD7-I. Precursors containing the mitochondria-targeting sequence at the N terminus are translated in the cytoplasm and then bind and extract PC from the cytoplasmic surfaces of the ER, Golgi apparatus, or plasma membranes. StarD7-I might also accept PC from other cytosolic PC-transporting proteins, such as StarD2/PC-TP and StarD10. The precursors binding to PC may be recognized by mitochondrial receptors, such as the preprotein translocase of the outer membrane complex, and then the presequence is cleaved by the mitochondrial-processing peptidase. PC would then be inserted into the outer mitochondrial membrane, because StarD7 is associated with the outer leaflet of mitochondria (Fig. 3).
Alternative pathways for the transport of PC to mitochondria independent of PC-transport proteins have been suggested. It is accepted that the phospholipids synthesized in the ER are transported to mitochondria through restricted membranes called mitochondria-associated membranes (MAMs), which are transient bridges from the ER to the outer mitochondrial membranes (16, 17). Recently, an ER-mitochondria tethering protein complex has been identified in yeast, and the phospholipid trafficking between the ER and mitochondria in the mutant cells was impaired (18). In this study, we could not find any significant reduction of PC transport to mitochondria by the knockdown of StarD7 (Fig. 5). One possible reason is that MAM may be important for PC transfer to mitochondria in steady state. In addition to MAM, cytosolic StarD2/PC-TP protein was also reported to transfer liposomal PC to mitochondria in vitro (19) and may facilitate the delivery of PC to mitochondria, because this protein could be translocated to mitochondria by phosphorylation in response to clofibrate (20). However, mice carrying a deletion in the gene encoding StarD2/PC-TP had a normal phenotype without any reported mitochondrial abnormalities (21). Recently, it was reported that brown adipocytes lacking StarD2/PC-TP showed enlarged and elongated mitochondria (22). Further study of the physiological importance and detailed mechanism of intracellular PC transport by StarD7 is necessary.
In the present study, we showed that overexpressed StarD7-I was distributed in mitochondria when HEAP-1 cells were plated at a low cellular density; however, the StarD7-I was distributed in the cytoplasm of cells plated at a high cellular density (Fig. 6). These results suggest that cell-cell contact might regulate the intracellular localization of StarD7-I. However, we could not determine the molecular mechanism responsible for the localization of StarD7-I. The intracellular localization of some proteins changes from the cytosol to the mitochondria in response to various stimuli. For example, the distribution of Bcl-2-associated X protein, Bax, changes from the cytoplasm to the mitochondria during apoptosis to activate the release of cytochrome c (23). Dd-TRAP1, a Dictyostelium homologue of tumor necrosis factor receptor-associated protein 1, is located in the cortex of cells growing at low density but is translocated to mitochondria when cell density is increased (24). We speculate that a post-translational modification of StarD7-I may change its localization. Further studies about the relationship between protein modifications and the subcellular localization of StarD7-I are being planned.
Mitochondrial PC has been reported to be involved in various enzymatic properties. For example, tafazzin, a mitochondrial enzyme that catalyzes PC-cardiolipin transacylation in an acyl-CoA-independent manner, specifically prefers PC that contains linoleoyl residues as a substrate (25). CoA synthase, which is localized in the outer mitochondrial membrane, was reported to be activated by PC (26). Thus, the present study may facilitate research regarding the biological functions and importance of mitochondrial PC.
Acknowledgments
We thank Dr. Takashi Namatame, Clinical Research Center, Dokkyo Medical University School of Medicine, for DNA sequencing, and the Research Support Center, Dokkyo Medical University School of Medicine, for allowing us to use their facilities.
This work was supported in part by Grant-in-Aid for Young Scientists (B) 20770091 from the Ministry of Education, Culture, Sports, Science and Technology, Japanese Government.
- ER
- endoplasmic reticulum
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PS
- phosphatidylserine
- SM
- sphingomyelin
- START
- steroidogenic acute regulatory protein-related lipid transfer
- StarD
- START domain-containing protein
- CERT
- ceramide transport protein
- NBD
- 7-nitrobenz-2-oxa-1,3-diazol-4-yl
- PC-TP
- phosphatidylcholine transfer protein
- MAM
- mitochondria-associated membrane
- GAPDH
- glyceraldehydes-3-phosphate dehydrogenase
- GFP
- green fluorescent protein
- EGFP
- enhanced GFP
- rh-PE
- lissamine rhodamine B-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt
- siRNA
- small interference RNA
- DMEM
- Dulbecco's modified Eagle's medium
- PBS
- phosphate-buffered saline
- CCCP
- carbonylcyanide-m-chlorophenylhydrazone.
REFERENCES
- 1.Voelker D. R. (2003) J. Lipid Res. 44, 441–449 [DOI] [PubMed] [Google Scholar]
- 2.Sprong H., van der Sluijs P., van Meer G. (2001) Nat. Rev. Mol. Cell Biol. 2, 504–513 [DOI] [PubMed] [Google Scholar]
- 3.D'Angelo G., Polishchuk E., Di Tullio G., Santoro M., Di Campli A., Godi A., West G., Bielawski J., Chuang C. C., van der Spoel A. C., Platt F. M., Hannun Y. A., Polishchuk R., Mattjus P., De Matteis M. A. (2007) Nature 449, 62–67 [DOI] [PubMed] [Google Scholar]
- 4.Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., Nishijima M. (2003) Nature 426, 803–809 [DOI] [PubMed] [Google Scholar]
- 5.Alpy F., Tomasetto C. (2005) J. Cell Sci. 118, 2791–2801 [DOI] [PubMed] [Google Scholar]
- 6.Durand S., Angeletti S., Genti-Raimondi S. (2004) Placenta 25, 37–44 [DOI] [PubMed] [Google Scholar]
- 7.Kanno K., Wu M. K., Scapa E. F., Roderick S. L., Cohen D. E. (2007) Biochim. Biophys. Acta 1771, 654–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olayioye M. A., Vehring S., Müller P., Herrmann A., Schiller J., Thiele C., Lindeman G. J., Visvader J. E., Pomorski T. (2005) J. Biol. Chem. 280, 27436–27442 [DOI] [PubMed] [Google Scholar]
- 9.Daum G. (1985) Biochim. Biophys. Acta 822, 1–42 [DOI] [PubMed] [Google Scholar]
- 10.Niwa H., Yamamura K., Miyazaki J. (1991) Gene 108, 193–199 [DOI] [PubMed] [Google Scholar]
- 11.Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 12.Nichols J. W., Pagano R. E. (1983) J. Biol. Chem. 258, 5368–5371 [PubMed] [Google Scholar]
- 13.Sleight R. G., Abanto M. N. (1989) J. Cell Sci. 93, 363–374 [DOI] [PubMed] [Google Scholar]
- 14.Guda C., Fahy E., Subramaniam S. (2004) Bioinformatics 20, 1785–1794 [DOI] [PubMed] [Google Scholar]
- 15.Bolender N., Sickmann A., Wagner R., Meisinger C., Pfanner N. (2008) EMBO Rep. 9, 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vance J. E. (2008) J. Lipid Res. 49, 1377–1387 [DOI] [PubMed] [Google Scholar]
- 17.Kuge O., Nishijima M. (2003) J. Biochem. 133, 397–403 [DOI] [PubMed] [Google Scholar]
- 18.Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S., Walter P. (2009) Science 325, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolay K., Hovius R., Bron R., Wirtz K., de Kruijff B. (1990) Biochim. Biophys. Acta 1025, 49–59 [DOI] [PubMed] [Google Scholar]
- 20.de Brouwer A. P., Westerman J., Kleinnijenhuis A., Bevers L. E., Roelofsen B., Wirtz K. W. (2002) Exp. Cell Res. 274, 100–111 [DOI] [PubMed] [Google Scholar]
- 21.van Helvoort A., de Brouwer A., Ottenhoff R., Brouwers J. F., Wijnholds J., Beijnen J. H., Rijneveld A., van der Poll T., van der Valk M. A., Majoor D., Voorhout W., Wirtz K. W., Elferink R. P., Borst P.(1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11501–11506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Won Kang H., Ribich S., Kim B. W., Hagen S. J., Bianco A. C., Cohen D. E. (2009) J. Lipid Res. 50, 2212–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou L., Chang D. C. (2008) J. Cell Sci. 121, 2186–2196 [DOI] [PubMed] [Google Scholar]
- 24.Morita T., Amagai A., Maeda Y. (2004) J. Cell Sci. 117, 5759–5770 [DOI] [PubMed] [Google Scholar]
- 25.Schlame M., Ren M. (2006) FEBS Lett. 580, 5450–5455 [DOI] [PubMed] [Google Scholar]
- 26.Zhyvoloup A., Nemazanyy I., Panasyuk G., Valovka T., Fenton T., Rebholz H., Wang M. L., Foxon R., Lyzogubov V., Usenko V., Kyyamova R., Gorbenko O., Matsuka G., Filonenko V., Gout I. T. (2003) J. Biol. Chem. 278, 50316–50321 [DOI] [PubMed] [Google Scholar]
- 27.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]