Abstract
DNA packaging in tailed bacteriophages and other viruses requires assembly of a complex molecular machine at a specific vertex of the procapsid. This machine is composed of the portal protein that provides a tunnel for DNA entry, an ATPase that fuels DNA translocation (large terminase subunit), and most frequently, a small terminase subunit. Here we characterized the interaction between the terminase ATPase subunit of bacteriophage SPP1 (gp2) and the procapsid portal vertex. We found, by affinity pulldown assays with purified proteins, that gp2 interacts with the portal protein, gp6, independently of the terminase small subunit gp1, DNA, or ATP. The gp2-procapsid interaction via the portal protein depends on gp2 concentration and requires the presence of divalent cations. Competition experiments showed that isolated gp6 can only inhibit gp2-procapsid interactions and DNA packaging at gp6:procapsid molar ratios above 10-fold. Assays with gp6 carrying mutations in distinct regions of its structure that affect the portal-induced stimulation of ATPase and DNA packaging revealed that none of these mutations impedes gp2-gp6 binding. Our results demonstrate that the SPP1 packaging ATPase binds directly to the portal and that the interaction is stronger with the portal embedded in procapsids. Identification of mutations in gp6 that allow for assembly of the ATPase-portal complex but impair DNA packaging support an intricate cross-talk between the two proteins for activity of the DNA translocation motor.
Keywords: Protein/Protein-Protein Interactions, ATPases, Viral DNA, Virus Assembly, Virus Structure, ATPase, DNA Packaging, SPP1 Bacteriophage, Molecular Motor, Viral Assembly
Introduction
A fundamental step in a virus life cycle is the encapsidation of the genome inside a protective protein shell. Some families of double-stranded DNA viruses, like tailed bacteriophages and herpes viruses, pack their genome into a preformed icosahedral procapsid to concentrations as high as 500 mg/ml (1–3). The molecular motor responsible for DNA packaging is among the most powerful biological machines described (4). Like many other molecular motors, this machine converts the chemical energy of ATP hydrolysis into mechanical movement of DNA. The requirements for DNA packaging have been identified in a number of viruses. Although details of this process differ among viruses, there are striking similarities, supporting a common strategy for motor assembly and functioning (reviewed in Refs. 5 and 6). The DNA-packaging motor assembles at a specific vertex of the procapsid, characterized by the presence of the portal protein. This protein is a cyclical oligomer possessing a central channel through which DNA translocation occurs (7–11). DNA translocation is powered by the energy of ATP hydrolysis carried out by the viral ATPase (or large terminase subunit), another essential component of the DNA-packaging motor that assembles at the portal vertex (12–16). In most cases, a third protein, the small terminase subunit, is also required for DNA encapsidation. Although the components of the DNA-packaging motor have been identified for several viruses and their function has been assigned, the nature of the packasome formed by interaction of the terminase with the prohead remains a central question of the DNA-packaging mechanism.
Bacillus subtilis phage SPP1 is a well recognized model system for viruses that package their DNA through a procapsid portal vertex. The SPP1 DNA-packaging reaction was reproduced in vitro and characterized using purified components (16). The packaging reaction requires substrate DNA, ATP, two terminase subunits (gp1and gp2), and procapsids with the portal protein, gp6. As in other phages, the large terminase subunit, gp2, has ATPase and endonuclease activities (17, 18). Although it is well established that the ATPase fuels DNA translocation, biochemical and genetic evidence also shows that gp1 and gp6 regulate the ATPase activity of gp2 and that a strict correlation exists between the portal-induced stimulation of the ATPase and DNA packaging (17–19). Enhancement of the ATPase activity of gp2 by the portal requires gp1 for a maximal effect and has a higher magnitude in the context of the procapsid (19). The SPP1 DNA-gp1-gp2 complex was hypothesized to dock at the portal vertex to assemble the machinery that pumps DNA to the procapsid interior (18). A detailed understanding of how these molecules build the functional motor is, however, still missing. Here we report that the SPP1 packaging ATPase binds directly to the portal protein, without requirement for gp1. Genetic evidence shows that this interaction is not sufficient for motor activity, indicating an intricate cross-talk between the ATPase and the portal proteins during DNA translocation to successfully accomplish viral genome encapsidation.
EXPERIMENTAL PROCEDURES
Enzymes and Reagents
Ultrapure acrylamide and isopropyl-1-thio-β-d-galactopyranoside were purchased from Euromedex. Agarose was from Bio-Rad. SYBR Gold was from Molecular Probes. Protein molecular weight markers and DNA restriction enzymes were from Biolabs. Proteinase K was from Roche Diagnostics. Lysozyme, DNase and dextran were from Sigma. ATP was purchased from Roche Applied Science.
Bacterial Strains, Bacteriophages, and Plasmids
SPP1 suppressor-sensitive mutants used were sus115, sus70, and sus70sus115 (20–22). B. subtilis HA101B (sup-3) and YB886 (sup°) were the permissive and non-permissive strains used for SPP1 multiplication (21). Bacterial and phage strains handling was as described previously (20–21). The Escherichia coli strain used for gp6 overproduction was XL-1 Blue (Stratagene). E. coli strains used for production of the terminase subunits gp2 and gp1 were BL21(DE3) and BL21(DE3)(pLysS), respectively (23). Plasmids pCB191 (24), pBT115 (21), and pREP4 (Qiagen) have been described. Plasmids encoding wild-type or mutant gp6 forms are derivatives of plasmid pHP13 (25) and have been described previously (19, 22).
DNA Purification
Plasmid DNA was purified with a plasmid purification kit from Qiagen. DNA concentration was determined using the molar extinction coefficient of 6500 m−1 × cm−1 at 260 nm. Analytical gel electrophoresis of plasmid DNA and restriction fragments was carried out in 0.8% (w/v) agarose/Tris borate-EDTA horizontal slab gels.
Protein Purification
gp1, gp2, and gp6 were purified as described previously (16, 19). Protein concentration was determined by the Bradford method (26) using bovine serum albumin as a standard. gp1, gp2, and gp6 concentrations were expressed as moles of protein decamers (27), monomers (17), and 13-mers (28), respectively.
Production and Purification of SPP1 Procapsids
Wild-type SPP1 procapsids, procapsids lacking gp6, or procapsids containing different gp6 mutant forms were produced and purified as described before (19). The molecular mass estimated for the procapsids was about 1.9 × 107 Da. The purity of the procapsid preparations was analyzed by SDS-PAGE, and the presence of gp6 was detected by Western blotting with an anti-gp6 antibody. Blots were developed using the ECL detection system (GE Healthcare). The quality of the different procapsid preparations was also checked by electron microscopy of negatively stained samples.
Pulldown Assays
Affinity pulldown assays were carried out using purified histidine-tagged gp2 and Co2+-coated magnetic beads (Dynabeads Talon, Invitrogen Dynal AS, Norway) according to the manufacturer's instructions. Unless stated otherwise, purified proteins or/and procapsids were incubated for 1 h at 30 °C in 50 mm Tris-HCl, pH 7.8, 50 mm NaCl, and 10 mm MgCl2. Samples were then mixed gently with the beads. After a 10-min incubation with gentle shaking, beads were subjected to a magnetic field, allowing bound material to be quickly and efficiently separated from the rest of the sample. Unbound material was then removed, and the beads were washed. Bead-bound material was released by directly adding a suitable volume of SDS-PAGE loading buffer and boiling the sample at 100 °C for 5–10 min. Proteins present in the beads were analyzed by SDS-PAGE and Western blotting using the appropriate rabbit polyclonal antisera and the ECL kit (GE Healthcare).
Antibody Production
Polyclonal rabbit antiserum was raised against purified gp2 according to standard protocols (29). The IgG fraction of the sera was purified by affinity chromatography with protein A-Sepharose 4B (GE Healthcare) as described by the suppliers.
DNA-packaging Reaction in Vitro
DNA-packaging reactions were performed using the SPP1 in vitro DNA-packaging system, as described before (16). Briefly, unless stated otherwise, a standard 20-μl reaction mixture contained 1 mm ATP, 1 nm DNA, 10 nm procapsids, and terminase proteins (1 μm gp2 monomers and 1 μm gp1 decamers) in 50 mm Tris-HCl, pH 7.8, 50 mm NaCl, 10 mm MgCl2, 10% dextran. Reaction mixtures were incubated for 1 h at 30 °C. DNase was then added at 20 μg/ml, and incubation continued for 10 min at 30 °C. Reactions were stopped by the addition of 50 mm EDTA and deproteinized. DNA was resolved by gel electrophoresis in 0.8% agarose gels and stained with SYBR Gold (Molecular Probes) according to the manufacturer's instructions.
RESULTS
gp2 Interacts Directly with the Isolated Portal Protein
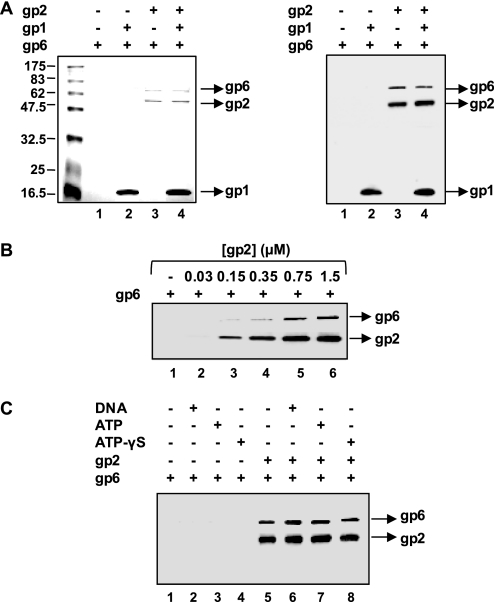
To analyze the network of interactions between the components of the SPP1 DNA-packaging machinery, pulldown assays were carried out with purified proteins. We first checked the capacity of purified gp2 carrying a histidine tag at its amino terminus (gp2-His, abbreviated from here on as gp2) to interact with the isolated portal protein, gp6. Tagged gp2 is fully functional, as revealed by previous in vivo and in vitro analysis (16, 17). The pulldown experiments were performed utilizing affinity of the histidine tag for cobalt-agarose magnetic beads, as described under “Experimental Procedures.” Preliminary control assays showed the expected binding of gp2 to beads, whereas gp6 alone was found in the flow-through (data not shown). As illustrated in Fig. 1A, showing Coomassie Blue-stained SDS-PAGE (left panel) and Western blot (right panel) analyses, incubation with gp2 led to association of gp6 to the bead fraction (compare lanes 1 with lanes 3 and 4). A rise in the amount of gp6 in the beads is observed with increasing doses of gp2, showing that gp2 binds to isolated gp6 in a concentration-dependent manner (Fig. 1B). The gp2-gp6 binding takes place in the absence of the terminase small subunit, gp1 (Fig. 1A, lane 3). Furthermore, the presence of gp1 in the reaction mixture did not result in significant changes in the amount of gp6 recruited to the beads (Fig. 1A, compare lanes 3 and 4). The same results were observed if preincubation of gp1 with gp2 occurred before gp6 addition (data not shown). It should be noted that gp1 alone binds strongly to the cobalt-agarose beads in an unspecific manner, being present mainly in the bead fraction (Fig. 1A, lane 2). This was also observed with other affinity matrices and various experimental conditions. Attempts to reduce the unspecific binding of gp1 did not give satisfactory results under conditions that did not disturb gp2-bead binding. Therefore, the gp2-gp1 physical interaction, which has been shown before (17, 24), is not addressed in these experiments. We then checked the effect of adding DNA or ATP in gp2-gp6 binding. No significant effect was observed with DNA or ATP concentrations identical to those used in the SPP1 in vitro DNA-packaging reactions (16) (Fig. 1C). Replacement of ATP by its poorly hydrolyzable analogue ATPγS3 did not cause any change in binding as well. Similar results were obtained in the absence or in the presence of gp1 (not shown). Overall these data show that gp2 is capable of binding isolated gp6 directly, without requirement for gp1, DNA, or ATP.
FIGURE 1.
Binding of gp2 to isolated gp6. A, recruitment of gp6 by gp2 analyzed by affinity pulldown. gp2 (500 nm) was incubated for 30 min at 30 °C with purified gp6 (80 nm) in the absence or in the presence of gp1 (500 nm). After pulldown (see “Experimental Procedures”), the proteins present in the bead fraction were identified by Coomassie Blue-stained SDS-PAGE (left panel) and Western blotting (right panel). Blots were reacted successively with anti-gp6, anti-gp1, and anti-gp2 antibodies. B, concentration dependence of gp2 on gp6 binding. gp6 (80 nm) was incubated with increasing doses of gp2 for 30 min at 30 °C before pulldown. Proteins present in the bead fraction were resolved by SDS-PAGE followed by Western blotting. Blots were incubated sequentially with anti-gp6 and anti-gp2 antibodies. C, effect of DNA and ATP on the gp2-gp6 interaction. gp2 (1 μm) was incubated with gp6 (80 nm) at 30 °C for 1 h in the absence or in the presence of DNA (1 nm), ATP (1 mm), or ATPγS (1 mm). After pulldown, the amounts of gp2 and gp6 present in the bead fraction were detected by Western blot.
gp2 Binds to Purified Procapsids Independently of gp1, DNA, or ATP
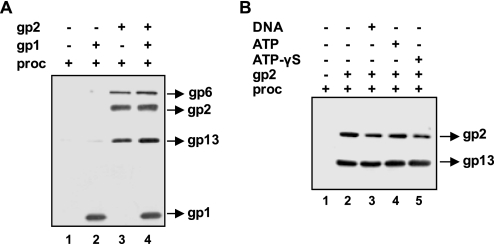
We next asked the question whether gp2 alone could interact with portal-containing SPP1 procapsids or whether this required the presence of the terminase small subunit. The interaction of gp2 with purified procapsids was checked by pulldown assays, as described above, followed by detection of the major capsid protein gp13 and gp6 to probe for procapsid components. As observed for isolated gp6, gp2 could recruit procapsids to the beads without requirement for gp1 DNA (Fig. 2A, compare lanes 1 and 3) or ATP (Fig. 2B). A slight increase in procapsid recruitment in the pulldown experiments could sometimes be observed in the presence of high gp1 concentrations (more than 500 nm) (Fig. 2A, lanes 3 and 4). However, gp1 can bind to the beads non-specifically (Figs. 1A and 2A, lane 2), which renders the interpretation of these observations difficult. The minor increase in procapsid binding in the pulldown experiments could be due either to a stimulation or/and to a stabilization of gp2-procapsid interaction by gp1 or simply to an additive effect due to potential gp1-procapsid interactions. We detected a gp1-procapsid association at high doses of gp1 in immunoprecipitation assays with anti-SPP1 or anti-gp1 antibodies. Identical results were obtained with wild-type and portal-less procapsids showing that co-immunoprecipitation is not due to an interaction between gp1 and the portal vertex (data not shown). It is likely therefore that the slight augmentation in the amount of procapsids recruited in the pulldown assays with gp2-bound beads in the presence of high amounts of gp1 (Fig. 2A and data not shown) is mainly due to some unspecific interactions between gp1, a basic protein, and the negatively charged procapsid lattice.
FIGURE 2.
Interaction of gp2 with purified procapsids. A, gp2-procapsid binding in the absence and in the presence of gp1. gp2 (1 μm) was incubated with procapsids (proc; 10 nm) for 1 h at 30 °C in the absence or in the presence of gp1 (500 nm). After pulldown, proteins bound to beads were detected by Western blot. Blots were successively reacted with anti-SPP1 (revealing the major capsid protein gp13), anti-gp6, anti-gp2, and anti-gp1 antibodies. B, effect of DNA, ATP, or ATPγS on gp2-procapsid binding. gp2 (1 μm) was incubated with procapsids for 1 h at 30 °C in the absence or in the presence of DNA (1 nm), ATP (1 mm), or ATPγS (1 mm). Blots of pulled-down material were successively reacted with anti-SPP1 and anti-gp2 antibodies.
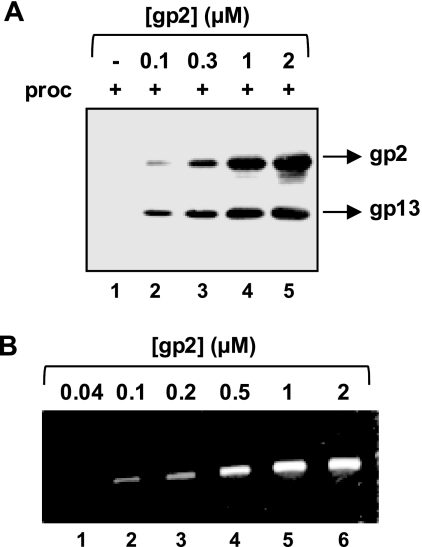
The addition of increasing concentrations of gp2 showed that the gp2-procapsid interaction occurs in a dose-dependent manner (Fig. 3A). This result correlated with the observed concentration-dependent effect of gp2 on DNA packaging in vitro (Fig. 3B).
FIGURE 3.
Effect of gp2 concentration on the gp2-procapsid interaction and on DNA packaging. A, dose-dependent effect of gp2 on procapsid binding. Procapsids (proc; 10 nm) were incubated in the absence (−) or in the presence of increasing doses of gp2 for 1 h followed by 10 min incubation with cobalt beads. The presence of procapsids in the bead fraction was checked by Western blot using anti-SPP1 and anti-gp2 antibodies. B, dose-dependent effect of gp2 on DNA packaging. DNA-packaging reactions in vitro were carried out for 1 h as described under “Experimental Procedures” using 10 nm procapsids and the gp2 concentrations indicated in the figure. The amount of packaged DNA was checked in a 0.8% agarose gel.
Effect of Magnesium on the gp2-Procapsid Interaction
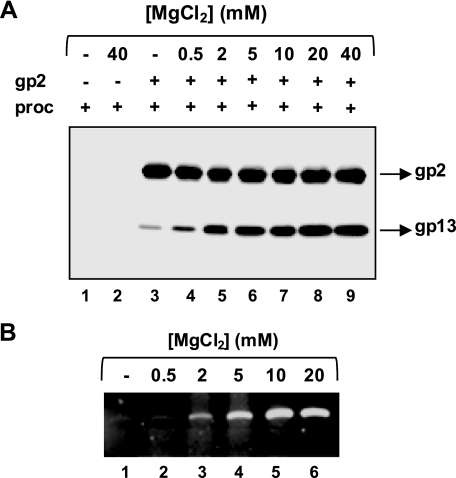
Because Mg2+ is a strict requirement for gp2 ATPase activity, we investigated whether it also plays a role in the gp2-procapsid interaction. As shown in Fig. 4A, Mg2+ is necessary for the interaction, and rising concentrations of the divalent cation led to an increase in gp2-procapsid binding, approaching a maximum at ∼10 mm. These data correlated well with the effect of MgCl2 concentrations on DNA packaging (Fig. 4B). Replacement of Mg2+ by Ca2+ (10 mm CaCl2) in the pulldown assays supported gp2-procapsid binding but precluded DNA packaging (data not shown).
FIGURE 4.
Effect of MgCl2 on gp2-procapsid binding and on DNA packaging. A, concentration-dependent effect of MgCl2 on the gp2-procapsid interaction. Procapsids (proc; 10 nm) and gp2 (1 μm) were incubated for 1 h in the absence (−) or in the presence of increasing doses of MgCl2, indicated above the gel lanes, before pulldown and Western blot. B, dose-dependent effect of gp2 on DNA packaging. DNA-packaging reactions in vitro were carried out as described under “Experimental Procedures.”
gp2-Procapsid Binding Is Mediated by the Portal Protein
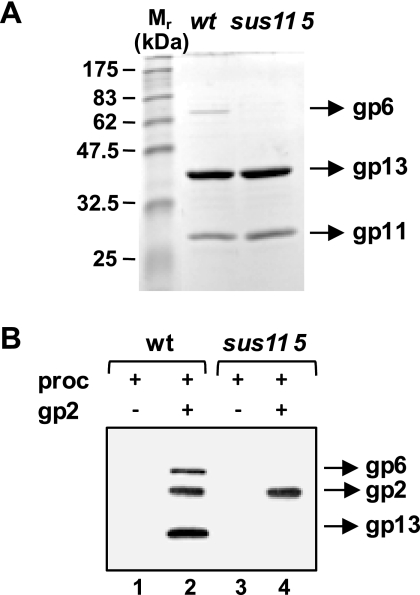
The portal vertex is believed to be the docking point for terminase binding. To examine whether the gp2-procapsid interaction occurs via the portal protein, portal-less procapsids were produced and purified (Fig. 5A), and their capacity to interact with gp2 was analyzed as well. As shown in Fig. 5B, procapsids without portal were not recruited by gp2, contrarily to wild-type structures (lanes 4 and 2, respectively). This finding demonstrates that gp2-procapsid binding is undeniably portal-mediated.
FIGURE 5.
Dependence of the gp2-procapsid interaction on the presence of gp6 at the procapsid portal vertex. A, SDS-PAGE analysis of wild-type (wt) and portal-lacking (sus115) SPP1 procapsids. The positions of the portal (gp6), the major head (gp13), and the scaffolding (gp11) proteins are indicated. The gel was stained with Coomassie Blue. M, molecular mass standards. B, interaction of gp2 with the two types of procapsids (proc). gp2 (1 μm) was incubated with procapsids (10 nm) before pulldown with cobalt beads. Procapsid proteins and gp2 bound to beads were detected by Western blot.
Competition of Isolated Portal Protein and Procapsis for gp2 Binding
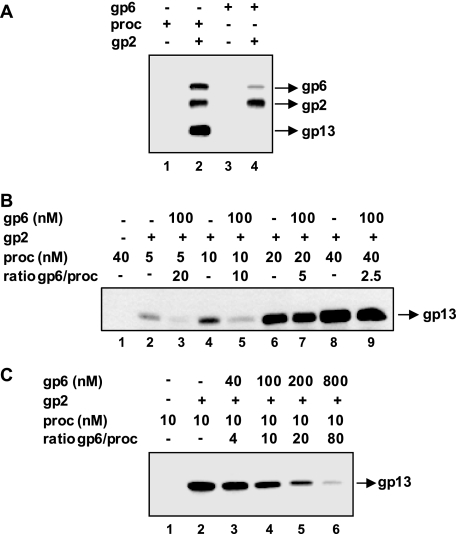
We then addressed the question of whether gp2 could discriminate between the isolated portal protein and the portal embedded in the procapsid structure. gp2 was first incubated with equimolar amounts of either purified gp6 or procapsids. gp2-gp6 binding was checked as previously. Although gp2 can recruit both forms of the protein, we consistently found a stronger signal for the portal incorporated in the procapsid (Fig. 6A, compare lanes 2 and 4). We have thus analyzed the competition of gp6-containing procapsids for the binding between isolated gp6 and gp2. Binding of increasing doses of procapsids to 100 nm gp2 that had been preincubated in the absence or in the presence of equimolar amounts of gp6 (gp2 (monomers):gp6 (13-mers) ratio of 1) was checked in pulldown assays. Procapsids could bind efficiently to gp2 preincubated with isolated gp6 when gp6 was at a molar excess of 2.5 or 5 relative to the procapsid input concentration (Fig. 6B, compare lanes 6 and 7 and lanes 8 and 9). Inhibition of procapsid-gp2 binding was only observed at higher gp6:procapsid ratios (Fig. 6B, lanes 3 and 5). We also analyzed binding of 10 nm procapsids to a large excess of gp2 (1 μm) preincubated with increasing concentrations of isolated gp6 (Fig. 6C). Inhibition of procapsid binding was observed at gp6 concentrations above 100 nm. The effect is observed at a concentration of gp2 monomers that is ∼10-fold higher than its interaction partners (isolated gp6 plus gp6 in the procapsid), a situation in which no competition was expected to occur. This result reveals that several gp2 monomers bind to a single gp6 molecule and/or the presence of a fraction of inactive gp2 protein in the reaction.
FIGURE 6.
Competition of the gp2-gp6 interaction between isolated portal and the portal protein embedded in the procapsid structure. A, binding of gp2 to portal-containing procapsids (proc) versus isolated portal. gp2 (1 μm) was incubated for 1 h with 10 nm of either procapsids or purified gp6 before being incubated with cobalt beads. After washing, the presence of gp6 in the bead fraction was detected by Western blot. B, competition of gp6-containing procapsids for the binding between gp2 and isolated gp6 13-mers. Increasing concentrations of procapsids (ranging from 5 to 40 nm) were added to gp2 (100 nm) that had been previously incubated with 100 nm gp6 for 30 min. Incubation proceeded for 30 min more before pulldown with cobalt beads. The presence of procapsids in the bead fraction was detected by Western blot. C, effect of increasing concentrations of purified portal protein on gp2-procapsid binding at high gp2 concentration. Procapsids (10 nm) were added to gp2 (1 μm) that had been previously incubated for 30 min without gp6 (−) or with various concentrations of purified gp6 as labeled above the gel lanes. Incubation proceeded for 30, min and procapsid binding to gp2 was then checked as in B.
Binding of gp2 to gp6 DNA-packaging Mutants
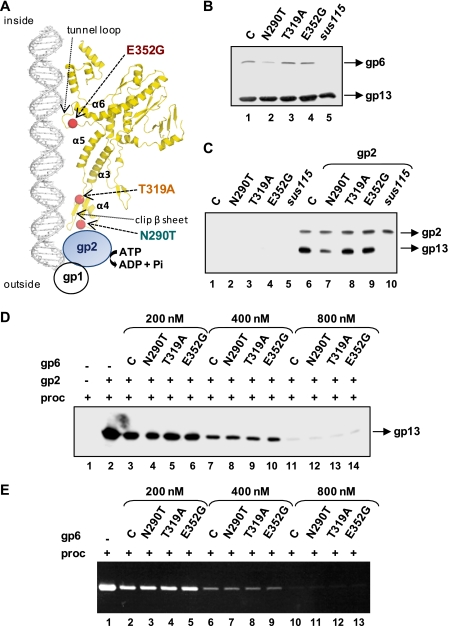
Previously we have identified and characterized single amino acid substitutions in the SPP1 portal protein that impair or block DNA packaging (19, 22, 30). Biochemical analysis of selected mutants presenting distinct phenotypes revealed that those mutations affected more or less severely the portal-induced stimulation of the ATPase activity. This effect was strictly correlated with the efficiency of DNA packaging into procapsids containing the mutant portals (19). Positioning of mutations in the now available crystallographic structure of the gp6 protein (Protein Data Bank (PDB) accession number 2JES) (11) showed that they affect residues at the putative terminase docking interface (N290T) or that they are buried inside the portal central channel (T319A and E352G) (Fig. 7A). These results did not allow discriminating whether the mutations affected the physical interaction between the motor components (assembly defect) or the motor mechanics during DNA translocation (motor activity defect). To address this question, procapsids carrying the mutant gp6 proteins were produced and purified (Fig. 7B), and their interaction with gp2 was examined in pulldown assays. Interestingly, gp2 could recruit all different procapsids, with the exception of the portal-less ones (sus115), as shown in Fig. 7C (lanes 6–10). The slightly small amount of procapsids carrying the gp6N290T that is pulled-down (Fig. 7C, lane 7) is likely due to the presence of a small fraction of portal-less procapsids in this population, as revealed by the lower ratio gp6:gp13 when compared with other procapsids (Fig. 7B). Similar results were obtained when the experiment was carried out in the absence or in the presence of gp1 (not shown). To confirm that all three gp6 mutant proteins were able to interact with gp2, we compared their capacity to inhibit the gp2-procapsid interaction and DNA packaging with the one verified with control gp6. gp2 (200 nm) was incubated with increasing concentrations of gp6 (200–800 nm) under conditions that inhibit procapsid binding (gp6:procapsid ratio ranging from 10 to 40). As shown in Fig. 7D, incubation of gp2 with all gp6 mutants caused inhibition of gp2-procapsid binding, to the same extent as verified for control gp6. The level of inhibition increased with the gp6:procapsid ratio in a similar fashion for the control and mutant proteins. Similar results were obtained concerning the inhibition of DNA packaging by the isolated portals, with identical effects observed with the mutant portals relatively to the control (Fig. 7E). Overall the data indicate that gp6N290T, gp6T319A, and gp6E352G proteins are not defective in gp2 binding.
FIGURE 7.
Impact of gp6 mutations that affect the ATPase activity on the gp2-procapsid interaction. A, positioning of a set of gp6 DNA-packaging mutations that differentially affect the terminase ATPase activity (19) in the gp6 crystallographic structure (PDB accession number 2JES) (11). A ribbon diagram of one gp6 subunit is shown. Structural elements of gp6 and the putative interface of interaction with the ATPase are indicated. B, analysis of protein content of procapsid preparations carrying distinct gp6 forms. All portals used in this experiment carry mutation sizX (E424K) in addition to the amino acid substitution indicated in the figure, as described (19, 22). Lane C, control. Procapsid content was characterized by Western blot. C, interaction between gp2 and the gp6 mutant procapsids assayed by pulldown. The different procapsid preparations shown in B were incubated for 1 h at 30 °C without (lanes 1–5) and with gp2 (1 μm) (lanes 6–10) followed by pulldown and Western blot. Lane C, control. D, comparison of the effect of purified gp6 mutant proteins and control gp6 on the interaction of gp2 with wild-type procapsids (proc). gp2 (200 nm) was preincubated with different concentrations of purified control (lane C) or mutant gp6 proteins for 30 min before further incubation of samples with wild-type procapsids (20 nm). Procapsid binding to gp2 was then checked as in C. E, effect of purified gp6 mutant proteins on in vitro DNA packaging. gp2 (200 nm) was preincubated with different amounts of purified control or mutant gp6 proteins for 30 min. Procapsids (20 nm), gp1, DNA, and ATP were then added to the reaction mixtures (see “Experimental Procedures”), and incubation continued for 30 more min. After DNase treatment, the amount of protected DNA was checked in a 0.8% agarose gel.
DISCUSSION
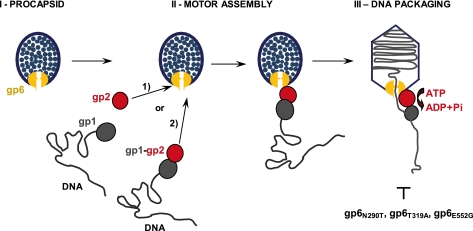
The mechanism by which viral DNA is packaged into a preformed procapsid is a fascinating but yet still unsolved biological problem. One of the key questions that remain to be addressed in molecular detail is the network of interactions between the substrate DNA, the terminase subunits, and the portal vertex to assemble the DNA-packaging motor. The first major outcome of this study is the finding that the SPP1 packaging ATPase, gp2, interacts directly with the portal protein, gp6, independently of the terminase small subunit gp1, DNA, or ATP (Figs. 1 and 8). This holds true both for the isolated portal and for the portal embedded in the procapsid structure (Figs. 1, 2, and 5). gp2 cannot interact with portal-less procapsids, demonstrating the specificity of the gp2-portal interaction (Fig. 5B). These results are in agreement with previous work suggesting that ATPase-portal interactions occur in phages T3, lambda, and T4 (14, 15, 31). In the case of phi29 phage, the interaction appears to be mediated by a pRNA, which binds directly to the portal (32, 33). The interaction of gp2 with the portal protein is dependent of the ATPase concentration (Figs. 1B and 3A). Interestingly, the effect of increasing gp2 concentrations on DNA packaging closely followed the result obtained with identical gp2 doses on the interaction with the portal vertex (Fig. 3). This strongly suggests that binding of the SPP1 ATPase to the portal plays a direct role in docking terminase to the procapsid, and hence, in the assembly of the macromolecular DNA translocation complex. We cannot distinguish at present whether (i) gp2 binds first to the procapsid followed by docking of the gp1-DNA complex, (ii) whether a preassembled gp2-gp1-DNA complex binds the portal vertex via gp2, or (iii) whether both assembly paths lead to formation of a functional motor (Fig. 8).
FIGURE 8.
Schematic representation of the SPP1 DNA-packaging motor assembly. gp6, gp2, and gp1 are represented in yellow, red, and gray, respectively. Assembly of the motor occurs by a direct gp6-gp2 interaction. It is presently not known whether isolated gp2 interacts with gp6 followed by gp1-DNA docking (pathway 1 in the figure) or whether a gp2-gp1-DNA complex binds to gp6 (pathway 2). The step of DNA packaging affected by single amino acid substitutions in gp6 studied in this work (Fig. 7) is shown.
Biochemical characterization of the ATPase-portal interaction showed that efficient binding of gp2 to the portal vertex requires the presence of divalent cations. Pulldown assays with increasing Mg2+ concentrations revealed that a maximal binding was reached around 10 mm Mg2+ (Fig. 4A), which is in the range of Mg2+ concentrations found in the B. subtilis cytoplasm (10–50 mm) under physiological conditions (34). Although gp2 is unable to hydrolyze ATP in the presence of Ca2+, (17), Mg2+ could be replaced by Ca2+ in the pulldown assays without loss of the gp2-gp6 binding affinity (data not shown). This result, together with the finding that gp2 does not require ATP for gp6 binding, indicates that the effect of Mg2+ on the gp2-gp6 interaction is not related to the gp2 ATPase activity. The dependence on the divalent cations Mg2+ or Ca2+ for efficient gp2-gp6 binding is most likely explained by a role on gp6 and/or gp2 structure that would adopt a conformation competent for interaction or on their physical interaction. Divalent cations were in fact shown to stabilize and lead to a more compact structure of SPP1 portal oligomers (35, 36). Potential Mg2+ binding sites have also been found in the recently solved gp2 nuclease domain structure (37) and in the ATPase and nuclease domains of the T4 large terminase subunit, gp17 (38–40). This correlates with the requirement for Mg2+ for both endonuclease and ATPase activities (17). Besides their role in promoting a gp6 structure that may be preferentially recognized by the ATPase, divalent cations may also have a structural effect on the ATPase itself that could favor its association with the portal and/or oligomerization around the portal vertex.
The second major output of the present work is the discovery that the gp2-portal physical interaction is stronger when the portal is embedded in the procapsid (Fig. 6). These results are in conformity with earlier DNA packaging in vitro experiments using crude cell extracts, which showed that the isolated portal protein did not inhibit DNA packaging in a competitive manner (41). The molecular basis for discrimination between the pool of free portal protein and the portal embedded in the procapsid structure during DNA packaging is now shown to be provided by the gp2-portal vertex interaction. This distinction is advantageous from an economical point of view by preventing premature and non-productive interactions between gp2 and the isolated portal co-existing in the cytoplasm of infected cells. A particular conformation of either the portal or the complete vertex structure could be involved in specific binding. A possible structural basis for the specificity of gp2 for the procapsid-embedded portal is the different oligomeric state of the portal protein. Although isolated gp6 is a cyclical 13-mer, in DNA-filled capsids, gp6 is a 12-mer ring embedded in the capsid vertex. This change was proposed to occur during procapsid assembly (42).
gp6 mutations that affect differently the portal-induced stimulation of the ATPase and DNA packaging are available (19). The novel possibility of analyzing the physical gp2-gp6 interaction provided an experimental framework to assess whether differences in the motor ATP hydrolysis rate caused by gp6 mutations are a consequence of a deficient gp2-gp6 binding. A weak protein-protein (gp2-gp6) interaction would have, in fact, an impact on all subsequent events engaging the complex, including ATPase activity and DNA packaging. The pulldown assays with procapsids carrying gp6 mutant proteins showed clearly that the gp6N290T, gp6T319A, and gp6E352G mutations do not impede gp2-gp6 binding. This holds true either for experiments performed in the absence of gp1 (Fig. 7C) or for experiments with gp1 present in the reaction mixture (data not shown) under conditions where DNA packaging is expected to occur. The capacity of mutant portals to interact with the terminase was further confirmed by competition experiments (Fig. 7, D and E). The finding that the portal mutations do not interfere with assembly of the ATPase-portal complex strongly suggests that they impair communication between gp2 and gp6 in the assembled packaging motor that is essential for its activity (Fig. 8). Residue Asn-290 is exposed at the putative interface of gp6 for ATPase binding, providing a simple explanation of how its substitution could affect cross-talk necessary for modulation of ATPase activity. Interestingly, Thr-319 and Glu-352 are found at different positions in the portal protein internal channel, inaccessible for gp2 direct interaction (Fig. 7A). Thr-319 is present at the extremity of an antiparallel β-sheet (the “clip β-sheet”; 11) that is also formed by Asn-290, found in another strand at the opposite end of the β-sheet. The T319A substitution is expected to disrupt neither the β-sheet nor the local structural organization of gp6, rendering its phenotype difficult to interpret mechanistically. Residue Glu-352 maps in the portal tunnel loop that protrudes to the internal channel (Fig. 7A). The gp6 subunit tunnel loops define the most constricted region of the portal channel. Motion of the loops and their contact with DNA were proposed to play a key role in DNA translocation (11). Mutation E352G abolishes hydrogen bonding to Ile-354, likely destabilizing the loop structure. The ensemble of the data suggests that the single amino acid substitutions under study have subtle effects in the gp6 structure allowing for normal docking of gp2 to the portal vertex but preventing conformational changes in gp6 that modulate gp2 hydrolysis of ATP. Changes involve tunnel loops, structural elements lining the internal portal channel, and the gp2 binding interface, arguing for their coordinated action with stimulation of gp2 ATPase activity. The mechanism likely involves motions of helix α-5 that connect the tunnel loop to the clip β-sheet (43). This far-distance communication can occur in the absence of DNA in the portal channel but is required for DNA packaging. Definition of its role on DNA translocation when the ATPase and the portal tunnel loops contact the double helix is of central importance to understanding the mechanics of this powerful viral nanomotor.
Acknowledgments
We thank Marie-Christine Vaney (Institut Pasteur) for advice on gp6 structure interpretation. We are indebted to Sandrine Brasilès (Unité de Virologie Moléculaire et Structurale, CNRS) for purified gp6N290T.
This work was supported by CNRS institutional funding and by Agence Nationale de la Recherche Grant ANR-06-BLAN-0168.
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
REFERENCES
- 1.Bazinet C., King J. (1985) Annu. Rev. Microbiol. 39, 109–129 [DOI] [PubMed] [Google Scholar]
- 2.Casjens S., Hendrix R. (1988) in The Bacteriophages (Calendar R. ed) Vol. 1, pp. 15–91, Plenum Press, New York [Google Scholar]
- 3.Black L. W. (1989) Annu. Rev. Microbiol. 43, 267–292 [DOI] [PubMed] [Google Scholar]
- 4.Smith D. E, Tans S. J., Smith S. B., Grimes S., Anderson D. L., Bustamante C. (2001) Nature 413, 748–752 [DOI] [PubMed] [Google Scholar]
- 5.Catalano C. E. (2005) in Viral Genome Packaging Machines: Genetics, Structure, and Mechanism (Catalano C. E. ed) pp. 1–4, Kluwer Academic/Plenium Publishers, NY [Google Scholar]
- 6.Rao V. B., Feiss M. (2008) Annu. Rev. Genet. 42, 647–681 [DOI] [PubMed] [Google Scholar]
- 7.Valpuesta J. M., Carrascosa J. L. (1994) Q. Rev. Biophys. 27, 107–155 [DOI] [PubMed] [Google Scholar]
- 8.Simpson A. A., Tao Y., Leiman P. G., Badasso M. O., He Y., Jardine P. J., Olson N. H., Morais M. C., Grimes S., Anderson D. L., Baker T. S., Rossmann M. G. (2000) Nature 408, 745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newcomb W. W., Juhas R. M., Thomsen D. R., Homa F. L., Burch A. D., Weller S. K., Brown J. C. (2001) J. Virol. 75, 10923–10932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agirrezabala X., Martín-Benito J., Valle M., González J. M., Valencia A., Valpuesta J. M., Carrascosa J. L. (2005) J. Mol. Biol. 347, 895–902 [DOI] [PubMed] [Google Scholar]
- 11.Lebedev A. A., Krause M. H., Isidro A. L., Vagin A. A., Orlova E. V., Turner J., Dodson E. J., Tavares P., Antson A. A. (2007) EMBO J. 26, 1984–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo P., Peterson C., Anderson D. (1987) J. Mol. Biol. 197, 219–228 [DOI] [PubMed] [Google Scholar]
- 13.Morita M., Tasaka M., Fujisawa H. (1993) Virology 193, 748–752 [DOI] [PubMed] [Google Scholar]
- 14.Yeo A., Feiss M. (1995) J. Mol. Biol. 245, 141–150 [DOI] [PubMed] [Google Scholar]
- 15.Lin H., Rao V. B., Black L. W. (1999) J. Mol. Biol. 289, 249–260 [DOI] [PubMed] [Google Scholar]
- 16.Oliveira L., Alonso J. C., Tavares P. (2005) J. Mol. Biol. 353, 529–539 [DOI] [PubMed] [Google Scholar]
- 17.Gual A., Camacho A. G., Alonso J. C. (2000) J. Biol. Chem. 275, 35311–35319 [DOI] [PubMed] [Google Scholar]
- 18.Camacho A. G., Gual A., Lurz R., Tavares P., Alonso J. C. (2003) J. Biol. Chem. 278, 23251–23259 [DOI] [PubMed] [Google Scholar]
- 19.Oliveira L., Henriques A. O., Tavares P. (2006) J. Biol. Chem. 281, 21914–21923 [DOI] [PubMed] [Google Scholar]
- 20.Tavares P., Santos M. A., Lurz R., Morelli G., de Lencastre H., Trautner T. A. (1992) J. Mol. Biol. 225, 81–92 [DOI] [PubMed] [Google Scholar]
- 21.Chai S., Bravo A., Lüder G., Nedlin A., Trautner T. A., Alonso J. C. (1992) J. Mol. Biol. 224, 87–102 [DOI] [PubMed] [Google Scholar]
- 22.Isidro A., Henriques A. O., Tavares P. (2004) Virology 322, 253–263 [DOI] [PubMed] [Google Scholar]
- 23.Studier F. W. (1991) J. Mol. Biol. 219, 37–44 [DOI] [PubMed] [Google Scholar]
- 24.Gual A., Alonso J. C. (1998) Virology 242, 279–287 [DOI] [PubMed] [Google Scholar]
- 25.Haima P., Bron S., Venema G. (1987) Mol. Gen. Genet. 209, 335–342 [DOI] [PubMed] [Google Scholar]
- 26.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 27.Chai S., Lurz R., Alonso J. C. (1995) J. Mol. Biol. 252, 386–398 [DOI] [PubMed] [Google Scholar]
- 28.Dube P., Tavares P., Lurz R., van Heel M. (1993) EMBO J. 12, 1303–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harlow E., Lane D. (1988) Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 30.Isidro A., Santos M. A., Henriques A. O., Tavares P. (2004) Mol. Microbiol. 51, 949–962 [DOI] [PubMed] [Google Scholar]
- 31.Morita M., Tasaka M., Fujisawa H. (1995) J. Mol. Biol. 245, 635–644 [DOI] [PubMed] [Google Scholar]
- 32.Koti J. S., Morais M. C., Rajagopal R., Owen B. A., McMurray C. T., Anderson D. L. (2008) J. Mol. Biol. 381, 1114–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morais M. C., Koti J. S., Bowman V. D., Reyes-Aldrete E., Anderson D. L., Rossmann M. G. (2008) Structure 16, 1267–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Médicis E. D., Paquette J., Gauthier J. J., Shapcott D. (1986) Appl. Environ. Microbiol. 52, 567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jekow P., Behlke J., Tichelaar W., Lurz R., Regalla M., Hinrichs W., Tavares P. (1999) Eur. J. Biochem. 264, 724–735 [DOI] [PubMed] [Google Scholar]
- 36.Orlova E. V., Gowen B., Dröge A., Stiege A., Weise F., Lurz R., van Heel M., Tavares P. (2003) EMBO J. 22, 1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smits C., Chechik M., Kovalevskiy O. V., Shevtsov M. B., Foster A. W., Alonso J. C., Antson A. A. (2009) EMBO reports 10, 592–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun S., Kondabagil K., Gentz P. M., Rossmann M. G., Rao V. B. (2007) Mol. Cell 25, 943–949 [DOI] [PubMed] [Google Scholar]
- 39.Sun S., Kondabagil K., Draper B., Alam T. I., Bowman V. D., Zhang Z., Hegde S., Fokine A., Rossmann M. G., Rao V. B. (2008) Cell 135, 1251–1262 [DOI] [PubMed] [Google Scholar]
- 40.Alam T. I., Draper B., Kondabagil K., Rentas F. J., Ghosh-Kumar M., Sun S., Rossmann M. G., Rao V. B. (2008) Mol. Microbiol. 69, 1180–1190 [DOI] [PubMed] [Google Scholar]
- 41.Dröge A., Tavares P. (2000) J. Mol. Biol. 296, 103–115 [DOI] [PubMed] [Google Scholar]
- 42.Lurz R., Orlova E. V., Günther D., Dube P., Dröge A., Weise F., van Heel M., Tavares P. (2001) J. Mol. Biol. 310, 1027–1037 [DOI] [PubMed] [Google Scholar]
- 43.Cuervo A., Vaney M. C., Antson A. A., Tavares P., Oliveira L. (2007) J. Biol. Chem. 282, 18907–18913 [DOI] [PubMed] [Google Scholar]