Abstract
Pro-lysyl oxidase is secreted as a 50-kDa proenzyme and is then cleaved to a 30-kDa mature enzyme (lysyl oxidase (LOX)) and an 18-kDa propeptide (lysyl oxidase propeptide (LOX-PP)). The presence of LOX-PP in the cell layers of phenotypically normal osteoblast cultures led us to investigate the effects of LOX-PP on osteoblast differentiation. Data indicate that LOX-PP inhibits terminal mineralization in primary calvaria osteoblast cultures when added at early stages of differentiation, with no effects seen when present at later stages. LOX-PP was found to inhibit serum- and FGF-2-stimulated DNA synthesis and FGF-2-stimulated cell growth. Enzyme-linked immunosorbent assay and Western blot analyses show that LOX-PP inhibits FGF-2-induced ERK1/2 phosphorylation, signaling events that mediate the FGF-2-induced proliferative response. LOX-PP inhibits FGF-2-stimulated phosphorylation of FRS2α and FGF-2-stimulated DNA synthesis, even after inhibition of sulfation of heparan sulfate proteoglycans. These data point to a LOX-PP target at or near the level of fibroblast growth factor receptor binding or activation. Ligand binding assays on osteoblast cell layers with 125I-FGF-2 demonstrate a concentration-dependent inhibition of FGF-2 binding to osteoblasts by LOX-PP. In vitro binding assays with recombinant fibroblast growth factor receptor protein revealed that LOX-PP inhibits FGF-2 binding in an uncompetitive manner. We propose a working model for the respective roles of LOX enzyme and LOX-PP in osteoblast phenotype development in which LOX-PP may act to inhibit the proliferative response possibly to allow cells to exit from the cell cycle and progress to the next stages of differentiation.
Keywords: Cell/Differentiation, Extracellular Matrix/Collagen, Extracellular Matrix/Glycosaminoglycans, Growth Factors, Protein/Connective Tissues, Protein/Multifunctional, Signal Transduction/Protein Kinases/MAP, Tissue/Organ Systems/Bone
Introduction
The process of bone formation is highly coordinated and has been divided into three identifiable stages as follows: proliferation, matrix maturation, and mineralization (1). Tightly controlled expression and action of growth factors and extracellular matrix molecules act in an autocrine and paracrine manner to promote osteoblast development and differentiation (2–4). The fibroblast growth factors (FGFs)2 are a family of structurally related proteins, with 23 members identified (5). The role of FGF-2 in bone formation is highlighted by genetic studies in which overexpression of FGF-2 induces abnormal long bone formation, although its knockdown inhibits bone formation and reduces bone mass (6, 7). Additionally, several mutations in FGF receptors have been implicated in the etiology of human skeletal dysplasias (5, 8). FGF-2 is a key mitogen for various types of bone cells, including precursor cells, bone marrow stromal cells, calvarial osteoblasts, and mature osteoblasts (9–12). FGF-2 contributes to the expansion of the pool of cells during the proliferative phase (9, 11, 13). Its role in the later stages of matrix maturation and mineralization is, however, controversial, with both positive (14–17) and negative effects described (18–21). Similar to effects of other mitogenic growth factors (22–25), chronic in vitro exposure of osteoblasts to FGF-2 stimulates proliferation and inhibits late stages of osteoblast differentiation, whereas an early pulse of FGF-2 exposure results in ultimately enhanced mineralization (10, 12, 26).
Lysyl oxidase is a critical enzyme in the normal biosynthesis of the extracellular matrix. It is synthesized and secreted as a 50-kDa proenzyme (pro-LOX) and is then processed to ∼30-kDa mature enzyme (LOX) and ∼18-kDa lysyl oxidase propeptide (LOX-PP) by extracellular procollagen C-proteinases encoded by the BMP1, Tll1, and Tll2 genes (27–29).
The importance of LOX enzyme in catalyzing the final enzyme reaction required for subsequent normal biosynthetic cross-linking of collagen and elastin precursors and its role in extracellular matrix production and maintaining correct bone phenotype are established (30–32). However, the biological functions of the released propeptide are less understood. The LOX gene was found to have tumor suppressor properties and is described as a “RAS-rescission gene.” In a study from our group, Palamakumbura et al. (33) mapped this “RAS rescission” activity of LOX to its propeptide domain. The expression of LOX-PP in Her-2/neu-driven breast cancer cells was then found to inhibit anchorage-independent growth and migration of cells, and LOX-PP was found to suppress the growth of Her-2/neu-driven tumors in a xenograft model (34). LOX-PP inhibits the phosphatidylinositol 3-kinase/AKT and the ERK1/2 MAP kinase pathways, as well as levels of downstream NF-κB and cyclin D1 in breast, pancreatic, and lung cancer cell lines (34, 35) and in prostate (36) and oral cancer cell lines (37). In addition, LOX-PP inhibits DNA synthesis in cultures of phenotypically normal, primary rat vascular smooth muscle cells (38).
We demonstrated previously the presence of LOX-PP in differentiating MC3T3-E1 osteoblast cultures (39, 40). Here, we investigate potential functions for this molecule in osteoblast cultures. We report an inhibition of osteoblast proliferation by LOX-PP and inhibition of important signaling intermediates activated by FGF-2. In addition, data indicate that one mechanism of action of LOX-PP is to inhibit FGF-2 binding to its high affinity FGF receptors. These studies suggest a possible biological role for LOX-PP in regulating osteoblast proliferation and point to the importance of both LOX enzyme and LOX-PP in bone formation.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant LOX-PP
Recombinant rat LOX-PP was generated and purified to homogeneity as described earlier (38). Detailed characterization of rLOX-PP will be published elsewhere.3
Primary Rat Calvaria Cell Culture
Calvaria were collected from 19-day-old CD IGS rat fetuses (Charles River Laboratories), and cells were isolated by trypsin/collagenase digestion (41). Briefly, calvaria were freed of adherent connective tissue and placed in digestion solution containing 0.175% trypsin (Invitrogen) and 1 mg/ml collagenase P (Roche Applied Science) in sterile PBS (containing calcium and magnesium chloride) at 37 °C and 5% CO2 in a fully humidified incubator. Three serial digestions were performed for 20, 20, and 90 min each. Cells released from the last digestion were collected by centrifugation and counted. 5.0 × 105 cells were plated in 10-cm culture plates and grown in media containing α-MEM supplemented with 10% fetal bovine serum (FBS), 1% nonessential amino acids, 100 units/ml penicillin, and 100 μg/ml streptomycin. Upon confluence, cells were passaged and plated in 24-well plates. For experiments where DNA synthesis was to be assayed, cells were plated at a density of 2.5 × 104 cells/well. For differentiation experiments, cells were plated at a density of 1.0 × 104 cells/well, and growth media were supplemented with 50 μg/ml ascorbic acid, 10 mm β-glycerophosphate, and 10 nm dexamethasone, to permit osteoblast differentiation and mineralization.
Alizarin Red Assay
Primary rat calvaria osteoblasts were cultured using differentiation media as described above. On day 14, culture media were removed; cells were washed twice with PBS and fixed in 10% phosphate-buffered formalin at room temperature for 30 min. Wells were then washed thoroughly with deionized water and air-dried overnight. To stain the mineral nodules, 1% alizarin red dye was added to each well for 10 min at room temperature and washed thoroughly under running water until the washes were colorless. Wells were then air-dried and photographed.
MC3T3-E1 Cell Culture
MC3T3-E1 subclone 4 (42) osteoblasts were obtained from American Type Culture Collection (ATCC). Cells were maintained in α-MEM supplemented with 10% FBS, 1% nonessential amino acids, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C and 5% CO2 in a fully humidified incubator. All experiments were carried out with cells of passage numbers 6–10. Unless otherwise noted, cell cultures were subjected to the following experimental protocol. MC3T3-E1 cells were plated at either 2.5 × 104 cells/well in 24-well plates or 8 × 104 cells/well in 6-well plates. Twenty four hours after plating, cells were transferred to serum-free medium containing 0.1% BSA for an additional 24 h. Cells were then treated as described in each experiment.
DNA Synthesis Assay
Rat calvaria cells or MC3T3-E1 cells were cultured as described above and placed in medium containing 0.1% BSA for 24 h. Cells were then treated with either serum (0–10%) or FGF-2 (0.25–10 ng/ml) or PBS with or without rLOX-PP or BSA or vehicle for 24 h. When indicated, 10 nm PD 173074 or DMSO was added 30 min prior to treatment. For the last 6 h of induction, [3H]thymidine (2 μCi) was added to each well. At the end of the incubation period, cells were washed three times in ice-cold PBS and then incubated with two washes of 5% trichloroacetic acid for 30 min on ice. Trichloroacetic acid was aspirated, and acid-insoluble precipitate was dissolved in 0.2 n NaOH, 0.5% SDS and subjected to liquid scintillation counting (Packard Tri-Carb 1500).
Cell Growth Assay
MC3T3-E1 cells were plated at a density of 2.0 × 104 cells/well in 24-well plates and cultured in 0.1% BSA as described above. Cells were treated with FGF-2 (1 ng/ml) or PBS with or without rLOX-PP (4 μg/ml) for 3 days. Cultures were harvested at 24-h intervals by washing twice in PBS, followed by fixation with 10% paraformaldehyde for 30 min at room temperature. Cells were then washed with PBS and then with water. Plates were air-dried overnight followed by staining with 0.1% crystal violet dye for 30 min under mild shaking. Plates were washed with water until the washes were colorless. Plates were then air-dried and cells counted in two randomly selected 1.7-mm2 fields per well, six wells per experimental group (n = 6) using Olympus MicroSuiteTM (Basic) software on a Zeiss Axiovert 200 microscope.
SDS-PAGE and Western Blotting
MC3T3-E1 cells grown in 6-well plates were cultured as described above and treated as indicated for each experiment. After treatments, cells were washed in ice-cold PBS and harvested in 200 μl of sample buffer containing 62.5 mm Tris-HCl, 2% SDS, 0.71 m β-mercaptoethanol, and 10% glycerol. Lysates were boiled for 5 min, and protein concentrations were determined with a NanoOrange® kit (Molecular Probes). Equal amounts of proteins from each sample were resolved by electrophoresis on 12% SDS-polyacrylamide gels and then transferred to polyvinylidene difluoride membranes overnight, followed by Western blotting. Membranes were blocked in 5% nonfat dry milk in TBST (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.5% Tween) at room temperature for 2 h. Blots were then incubated with primary antibody (1:1000) overnight in 5% milk/TBST. Primary antibodies employed were anti-phospho-p44/42 MAPK (catalog no. 9101S, Cell Signaling), anti-p44/42 MAPK (catalog no. 9102, Cell Signaling), anti-phospho-FRS2α (Tyr-436, catalog no. 3861S, Cell Signaling), and anti-β actin (catalog no. CP01, Calbiochem). Membranes were washed three times each for 15 min in TBST followed by incubation with the appropriate horseradish peroxidase-coupled secondary antibody for 1.5 h at room temperature. Membranes were washed and visualized with the enhanced chemiluminescent reagent (ECL, Amersham Biosciences) and x-ray film. Membranes were subsequently stripped using Restore Western Stripping Solution (Pierce) and then re-probed for loading controls as indicated. Radiographs were quantified using the Versadoc 3000 imaging system with the Quantity One software (Bio-Rad).
ERK1/2 FACETM ELISA
Fast-activated cell-based ELISA (FACETM, Active Motif) was used to analyze ERK1/2 activation in MC3T3-E1 cells, according to the manufacturer's instructions. Briefly, cells were grown in 96-well plates and then placed in 0.1% BSA and induced with FGF-2 (1 ng/ml) with or without rLOX-PP (4 μg/ml) for the indicated periods of time. Cells were rapidly fixed with 4% formaldehyde solution for 20 min and were then washed and blocked for 1 h, followed by incubation with primary antibodies against phospho-ERK1/2 (Thr-202/Tyr-204 and Thr-185/Tyr-187) for 1 h at room temperature. Wells were washed again and incubated with horseradish peroxidase-conjugated secondary antibody for an additional hour at room temperature. Wells were washed and then incubated with developing solution for 10 min. Absorbance at 450 nm was measured using an ELISA plate reader (Tecan). The cells were then washed and stained with crystal violet for 30 min. Bound dye was eluted with 1% SDS, and its absorbance was measured at 595 nm. Data are expressed as the ratio of A450/A595.
Inhibition of HSPG Sulfation with Sodium Chlorate
MC3T3-E1 cells were grown in 100-mm culture dishes until 80% confluent. Cells were then cultured in α-MEM containing 0.2% FBS (without penicillin and streptomycin) with or without 50 mm sodium chlorate for an additional 24 h to inhibit ATP sulfurylase and the consequent formation of adenosine 3′-phosphate,5′-phosphosulfate, which is required for sulfation of glycosaminoglycans (43). Cells were washed twice in PBS and treated with trypsin/EDTA for 5 min and then re-plated in 24-well plates at a density of 7 × 104 cells/well. Cells were allowed to attach for 2.5 h after which they were treated with FGF-2, PBS, rLOX-PP, or LOX-PP vehicle (water) as indicated. Treatment volumes included 0.1% or less of the total volume of media. Cells were then assayed for DNA synthesis after 24 h, as described above. Experimental groups were the following: the “chlorate” group contained 50 mm sodium chlorate during all the experimental treatments; the “untreated control” group was subjected to all experimental manipulations but without sodium chlorate; the “recovered” group consisted of cells that were exposed to chlorate for the first 24-h period, but chlorate was eliminated during the re-plating of cells and the 24-h FGF-2 treatment periods.
FGF Radiolabeling
125I-FGF-2 was prepared using a modified Bolton-Hunter procedure (44). Briefly, 1 mCi of 125I-labeled Bolton-Hunter reagent (PerkinElmer Life Sciences), in anhydrous benzene, was dried under a gentle stream of nitrogen gas. 10 μg of carrier-free recombinant human FGF-2 (PeproTech), reconstituted in 100 mm sodium phosphate buffer, pH 8.5, was added to the vial in a volume of 35 μl, and the reaction was allowed to progress for 2.5 h on ice. The reaction was then quenched using 200 μl of 0.2 m glycine for 45 min on ice. The volume of the reaction was brought up to 500 μl with column equilibration buffer (50 mm Tris, pH 7.5, 1 mm dithiothreitol, 0.3 m NaCl, 0.05% gelatin). Trichloroacetic acid precipitation was performed on an aliquot to determine the specific radioactivity of labeled FGF-2 (activity present in the precipitate). To separate the labeled FGF-2 from low molecular weight reagents, the entire sample was applied to a pre-equilibrated, PD-10 Sephadex G-25M column (GE Healthcare). The void fractions containing 125I-FGF-2 peak were pooled. The specific activity was typically ∼29.4 μCi/μg. Aliquots of prepared stocks were stored at −80 °C for no longer than 8 weeks.
Radioligand Binding Assays
MC3T3-E1 cells were plated at 2.5 × 104 cells/well of a 24-well plate. At 90% confluence, the medium was changed to serum-free medium containing 0.1% BSA. The following day, equilibrium binding assays were performed as described earlier (44, 45). Cells were washed three times with ice-cold binding buffer (α-MEM, 25 mm HEPES, 0.1% BSA) and then equilibrated with fresh binding buffer for 10 min in a final volume of 500 μl. Vehicle or rLOX-PP was then added for an additional 15 min, before incubating with radiolabeled 125I-FGF-2 (1 ng/ml) for 2 h at 4 °C. At the end of the incubation period, cells were washed three times in cold binding buffer. Bound 125I-FGF-2 was released using 1 n NaOH and quantified using a Cobra II auto-gamma 5005 counter. Excess nonlabeled (500-fold) FGF-2 in column equilibration buffer was added to parallel wells to analyze nonspecific binding.
In Vitro Binding Assays
In vitro binding assays were performed using recombinant chimeric FGFR1-β (IIIc) fused to the Fc domain of human IgG (R & D Systems) immobilized onto Reacti-Bind protein A/G-coated 8-well strip plates (Pierce) via the Fc domain of the fusion protein, so as to preserve the availability of the receptor for ligand binding. Briefly, 10 ng of FGFR1-Fc reconstituted in PBS, 0.1% BSA was incubated in ice-cold binding buffer (α-MEM, 25 mm HEPES, 0.1% BSA) for 2 h to bind to each well. Following removal of unbound FGFR1, the indicated wells were pretreated with rLOX-PP (1–16 μg/ml) or vehicle for 15 min and then incubated with increasing concentrations of 125I-FGF-2 (1–50 nm; total reaction volume = 100 μl). The binding reaction was allowed to progress to equilibrium at 4 °C for 2 h. After washing the unbound ligand, the radioactivity retained in each well was measured using a Cobra II auto-gamma 5005 counter. Nonspecific binding was determined by a parallel set of experiments in which 1500-fold excess unlabeled FGF-2 was added during binding. Nonspecific binding was subtracted from the total observed binding, and the data were analyzed using a single site binding model, according to Equation 1,
where [bound FGF-2] and [free FGF-2] denote the concentrations of bound and free ligand, respectively; Bmax indicates the maximum number of receptor binding sites, and Kd indicates the dissociation equilibrium constant. Data fit was conducted in KaleidaGraph 3.6 (Synergy Software).
RESULTS
Addition of rLOX-PP Early during Osteoblast Differentiation Inhibits Mineralization
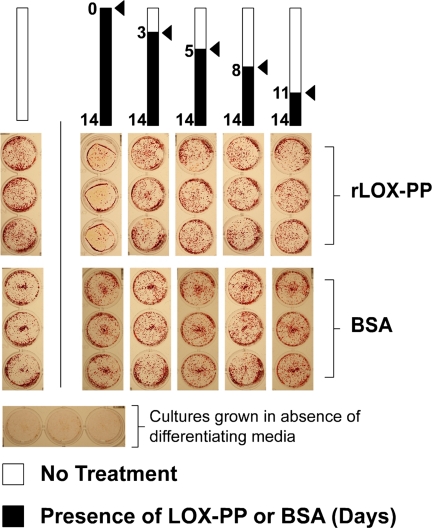
The presence of LOX-PP in differentiating osteoblast cultures has been reported previously (39, 40). To investigate whether LOX-PP has the potential to participate in osteoblast development, we tested the effects of rLOX-PP on differentiating osteoblast cultures. Primary rat calvaria osteoblasts were induced to differentiate in α-MEM supplemented with ascorbate, β-glycerophosphate, and dexamethasone. rLOX-PP or BSA was added beginning at different times (days 0, 3, 5, 8, and 11) and was maintained throughout the remainder of the 14-day experimental period as indicated (Fig. 1). Cultures were fixed and stained with alizarin red to visualize mineralized nodule formation. The presence of rLOX-PP beginning on day 0 of differentiation results in a nearly complete inhibition of mineralization, although its addition at later time points does not have any apparent effect on mineralization compared with its untreated control grown on the same 24-well plate (Fig. 1). In contrast, the presence of BSA (4 μg/ml) has no effect on mineral nodule formation compared with its respective untreated control grown on the same 24-well plate.
FIGURE 1.
Early addition of rLOX-PP inhibits mineralization of osteoblast cultures. Primary rat calvaria osteoblasts were cultured in differentiating medium and treated with either rLOX-PP or BSA control (4 μg/ml) beginning on the days indicated and continuing for the remainder of the experiment (black bars). After 14 days of culture, cells were fixed and stained with alizarin red to visualize mineralized nodules. The figure shows one of two separate experiments with the same outcomes.
rLOX-PP Inhibits Serum-induced Osteoblast DNA Synthesis
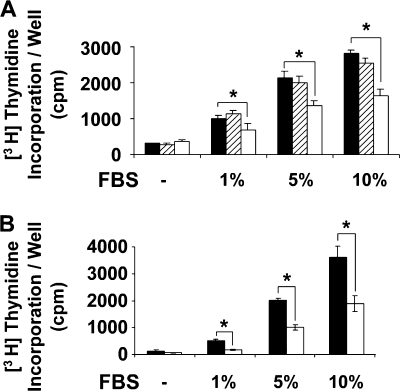
Because rLOX-PP inhibits mineralization of osteoblasts only when present during early stages of differentiation (Fig. 1), we reasoned that rLOX-PP might inhibit osteoblast proliferation. Hence, we tested the effect of LOX-PP on serum-stimulated DNA synthesis. Cells were stimulated for 24 h with increasing concentrations of FBS (0–10%) in the absence or presence of rLOX-PP (4 μg/ml). As seen in Fig. 2A, increasing FBS concentrations result in increased DNA synthesis that is dose-dependent. In the presence of rLOX-PP, this stimulation is significantly inhibited in all groups by at least 50%. Similar results were obtained with phenotypically the normal mouse osteoblastic MC3T3-E1 cell line (Fig. 2B).
FIGURE 2.
rLOX-PP inhibits fetal bovine serum-induced DNA synthesis in osteoblasts. Primary rat calvaria osteoblasts (A) or murine MC3T3-E1 cells (B) were grown in 24-well plates until 80% confluent and then placed in 0.1% BSA medium for 24 h. Cells were then treated with the indicated concentrations of serum (FBS) in the presence of 4 μg/ml rLOX-PP (white bars) or BSA control (hatched bars) or vehicle (solid bars) for a total of 24 h. Incorporation of [3H]thymidine into DNA was measured by trichloroacetic acid precipitation. Data shown are mean counts/min values/well ± S.D. from one experiment (n = 3, *, p < 0.05). This experiment was performed two times with similar results.
FGF Signaling Is a Significant Mediator of Serum-stimulated DNA Synthesis
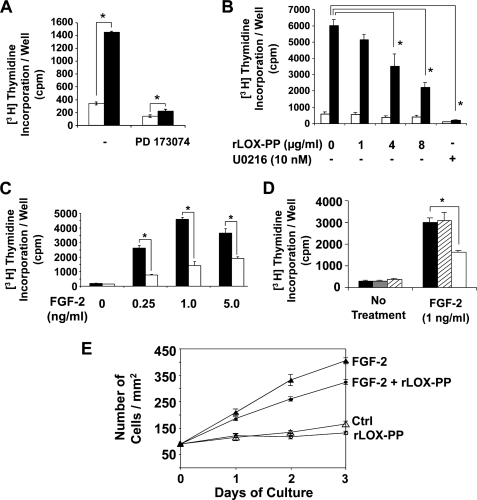
Serum either contains or induces cells to produce a variety of growth factors that act as mitogens in a paracrine or autocrine manner. The importance of FGF signaling in osteoblasts is demonstrated in a study by Fakhry et al. (10), where the expression of a dominant negative FGF receptor in calvaria osteoblasts was found to inhibit basal proliferation by more than 50%. We therefore inhibited FGF receptor activity by pretreating serum-deprived MC3T3-E1 cells with 10 nm PD 173074, a pharmacological inhibitor of FGFR activity, or DMSO (vehicle) control. This concentration of PD 173074 is known to inhibit FGF receptor tyrosine kinase activity (46). Thirty minutes after treatment, cells were treated with 1% serum for a period of 24 h, and DNA synthesis was measured as described earlier. Fig. 3A shows that in the presence of the inhibitor, serum addition results in only a modest increase in DNA synthesis (∼1.5-fold), as opposed to the strong induction in the control group without the inhibitor (∼4.2-fold). Hence, serum-induced DNA synthesis is largely blocked in the presence of a pharmacological inhibitor of FGF receptors, suggesting that FGF signaling mediates the serum-induced proliferative response of osteoblasts to a significant degree.
FIGURE 3.
Serum stimulation of DNA synthesis in osteoblasts depends in part on FGF signaling (A), and rLOX-PP inhibits FGF-2-induced DNA synthesis (B–D) and proliferation of osteoblasts (E). MC3T3-E1 cells were plated in 24-well plates (2.5 × 104 cells/well), and after 24 h cells were placed in medium containing 0.1% BSA medium without serum. A, after 24 h, cells were treated with either 10 nm PD 173074 (FGFR inhibitor) or DMSO (vehicle) for 30 min and then treated with 1% FBS (black bars) or serum-free medium (white bars) for an additional 24 h. [3H]Thymidine was added for the last 6 h, and its incorporation into DNA was measured by trichloroacetic acid precipitation. Data are mean counts/min values per well ± S.D. (n = 3, *, p < 0.05). After 24 h, cells were treated for an additional 24 h with FGF-2 (1 ng/ml) (black bars) or PBS (white bars) in the presence of increasing concentrations of rLOX-PP (0–8 μg/ml) or 10 nm U0216 (MEK inhibitor) or vehicle (B) or with increasing concentrations of FGF-2 (0–5 ng/ml) in the presence of 4 μg/ml rLOX-PP (white bars) or vehicle (black bars) (C). [3H]Thymidine was added for the last 6 h, and levels of radioactive thymidine incorporated into DNA were measured by trichloroacetic acid precipitation. Data are from one experiment and are mean counts/min values/well ± S.D. (n = 3, *, p < 0.05). Experiments were performed three times with the same outcomes. D, primary rat calvaria osteoblasts were treated with FGF-2 (1 ng/ml) in the presence of 4 μg/ml rLOX-PP (white bars), BSA control (hatched bars), or vehicle (solid bars), and DNA synthesis was assessed as described above. Data are from one experiment and are mean counts/min values/well ± S.D. (n = 3, *, p < 0.05). E, MC3T3-E1 cells were plated in 24-well plates (2.0 × 104 cells/well) for 24 h and then placed in 0.1% BSA medium without serum. After 24 h, they were treated with FGF-2 (1 ng/ml) or vehicle (serum-free control medium) for a total of 3 days in the presence of 4 μg/ml rLOX-PP or vehicle. Cultures were harvested at 24-h intervals and fixed in 10% paraformaldehyde and stained using 0.1% crystal violet dye. Cells present in area of 1.75 mm2 were counted with a Zeiss Axiovert 200 microscope and Olympus MicroSuiteTM (Basic) software. Two randomly selected areas were used from each well, and six wells were used per experimental condition (n = 6). Data are mean number of cells/mm2 ± S.E. Two-way analysis of variance indicates significant inhibition in cell numbers in the presence of rLOX-PP in FGF-2-treated cells (p < 0.001) and not in the absence of FGF-2 (p > 0.05). Ctrl, control.
rLOX-PP Inhibits FGF-2-induced DNA Synthesis and Cell Growth
The effect of rLOX-PP on FGF-2-induced DNA synthesis of osteoblast cultures was next determined. Cells were plated, and after 24 h the medium was changed to α-MEM containing 0.1% BSA (without serum), and cells were grown for an additional 24 h. Cells were then treated with recombinant human FGF-2 (1 ng/ml) in the presence of absence of rLOX-PP for 24 h, and DNA synthesis was measured. Data show that FGF-2 alone resulted in an 8-fold increase in DNA synthesis, which is inhibited by increasing levels of rLOX-PP in MC3T3-E1 cultures (Fig. 3B). Data in Fig. 3C show that rLOX-PP inhibits DNA synthesis over a range of FGF-2 concentrations. Inhibition of FGF-2-induced DNA synthesis was also observed in primary rat calvaria osteoblasts (Fig. 3D), although equal amounts of BSA have no inhibitory effects.
To establish that LOX-PP inhibition of DNA synthesis results in an inhibition of cell culture growth, MC3T3-E1 cultures were serum-deprived for 24 h and then treated with FGF-2 (1 ng/ml), in the presence or absence of rLOX-PP (4 μg/ml). Cultures (n = 6) were harvested, stained, and counted at 24-h intervals for 72 h. Fig. 3E shows that 4 μg/ml rLOX-PP significantly inhibits the FGF-2-induced increase in cell numbers (p < 0.001), although LOX-PP alone does not affect cell culture growth (p > 0.05, two-way analysis of variance).
rLOX-PP Inhibits FGF-2-induced Phosphorylation of ERK1/2 and FRS2α
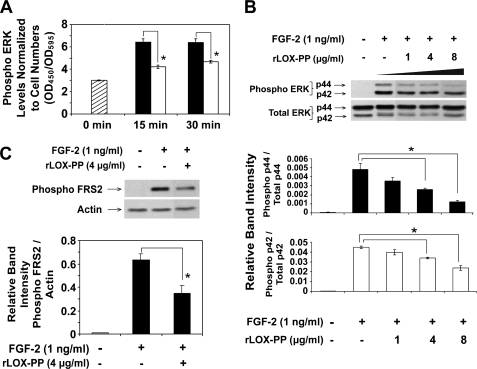
We next sought to identify a mechanism by which LOX-PP inhibits FGF-2-stimulated DNA synthesis and cell proliferation. ERK1/2 MAP kinases are important signaling intermediates involved in FGF-2-induced DNA synthesis (10, 47, 48). Because earlier studies have demonstrated that LOX-PP inhibits ERK1/2 phosphorylation (34, 35, 49), we tested the effect of LOX-PP on FGF-2-induced ERK1/2 phosphorylation in MC3T3-E1 cells first with an ELISA. Cells were seeded and grown in 96-well plates and serum-deprived for 24 h followed by treatment with FGF-2 (1 ng/ml) in the presence or absence or rLOX-PP (4 μg/ml). Cells were then fixed at the indicated time points and probed with phospho-specific antibodies against ERK1/2 and subsequently with crystal violet for normalization as described under “Experimental Procedures.” Data (Fig. 4A) show that rLOX-PP (4 μg/ml) inhibits FGF-2-induced ERK1/2 phosphorylation over base-line levels by nearly 50% at 15 and 30 min following induction.
FIGURE 4.
rLOX-PP inhibits FGF-2-induced ERK1/2 and FRS2α phosphorylation determined by ELISA (A) and Western blotting (B). The effect of rLOX-PP on FGF-2-induced ERK1/2 activation was first analyzed using the FACETM (Active Motif). A, MC3T3-E1 cells were grown in 96-well plates for 24 h in medium containing 0.1% BSA. Cells were induced with FGF-2 (1 ng/ml) in the presence of 4 μg/ml rLOX-PP (white bars) or vehicle (black bars) for 0–30 min, after which the cells were fixed and later probed with primary antibodies against phospho-ERK1/2. Levels of phosphoprotein were determined colorimetrically (A450). Wells were subsequently stained with crystal violet to allow for normalization (A595). Bars indicate mean readings (A450/A595) for each group ± S.E. (n = 4, *, p < 0.05). In Western blotting experiments (B and C), MC3T3-E1 cells were plated in 6-well plates (8 × 104 cells/well), and after 24 h, cells were placed in medium containing 0.1% BSA for 24 h. Cells were then treated with FGF-2 (1 ng/ml) in the presence of 0–8 μg/ml rLOX-PP for 20 min (B) or 4 μg/ml rLOX-PP for 5 min (C). Cell lysates were collected in SDS-PAGE sample buffer and subjected to SDS-PAGE followed by Western blot analysis. Upper panels show representative blots probed with antibody specific for phosphorylated ERK1/2 (B) or phosphorylated FRS2α (Tyr-436) and for total ERK1/2 and β-actin for normalization (C). The lower panel shows mean values obtained from densitometric analysis of blots ± S.D. from one experiment (n = 3, *, p < 0.05). This experiment was performed twice with the same outcome.
To confirm these findings, MC3T3-E1 cells were next treated with FGF-2 (1 ng/ml) for 20 min, in the absence or presence of increasing concentrations of rLOX-PP (1–8 μg/ml). Cell lysates were collected and subjected to SDS-PAGE and Western blotting using phospho-specific antibodies against ERK1/2. A dose-dependent inhibition of ERK phosphorylation by rLOX-PP was observed (Fig. 4B). Moreover, the dose response of rLOX-PP inhibition of FGF-2-stimulated ERK1/2 activation is similar to the dose response for rLOX-PP inhibition of FGF-2-stimulated DNA synthesis (Fig. 3B and Fig. 4B).
FRS2 is a substrate for FGFRs, thereby acting immediately downstream of the FGF receptors (50). It serves as an adaptor protein that relays signals from the phosphorylated and active FGFRs to GRB-SOS-SHP complexes that activate RAS and ultimately leads to activation of ERK1/2 MAP kinases. MC3T3-E1 cells in serum-free medium containing 0.1% BSA were treated with FGF-2 (1 ng/ml) for 5 min, in the presence or absence of rLOX-PP (4 μg/ml). Cell lysates were collected and subjected to Western blot analysis using antibodies against phosphorylated FRS2α (Tyr-436). A 50% inhibition in the levels of FGF-2-induced phosphorylation of FRS2α was observed in the presence of rLOX-PP (Fig. 4C).
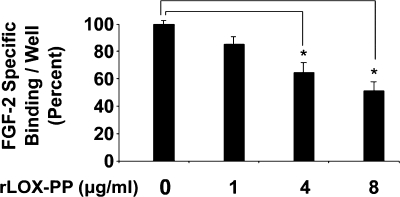
rLOX-PP Inhibits Binding of FGF-2 to MC3T3-E1 Cells
The finding that LOX-PP can inhibit FGF-2-activated FRS2α phosphorylation points to an inhibition at the level of FGF receptor function. The effect of LOX-PP on binding of FGF-2 to MC3T3-E1 monolayers was therefore tested. Osteoblasts were exposed to 125I-FGF-2 in the presence of varying concentrations of rLOX-PP (0–8 μg/ml) for 2 h at 4 °C. Unbound ligand was washed out, and the bound radiolabeled ligand was quantified. Specific binding was assessed by competing with excess, nonlabeled FGF-2. A dose-dependent inhibition of specific binding of FGF-2 to osteoblasts was observed in the presence of increasing concentrations of rLOX-PP (Fig. 5). Moreover, the dose response for rLOX-PP inhibition of FGF-2 binding observed here is similar to the dose response of rLOX-PP inhibition of FGF-2-induced DNA synthesis and ERK activation (Figs. 3 and 4).
FIGURE 5.
rLOX-PP inhibits FGF-2 binding to MC3T3-E1 osteoblast cell layers. Preconfluent MC3T3-E1 cells were grown in 24-well plates (2.5 × 104 cells/well) in medium containing 0.1% BSA for 24 h. An equilibrium binding assay was then performed as described under “Experimental Procedures” with 125I-FGF-2 in the presence or absence of increasing concentrations of rLOX-PP (0–8 μg/ml). Wells were then washed, and bound 125I-FGF-2 was measured. Specific binding was assessed using 500-fold excess nonlabeled FGF-2. Data are expressed as percent of FGF-2 bound/well measured in the absence of rLOX-PP. Data from three separate experiments were pooled (n = 10, *, p < 0.05).
rLOX-PP Inhibits Signaling Induced by FGF-FGFR Interactions
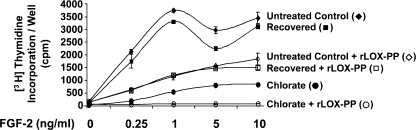
FGF-2 binds in a ternary complex to high affinity FGF receptors as well as to cell surface heparan sulfate proteoglycans (51, 52). It is therefore possible that LOX-PP inhibits FGF-HSPG interactions that are required for optimal FGFR activation. Alternatively, LOX-PP could directly inhibit FGF-FGFR binding independent of HSPGs. To determine whether the observed inhibition of FGF binding to cells is due primarily to inhibition of FGF-FGFR interactions or FGF-HSPG interactions, cells were treated with sodium chlorate to inhibit HSPG sulfation. Without the necessary sulfations, HSPGs cannot participate in the FGF-FGFR-HSPG ternary complex (12, 53–55). Such a treatment results in diminished FGF-2-induced cellular responses, enabling analysis of FGF-2 activity directly mediated by its high affinity receptors in the absence of functional HSPGs.
MC3T3-E1 cells were pretreated with 50 mm sodium chlorate for a period of 24 h before testing the ability of FGF-2 to stimulate DNA synthesis. Moreover, to ensure that residual sulfated glycosaminoglycans are eliminated, cells were trypsinized to cleave cell surface HSPGs and re-plated prior to treatment (53). Cells were permitted to adhere for a period of 2.5 h, following which they were treated with increasing concentrations of FGF-2 (0–10 ng/ml) in the presence or absence of rLOX-PP (4 μg/ml) for a period of 24 h. [3H]Thymidine was added for the last 6 h of treatment and its incorporation into DNA was assessed. The chlorate group was exposed to 50 mm chlorate during all of these steps. Chlorate was not used in the untreated control group, but cells were subjected to all the steps indicated. To ensure that the chlorate treatment protocol did not affect cells irreversibly, cells were treated with chlorate initially for the 24-h period and were replated in the absence of sodium chlorate and treated with FGF-2 (recovered group). Because HSPGs undergo a rapid turnover (56), these cells are expected to mimic the untreated control cells (grown without chlorate) in their response to FGF-2 and LOX-PP.
Fig. 6 shows that chlorate treatment of cells inhibits DNA synthesis induced by FGF-2, as expected (chlorate group compared with untreated control group). Interestingly, rLOX-PP further inhibits DNA synthesis induced by FGF-2 fully to base-line levels in the chlorate group, indicating a potent inhibition by LOX-PP of the signaling events induced by FGF-FGFR interactions independent of HSPGs. The recovered cells closely follow the untreated control cells indicating the reversibility of the chlorate effect.
FIGURE 6.
rLOX-PP inhibits signaling induced by FGF-FGFR. Cells were grown in the absence (untreated control group) or presence (chlorate and recovered groups) of 50 mm sodium chlorate for 24 h to inhibit the sulfation of HSPGs. Cells were then trypsinized and re-plated in the presence (chlorate group) or absence (recovered and untreated control groups) of 50 mm sodium chlorate (see “Experimental Procedures”). Cells were allowed to attach for 2.5 h and then treated with FGF-2 (0–10 ng/ml) in the presence or absence or rLOX-PP (4 μg/ml) for 24 h. The DNA synthesis assay was used to determine DNA synthesis (“Experimental Procedures”). Data are mean counts/min values for each group ± S.D. from one experiment. Treatment with rLOX-PP shows statistically significant decreases in all groups where present, compared with respective controls, at all concentrations tested, n = 3 (p < 0.005). This experiment was performed twice with similar results.
Increasing the concentration of FGF-2 beyond 1 ng/ml did not continuously increase DNA synthesis in control cells (Fig. 6) indicating a peak in FGF-2 response at this concentration. Such concentration-dependent effects of FGF-2 have been described before in osteoblasts as well as other cell types in the presence of the normal complement of HSPGs (10, 11, 45). In the chlorate treatment group, however, an increase in DNA synthesis was observed with increasing concentrations of FGF-2 beyond 1 ng/ml, again consistent with other studies (45), validating these experiments. Most important, in the presence of rLOX-PP, DNA synthesis is fully inhibited at all concentrations of FGF-2 in the chlorate-treated cells (Fig. 6). Thus, rLOX-PP fully inhibits the induction of DNA synthesis by FGF-2 via its high affinity receptors in the absence of HSPGs. Taken together with data obtained in the binding experiments (Fig. 5), these results suggest that rLOX-PP inhibits FGF-2 binding to its high affinity FGF receptors.
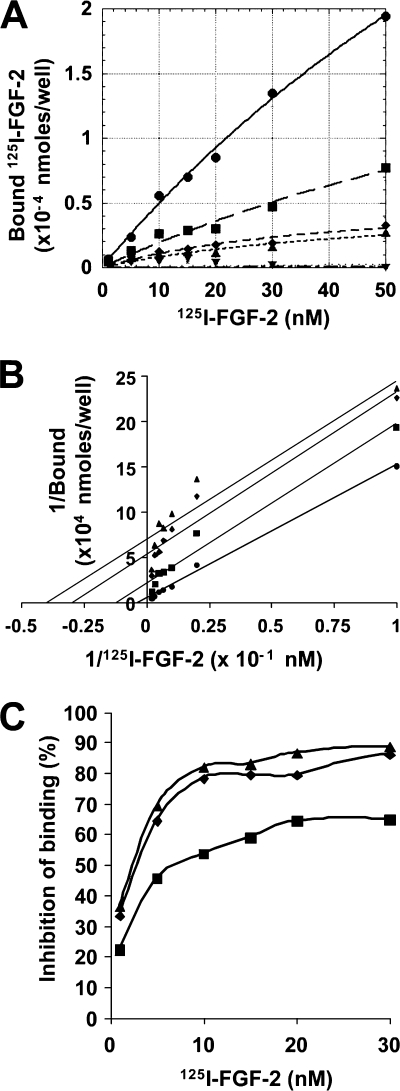
rLOX-PP Inhibits in Vitro Binding of FGF-2 to FGFR
To determine whether LOX-PP indeed inhibits binding of FGF to its high affinity receptors, in vitro binding assays were performed with recombinant FGFR and 125I-FGF-2. Briefly, chimeric FGFR molecules, which contain the extracellular domain of FGFR1-β (IIIc) fused to the Fc domain of human IgG, were absorbed onto protein A/G-coated wells and were incubated with 125I-FGF-2 (1–50 nm) in the presence of increasing concentrations of rLOX-PP. The binding reaction was allowed to progress at 4 °C for 2 h, after which the wells were washed, and the radioactivity retained was measured. The data shown in Fig. 7A were analyzed using a simple single site model for the interaction between FGF-2 and FGFR1 (see under “Experimental Procedures”). Data showed that in the presence of increasing concentrations of LOX-PP, both the Kd value for FGF-2 and the maximum binding for FGF-2 decrease, consistent with uncompetitive binding. A double-reciprocal plot (Fig. 7B) resulted in parallel lines, consistent with uncompetitive kinetics. Uncompetitive kinetics of LOX-PP inhibition of FGF-2-FGFR1 binding is further confirmed in Fig. 7C, in which the binding of FGF-2 is inhibited to a greater degree at higher concentrations of FGF-2 than at lower concentrations (57). These analyses suggest that LOX-PP forms a trivalent complex with FGF-2 and FGFR1 and thereby inhibits FGFR1 activity. Such a mechanism of action is consistent with LOX-PP attenuation of FGF-2 binding and signaling that is less than 100% inhibition, as is seen throughout this study. Such observations are characteristic of uncompetitive inhibitors of receptors (58).
FIGURE 7.
rLOX-PP inhibits binding of FGF-2 to high affinity FGF receptors. Equilibrium binding analyses were performed with FGFR1-β (IIIc)-Fc molecules prebound to protein A/G-coated plates and 125I-FGF-2 in concentrations ranging between 1 and 50 nm in the presence or absence of rLOX-PP (0–16 μg/ml). The binding reaction was allowed to progress at 4 °C for 2 h, after which free and bound ligands in each well were measured. A 1500-fold excess nonlabeled FGF-2 was used to determine nonspecific binding. This experiment was performed twice with similar results. A, data represent mean values for specific bound FGF-2 plotted against total free ligand ± S.E. (n = 3) either in the absence (●) or in the presence of 2 μg/ml (■), 4 μg/ml (♦), 8 μg/ml (▴), and 16 μg/ml (▾) rLOX-PP. B, double-reciprocal (Lineweaver-Burke) plot of data representing 1/[FGF-2] (x axis) against 1/bound (y axis). Correlation coefficients for each group are as follows: FGF alone (●) R2 = 0.99; rLOX-PP 2 μg/ml (■) R2 = 0.97; 4 μg/ml (♦) R2 = 0.93; and 8 μg/ml (▴) R2 = 0.89. C, FGF-2 concentration versus percent inhibition at different concentrations of rLOX-PP as follows: 2 μg/ml (■), 4 μg/ml (♦), and 8 μg/ml (▴).
DISCUSSION
Two transition points have been suggested to occur in the normal differentiation of osteoblasts (1, 59). The first occurs when cells become less proliferative down-regulating genes important for cell cycle and cell growth control, while up-regulating those for extracellular matrix production and maturation. The second transition point occurs at the onset of mineralization. Although proliferation is a primary requirement for osteoblast development, an exit from the cell cycle with a concomitant commitment to a phenotypic lineage is equally imperative. Deregulation of this process is detrimental for bone formation as is apparent from observations in osteosarcoma cells in which cell cycle-dependent events are deregulated (60).
Here, we show that rLOX-PP inhibits the mineralization of primary calvaria osteoblast cultures when present during the early proliferative stages of osteoblast differentiation, suggesting that LOX-PP inhibits osteoblast proliferation. In accordance with this hypothesis, data show that rLOX-PP inhibits serum-induced DNA synthesis in primary calvaria osteoblasts and in the phenotypically normal murine osteoblast MC3T3-E1 cell line. These findings may have important implications regarding the roles of both LOX enzyme and LOX-PP in the control of osteoblast differentiation and function, as outlined below.
We have previously demonstrated that levels of pro-LOX mRNA are initially low and increase through the early phases of osteoblast development (39). Immunohistochemistry and Western blot analysis of differentiating MC3T3-E1 cultures using anti-LOX-PP antibody showed maximum immunoreactivity at time points that correspond to the end of the proliferative phase of osteoblast differentiation (39). Initial low levels of LOX-PP during this early phase may help to permit the necessary proliferation of osteoblasts required for ultimate bone formation, although time-dependent increasing levels of LOX-PP are suggested to inhibit proliferation and permit subsequent stages of differentiation.
Data presented here are not sufficient to identify a role of LOX-PP in the later stages of differentiation. Although we see no apparent effects for the addition of rLOX-PP after day 3 in mineralizing cultures, these cells may have accumulated high levels of endogenous LOX-PP in the matrix by this time (39), rendering the added recombinant protein redundant. Although knockdown studies seemingly could help to address the role of LOX-PP in later stages of differentiation, LOX gene knockdown results in the loss of both mature LOX enzyme and LOX-PP. LOX enzyme itself is crucial for collagen cross-linking and osteoblast phenotype development and mineralization (61). Complex outcomes in the phenotype of murine LOX−/− calvaria osteoblasts have been reported recently (62) and result from the loss of several functions that depend on production of the LOX proenzyme, LOX-PP, and mature LOX. Ongoing experiments are investigating possible functions for LOX-PP itself at later stages of osteoblast differentiation.
The growth inhibitory effects of LOX-PP on osteoblast cultures are in line with recent studies that demonstrate growth inhibitory activity of LOX-PP in some transformed cell lines and in primary vascular cells (33, 34, 38). In vascular smooth muscle cells, tumor necrosis factor-α signaling is inhibited (38), although in breast cancer cells RAS-dependent pathways and FAK activation and haptotaxis are inhibited (63). Taken together, these findings suggest that LOX-PP has more than one molecular target and mechanism of action. Several mitogens have been shown to induce the proliferation of osteoblasts. Of these, the FGFs have been found to play a central role in bone formation and osteoblast function (64, 65) and are potent inducers of osteoblast proliferation (9–11, 26, 66). Here, we demonstrate that a major mode of action of LOX-PP is to inhibit FGF-2-induced DNA synthesis and cell growth in developing normal primary and MC3T3-E1 osteoblasts.
FGFs activate a range of signaling molecules leading to cell proliferation, mediated largely by the RAS-RAF-MEK-ERK MAP kinase pathway (10, 47, 48). Here, rLOX-PP was found to inhibit FGF-2-induced ERK phosphorylation in MC3T3-E1 cells. Although most receptor tyrosine kinases activate signaling via the small adaptor protein GRB2 (67), FGFRs cannot directly bind GRB2 (50). Instead, FRS2 (fibroblast growth factor receptor substrate-2), serves as a link between the FGFRs and the downstream RAS-MAP kinase pathway (68). Data presented here show that rLOX-PP inhibits FGF-2 activation of FRS2α. Because FRS2α acts immediately downstream of the FGF receptors, we investigated whether LOX-PP may act at the level of these receptors. Fig. 5 shows that LOX-PP inhibits FGF-2 binding to cell layers in a dose-dependent manner.
FGFs bind to high affinity FGF receptors as well as cell surface HSPGs (69–71) in a ternary complex (51, 52). Although the molecular details of this interaction are not fully characterized, there is general agreement that HSPGs increase the affinity of FGFs to their receptors (72). Hence, we wished to investigate whether the inhibition of binding by LOX-PP was due to an inhibition of FGF-FGFR interactions or FGF-HSPG interactions. Studies indicate that nonsulfated HSPGs cannot interact with the FGF-FGFR complex (12, 53–55). Inhibition of HSPG sulfation as presented here points to a mechanism in which LOX-PP inhibits the interaction between FGF and its high affinity receptors independent of HSPGs. Moreover, in vitro ligand binding assays performed with radiolabeled FGF-2 and rFGFR1 show that LOX-PP indeed inhibits FGF-2 binding to FGF receptor chimera. Specifically, LOX-PP decreased the total available binding sites while at the same time decreasing the dissociation constant, suggestive of uncompetitive inhibition. Uncompetitive inhibition implies that LOX-PP interacts with a complex of the receptor and the ligand, resulting in decreased receptor activation. Further protein-protein interaction studies are needed to fully understand this interesting mechanism of action of LOX-PP.
Studies demonstrate that the stimulatory effect of FGF-2 on bone formation lies primarily in its ability to increase osteogenic cell populations (73–75). Additionally, FGF signaling is important in the growth and proliferation of osteoblasts in vivo and in vitro (76–78). Hence, although there is general consensus that FGF has a positive, inductive effect on osteoblast proliferation (9–11, 26, 66), the role of FGF in the later stages of differentiation is controversial. FGF-2 has been shown to up-regulate Runx2, osteocalcin, and fibronectin (14–17). At the same time FGF-2 down-regulates transcripts of type 1 collagen, lysyl oxidase, as well as alkaline phosphatase (18–21), molecules that are important at later stages of differentiation.
This variety of FGF-2 effects may be due to stage-specific effects on osteoblast differentiation (10, 79). Studies suggest that prolonged exposure to FGF-2 is unfavorable to normal bone mineralization, although short term exposure is beneficial (10, 12, 26). Hence, although the proliferative effect of FGF-2 is necessary, its favorable effect is temporal, and its actions need to be regulated, specifically beyond the first transition point. In vivo, bone cells may be able to regulate the actions of FGF-2 by either temporally regulating levels of the growth factor and/or expression of their receptors (11). Other cell surface, ECM, and intracellular molecules influence FGF-2 activity, such as the expression of diverse cell surface HSPGs and their differential modifications (12, 53, 54). For example, Jackson et al. (53) showed that heparan sulfate harvested from differentiating MC3T3-E1 cultures, when re-applied to proliferating MC3T3-E1 cultures, results in inhibition of FGF-2 mitogenic potential, indicating that these cultures are regulating the production of heparan sulfate molecules capable of antagonizing FGF-2 signaling. Our data suggest that LOX-PP could potentially similarly modulate FGF-2-induced proliferative cues.
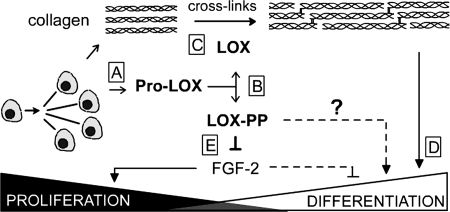
Hence, we propose a working model for the roles of both LOX-PP and LOX enzyme in osteoblast differentiation (Fig. 8). As the cells progress through the proliferative stage, they increasingly synthesize pro-LOX (Fig. 8A), which is cleaved by extracellular procollagen C-proteinases, hence accumulating LOX-PP and LOX enzyme (Fig. 8B) (80). The LOX enzyme is required for the formation of cross-links in collagen molecules (Fig. 8C), which contribute to the formation of a mature, mineralized osteoblast matrix (Fig. 8D) (31, 32, 39). At the same time, data suggest that LOX-PP may function to inhibit proliferation of osteoblasts beyond the first transition point, in part by regulating FGF-2-induced proliferation of cells (Fig. 8E). Such an inhibition would facilitate the entry of cells into subsequent stages of osteoblast differentiation.
FIGURE 8.
Working model for the respective roles of LOX-PP and LOX enzyme in osteoblast differentiation. Proliferating osteoblasts synthesize pro-LOX (A), which is cleaved extracellularly into the mature LOX enzyme and a propeptide, LOX-PP (B). The mature LOX enzyme catalyzes the final enzymatic step required for formation of normal cross-links in collagen molecules (C), hence contributing to the formation of a mature, functional osteoblast matrix (D). LOX-PP inhibits FGF-2-induced signaling and proliferation (E). Because the levels of LOX-PP peak around the first transition point, its ability to inhibit osteoblast proliferation could facilitate the entry of cells into the later stages of differentiation.
Acknowledgments
We acknowledge the expert technical assistance of Danielle N. Stephens and JoAnn Buczek-Thomas.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DE14066 from NIDCR and R01 HL56200 from NHLBI. This work was also supported by Department of Defense Award W81XWH-08-1-0349.
S. R. Vora, Y. Guo, D. N. Stephens, E. Salih, E. D. Vu, K. H. Kirsch, G. E. Sonenshein, and P. C. Trackman, manuscript in preparation.
- FGF
- fibroblast growth factor
- FGFR
- FGF receptor
- ELISA
- enzyme-linked immunosorbent assay
- HSPG
- heparan sulfate proteoglycan
- LOX-PP
- lysyl oxidase propeptide
- LOX
- lysyl oxidase
- α-MEM
- α-minimal essential medium
- FBS
- fetal bovine serum
- BSA
- bovine serum albumin
- PBS
- phosphate-buffered saline
- ERK
- extracellular signal-regulated kinase
- MAP
- mitogen-activated protein
- r
- recombinant.
REFERENCES
- 1.Stein G. S., Lian J. B., Owen T. A. (1990) FASEB J. 4, 3111–3123 [DOI] [PubMed] [Google Scholar]
- 2.Canalis E., McCarthy T. L., Centrella M. (1991) Annu. Rev. Med. 42, 17–24 [DOI] [PubMed] [Google Scholar]
- 3.Stein G. S., Lian J. B., Stein J. L., Van Wijnen A. J., Montecino M. (1996) Physiol. Rev. 76, 593–629 [DOI] [PubMed] [Google Scholar]
- 4.Baylink D. J., Finkelman R. D., Mohan S. (1993) J. Bone Miner. Res. 8, S565–S572 [DOI] [PubMed] [Google Scholar]
- 5.Ornitz D. M., Marie P. J. (2002) Genes Dev. 16, 1446–1465 [DOI] [PubMed] [Google Scholar]
- 6.Coffin J. D., Florkiewicz R. Z., Neumann J., Mort-Hopkins T., Dorn G. W., 2nd, Lightfoot P., German R., Howles P. N., Kier A., O'Toole B. A., et al. (1995) Mol. Biol. Cell 6, 1861–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montero A., Okada Y., Tomita M., Ito M., Tsurukami H., Nakamura T., Doetschman T., Coffin J. D., Hurley M. M. (2000) J. Clin. Invest. 105, 1085–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muenke M., Schell U. (1995) Trends Genet. 11, 308–313 [DOI] [PubMed] [Google Scholar]
- 9.Canalis E., Centrella M., McCarthy T. (1988) J. Clin. Invest. 81, 1572–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fakhry A., Ratisoontorn C., Vedhachalam C., Salhab I., Koyama E., Leboy P., Pacifici M., Kirschner R. E., Nah H. D. (2005) Bone 36, 254–266 [DOI] [PubMed] [Google Scholar]
- 11.Berrada S., Lefebvre F., Harmand M. F. (1995) In Vitro Cell Dev. Biol. Anim. 31, 698–702 [DOI] [PubMed] [Google Scholar]
- 12.Ling L., Murali S., Dombrowski C., Haupt L. M., Stein G. S., van Wijnen A. J., Nurcombe V., Cool S. M. (2006) J. Cell Physiol. 209, 811–825 [DOI] [PubMed] [Google Scholar]
- 13.De Luca F., Baron J. (1999) Trends Endocrinol. Metab. 10, 61–65 [DOI] [PubMed] [Google Scholar]
- 14.Xiao G., Jiang D., Gopalakrishnan R., Franceschi R. T. (2002) J. Biol. Chem. 277, 36181–36187 [DOI] [PubMed] [Google Scholar]
- 15.Boudreaux J. M., Towler D. A. (1996) J. Biol. Chem. 271, 7508–7515 [DOI] [PubMed] [Google Scholar]
- 16.Tang C. H., Yang R. S., Chen Y. F., Fu W. M. (2007) J. Cell Physiol. 211, 45–55 [DOI] [PubMed] [Google Scholar]
- 17.Xiao G., Jiang D., Thomas P., Benson M. D., Guan K., Karsenty G., Franceschi R. T. (2000) J. Biol. Chem. 275, 4453–4459 [DOI] [PubMed] [Google Scholar]
- 18.Hurley M. M., Abreu C., Harrison J. R., Lichtler A. C., Raisz L. G., Kream B. E. (1993) J. Biol. Chem. 268, 5588–5593 [PubMed] [Google Scholar]
- 19.Feres-Filho E. J., Menassa G. B., Trackman P. C. (1996) J. Biol. Chem. 271, 6411–6416 [DOI] [PubMed] [Google Scholar]
- 20.Fang M. A., Glackin C. A., Sadhu A., McDougall S. (2001) J. Cell. Biochem. 80, 550–559 [PubMed] [Google Scholar]
- 21.Rodan S. B., Wesolowski G., Yoon K., Rodan G. A. (1989) J. Biol. Chem. 264, 19934–19941 [PubMed] [Google Scholar]
- 22.Hsieh S. C., Graves D. T. (1998) J. Cell. Biochem. 69, 169–180 [DOI] [PubMed] [Google Scholar]
- 23.Janssens K., ten Dijke P., Janssens S., Van Hul W. (2005) Endocr. Rev. 26, 743–774 [DOI] [PubMed] [Google Scholar]
- 24.Alliston T., Choy L., Ducy P., Karsenty G., Derynck R. (2001) EMBO J. 20, 2254–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeda S., Hayashi M., Komiya S., Imamura T., Miyazono K. (2004) EMBO J. 23, 552–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalajzic I., Kalajzic Z., Hurley M. M., Lichtler A. C., Rowe D. W. (2003) J. Cell. Biochem. 88, 1168–1176 [DOI] [PubMed] [Google Scholar]
- 27.Uzel M. I., Scott I. C., Babakhanlou-Chase H., Palamakumbura A. H., Pappano W. N., Hong H. H., Greenspan D. S., Trackman P. C. (2001) J. Biol. Chem. 276, 22537–22543 [DOI] [PubMed] [Google Scholar]
- 28.Trackman P. C., Bedell-Hogan D., Tang J., Kagan H. M. (1992) J. Biol. Chem. 267, 8666–8671 [PubMed] [Google Scholar]
- 29.Cronshaw A. D., Fothergill-Gilmore L. A., Hulmes D. J. (1995) Biochem. J. 306, 279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oxlund H., Barckman M., Ortoft G., Andreassen T. T. (1995) Bone 17, S365–S371 [DOI] [PubMed] [Google Scholar]
- 31.Knott L., Bailey A. J. (1998) Bone 22, 181–187 [DOI] [PubMed] [Google Scholar]
- 32.Trackman P. C. (2005) J. Cell. Biochem. 96, 927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palamakumbura A. H., Jeay S., Guo Y., Pischon N., Sommer P., Sonenshein G. E., Trackman P. C. (2004) J. Biol. Chem. 279, 40593–40600 [DOI] [PubMed] [Google Scholar]
- 34.Min C., Kirsch K. H., Zhao Y., Jeay S., Palamakumbura A. H., Trackman P. C., Sonenshein G. E. (2007) Cancer Res. 67, 1105–1112 [DOI] [PubMed] [Google Scholar]
- 35.Wu M., Min C., Wang X., Yu Z., Kirsch K. H., Trackman P. C., Sonenshein G. E. (2007) Cancer Res. 67, 6278–6285 [DOI] [PubMed] [Google Scholar]
- 36.Palamakumbura A. H., Vora S. R., Nugent M. A., Kirsch K. H., Sonenshein G. E., Trackman P. C. (2009) Oncogene 28, 3390–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palamakumbura A. H., Vora S. R., Sonenshein G. E. (2008) J. Dent. Res. 87, 854 [Google Scholar]
- 38.Hurtado P. A., Vora S., Sume S. S., Yang D., St Hilaire C., Guo Y., Palamakumbura A. H., Schreiber B. M., Ravid K., Trackman P. C. (2008) Biochem. Biophys. Res. Commun. 366, 156–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong H. H., Pischon N., Santana R. B., Palamakumbura A. H., Chase H. B., Gantz D., Guo Y., Uzel M. I., Ma D., Trackman P. C. (2004) J. Cell Physiol. 200, 53–62 [DOI] [PubMed] [Google Scholar]
- 40.Guo Y., Pischon N., Palamakumbura A. H., Trackman P. C. (2007) Am. J. Physiol. Cell Physiol. 292, C2095–C2102 [DOI] [PubMed] [Google Scholar]
- 41.Bellows C. G., Aubin J. E., Heersche J. N., Antosz M. E. (1986) Calcif. Tissue Int. 38, 143–154 [DOI] [PubMed] [Google Scholar]
- 42.Wang D., Christensen K., Chawla K., Xiao G., Krebsbach P. H., Franceschi R. T. (1999) J. Bone Miner. Res. 14, 893–903 [DOI] [PubMed] [Google Scholar]
- 43.Baeuerle P. A., Huttner W. B. (1986) Biochem. Biophys. Res. Commun. 141, 870–877 [DOI] [PubMed] [Google Scholar]
- 44.Nugent M. A., Edelman E. R. (1992) Biochemistry 31, 8876–8883 [DOI] [PubMed] [Google Scholar]
- 45.Fannon M., Nugent M. A. (1996) J. Biol. Chem. 271, 17949–17956 [DOI] [PubMed] [Google Scholar]
- 46.Mohammadi M., Froum S., Hamby J. M., Schroeder M. C., Panek R. L., Lu G. H., Eliseenkova A. V., Green D., Schlessinger J., Hubbard S. R. (1998) EMBO J. 17, 5896–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai C. F., Chaudhary L., Fausto A., Halstead L. R., Ory D. S., Avioli L. V., Cheng S. L. (2001) J. Biol. Chem. 276, 14443–14450 [DOI] [PubMed] [Google Scholar]
- 48.Kim H. J., Lee M. H., Park H. S., Park M. H., Lee S. W., Kim S. Y., Choi J. Y., Shin H. I., Kim H. J., Ryoo H. M. (2003) Dev. Dyn 227, 335–346 [DOI] [PubMed] [Google Scholar]
- 49.Jeay S., Pianetti S., Kagan H. M., Sonenshein G. E. (2003) Mol. Cell. Biol. 23, 2251–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kouhara H., Hadari Y. R., Spivak-Kroizman T., Schilling J., Bar-Sagi D., Lax I., Schlessinger J. (1997) Cell 89, 693–702 [DOI] [PubMed] [Google Scholar]
- 51.Kan M., Wang F., Xu J., Crabb J. W., Hou J., McKeehan W. L. (1993) Science 259, 1918–1921 [DOI] [PubMed] [Google Scholar]
- 52.Pantoliano M. W., Horlick R. A., Springer B. A., Van Dyk D. E., Tobery T., Wetmore D. R., Lear J. D., Nahapetian A. T., Bradley J. D., Sisk W. P. (1994) Biochemistry 33, 10229–10248 [DOI] [PubMed] [Google Scholar]
- 53.Jackson R. A., Murali S., van Wijnen A. J., Stein G. S., Nurcombe V., Cool S. M. (2007) J. Cell Physiol. 210, 38–50 [DOI] [PubMed] [Google Scholar]
- 54.Lundin L., Larsson H., Kreuger J., Kanda S., Lindahl U., Salmivirta M., Claesson-Welsh L. (2000) J. Biol. Chem. 275, 24653–24660 [DOI] [PubMed] [Google Scholar]
- 55.Wang S., Ai X., Freeman S. D., Pownall M. E., Lu Q., Kessler D. S., Emerson C. P., Jr. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4833–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yanagishita M., Hascall V. C. (1992) J. Biol. Chem. 267, 9451–9454 [PubMed] [Google Scholar]
- 57.Lipton S. A. (2006) Nat. Rev. Drug. Discov. 5, 160–170 [DOI] [PubMed] [Google Scholar]
- 58.Chen H. S., Pellegrini J. W., Aggarwal S. K., Lei S. Z., Warach S., Jensen F. E., Lipton S. A. (1992) J. Neurosci. 12, 4427–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lian J. B., Stein G. S. (1995) Iowa Orthop. J. 15, 118–140 [PMC free article] [PubMed] [Google Scholar]
- 60.Holthuis J., Owen T. A., van Wijnen A. J., Wright K. L., Ramsey-Ewing A., Kennedy M. B., Carter R., Cosenza S. C., Soprano K. J., Lian J. B., et al. (1990) Science 247, 1454–1457 [DOI] [PubMed] [Google Scholar]
- 61.Gerstenfeld L. C., Riva A., Hodgens K., Eyre D. R., Landis W. J. (1993) J. Bone Miner. Res. 8, 1031–1043 [DOI] [PubMed] [Google Scholar]
- 62.Pischon N., Mäki J. M., Weisshaupt P., Heng N., Palamakumbura A. H., N′Guessan P., Ding A., Radlanski R., Renz H., Bronckers T. A., Myllyharju J., Kielbassa A. M., Kleber B. M., Bernimoulin J. P., Trackman P. C. (2009) Calcif. Tissue Int. 85, 119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao Y., Min C., Vora S. R., Trackman P. C., Sonenshein G. E., Kirsch K. H. (2009) J. Biol. Chem. 284, 1385–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ornitz D. M., Itoh N. (2001) Genome Biol. 2, REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hauschka P. V., Mavrakos A. E., Iafrati M. D., Doleman S. E., Klagsbrun M. (1986) J. Biol. Chem. 261, 12665–12674 [PubMed] [Google Scholar]
- 66.Jackson R. A., Nurcombe V., Cool S. M. (2006) Gene 379, 79–91 [DOI] [PubMed] [Google Scholar]
- 67.Schlessinger J. (1994) Curr. Opin. Genet. Dev. 4, 25–30 [DOI] [PubMed] [Google Scholar]
- 68.Gotoh N. (2008) Cancer Sci. 99, 1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moscatelli D. (1987) J. Cell Physiol. 131, 123–130 [DOI] [PubMed] [Google Scholar]
- 70.Friesel R., Burgess W. H., Mehlman T., Maciag T. (1986) J. Biol. Chem. 261, 7581–7584 [PubMed] [Google Scholar]
- 71.Yayon A., Klagsbrun M., Esko J. D., Leder P., Ornitz D. M. (1991) Cell 64, 841–848 [DOI] [PubMed] [Google Scholar]
- 72.Roghani M., Mansukhani A., Dell'Era P., Bellosta P., Basilico C., Rifkin D. B., Moscatelli D. (1994) J. Biol. Chem. 269, 3976–3984 [PubMed] [Google Scholar]
- 73.Tanaka H., Ogasa H., Barnes J., Liang C. T. (1999) Mol. Cell. Endocrinol. 150, 1–10 [DOI] [PubMed] [Google Scholar]
- 74.Okazaki H., Kurokawa T., Nakamura K., Matsushita T., Mamada K., Kawaguchi H. (1999) Calcif. Tissue Int. 64, 542–546 [DOI] [PubMed] [Google Scholar]
- 75.Martin I., Muraglia A., Campanile G., Cancedda R., Quarto R. (1997) Endocrinology 138, 4456–4462 [DOI] [PubMed] [Google Scholar]
- 76.Yu K., Xu J., Liu Z., Sosic D., Shao J., Olson E. N., Towler D. A., Ornitz D. M. (2003) Development 130, 3063–3074 [DOI] [PubMed] [Google Scholar]
- 77.Li C. F., Hughes-Fulford M. (2006) J. Bone Miner. Res. 21, 946–955 [DOI] [PubMed] [Google Scholar]
- 78.Mansukhani A., Bellosta P., Sahni M., Basilico C. R. (2000) J. Cell Biol. 149, 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Debiais F., Hott M., Graulet A. M., Marie P. J. (1998) J. Bone Miner. Res. 13, 645–654 [DOI] [PubMed] [Google Scholar]
- 80.Hong H. H., Uzel M. I., Duan C., Sheff M. C., Trackman P. C. (1999) Lab. Invest. 79, 1655–1667 [PubMed] [Google Scholar]