Abstract
A member of the sirtuin family of NAD+-dependent deacetylases, SIRT3, is located in mammalian mitochondria and is important for regulation of mitochondrial metabolism, cell survival, and longevity. In this study, MRPL10 (mitochondrial ribosomal protein L10) was identified as the major acetylated protein in the mitochondrial ribosome. Ribosome-associated SIRT3 was found to be responsible for deacetylation of MRPL10 in an NAD+-dependent manner. We mapped the acetylated Lys residues by tandem mass spectrometry and determined the role of these residues in acetylation of MRPL10 by site-directed mutagenesis. Furthermore, we observed that the increased acetylation of MRPL10 led to an increase in translational activity of mitochondrial ribosomes in Sirt3−/− mice. In a similar manner, ectopic expression and knockdown of SIRT3 in C2C12 cells resulted in the suppression and enhancement of mitochondrial protein synthesis, respectively. Our findings constitute the first evidence for the regulation of mitochondrial protein synthesis by the reversible acetylation of the mitochondrial ribosome and characterize MRPL10 as a novel substrate of the NAD+-dependent deacetylase, SIRT3.
Keywords: Protein/Post-translational Modification, Protein/Synthesis, Proteomics, RNA/Ribosome Function, Subcellular Organelles/Mitochondria, Translation, Translation/Regulation Eukaryote, Acetylation, Mitochondrial Ribosomal Protein MRPL10, SIRT3
Introduction
Mitochondria produce over 90% of the energy used by mammalian cells through the process of oxidative phosphorylation. Reversible acetylation regulates many biological processes, including mitochondrial energy metabolism (1–6). Although the enzymes involved in the acetylation of mitochondrial proteins are not known, members of the class III histone deacetylases (sirtuins), SIRT3, SIRT4, and SIRT5, have been found to reside in mitochondria (6–8). Sirtuins are homologs of the yeast SIR2 (silent mating type information regulation 2) gene and use NAD+ as a cosubstrate (9–11). Both SIRT3 and SIRT4 are required to maintain cell survival after genotoxic stress in a NAD+-dependent manner (12, 13). Genetic variations in the human Sirt3 gene have also been linked to longevity (12, 13). We have previously shown that SIRT3 expression in adipose tissue is increased by caloric restriction and cold exposure (1, 14). Mitochondrial acetyl-CoA synthetase 2 and glutamate dehydrogenase are the two key metabolic enzymes regulated through deacetylation by SIRT3 (3, 6, 15). Thus, SIRT3 and SIRT4 modulate mitochondrial function in response to its [NADH]/[NAD+] ratio by regulating the activity of key metabolic enzymes.
In addition to metabolic enzymes, nucleus-encoded subunits of the electron transport chain complexes were found to be acetylated (1). In fact, Complex I subunit NDUFA9 is a SIRT3 substrate, and acetylation/deacetylation of Complex I is proposed to regulate and maintain basal ATP levels in mammalian mitochondria (16).
Thirteen of the essential protein components of the electron transport chain, as well as ATP synthase, are the products of genes present in mitochondrial DNA. The synthesis of these proteins is carried out by mitochondrial ribosomes within this organelle. We and others have previously identified 77 mammalian mitochondrial ribosomal proteins, of which 29 are in the small subunit and 48 are in the large subunit (17–21). About half of these proteins have homologs in bacterial ribosomes, whereas the remainders represent new classes of ribosomal proteins. However, we have observed that the functional core of the mitochondrial ribosome, essential for protein synthesis, was conserved in the cryoelectron microscopy reconstruction studies (22).
Mammalian mitochondrial ribosomal proteins are all nucleus-encoded, and some of them have been mapped to regions associated with disorders of mitochondrial energy metabolism (23). Alterations in expression levels and mutations of these ribosomal proteins affect mitochondrial protein synthesis, cell growth, and apoptosis (24–28). Some of the ribosomal proteins with bacterial homologs, such as MRPS12, MRPS16, and MRPL12, have been shown to be essential to support protein synthesis in mitochondria (24, 29–31).
In the present study, we demonstrate for the first time the acetylation of a mitochondrial ribosomal protein, MRPL10 (mitochondrial ribosomal protein L10), and its deacetylation by the NAD+-dependent deacetylase SIRT3. Using various biochemical and proteomics techniques, we also show that SIRT3 interacts with the mitochondrial ribosome. We propose that mitochondrial protein synthesis is regulated by reversible acetylation of MRPL10 and that the NAD+-dependent SIRT3 stimulates deacetylation of MRPL10, consequently regulating protein synthesis in mammalian mitochondria.
EXPERIMENTAL PROCEDURES
Sirt3 Knock-out Mice
Mice in which the Sirt3 gene was targeted by gene trapping were obtained from the Texas Institute for Genomic Medicine (Houston, TX). Briefly, these mice were created by generating embryonic stem cells (Omnibank number OST341297) bearing a retroviral promoter trap that functionally inactivates one allele of the Sirt3 gene, as described previously (32). Sequence analysis indicated that retroviral insertion occurred in the intron preceding coding exon 1 (accession number NM_022433). Targeted 129/SvEvBrd embryonic stem cells were injected into C57BL/6 albino blastocysts. The chimeras (129/SvEvBrd) were then crossed with C57BL/6 albinos to produce heterozygotes. Heterozygotes were then mated, and the offspring were genotyped using PCR, containing two primers flanking the trapping cassette insertion site TG0003–5′ (ATCTCGCAGATAGGCTATCAGC) and TG0003–3′ (AACCACGTAACCTTACCCAAGG), as well as a third primer, LTR rev, a reverse primer located at the 5′-end of the trapping cassette (ATAAACCCTCTTGCAGTTGCATC). Primer pair TG0003-5′ and TG0003-3′ amplifies a 336-bp fragment from the wild type allele, whereas primer pair TG0003–5′ and LTR rev amplifies a 160-bp fragment from the knock-out allele.
Plasmid Constructs
The mouse full-length MRPL10 coding sequence was amplified by reverse transcription-PCR, using mouse muscle RNA and the primer pair 5′-CCGGAATTCCGAACTTCCTGTAGCG-3′ and 5′-CTCGAGGGCATCTGGAGCAGGATCG-3′. The full-length human MRPL19 mRNA was amplified from Jurkat cell lines using primers 5′-AAACGGGGATCCATGGCGGCCTGCATTGCAACG-3′ and 5′-AGACGGTCGAGTTACTTATCGTCGTCATCCTTGTAATCAGACCTTTTCGACGCTTCAAT-3′. For transient expression, the DNA fragments from MRPL10 and MRPL19 were digested with EcorI/XhoI and BamHI/XhoI, respectively, and then inserted into the pcDNA3.1-MycHis and pcDNA-FLAG vectors that were derived from pcDNA3.1(+) to yield C-terminal FLAG-tagged constructs (Invitrogen). To generate stable cell lines overexpressing MRPL10, the MRPL10 cDNA was first cloned into pLNCX2 (Clontech) by using BglII and SalI. RetroPackTMPT67 cells were transfected with a retroviral expression vector, pLNCX2, to obtain retrovirus particles. MRPL10 mutants were created by site-directed mutagenesis (QuikChange® II site-directed mutagenesis kits, Stratagene, La Jolla, CA) at Lys124, Lys162, and Lys196. The primers for K124A (AAG to GCG) forward (5′-cacaagatcctgatgGCggtcttccccaaccag) and reverse (5′-ctggttggggaagaccGCcatcaggatcttgtg); K162A (AAG to GCG) forward (5′-gaagagcccaaggtcGCggagatggtacgg) and reverse (5′-ccgtaccatctccGCgaccttgggctcttc); and K196A (AAG to GCG) forward (5′-ggctttatcaactactccGCgctccccagcctgccc) and reverse (5′-gggcaggctggggagcGCggagtagttgataaagcc) were utilized for the PCR amplification, respectively. Mutated sequences are capitalized. Mouse NDUFA7 cDNA was amplified by reverse transcription-PCR, using the primer pair 5′-CGGGATCCGGAAGGAATATGGCGTC-3′ and 5′-GCTCTAGACAGGTATGGCTGGTCCTTG-3′ and inserted into BamHI and XbaI sites of pcDNA3.1-MycHis. The pcDNA3.1-SIRT3-FLAG, pBabe-SIRT3-puro, and pBabe-SIRT3N87A-puro were described previously (14). To generate pcDNA3.1-SIRT3N87A-FLAG, SIRT3N87A (7) was used as a template to be PCR-amplified with the same primers used to generate SIRT3-FLAG (14) and cloned into pCR-Blunt II-TOPO (Invitrogen). The SIRT3N87A-FLAG fragment was then excised out with HindIII/XhoI and ligated into pcDNA3.1(+). The human SIRT3-FLAG expression vector was kindly provided by Eric Verdin (University of California, San Francisco). To prepare SIRT5 with a C-terminal FLAG tag, the mouse SIRT5 coding sequence was amplified from mouse brown adipose tissue by PCR with the primer pair 5′-GAATGATCAATGCGACCTCTCCTGATTGC-3′ and 5′-ATAGAATTCAGAAGTCCTTTCAGTTTCATG-3′. The DNA fragment was then digested with BcII/EcoRI and inserted into BamHI/EcoRI sites of pcDNA3.1(+) containing a FLAG tag sequence (5′-GAATTCGACTACAAAGACGATGACGACAAGTAACTCGAG-3′) between EcoRI/XhoI sites. To produce a GST-SIRT3 fusion protein, the full-length SIRT3 was amplified by the primer pair 5′-AAAAGCTTGTGGGGGCCGGCATCAGCA-3′ and 5′- TGATCTGCTGTGTGACTTCC-3′ and inserted into pCR-Blunt II-TOPO. The SIRT3 coding sequence was then excised out with HindIII/EcoRI, treated with mung bean nuclease, and inserted into the SmaI site of pGEX-2T (Amersham Biosciences). To establish a series of SIRT3 deletion mutants, the SIRT3 mutants generated by using restriction endonucleases were inserted in frame into pGEX-4T-1 vector. All expression constructs were verified by DNA sequencing.
Cell Culture
HeLa, human embryonic kidney 293 (HEK293),4 BOSC23, RetroPackTMPT67, and C2C12 cells were cultured in Dulbecco's modified Eagle's medium (Hyclone) supplemented with 10% bovine calf serum (Hyclone), 100 IU/ml penicillin, and 100 μg/ml streptomycin at 37 °C and 5% CO2. HEK293 cells were transfected as indicated (33), and the transfected cells were lysed in cell lysis buffer (50 mm Tris-HCl, pH 8.0, 50 mm KCl, 20 mm NaF, 1 mm Na3VO4, 5 mm EDTA, and 0.5% Nonidet P-40) supplemented with a protease inhibitor mixture (Roche Applied Science).
RNA Interference
C2C12 cells with a SIRT3 shRNA knockdown were generated using a lentivirus-mediated delivery system as reported (34). The DNA cassette used to generate shRNA against SIRT3 mRNA in this experiment was as follows: tcgagaaaaagccatctttgaacttggcttctcttgaaagccaagttcaaagatgg (the 18-nucleotide sense and reverse complementary targeting sequence are underlined). Briefly, double-stranded oligonucleotides were inserted into the BbsI/XhoI site of pBS-hU6–1 vector. The cloned oligonucleotide sequence together with an upstream human U6 promoter were excised with XbaI/XhoI and then subcloned into FG12 vector, which contains an enhanced green fluorescent protein marker for cell tracking. FG12-hU6-siRNA lentiviral vectors were transfected into 293T cells together with three packaging plasmids: pMDLg/pRRE, CMV-VSVG, and RSV-Rev. The recombinant lentiviruses produced from the transfected 293T cells were used to infect C2C12 cells. Seventy-two hours after infection, transduced C2C12 cells were sorted by flow cytometry based on enhanced green fluorescent protein expression.
Mitochondrial Ribosome Preparation
Preparation of mitochondrial ribosomes from bovine liver was adapted from previously described methods (19, 21, 35, 36). In order to preserve the phosphorylation and acetylation status of ribosomal proteins, phosphatase inhibitors (2 mm imizadole, 1 mm Na3VO4, 1.15 mm Na2MoO4·2H2O, 1 mm NaF, 4 mm Na2C4H4O6·2H2O) and deacetylase inhibitor (1 mm sodium butyrate) were added during the homogenization process. Crude ribosomes prepared at 0.2, 0.4, and 1.6% Triton X-100 concentrations were loaded onto 10–30% sucrose gradients and fractionated to isolate mitochondrial 55 S ribosomes (16, 18, 34). Purified ribosomes were sedimented by ultracentrifugation at 40,000 rpm for 5 h in a Beckman Type 40 rotor. Acetone pellets of ribosome preparations (∼1.8 A260 units) were resuspended in two-dimensional lysis buffer consisting of 9.8 m urea, 2% (w/v) Nonidet P-40, 2% ampholytes, pI 3–10 and 8–10, and 100 mm dithiothreitol. The samples were loaded on non-equilibrium pH gradient electrophoresis tube gels and equilibrated in buffer containing 60 mm Tris-HCl, pH 6.8, 2% SDS, 100 mm dithiothreitol, and 10% glycerol. The 14% second dimension gel was stained with Coomassie Blue, and the protein spots corresponding to the acetylated ribosomal proteins were excised based on their locations determined by Western blot analysis. To complement this, in-gel and in-solution tryptic digestions were carried out using ribosomes prepared at different Triton X-100 concentrations and analyzed by LC-MS/MS to identify acetylated ribosomal proteins and the other ribosome-associated proteins as described in the supplemental material.
Yeast Two-hybrid Interaction Screening
To search for SIRT3-interacting proteins, yeast two-hybrid screening was performed using the MatchmakerTM two-hybrid system (Clontech) according to the manufacturer's instructions. Briefly, SIRT3 cDNA was inserted into the pGBKT7 vector and transformed in yeast, which was mated with pretransformed yeast carrying human placental cDNAs. Positive clones were identified by staining for β-galactosidase activity, and the encoded human cDNAs were identified by sequencing.
GST Pull-down Assay
All pGEX-constructs were transformed into the BL21(DE3) strain of Escherichia coli, and the fusion proteins were induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside at 30 °C for 3 h. The bacteria were collected by centrifugation, and the resulting pellets were lysed with bacterial lysis buffer (20 mm Tris-HCl, pH 8.0, 5 mm EDTA, 0.1% Triton X-100, and 2% glycerol). Fusion proteins were purified by affinity chromatography, using glutathione-agarose beads (Sigma), and analyzed by SDS-PAGE. Four μg of GST, GST-SIRT3, or GST-SIRT3 deletion mutants were immobilized on glutathione-agarose. The beads were incubated with 500 μg of total protein from transfected HEK293 cells at 4 °C for 2 h. The agarose beads were washed three times in cell lysis buffer, and proteins were eluted in SDS sample buffer. For the purification of bacterially expressed SIRT3 or SIRT3N87A, about 10 μg of GST fusion protein after purification on glutathione-agarose beads was digested with thrombin (2 units) at room temperature for 2 h, and the released SIRT3 or SIRT3N87A was collected in the supernatant after centrifugation.
Immunoprecipitation and Immunoblotting
For co-immunoprecipitation assays, lysates of HEK293 cells expressing proteins by transient transfection were incubated with anti-FLAG-agarose beads (Sigma) at 4 °C overnight or with Talon metal affinity resin (BD Biosciences) at 4 °C for 2 h. Beads were washed three times with lysis buffer, and proteins were released from beads in SDS-sample buffer and analyzed by immunoblotting.
For immunoblotting, protein samples for either one-dimensional or two-dimensional analysis were loaded onto SDS-polyacrylamide gels and transferred to a polyvinylidene difluoride membrane or nitrocellulose membrane. The blot was probed with a monoclonal MRPS29 (DAP3) antibody at a 1:5000 dilution (Transduction Laboratories), a monoclonal Hsp60 antibody at a 1:4000 dilution (Transduction Laboratories), polyclonal mouse and human MRPL10 antibodies at a 1:3000 dilution, a polyclonal human MRPL41 antibody at 1:3000, a polyclonal anti-SIRT3 antiserum (against C-terminal domain of murine SIRT3) at 1:3000, a mouse monoclonal anti-FLAG M2-peroxidase at 1:3000 (Sigma), a monoclonal anti-N-acetyl lysine antibody at 1:3000 (Cell Signaling Technology Inc.), or a mouse monoclonal anti-Myc antibody at 1:4000 (Clontech). Total oxidative phosphorylation human antibody mixture (MitoProfile® Total OXPHOS Rodent WB Antibody Cocktail, Mitosciences Inc, Eugene, OR) was used at a 1:3000 dilution. The secondary antibody was ImmunoPure antibody goat anti-mouse IgG (Pierce) at a 1:5000 dilution; goat anti-rabbit IgG at a 1:1000 dilution; or Affinipure rabbit anti-mouse IgG, rabbit anti-goat IgG, or goat anti-rabbit IgG (Jackson ImmunoResearch), all at a 1:10,000 dilution, followed by development with the SuperSignal West Pico Chemiluminescent Substrate (Pierce) according to the protocol provided by the manufacturer.
In Vitro Deacetylation Assays
Deacetylation of ribosomal proteins was performed in the presence of NAD+ as a substrate for the deacetylase using previously described methods (11, 37). For this reaction, 0.1 or 0.2 A260 units of sucrose gradient-purified or crude 55 S ribosomes, respectively, prepared in 0.2% Triton X-100, were incubated with 3 mm NAD+ for 30 min at 37 °C in the presence and absence of 0.25 μg of recombinant mouse SIRT3. Acetylated/deactylated ribosome samples were analyzed by immunoblotting using rabbit polyclonal anti-N-acetyl lysine antibody.
For deacetylation of MRPL10, HEK293 cells were first transfected with FLAG-tagged MRPL10, and cell extracts were immunoprecipitated with anti-FLAG-agarose beads. Then the immunoprecipitated MRPL10 was incubated with or without 0.3 μg of SIRT3 or SIRT3N87A in 50 mm Tris-HCl, pH 8.0, 4 mm MgCl2, 50 mm NaCl, 0.5 mm dithiothreitol for 30 min to 2 h at 30 °C in the presence or absence of 5 mm NAD+ or 10 mm nicotinamide. After incubation, the level of MRPL10 acetylation was determined by immunoblotting analysis as described above.
Mouse Mitochondrial Ribosome Isolation and Translation Assays
To assess the mitochondrial protein synthesis, pulse-labeling experiments with [35S]methionine were performed according to a protocol described previously (38). About 2 × 106 cells of the desired type were plated on 10-cm cell culture dishes, incubated overnight, washed with methionine-free DMEM, and then incubated for 7 min at 37 °C in 4 ml of the same medium containing a 100 μg/ml concentration of the cytoplasmic translational inhibitor emetine. Thereafter, [35S]methionine (0.2 mCi (1175 Ci/mmol)) was added, and the cells were incubated for 30 min. The labeled cells were lysed, and 70 μg of protein were electrophoresed through an SDS-polyacrylamide gel (15–20% exponential gradient). The intensities of the bands were quantified by phosphorimaging analyses.
Liver tissue obtained from Sirt3+/+, Sirt3+/−, and Sirt3−/− mice was resuspended in an isotonic mitochondrial buffer (210 mm mannitol, 70 mm sucrose, 1 mm EDTA, 10 mm HEPES-KOH, pH 7.5), supplemented with protease inhibitors (1 mm phenylmethylsulfonyl fluoride, 50 μg/ml leupeptin), and then homogenized in a Dounce homogenizer on ice. The suspension was centrifuged at 400 × g at 4 °C. This procedure was repeated twice, and supernatants were centrifuged at 10,000 × g at 4 °C for 10 min to pellet mitochondria. Mitochondria were lysed in a buffer containing 0.26 m sucrose, 20 mm Tris-HCl, pH 7.6, 40 mm KCl, 20 mm MgCl2, 0.8 mm EDTA, 0.05 mm spermine, 0.05 mm spermidine, 6 mm β-mercaptoethanol, and 1.6% Triton X-100, and the lysates were loaded onto 34% sucrose cushions and centrifuged at 100,000 × g at 4 °C for 16 h. The crude ribosome pellets were resuspended in 20 mm Tris-HCl, pH 7.6, 40 mm KCl, 20 mm MgCl2, and 1 mm dithiothreitol. EF-Tumt and EF-Gmt were prepared from the recombinant proteins (supplemental material). In vitro translation assays were performed in 100-μl reactions containing 50 mm Tris-HCl, pH 7.8, 1 mm dithiothreitol, 0.1 mm spermine, 40 mm KCl, 7.5 mm MgCl2, 2.5 mm phosphoenolpyruvate, 0.18 units of pyruvate kinase, 0.5 mm GTP, 50 units of RNasin Plus, 12.5 μg/ml poly(U), 20 pmol of [14C]Phe-tRNA, 0.15 μm EF-Tumt, 1 μg of EF-Gmt, and varying amounts of mitochondrial ribosomes obtained from mouse mitochondria. The reaction mixtures were incubated at 37 °C for 15 min and terminated by the addition of cold 5% trichloroacetic acid, followed by incubation at 90 °C for 10 min. The in vitro translated 14C-labeled poly(Phe) was collected on nitrocellulose filter membranes and quantified using a liquid scintillation counter.
RESULTS AND DISCUSSION
Mitochondrial Ribosomal Protein MRPL10 Is Acetylated and SIRT3 Is Responsible for Its Deacetylation
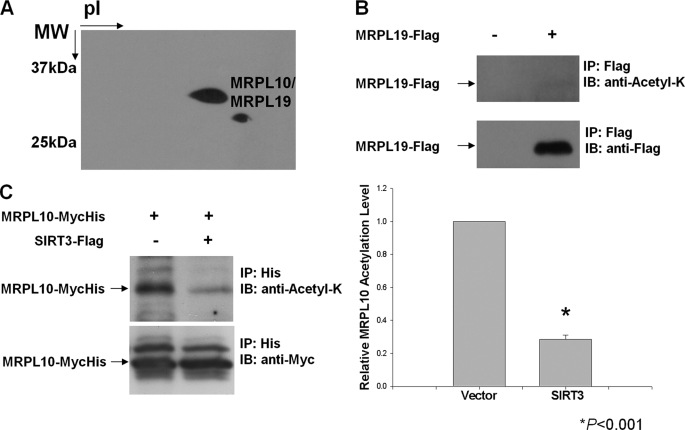
A combination of two-dimensional gel separation and capillary LC-MS/MS analyses has been successfully used in the identification of ribosomal proteins and their post-translational modifications in our laboratory (28, 39–42). Acetylation of several components of the translational machinery in bacteria either at N-terminal amino groups or at ϵ-amino groups of Lys residues has been reported previously (43). To determine the acetylated proteins of mammalian mitochondrial ribosomes, 55 S ribosomes were purified from bovine liver using previously described methods (35, 36). Ribosomal proteins were separated by two-dimensional gel electrophoresis, and acetylated ribosomal proteins were identified by immunoblotting using anti-N-acetyl lysine antibody (Fig. 1A). Protein bands corresponding to acetylated proteins detected in the gel were excised, digested in-gel with trypsin, and analyzed by capillary LC-MS/MS for identification (supplemental material). The mass spectrometric analyses of the two-dimensional gel spots revealed the presence of the two mitochondrial ribosomal proteins, MRPL10 (29.3 kDa) and MRPL19 (33.3 kDa) (Fig. 1A and Table 1). Clearly, after the cleavage of the N-terminal signal sequences during their translocation into mitochondria, these two proteins have almost the same pI and molecular weight values because they were detected in the same gel spot in the two-dimensional gel analysis (Fig. 1A). The existence of the two mitochondrial ribosomal proteins in the same spot suggests the possibility for acetylation of both MRPL10 and MRPL19 in the ribosome; however, no acetylated peptides were detected in the mass spectrometric analysis of tryptic peptides obtained from the in-gel digestion of these spots, possibly because of the high arginine and lysine content of MRPL10 and MRPL19. MRPL10 was also found to be phosphorylated in mitochondrial ribosomes, and this modification interferes with the detection of acetylated peptides, especially if the modification sites are in close proximity to each other (42). In addition, the less prominent acetylation signal observed in two-dimensional gel immunoblotting corresponded to a faint Coomassie-stained protein spot, which did not lead to the identification of a protein in mass spectrometric analysis. It is therefore possible that this second acetylated protein signal is due to proteolytic degradation of acetylated MRPL10, which occurred during the lengthy process of mitochondrial ribosome purification.
FIGURE 1.
Detection of acetylated mitochondrial ribosomal proteins. Acetylated proteins found in ribosomes purified from bovine mitochondria were detected by immunoblot (IB) analysis using anti-N-acetyl lysine antibody (anti-Acetyl-K). A, approximately 1.8 A260 units of purified bovine 55 S ribosomes were separated on two-dimensional non-equilibrium pH gradient electrophoresis gels, and acetylated proteins were detected with anti-N-acetyl lysine antibody. Protein spots corresponding to the acetylated ribosomal proteins were identified by mass spectrometry. B and C, HEK293 cells were transfected with FLAG-tagged MRPL19 (B) and MycHis-tagged MRPL10 alone or together with SIRT3 (C). The tagged proteins were enriched by affinity chromatography or immunoprecipitation (IP), and MRPL10 and MRPL19 acetylation levels were detected by immunoblotting with anti-N-acetyl lysine antibody. *, p < 0.001.
TABLE 1.
Peptides detected from tryptic digests of acetylated band detected in mammalian mitochondrial ribosomal proteins by LC-MS/MS analysis
Collision-induced dissociation spectra of peptides were searched by MASCOT as described in the supplemental materials, and the spectra for each peptide are provided in supplemental Table 1. Modifications of peptides by acetylation (ac), phosphorylation (p) and oxidation (o) are shown for each peptide.
| Peptide sequence | m/z | Mr (experimental) | Score |
|---|---|---|---|
| MRPL10 | |||
| QKLoMAVTEYIAPKPVVNPR | 725.1 | 2172.3 | 39 |
| QacKLoMAVTEYIAPKPVVNPR | 738.8 | 2213.3 | 22 |
| LoMAVTEYIAPKPVVNPR | 958.5 | 1915.1 | 72 |
| oMIAVCQNVAoMSAEDK | 822.5 | 1642.9 | 96 |
| VFPNQILKPFLEDSK | 888.4 | 1774.8 | 69 |
| YQNLLPLFVGHNLLLVSEEPK | 1213.1 | 2424.1 | 96 |
| VKEoMVRILK | 567.7 | 1132.3 | 21 |
| VacKEoMVRILK | 588.5 | 1173.5 | 15 |
| ILacKpSVPFLPLLGGCIDDTILSR | 858.0 | 2571.0 | 47 |
| QGFINYpSacKLPpSLALAQGELVGGLpTLLTAR | 1104.6 | 3310.9 | 54 |
| LPSLALAQGELVGGLTLLTAR | 1048.2 | 2094.3 | 145 |
| THSLLQHHPLQLTALLDQYAR | 1229.3 | 2455.8 | 78 |
| QQLEGDPVVPASAQPDPPNPVQDS | 829.8 | 2486.3 | 83 |
| MRPL19 | |||
| FLSPEFIPPR | 602.4 | 1202.8 | 79 |
| ILHIPEFYVGSILR | 829.4 | 1656.8 | 68 |
| LDDSLLYLR | 555.2 | 1108.4 | 87 |
| DALPEYSTFDMNMKPVAQEPSR | 843.5 | 2527.3 | 80 |
| WSQPWLEFDoMMR | 822.1 | 1642.2 | 62 |
| IEAAIWNEIEASK | 737.7 | 1473.5 | 91 |
To confirm the acetylation status of these two mitochondrial ribosomal proteins, another approach was taken. First, coding sequences for MRPL10 and MRPL19 were inserted into pcDNA3-MycHis and pcDNA3-FLAG vectors, respectively. MRPL10 and MRPL19 were then expressed in HEK293 cells and immunoprecipitated by Myc-His or FLAG tags. The acetylation status of these proteins was examined by immunoblotting with anti-N-acetyl lysine antibody. No significant acetylation of MRPL19 was detected, whereas MRPL10 was found to be clearly acetylated in a parallel experiment (Fig. 1, B and C). Although there are no known protein acetyl transferases in mitochondria, three NAD+-dependent deacetylases, SIRT3, SIRT4, and SIRT5, are localized to the mitochondria (6–8). Given that the SIRT3 has a known protein deacetylase activity, FLAG-tagged SIRT3 was also co-transfected with MRPL10. Its co-expression reduced the acetylation of MRPL10 significantly, as shown in Fig. 1C. These observations allowed us to conclude that the acetylated protein spots detected in the immunoblotting analysis of mitochondrial ribosome preparations were primarily from acetylated MRPL10 and possibly its proteolytic products. For this reason, MRPL10 has been designated as the major acetylated protein of the 55 S ribosome, and its reversible acetylation could have a role in the regulation of mitochondrial translation.
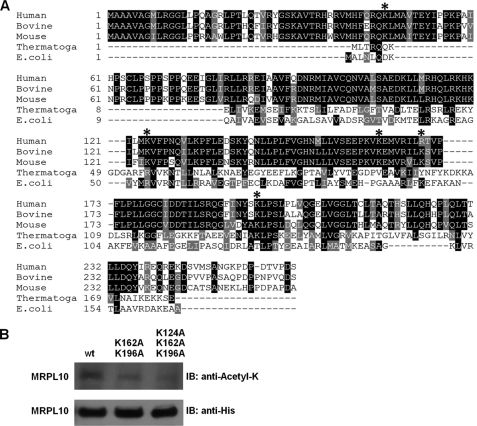
For further confirmation of MRPL10 acetylation and mapping of the acetylated Lys residues, mitochondrial ribosomal proteins isolated from bovine 55 S ribosomes were separated on reverse phase HPLC, and tryptic digests of the band(s) corresponding to the acetylated MRPL10 were analyzed by LC-MS/MS (supplemental Fig. S1 and Table 1). Acetylated and non-acetylated forms of MRPL10 were well separated on the C4-reverse phase column, and the acetylated form of MRPL10 was eluted in the later fraction as expected due to the neutralization of positive charges on Lys residues by more hydrophobic acetyl groups. In the immunoblotting analysis of HPLC fractions by anti-MRPL10 and anti-N-acetyl lysine antibodies, several different forms of MRPL10 were observed. This observation might be the result of differential post-translational modification(s) of MRPL10, such as acetylation and phosphorylation (supplemental Fig. S1). Depending on the relative signal intensities of acetylated and non-acetylated MRPL10, acetylated MRPL10 can be estimated to be 30% of the total MRPL10 found in the dissociated 39 S subunits and 55 S ribosomes (supplemental Fig. S1). However, this estimation is based on recognition of all of these different forms of MRPL10 by anti-MRPL10 antibody equally (supplemental Fig. S1). In addition to confirming acetylation of MRPL10, we also performed LC-MS/MS analysis to map the acetylated Lys residues from the tryptic digests of acetylated MRPL10 fractions. This analysis enabled us to map several highly conserved Lys residues (lysines 46, 124, 162, 169, and 196) as acetylated in bovine 55 S ribosomes (Table 1 and supplemental Table S1). The majority of these Lys residues are highly conserved in human, bovine, and mouse MRPL10 proteins (Fig. 2A).
FIGURE 2.
Role of Lys124, Lys162, and Lys196 in acetylation of MRPL10. A, primary sequence alignment of MRPL10 homologs from different species. The bovine (XP_592952), human (NP_660298), and mouse (NP_080430) mitochondrial ribosomal MRPL10 proteins were aligned with Thermotoga maritimi (Thermotoga) (NP_228266) and E. coli (AAC43083) L10 proteins. *, acetylated Lys residues detected in the LC-MS/MS analysis. The alignment was created with the ClustalW program in Biology Workbench and displayed in BOXSHADE. B, approximately 20 μg of affinity-enriched lysates obtained from HeLa cells stably expressing His-tagged wild type (wt), double (K162A and K196A), and triple (K124A, K162A and K196A) mutants were loaded on 12% SDS-PAGE and probed with anti-N-acetyl lysine and anti-His tag antibodies. IB, immunoblot.
To investigate the contribution of highly conserved Lys residues in acetylation of MRPL10, wild type, double mutants (K162A and K196A), or triple mutants (K124A, K162A, and K196A) of MRPL10 were stably expressed in HeLa cells. Acetylation and expression of these MRPL10 proteins were detected by anti-N-acetyl lysine and anti-His antibodies, respectively, after enriching the proteins by His tag affinity chromatography (Fig. 2B). As confirmed by immunoblotting analysis, overexpressed MRPL10 is acetylated, and the acetylation status of MRPL10 gradually declined in both double and triple Lys → Ala mutants (Fig. 2B). In the triple mutant, only three of the four acetylated lysines detected by mass spectrometry were mutated to Ala; therefore, it is possible that another acetylated lysine residue(s) contributes to the residual acetylation signal observed in the immunoblot (Fig. 2B).
NAD+-dependent Deacteylase-SIRT3 Is Associated with the 55 S Mitochondrial Ribosome and Interacts Directly with MRPL10
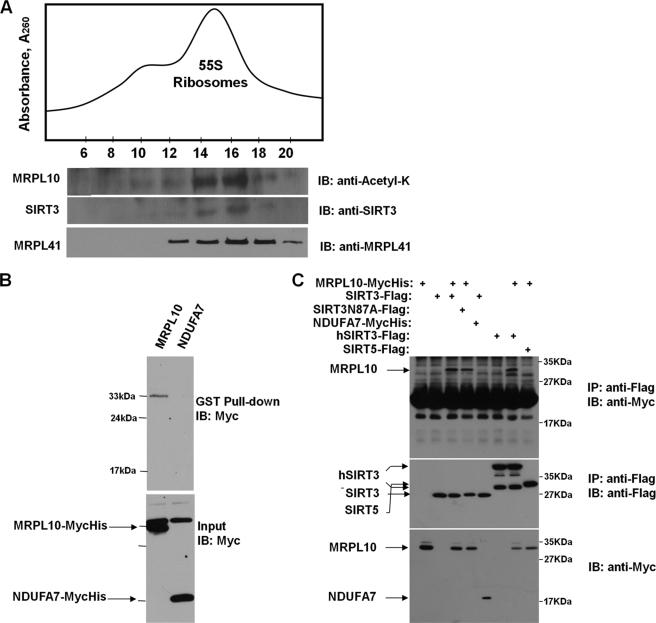
There are no protein acetyltransferases identified in mammalian mitochondria to date. Thus far, three mitochondrially localized deacetylases, SIRT3, SIRT4, and SIRT5, have been identified. In particular, SIRT3 possesses NAD+-dependent deacetylase activity, and SIRT3-dependent deacetylation of several metabolic enzymes and complex I subunit NDUFA9 was demonstrated in mammalian mitochondria (6, 15, 16). The presence of acetylated MRPL10 in the mitochondrial ribosome and deacetylation of ectopically expressed MRPL10 by SIRT3 (Fig. 1, A and C) prompted us to search for enzymes responsible for this reversible post-translational modification. To determine whether any of the deacetylases mentioned above are associated with ribosomes, crude mitochondrial ribosomes were isolated at different salt and non-ionic detergent concentrations to preserve their interactions with associated proteins. They were then fractionated on 10–30% linear sucrose gradients (Fig. 3A). To determine the location of ribosomes and ribosome-associated proteins, immunoblots of sucrose gradient fractions were probed with antibodies to N-acetyl lysine, MRPL41, and SIRT3 (Fig. 3A). The SIRT3-containing fractions (12–18) overlapped with the acetylated MRPL10, another large subunit ribosomal protein MRPL41 (Fig. 3A), and a small subunit protein MRPS29 (data not shown). To further verify association of SIRT3 with mitochondrial ribosomes, ribosome preparations were treated with RNase A and loaded on 10–30% gradients (supplemental Fig. S2). RNase A treatment only digests rRNAs of 55 S ribosomes. Proteins physically associated with the rRNA were spread through the gradient, and no significant amounts of SIRT3 or MRPL10 were retained in the gradients (supplemental Fig. S2).
FIGURE 3.
Interactions between SIRT3 and mitochondrial 55 S ribosomes and MRPL10. A, crude mitochondrial ribosomes were loaded on to 10–30% linear sucrose gradients to sediment 55 S ribosomes. To demonstrate the co-sedimentation of SIRT3 with the 55 S ribosome, immunoblot (IB) analyses were performed with anti-N-acetyl lysine antibody detecting the acetylated MRPL10, as well as anti-SIRT3 anti-MRPL41 antibodies, after separating 30 μl of each fraction on 12% SDS-PAGE. B, lysates prepared from HEK293 cells transfected with MycHis-tagged MRPL10 or NDUFA7 were incubated with recombinant GST-SIRT3 fusion protein immobilized on the glutathione-conjugated agarose beads. The proteins associated with the beads were analyzed by immunoblotting using anti-Myc antibody. C, HEK293 cells were transfected with MycHis-tagged MRPL10 or NDUFA7 with or without FLAG-tagged murine SIRT3, SIRT3N87A mutant, human SIRT3, or murine SIRT5, as indicated. The cell lysates were immunoprecipitated (IP) with anti-FLAG-agarose beads and detected by immunoblotting with anti-Myc or anti-FLAG antibodies.
In our analysis, we have found the association of SIRT3 with the mitochondrial ribosome only at low detergent and ionic conditions, implying a possibly transient interaction. In addition to immunoblotting analysis, in-gel proteolytic digestion and mass spectrometric analysis of the protein band detected with the SIRT3 antibody confirmed the association of SIRT3 with the ribosome. Peptide sequence matches obtained from the LC-MS/MS analysis of the in-gel digested SIRT3 band are presented in Table 2. The other mitochondrial deacetylases, SIRT4 and SIRT5, were not detected in immunoblotting analysis performed with SIRT4 and SIRT5 antibodies or in LC-MS/MS analysis of ribosome preparations (data not shown); therefore, we concluded that SIRT3 was the only mitochondrial deacetylase associated with the ribosome.
TABLE 2.
Peptides detected from tryptic digests of mitochondrial ribosome-associated bovine SIRT3 by LC-MS/MS analysis
Spectra for each peptide are provided in supplemental Table 1.
| Peptide sequence | m/z | Mr (experimental) | Score |
|---|---|---|---|
| KFLLQDIAELIK | 717.5 | 1433.0 | 101 |
| LYTQNIDGLER | 662.8 | 1323.5 | 73 |
| LVEAHGSLASATCTVCR | 860.2 | 1718.3 | 56 |
| DVAQLGDVVHGVK | 669.5 | 1337.1 | 63 |
| LVELLGWTDDIQDLIQR | 1015.7 | 2029.3 | 104 |
In a parallel approach to identify proteins interacting with SIRT3, we carried out yeast two-hybrid screening and identified MRPL10 as one of the SIRT3 interacting mitochondrial proteins (data not shown). To confirm and characterize the interaction between SIRT3 and MRPL10 further, MycHis-tagged MRPL10 or empty vector were transfected into HEK293 cells. The resulting cell lysates were incubated with SIRT3-GST fusion protein immobilized on glutathione-conjugated agarose beads to demonstrate their interactions in vitro. As shown in Fig. 3B, MRPL10, but not the mitochondrial complex I subunit NDUFA7, which was also identified in the yeast two-hybrid screening, co-purified with SIRT3 in GST pull-down. This observation indicates that there is a specific interaction between MRPL10 and SIRT3. The MRPL10-SIRT3 interaction in mammalian cells was further confirmed by a co-immunoprecipitation assay. In this experiment, HEK293 cells were transfected with MycHis-tagged MRPL10, murine SIRT3, the SIRT3 deacetylase mutant carrying the N87A mutation (14), NDUFA7, human SIRT3, or murine SIRT5 (Fig. 3C). The SIRT3, either from mice or humans, specifically interacted with MRPL10; however, SIRT5 was unable to bind to MRPL10. In addition, the SIRT3 mutant (N87A) lacking the deacetylase activity still interacted with MRPL10, indicating that this interaction is independent of SIRT3 enzymatic activity (Fig. 3C). These results suggest that the interaction between MRPL10 and SIRT3 is highly specific. This was not surprising because SIRT3, but not mitochondrial SIRT5 (data not shown), was identified as the ribosome-associated deacetylase in our proteomics analysis of the 55 S ribosome. Our findings are also in agreement with a recent report that deficiency of SIRT3, but not SIRT4 or SIRT5, affects mitochondrial protein deacetylation (15).
SIRT3 Interacts with the N-terminal Domain of MRPL10
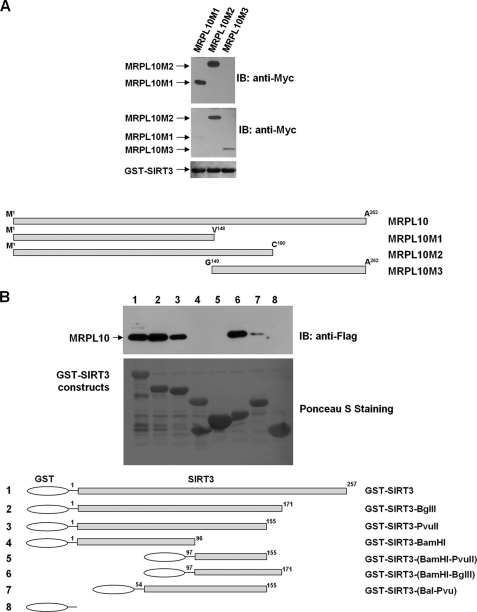
Similar to its bacterial and archaeal counterparts, MRPL10 is located in the L7/L12 stalk of the ribosomal large subunit, interacting with MRPL11 and possibly multiple copies of MRPL12 in the mitochondrial ribosome. To provide insight into ribosome and/or MRPL10-SIRT3 interactions, several different deletion constructs of MRPL10 and SIRT3 were generated. First, to map SIRT3 binding domain(s) in MRPL10, three deletion mutants were constructed and tested in GST-SIRT3 pull-down experiments (Fig. 4A). In these experiments, only MRPL10M1 and M2 constructs were pulled down with the GST-SIRT3 construct, as shown in the upper panel of the immunoblotting analysis performed with anti-Myc antibody (Fig. 4A). The first 148 amino acid (aa) residues of the N-terminal domain of MRPL10 are sufficient for binding SIRT3. Lack of this region severely disrupted the L10 interaction with SIRT3 (Fig. 4A).
FIGURE 4.
Domains mediating the interaction between MRPL10 and SIRT3. A, GST-pull-down assays were performed using GST-SIRT3 fusion protein and three different in vitro translated MycHis-MRPL10 constructs, MRPL10M1, -M2, and -M3. MRPL10 proteins interacting with SIRT3 (top) and the expression levels of these MRPL10 proteins used for GST pull-down (middle) were probed using anti-Myc antibody. The GST-SIRT3 protein input (bottom) for each pull-down assay was visualized by Ponceau S staining of the membrane. B, GST-pull down assays were performed using FLAG-tagged MRPL10 expressed in HEK293 cells and various recombinant GST-SIRT3 constructs. MRPL10 associated with GST-SIRT3 was analyzed by immunoblotting using anti-FLAG antibody, and the GST fusion proteins were visualized by Ponceau S staining of the membrane. IB, immunoblot.
Similarly, to determine the MRPL10 binding domain(s) in SIRT3, six different constructs of SIRT3 were generated (Fig. 4B). Deletion of 86 aa residues at the C terminus (aa 172–257) of SIRT3 did not affect the interaction between SIRT3 and MRPL10 (lane 2). Further deletion of 16 additional aa residues slightly decreased the interaction, suggesting a potential MRPL10 binding site between aa 155 and 171 (lane 3). In addition, neither residues 1–96 nor 97–155 displayed no binding to MRPL10 (lanes 4 and 5), whereas residues 97–171 strongly interacted with MRPL10 (lane 6), confirming the existence of a binding domain between residues 156 and 171. Because residues 1–155 still interacted with MRPL10 (lane 3), but neither residues 1–96 (lane 4) nor residues 97–155 (lane 5) did, it is likely that there is another interaction domain spanning the region around residue 96 in SIRT3. This was confirmed by a weak interaction between MRPL10 and the SIRT3 fragment containing residues aa 54–155 (lane 7) (Fig. 4B). Therefore, we conclude that there is a major MRPL10 binding domain around residues 156–171 and a minor binding site around residue 96.
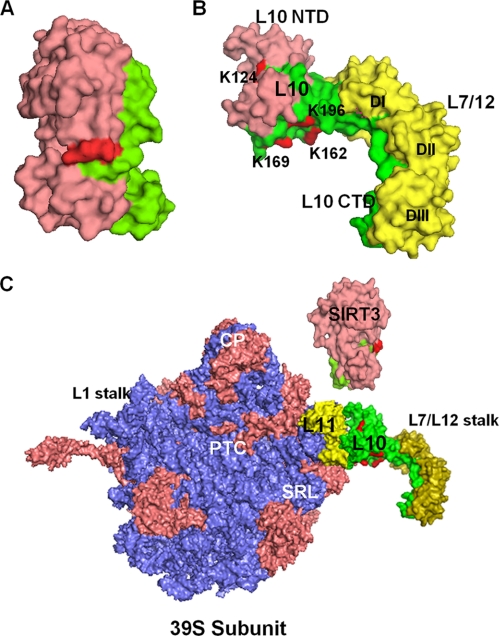
The SIRT3 and MRPL10 interaction domains were also evaluated using a structural model based on the coordinates of the human SIRT3 and the Thermotoga maritimi L10-L7/L12 complex (Fig. 5, A and B) (44). As illustrated in Fig. 5B, the region in the first 148 residues of MRPL10 (marked in pink) required for SIRT3 binding is probably the most accessible region for this interaction because MRPL10 also interacts with MRPL11 and MRPL12 to form the mitochondrial L7/L12 stalk. The C-terminal helix 8 of Thermotoga L10 binds three L7/L12 dimers (two L7/L12 dimers in E. coli), and the N-terminal globular domain interacts with the N-terminal domain of L11 and SIRT3 (Fig. 5B). Regions of MRPL10 and SIRT3 responsible for binding to each other and their interactions with the rest of the ribosome are in agreement with the idea of a transient interaction between SIRT3 and the mitochondrial ribosome. Moreover, acetylated Lys residues in MRPL10 are located near the SIRT3 binding domain, and reversible acetylation/deacetylation of these residues could be involved in regulation of ribosomal function by NAD+-dependent SIRT3 (Fig. 5, B and C). The L7/L12 stalk is known to be essential and to be the most highly regulated and flexible region of the large subunit due to its interaction with elongation, initiation, and release factors at different stages of translation (45–47). For this reason, it is plausible to suggest that the mitochondrial translational machinery might be regulated by reversible acetylation of the L7/L12 stalk protein MRPL10.
FIGURE 5.
Structural model of the SIRT3 and MRPL10 interactions in the ribosomal L7/L12 stalk. A, crystal structure model of the human SIRT3 (Protein Data Bank code 3GLU) representing the MRP-L10 (green surface) interaction site and the Acecs2 peptide at the active site (red). B, structure of the L10-L7/L12 complex (Protein Data Bank code 1ZAX) from T. maritimi was used to model L7/L12 stalk in mitochondria. In the model, MRPL10 was colored pink (to represent the SIRT3 binding site) and green, and the conserved Lys residues found to be acetylated in bovine MRPL10 (shown by asterisks in Fig. 2A) were colored red. The L7/L12 dimers (DI, DII, and DIII) were colored yellow. C, models of the human SIRT3 and T. maritimi L10-L7/L12 complex were used to represent their possible interactions with 55 S ribosomes using coordinates from the E. coli 50 S subunit (Protein Data Bank code 2AW4). The 50 S ribosomal rRNAs, L10, and SIRT3 were colored blue, green, and pink, respectively. The other functional regions, such as peptidyltransferase center (PTC), central protuberance (CP), sarcin-ricin loop (SRL), L1, and L7/L12 stalks of the large subunit and ribosomal proteins (salmon) are labeled in the model. NTD, N-terminal domain; CTD, C-terminal domain. The structural model was generated by PyMol software (DeLano Scientific LLC).
Recombinant and Ribosome-associated Endogenous NAD+-dependent SIRT3 Deacetylates MRPL10
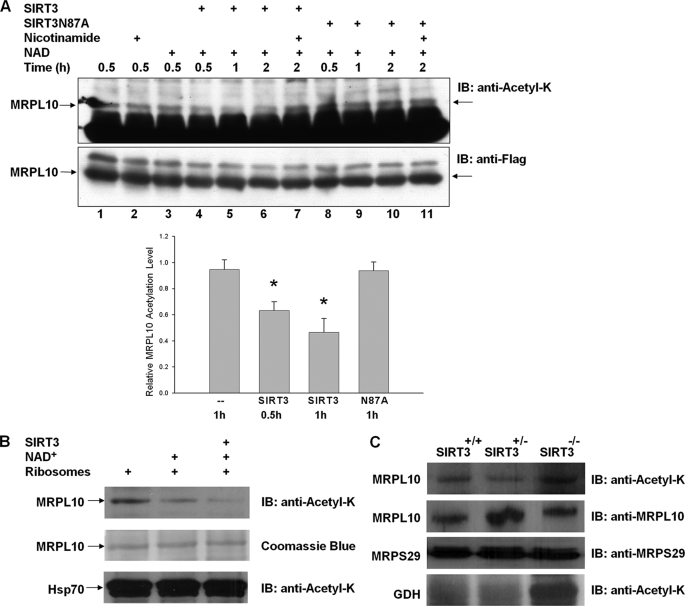
Given that the ribosomal protein MRPL10 is acetylated and interacts directly with the NAD+-dependent deacetylase SIRT3, we also investigated the effects of SIRT3 on deacetylation of MRPL10 in in vivo and in vitro assays. We have already shown that co-expression of SIRT3 in HEK293 cells with MRPL10 decreased MRPL10 acetylation (Fig. 1C). To further confirm this effect of SIRT3 on MRPL10, in vitro deacetylation assays were carried out using ectopically expressed FLAG-tagged MRPL10, purified with anti-FLAG-agarose beads in the presence of the recombinant SIRT3, SIRT3N87A, nicotinamide, or NAD+ (Fig. 6A). As shown in Fig. 6A, MRPL10 was acetylated (Fig. 6A, lane 1) and its incubation with nicotinamide or NAD+ did not affect the acetylation level (lanes 2 and 3), possibly due to the absence of endogenous deacetylase that co-immunoprecipitated with MRPL10. When incubated with SIRT3 and NAD+, the acetylation level of MRPL10 decreased significantly; however, deacetylation of MRPL10 was inhibited by the general NAD+-dependent deacetylase inhibitor nicotinamide (lanes 4–7). These results are consistent with the inhibition of the NAD+-dependent deacetylase activity of SIRT3 by nicotinamide. Moreover, the inactive deacetylase mutant, SIRT3N87A, was not effective in the deacetylation of MRPL10 in the presence or absence of NAD+ or nicotinamide (lanes 8–11).
FIGURE 6.
Deacetylation of MRPL10 by NAD+-dependent deacetylase, SIRT3. A, MRPL10 protein was produced by transfecting FLAG-tagged MRPL10 into HEK293 cells and then immunoprecipitated with anti-FLAG-agarose beads. Purified MRPL10 was then incubated with or without recombinant SIRT3 or SIRT3 mutant, SIRT3N87A, together with 10 mm nicotinamide or 5 mm NAD+, as indicated. The acetylation of MRPL10 was detected by immunoblotting with anti-N-acetyl lysine. *, p < 0.005. B, in vitro deacetylation reactions of about 0.1 A260 units of 55 S bovine mitochondrial ribosomes were performed in the presence of 3 mm NAD+ and 0.2 μg of recombinant SIRT3 as labeled and detected by immunoblotting (IB) analysis probed with anti-N-acetyl lysine antibody. The arrows indicate the specific deacetylation of MRPL10 but not the acetylated Hsp70 sedimented with ribosomes by endogenous and recombinant SIRT3 in the presence of 3 mm NAD+. C, mitochondrial ribosomes were prepared as described under “Experimental Procedures” from Sirt3−/−, Sirt3+/−, and Sirt3+/+ mouse liver, and the acetylation of ribosomal protein MRPL10 was detected by immunoblot analysis probed with anti-N-acetyl lysine antibody. As a control for acetylation of glutamate dehydrogenase in the absence of SIRT3 and equal loading, immunoblots were developed with anti-N-acetyl lysine and mouse MRPL10 and MRPS29 antibodies.
In addition to in vitro deacetylation assays performed with recombinant SIRT3 and MRPL10, similar assays were performed using purified bovine mitochondrial ribosomes containing acetylated MRPL10 as the SIRT3 substrate. The NAD+-dependent deacetylation of ribosome-associated MRPL10 by recombinant SIRT3 was detected by immunoblot analysis with anti-N-acetyl lysine antibody (Fig. 6B). In the presence of NAD+, the acetylation level of the protein band containing MRPL10 significantly decreased even without the addition of recombinant SIRT3, presumably due to the catalytic activity of the ribosome-associated endogenous SIRT3. This observation also confirmed the presence of ribosome-associated SIRT3 in the mitochondrial ribosome preparations, as detected by immunoblotting and mass spectrometric analysis (Fig. 3A and Table 2). Furthermore, the addition of the recombinant SIRT3 to the in vitro deacetylation assay further decreased the acetylation level of MRPL10. In contrast, acetylated Hsp70 (around 75 kDa), which co-sedimented with the ribosome, was neither deacetylated by an endogenous deacetylase nor by the recombinant SIRT3 in vitro.
To evaluate whether SIRT3 was the major deacetylase responsible for deacetylation of MRPL10, we also evaluated the acetylation level of MRPL10 in a SIRT3 knock out (Sirt3−/−) mouse. Analysis of mitochondrial ribosomes isolated from Sirt3−/−, Sirt3+/+, and Sirt3+/− mice revealed that the MRPL10 in the Sirt3−/− strain was more heavily acetylated than in either of the other strains (Fig. 6C). In agreement with Lombard et al. (15), we also detected hyperacetylation of mitochondrial glutamate dehydrogenase using two-dimensional gel electrophoresis and capillary LC-MS/MS analysis in SIRT3-knock-out mouse mitochondria (Fig. 6D) (data not shown). Protein blots were also probed with anti-mouse MRPL10 and MRPS29 antibodies to ensure equal loading of mitochondrial ribosomal proteins (Fig. 6C). Data obtained from deacetylation of ectopically expressed and endogenous ribosome-associated MRPL10 strongly suggest that the mitochondrial SIRT3 is associated with the mitochondrial ribosome and involved in the specific deacetylation of MRPL10 on the mitochondrial ribosome. Moreover, in the absence of SIRT3, the degree of MRPL10 acetylation was increased compared with the MRPL10 isolated from wild type and heterozygote mice expressing SIRT3.
Ribosomal Protein Acetylation Promotes Mitochondrial Protein Synthesis
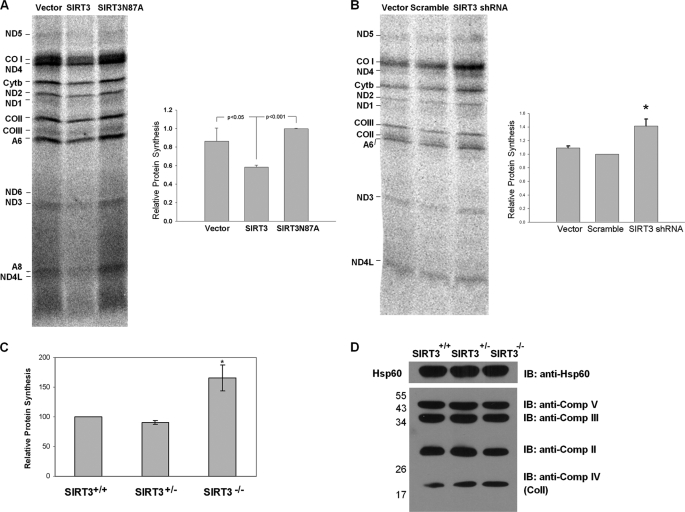
The mitochondrial translation machinery is responsible for the synthesis of 13 mitochondrially encoded proteins essential for oxidative phosphorylation: subunits of Complex I (ND1, ND2, ND3, ND4, ND4L, ND5, and ND6), cytochrome b, Complex IV (COI, COII, and COIII), and Complex V (ATP6 and ATP8). Using a well established in vivo labeling method for monitoring the synthesis of these mitochondrial proteins in mammalian cultures (38), we examined the role of reversible acetylation on mitochondrial protein synthesis in cells ectopically expressing SIRT3. Changes in the expression of mitochondrially encoded proteins were determined in C2C12 cells constitutively expressing murine SIRT3 or the SIRT3-N87A deacetylase mutant. In these assays, only de novo synthesized mitochondrial proteins were labeled with [35S]methionine in the presence of emetine, an inhibitor of cytoplasmic protein synthesis. In C2C12 cells expressing SIRT3, incorporation of [35S]methionine into 13 mitochondrially encoded proteins was decreased by about 20–25%, whereas there was no effect on the synthesis of mitochondrial proteins in the cells expressing the SIRT3 N87A deacetylase mutant (Fig. 7A). The decreased mitochondrial protein synthesis in SIRT3-expressing cells was neither due to a decrease in number of mitochondria as determined by quantitative reverse transcription-PCR of mitochondrial DNA nor due to mitochondrial mRNA levels (supplemental Fig. S3). In fact, SIRT3 expression slightly increased the levels of mitochondrial mRNA (supplemental Fig. S3). To further confirm the effect of SIRT3 on mitochondrial protein synthesis, C2C12 cells with stable expression of a short hairpin RNA against murine SIRT3 or a scrambled shRNA were tested for mitochondrial protein synthesis. As shown in Fig. 7B, in the SIRT3 knockdown C2C12 cells, the incorporation of [35S]methionine into all mitochondrially encoded proteins was significantly increased (about 1.4-fold) compared with the cells expressing the scrambled shRNA. These observations suggest that the expression of SIRT3 causes deacetylation of MRPL10 and down-regulates the synthesis of mitochondrial proteins.
FIGURE 7.
Role of ribosome acetylation/deacetylation in mitochondrial translation. A, SIRT3 expression decreases mitochondrial protein synthesis in C2C12 cells. Mitochondrial DNA-encoded protein synthesis in C2C12 cells stably expressing vector alone, SIRT3, or SIRT3N87A. The cells were exposed to [35S]methionine in the presence of a cytosolic translation inhibitor, emetine. A representative electrophoretic pattern of newly synthesized translational products is presented. ND1, -2, -3, -4, -4L, -5, and -6, subunits of NADH dehydrogenase 1, 2, 3, 4, 4L, 5, and 6, respectively; Cytb, apocytochrome b; COI, -II, and -III, subunits I, II, and III, respectively; A6 and A8, subunits 6 and 8, respectively, of the H+-ATPase. The combined intensities of 12 mitochondrial DNA-encoded proteins from each lane were used as an indicator of mitochondrial protein synthesis. The graph is a quantification of three independent pulse-labeling experiments. B, SIRT3 knockdown increases mitochondrial protein synthesis in C2C12 cells. Mitochondrial protein synthesis in C2C12 cells stably expressing vector alone, scrambled shRNA, or SIRT3 shRNA were measured by pulse-labeling experiments as described above. The combined intensities of 12 mitochondrial DNA-encoded proteins from each lane were used as an indicator of mitochondrial protein synthesis. C, acetylated ribosomes promote mitochondrial protein synthesis in vitro. Mitochondrial ribosomes (0.05–0.1 A260 units) isolated from Sirt3+/+, Sirt3+/−, and Sirt3−/− mouse liver mitochondria were used in the poly(U)-directed in vitro translation assays described under “Experimental Procedures.” *, p < 0.05. D, immunoblotting (IB) analysis of oxidative phosphorylation complexes obtained from Sirt3+/+, Sirt3+/−, and Sirt3−/− mouse liver mitochondria. Approximately 50 μg of mitochondrial lysate from each sample was separated on 12% SDS-PAGE, and immunoblot analysis was performed with Hsp60 antibody and total oxidative phosphorylation complex antibody mixture containing Complex I subunit NDUFB8, Complex II subunit of 30 kDa, Complex III subunit Core 2, Complex IV subunit II (COII), and ATP synthase subunit α antibodies.
We also performed poly(U)-directed polymerization assays to monitor mitochondrial ribosome activity as a function of the acetylation status of MRPL10. This assay is one of the primary techniques in assessing the interactions of mitochondrial elongation factors with the L7/L12 stalk of the ribosome (48, 49). To test the role of reversible acetylation on mitochondrial ribosome activity using this in vitro assay, we first isolated ribosomes from SIRT3 knock-out (Sirt3−/−), wild-type (Sirt3+/+), and heterozygote (Sirt3+/−) mouse liver mitochondria and performed poly(U)-directed poly(phenylalanine) synthesis in the presence of [14C]tRNAPhe and mammalian mitochondrial elongation factors EF-Tumt and EF-G1mt (49). To ensure that equal amounts of mitochondrial ribosomes were used in each in vitro translation assay, ribosomes were quantified using A260 measurements and immunoblotting using MRPS29 antibody as equal loading control (Fig. 6D). In these assays, mitochondrial ribosomes isolated from Sirt3−/− mice had 50–60% higher translational activity compared with those from Sirt3+/+ or Sirt3+/− mice (Fig. 7C). Because nicotinamide is a general inhibitor for sirtuins, including SIRT3, mitochondrial ribosomes prepared in the presence or absence of nicotinamide and cytoplasmic translation inhibitor emetine were also examined by poly(U)-dependent translation assays (supplemental Fig. S4). In these assays, we first verified the increased acetylation of mitochondrial ribosomes prepared in the presence of nicotinamide and tested the translation activity of these ribosomes. In agreement with the increased activity obtained in Sirt3−/− mice, the translation activity of mitochondrial ribosomes from nicotinamide-treated mitochondria was significantly more active, and this activity was not inhibited by cytoplasmic translation inhibitor emetine (supplemental Fig. S4).
We have also observed increased expression of mitochondrially encoded cytochrome c oxidase subunit II (COII) compared with the nucleus-encoded components of the oxidative phosphorylation complexes in wild-type, heterozygous, and SIRT3 knock-out mouse mitochondria (Fig. 7D). The increased acetylation and protein synthesis activity of ribosomes isolated from Sirt3−/− mice and nicotinamide-treated bovine mitochondria indicate that ribosomes containing acetylated MRPL10 are more active in synthesis of mitochondrial proteins.
Conclusions
Mitochondria are required for the production of more than 90% of the ATP required for survival of eukaryotic cells. However, slower metabolic rates and a reduction of oxidative phosphorylation and ATP synthesis have been associated with extended life span and slower aging. In fact, reduced expression of oxidative phosphorylation components by RNA interference in Caenorhabditis elegans decreases ATP production and oxygen consumption (50). Reduction of caloric intake induces expression of mitochondrial SIRT3 and causes similar effects by deacetylating mitochondrial proteins in an NAD+-dependent fashion (13). Similarly, the specific deacetylation of MRPL10 by the ribosome-associated deacetylase SIRT3 may play a pivotal role in coordinating the activity of the mitochondrial protein synthesis machinery to the [NADH]/[NAD+] ratio in mammalian mitochondria. This idea is supported by our findings suggesting that acetylated MRPL10 and/or ribosomes from SIRT3 knock-out mice are more active in protein synthesis, leading to an increase in the expression of mitochondrially encoded components of oxidative phosphorylation. Deacetylation of MRPL10 and/or ribosomes may impair protein synthesis and reduce the rate of oxidative phosphorylation by lowering the expression of essential proteins involved in oxidative phosphorylation.
Supplementary Material
Acknowledgments
We thank Margaret Nguyen for technical assistance and Eric Verdin for providing the human SIRT3-FLAG clone. We thank Robert Schlegel and Joseph Reese for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AG025223 (to Y. B.), R01DK075978 (to Q. T.), and R01GM071034 (to E. C. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
- HEK293 cells
- human embryonic kidney 293 cells
- LC
- liquid chromatography
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry
- HPLC
- high performance liquid chromatography
- sirtuin
- silent information regulator two (SIR2) homolog
- GST
- glutathione S-transferase
- aa
- amino acid(s).
REFERENCES
- 1.Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 2.Jackson P. J., Harris D. A. (1986) Biochem. J. 235, 577–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hallows W. C., Lee S., Denu J. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinardo M. M., Musicco C., Fracasso F., Milella F., Gadaleta M. N., Gadaleta G., Cantatore P. (2003) Biochem. Biophys. Res. Commun. 301, 187–191 [DOI] [PubMed] [Google Scholar]
- 5.Gerhart-Hines Z., Rodgers J. T., Bare O., Lerin C., Kim S. H., Mostoslavsky R., Alt F. W., Wu Z., Puigserver P. (2007) EMBO J. 26, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwer B., Bunkenborg J., Verdin R. O., Andersen J. S., Verdin E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10224–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michishita E., Park J. Y., Burneskis J. M., Barrett J. C., Horikawa I. (2005) Mol. Biol. Cell 16, 4623–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Onyango P., Celic I., McCaffery J. M., Boeke J. D., Feinberg A. P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13653–13658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000) Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 10.Smith J. S., Brachmann C. B., Celic I., Kenna M. A., Muhammad S., Starai V. J., Avalos J. L., Escalante-Semerena J. C., Grubmeyer C., Wolberger C., Boeke J. D. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landry J., Sutton A., Tafrov S. T., Heller R. C., Stebbins J., Pillus L., Sternglanz R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5807–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rose G., Dato S., Altomare K., Bellizzi D., Garasto S., Greco V., Passarino G., Feraco E., Mari V., Barbi C., BonaFe M., Franceschi C., Tan Q., Boiko S., Yashin A. I., De Benedictis G. (2003) Exp. Gerontol. 38, 1065–1070 [DOI] [PubMed] [Google Scholar]
- 13.Yang H., Yang T., Baur J. A., Perez E., Matsui T., Carmona J. J., Lamming D. W., Souza-Pinto N. C., Bohr V. A., Rosenzweig A., de Cabo R., Sauve A. A., Sinclair D. A. (2007) Cell 130, 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi T., Wang F., Stieren E., Tong Q. (2005) J. Biol. Chem. 280, 13560–13567 [DOI] [PubMed] [Google Scholar]
- 15.Lombard D. B., Alt F. W., Cheng H. L., Bunkenborg J., Streeper R. S., Mostoslavsky R., Kim J., Yancopoulos G., Valenzuela D., Murphy A., Yang Y., Chen Y., Hirschey M. D., Bronson R. T., Haigis M., Guarente L. P., Farese R. V., Jr., Weissman S., Verdin E., Schwer B. (2007) Mol. Cell. Biol. 27, 8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn B. H., Kim H. S., Song S., Lee I. H., Liu J., Vassilopoulos A., Deng C. X., Finkel T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14447–14452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Brien T. W., Fiesler S. E., Denslow N. D., Thiede B., Wittmann-Liebold B., Mougey E. B., Sylvester J. E., Graack H. R. (1999) J. Biol. Chem. 274, 36043–36051 [DOI] [PubMed] [Google Scholar]
- 18.Koc E. C., Burkhart W., Blackburn K., Moseley A., Koc H., Spremulli L. L. (2000) J. Biol. Chem. 275, 32585–32591 [DOI] [PubMed] [Google Scholar]
- 19.Cavdar Koc E., Burkhart W., Blackburn K., Moseley A., Spremulli L. L. (2001) J. Biol. Chem. 276, 19363–19374 [DOI] [PubMed] [Google Scholar]
- 20.Suzuki T., Terasaki M., Takemoto-Hori C., Hanada T., Ueda T., Wada A., Watanabe K. (2001) J. Biol. Chem. 276, 21724–21736 [DOI] [PubMed] [Google Scholar]
- 21.Koc E. C., Burkhart W., Blackburn K., Moyer M. B., Schlatzer D. M., Moseley A., Spremulli L. L. (2001) J. Biol. Chem. 276, 43958–43969 [DOI] [PubMed] [Google Scholar]
- 22.Sharma M. R., Koc E. C., Datta P. P., Booth T. M., Spremulli L. L., Agrawal R. K. (2003) Cell 115, 97–108 [DOI] [PubMed] [Google Scholar]
- 23.O'Brien T. W., O'Brien B. J., Norman R. A. (2005) Gene 354, 147–151 [DOI] [PubMed] [Google Scholar]
- 24.Miller C., Saada A., Shaul N., Shabtai N., Ben-Shalom E., Shaag A., Hershkovitz E., Elpeleg O. (2004) Ann. Neurol. 56, 734–738 [DOI] [PubMed] [Google Scholar]
- 25.Kissil J. L., Cohen O., Raveh T., Kimchi A. (1999) EMBO J. 18, 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chintharlapalli S. R., Jasti M., Malladi S., Parsa K. V., Ballestero R. P., González-García M. (2005) J. Cell Biochem. 94, 611–626 [DOI] [PubMed] [Google Scholar]
- 27.Yoo Y. A., Kim M. J., Park J. K., Chung Y. M., Lee J. H., Chi S. G., Kim J. S., Yoo Y. D. (2005) Mol. Cell Biol. 25, 6603–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller J. L., Koc H., Koc E. C. (2008) Protein Sci. 17, 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z., Cotney J., Shadel G. S. (2007) J. Biol. Chem. 282, 12610–12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emdadul Haque M., Grasso D., Miller C., Spremulli L. L., Saada A. (2008) Mitochondrion 8, 254–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zanotto E., Lehtonen V., Jacobs H. T. (2008) Biochim. Biophys. Acta 1783, 2352–2362 [DOI] [PubMed] [Google Scholar]
- 32.Zambrowicz B. P., Friedrich G. A., Buxton E. C., Lilleberg S. L., Person C., Sands A. T. (1998) Nature 392, 608–611 [DOI] [PubMed] [Google Scholar]
- 33.Jordan M., Schallhorn A., Wurm F. M. (1996) Nucleic Acids Res. 24, 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao Z. H., Becker L. B., Vanden Hoek T. L., Schumacker P. T., Li C. Q., Zhao D., Wojcik K., Anderson T., Qin Y., Dey L., Yuan C. S. (2003) Pharmacol. Res. 47, 463–469 [DOI] [PubMed] [Google Scholar]
- 35.Matthews D. E., Hessler R. A., Denslow N. D., Edwards J. S., O'Brien T. W. (1982) J. Biol. Chem. 257, 8788–8794 [PubMed] [Google Scholar]
- 36.Spremulli L. L. (2007) Methods Mol. Biol. 372, 265–275 [DOI] [PubMed] [Google Scholar]
- 37.Landry J., Sternglanz R. (2003) Methods 31, 33–39 [DOI] [PubMed] [Google Scholar]
- 38.Chomyn A. (1996) Methods Enzymol. 264, 197–211 [DOI] [PubMed] [Google Scholar]
- 39.Cavdar Koc E., Blackburn K., Burkhart W., Spremulli L. L. (1999) Biochem. Biophys. Res. Commun. 266, 141–146 [DOI] [PubMed] [Google Scholar]
- 40.Koc E. C., Burkhart W., Blackburn K., Koc H., Moseley A., Spremulli L. L. (2001) Protein Sci. 10, 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soung G. Y., Miller J. L., Koc H., Koc E. C. (2009) J. Proteome Res. 8, 3390–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller J. L., Cimen H., Koc H., Koc E. C. (2009) J. Proteome Res. 8, 4789–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J., Sprung R., Pei J., Tan X., Kim S., Zhu H., Liu C. F., Grishin N. V., Zhao Y. (2009) Mol. Cell Proteomics 8, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin L., Galonek H., Israelian K., Choy W., Morrison M., Xia Y., Wang X., Xu Y., Yang Y., Smith J. J., Hoffmann E., Carney D. P., Perni R. B., Jirousek M. R., Bemis J. E., Milne J. C., Sinclair D. A., Westphal C. H. (2009) Protein Sci. 18, 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahl M. C., Möller W. (2002) Curr. Protein Pept. Sci. 3, 93–106 [DOI] [PubMed] [Google Scholar]
- 46.Diaconu M., Kothe U., Schlünzen F., Fischer N., Harms J. M., Tonevitsky A. G., Stark H., Rodnina M. V., Wahl M. C. (2005) Cell 121, 991–1004 [DOI] [PubMed] [Google Scholar]
- 47.Allen G. S., Zavialov A., Gursky R., Ehrenberg M., Frank J. (2005) Cell 121, 703–712 [DOI] [PubMed] [Google Scholar]
- 48.Woriax V. L., Bullard J. M., Ma L., Yokogawa T., Spremulli L. L. (1997) Biochim. Biophys. Acta 1352, 91–101 [DOI] [PubMed] [Google Scholar]
- 49.Hunter S. E., Spremulli L. L. (2004) Biochemistry 43, 6917–6927 [DOI] [PubMed] [Google Scholar]
- 50.Lee S. S., Lee R. Y., Fraser A. G., Kamath R. S., Ahringer J., Ruvkun G. (2003) Nat. Genet. 33, 40–48 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.