Abstract
Here we describe features of the first non-mammalian T-type calcium channel (LCav3) expressed in vitro. This molluscan channel possesses combined biophysical properties that are reminiscent of all mammalian T-type channels. It exhibits T-type features such as “transient” kinetics, but the “tiny” label, usually associated with Ba2+ conductance, is hard to reconcile with the “bigness” of this channel in many respects. LCav3 is 25% larger than any voltage-gated ion channel expressed to date. It codes for a massive, 322-kDa protein that conducts large macroscopic currents in vitro. LCav3 is also the most abundant Ca2+ channel transcript in the snail nervous system. A window current at typical resting potentials appears to be at least as large as that reported for mammalian channels. This distant gene provides a unique perspective to analyze the structural, functional, drug binding, and evolutionary aspects of T-type channels.
Keywords: Calcium Channels, Membrane Biophysics, Neuroscience, RNA Abundance, Site-directed mutagenesis, T-type Channels, Invertebrates, Patch Clamp Electrophysiology, Site-directed Mutagenesis
Introduction
T-type calcium channels open in response to slight depolarizations in the low voltage range. Paradoxically, they are also recruited after membrane hyperpolarization as occurs during rebound burst firing (1). A window current of T-type channels is a feature that permits Ca2+ entry at rest (2) and contributes to differentiation and growth promoting functions in both excitable and non-excitable cells (3). T-type channels are also a leading pharmaceutical drug target and are implicated in a wide range of conditions such as epilepsy, pain, hypertension, cancer, and mental disorders (4).
T-type Ca2+ currents were first measured in starfish eggs using a two-electrode voltage clamp (5). Currents conducted by “Channel I” were evoked by small depolarizations (low voltage-activated), visible as a small hump in a current amplitude versus test potential plot, appearing inconsequential beside the Channel II currents elicited by larger depolarizations (high voltage-activated). Ca2+ channel types would be discriminated further by Tsien and co-workers (6) on the basis of properties where Ba2+ is the charge carrier. High voltage-activated L-type channels have a large unitary Ba2+ conductance with long-lasting openings, N-type (or non-L-type) channels are typically associated with neurons of intermediate unitary conductance, and the low voltage-activated, T-type channels produce transient currents that are of tiny unitary conductance in Ba2+ and close slowly upon membrane repolarization, producing a slowly deactivating tail current (6).
T-type channels remain as the least understood among the Ca2+ channel families. Although most of the 10 mammalian Ca2+ channel genes were characterized in the late 1980s, an additional decade was required for a description of the three T-type genes, Cav3.1 (α1G), Cav3.2 (α1H), and Cav3.3 (α1I) (7). Progress in understanding T-type channel functions continues to be hampered by the lack of highly selective blockers that discriminate between Cav3 channel types or separate Cav3 channels from related L-type (Cav1) and non-L-type (Cav2) Ca2+ channels, which usually produce more robust Ca2+ entry into the same cells (7).
Here we describe the in vitro expression characteristics of the first non-mammalian, T-type channel, LCav3, cloned from the pond snail, Lymnaea stagnalis. This structurally distant channel has quintessential features of T-types such as transient kinetics. LCav3 is big in many respects, such as its protein size; it expresses large macroscopic currents in human cells, it is the most abundant Ca2+ channel transcript in the snail nervous system, and it generates window currents that appear to be at least as large as those reported for mammalian channels. LCav3 provides a unique perspective to analyze the structure, function, and drug binding of T-type channels and serves as a useful surrogate in residue swapping experiments. Searches for the fundamental mechanisms that regulate this singleton invertebrate T-type channel will be facilitated by the simple molluscan preparation, where accessible and identified neurons underlying well described behaviors can be studied in isolated Lymnaea neurons, cultured synapses, or within intact, identified networks in situ. Also, LCav3 provides nourishment for evolutionary speculation. Although the first gastropods (500 million years ago) are likely quite distant from this ancestral branch point, the extant snail homolog, LCav3, is reminiscent of the gene that predates the speciation that led to the emergence of the three distinct, mammalian T-type channel genes.
EXPERIMENTAL PROCEDURES
Cloning and Sequencing of LCav3
The complete open reading frame for LCav3 was determined from at least three independent, overlapping DNA fragments from the PCR screening of L. stagnalis central nervous system λZAP cDNA libraries to generate a consensus gene. The full-length 9031-bp cDNA transcript is available in DDBJ/EMBL/GenBankTM databases under accession no. AF484084 and replaced a previous partial coding sequence entry of 5991 bp. The final clone was assembled from four overlapping PCR with sticky ends (numbered by cDNA transcript positions), XhoI-SpeI (209–2865), SpeI-SalI (2812–4544), SalI-MluI (4503–6874), and MluI-BamHI (6850–8869). Silent mutations were created in a Kozak consensus sequence upstream of the start codon (209–211), an MluI site (6858–6863), and several hairpin structures thought to interfere with the site-directed mutagenesis reaction (5225–5285). The full-length LCav3 coding sequence was assembled between XhoI and BamHI sites in bicistronic vector pIRES2-EGFP2 (Clontech). Low frequency of positive recombinants during cloning and the slow rate of growth of the full-length plasmid in bacteria (five full days before a colony appears on a bacterial plate after transformation) suggest that the plasmid insert is toxic to bacteria.
Transfections
HEK-293T cells (M. Calos, Stanford University) were cultured in Dulbecco's modified Eagle's medium (Sigma) with 10% fetal bovine serum (Sigma) and supplemented with 0.5% (v/v) penicillin-streptomycin solution (Sigma). For electrophysiology, 6 μg of the LCav3 pIRES2-EGFP construct was transfected into cells at 40–50% confluency using the standard Ca2+ phosphate transfection method. After overnight transfection, the cells were washed twice with culture media and incubated at 28 °C in a humidified, 5% CO2 chamber for 3 days. After incubation, cells were detached using a trypsin-EDTA solution (Sigma), plated at 10% confluency onto glass coverslips, and incubated at 37 °C for 4 h because adhesion to the glass substrate requires warmer temperatures (8).
Whole Cell Patch Clamp Recordings
Whole-cell recordings were carried out at 23 °C using either a 5 mm external Ca2+ solution (5 mm CaCl2, 166 mm tetraethylammonium chloride, 10 mm HEPES, pH 7.4) or 5 mm external Ba2+ solution (5 mm BaCl2, 166 mm tetraethylammonium chloride, 10 mm HEPES pH 7.4) and an internal solution consisting of 125 mm CsCl, 10 mm EGTA, 2 mm CaCl2, 1 mm MgCl2, 4 mm MgATP, 0.3 mm Tris-GTP, and 10 mm HEPES, pH 7.2. Recordings were obtained using an Axopatch 200B amplifier, sampled to a PC through a Digidata 1440a A/D converter. Data were filtered at 2 kHz and digitized at 5 kHz and acquired using pCLAMP 10.1 software (Molecular Devices). The pipette resistance was maintained between 3 and 5 megaohms, and the typical access resistance was between 4 and 5 megaohms. Only recordings with minimal leak (<10%) and small current sizes (<2 nA) were used for analysis, and offline leak subtraction was carried out using the Clampfit 10.1 software (Molecular Devices). Series resistance was compensated to 70% (prediction and correction; 10-μs lag). A gravity flow system was used to perfuse 5 mm Ca2+- or Ba2+-containing extracellular solution or 5 mm Ca2+ external solution containing solubilized Ni2+ (Sigma) or mibefradil (Sigma).
Data Analysis
Ca2+ current activation curves were constructed by converting the peak current values from each current-voltage relationship data set to conductance using the equation gCa = Ipeak/(Vcommand − ECa), where Ipeak is the peak current, Vcommand is the command pulse potential, gCa is the calcium conductane, and ECa is the Ca2+ reversal potential as determined by linear extrapolation of the current values in the ascending portion of the current-voltage relationships. Conductance values were then normalized and individually fitted with the Boltzmann equation, g/gmax = (1 + (exp(−Vcommand − V½)/K)) − 1, where g is the peak conductance, gmax is the maximal peak Ca2+ conductance, Vcommand is the conditioning potential, V½ is the half-maximal activation, and k is the activation slope factor. The steady-state inactivation curves were constructed by plotting normalized current (peak test pulse current/peak prepulse current) as a function of the inactivating potential. The data were fitted with a Boltzmann equation, I/Imax = (1 + exp((Vinact − V½/k)) − 1, where I is the peak test pulse current, Imax is the peak test pulse current when the conditioning pulse was −110 mV, Vinact and V½ are the conditioning potential and the half-maximal inactivation, respectively, and k is the inactivation slope factor. Kinetics of activation, inactivation, and deactivation were determined by fitting monoexponential functions over the growing or decaying phases of each current trace using the software Clampfit 10.1.
Antibody Production
LCav3 I-II linker coding sequence (1976–2575 bp) was PCR-amplified and cloned into the bacterial protein expression vector pET-22b(+) (Novagen) via NdeI and XhoI restriction sites. Peptide expression was induced by 1 mm isopropyl 1-thio-β-d-galactopyranoside in RosettaTM (DE3) cells (Novagen) transformed with the pET22b(+) plasmid-containing construct. Supernatant of lysed bacterial cells containing His6 LCav3 I-II linker expression was run and washed through a column containing Ni2+-charged His·Bind® resin (Novagen), then eluted off of the beads, dialyzed, and quantified using the Bradford assay. A rabbit was injected three times with recombinant proteins emulsified with Freund's complete adjuvant for the first injection and Freund's incomplete adjuvant (Sigma) for the subsequent injections. IgG rabbit antiserum was tested for immune reactivity with the antigen by Western blotting.
Immunolabeling of Recombinant LCav3 in HEK-293T Cells
HEK-293T cells were transfected with either 8 μg of LCav3 in pIRES2-EGFP alone or with 3.2 μg of LCav1 α1 subunit in pIRES2-EGFP (9) plus 2.4 μg of the rat β1 subunit in pMT2 and 2.4 μg of rat α2δ in pMT2. Cells were washed after transfection and incubated at 28 °C for 1 week before trypsinization and plating onto glass coverslips. Cells were then fixed with 1% paraformaldehyde in PBS overnight at 4 °C, washed twice with PBS, then permeabilized using phosphate-buffered saline containing 0.2% Tween 20 (PBS-T) for 10 min at room temperature. 1:500 primary antibody or preimmune serum was applied to preblocked cells overnight in PBS-T containing 3% bovine serum albumin. 3×-washed cells were incubated with 1:1000 diluted AlexaFluor 594 goat anti-rabbit secondary antibody in PBS-T containing 3% bovine serum albumin for 1 h at 23 °C, washed 4×, and imaged at 40× magnification with a Zeiss AxioObserver Z1 inverted epifluorescent microscope to detect AlexaFluor 594 antibody and EGFP. Images were captured using Zeiss AxioVision software, an brightness/contrast was adjusted using Adobe Photoshop.
Southern Blot
15-μg aliquots of genomic DNA isolated from Lymnaea tissue were digested with EcoRV, HindIII, EcoRI, and XhoI, and DNA fragments were separated through a 1% agarose gel. Digested DNA was transferred onto a positively charged nylon membrane (Roche Applied Science), and a standard hybridization procedure was carried out using manufacturer's instructions (EasyHyb, Roche Applied Science). Membranes were probed with a gel-purified 597-bp PCR product of the LCav3 gene (1979–2575 bp) incorporated with DIG-11-dUTP (Roche Applied Science). The probe was localized on the membrane using anti-digoxigenin alkaline phosphatase-conjugated antibody (Roche Applied Science; 1:5000 dilution) and color substrate solution nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Roche Applied Science).
Semiquantitative RT-PCR
RNA was extracted from the central nervous system of L. stagnalis using Tri Reagent (Sigma) and optimized from standard methods (10). RNA was treated with DNase (Fermentas) to remove contaminant DNA for RT-PCR analysis. RNA was quantified by spectrophotometry and visualized by gel electrophoresis to confirm the lack of degradation. First-strand cDNA were synthesized from RNA at 54 °C for 80 min and 70 °C for 15 min using either oligo-dT or random hexamer primers and Superscript III reverse transcriptase (Invitrogen) and controls were prepared that lacked reverse transcriptase. Primers used for cDNA amplification were designed to have similar melting temperatures, minimal secondary structure, and amplified fragments of similar sizes (500–600 bp). PCR primers spanned sequences from the following genes (GenBankTM accession nos.: cDNA transcript positions): Lactin (DQ206431: 38–628), LNALCN (AF484086, 3149–3761), LCav1 (AF484079: 5421–6103), LCav2 (AF484082: 5427–6095), LCav3 (AF484084: 7816–8546). PCR products generated after 25 cycles in the thermocycler were imaged in ethidium bromide-stained gels using a gel documentation system (Alpha Innotech) under UV light. Densinometric analysis of DNA band intensity was performed using Automatic Image Capture software (Alpha Innotech).
RESULTS
Identity of an Invertebrate T-type Channel (LCav3)
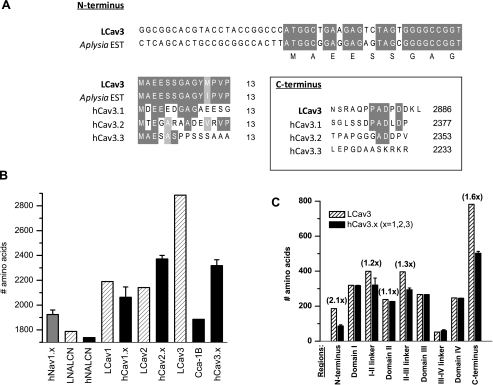
A novel invertebrate T-type channel transcript (9031 bp) was assembled from cDNA derived from the central nervous system of the freshwater pond snail, L. stagnalis, with a coding region that starts as an almost perfect match to an Expressed Sequence Tag data base entry from the marine snail Aplysia californica (accession no. EB302921). The LCav3 open reading frame predicts a 2886-amino acid protein, with an estimated ∼322-kDa molecular mass. The start and end of the LCav3 amino acid sequence also resemble those of the human Cav3.1 channel (Fig. 1A). Consistent with other related cation channels of this type, LCav3 has four repeat domains (DI to DIV) with each domain containing six membrane-spanning segments similar to the voltage-gated K channels (Fig. 3A). LCav3 is the largest voltage-gated ion channel expressed to date, being 25% larger than the mammalian T-types, 50% larger than a T-type homologue from Caenorhabditis elegans (cca-1b), and significantly larger than other Ca2+ channels (Cav1, Cav2), sodium channels (Nav), and NALCN (Fig. 1B). The extra size is mostly due to long cytoplasmic N and C termini and the cytoplasmic I-II and II-III linkers (Fig. 1C).
FIGURE 1.
Full-length snail LCav3 is the largest identified voltage-gated ion channel expressed to date. It is coded by a 9031-bp cDNA transcript that forms a 2886-amino acid protein with a molecular mass of 322 kDa. A, the N terminus closely matches with a putative start site derived from marine snail A. californica EST (EB302921) and slightly resembles the N and C termini of human Cav3.1–3.3. B, LCav3 is 1.25× larger than human Cav3 channels and 1.5× larger than nematode T-type, cca-1B, and all other four repeat ion channels. C, LCav3 is larger than human Cav3 channels in the N and C terminus and also the I-II and II-III cytoplasmic linkers.
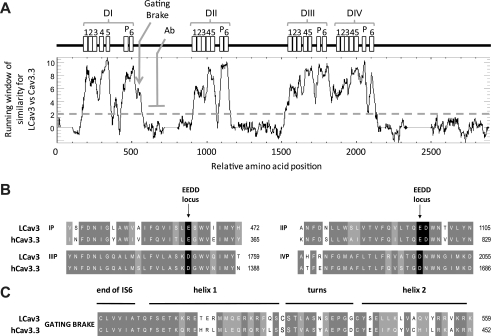
FIGURE 3.
Running window of similarity (A) and alignments (B and C) between amino acid sequences of distant T-type channel homologs (snail LCav3 and human Cav3.3) reveal that the invariant structures for T-type channels are harbored in six membrane-spanning segments in all four domains (I, II, III, and IV), including an ion conducting pore (S5-P-loop-S6) and voltage sensor (S1-S4). Illustrated is the position in the I-II linker where LCav3 polyclonal antibody (Ab) was generated in rabbits against a 200-amino acid peptide. B, shown is amino acid sequence alignment of the re-entrant P-loop located between S5 and S6 of each of the four domains illustrating the signature sequence (EEDD locus) that influences Ca2+ ion permeation and selectivity. The conserved aspartate residue (1097 in LCav3) in a position downstream of the selectivity filter glutamate residue is positioned to attract incoming Ca2+ ions to the pore.3 LCav3 contains a neutral isoleucine in the outer pore at position 468 where mammalian T-type channels have a negatively charged residue (Glu or Asp) that influence pore blocking drugs. C, alignment of the cytoplasmic gating brake in proximal I-II linker is shown. The gating brake is thought to prevent T-type channel gating at more hyperpolarized potentials.
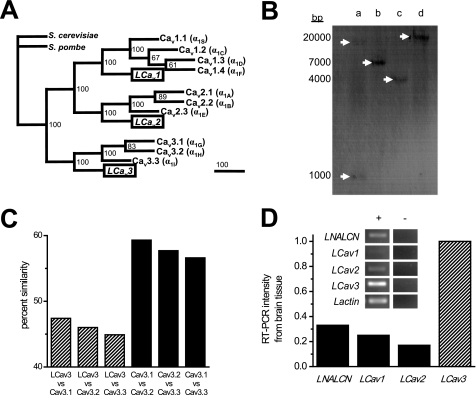
Snails, like most invertebrates, appear to have branched before the genomic duplication events that led to the expansion of gene isoforms and as such bear only single representatives for each of the three Ca2+ channel gene families (Cav1, Cav2, and Cav3) compared with the 10 different Ca2+ channel genes in mammals (Fig. 2A). The snail T-type gene diverges dramatically in amino acid sequence from the mammalian genes (mean identity/similarity = 37.6/46.1), whereas the mammalian homologues are clustered closer together (mean identity/similarity = 50.7/57.9) (Fig. 2C). The single copy nature of the LCav3 gene in the genome is evidenced by Southern blotting, producing a single banding hybridization pattern, except for a gel lane where the restriction enzyme (EcoRV) cuts the genomic DNA within the probe sequence, leading to two bands of weaker intensity (lane a, Fig. 2B). The LCav3 transcript is more abundant than other Ca2+ channels (LCav1 and LCav2) or LNALCN in central nervous system tissue as measured by semiquantitative RT-PCR (Fig. 2D). The higher expression of LCav3 compared with these others was also found by single cell, quantitative real-time PCR of individual VD4 neurons in a previous study using different primer sets (see Spafford et al. (11) and Fig. 2).
FIGURE 2.
Singleton, snail T-type Ca2+ channel gene is distantly related to vertebrate homologs and is the most abundant Ca2+ channel transcript in the snail brain. A, shown is the most parsimonious gene tree generated using multiple aligned sequences, analyzed in PAUP4.0 (D. L. Swofford) and illustrated with TreeView (R. D. M. Page). Sequences include official human sequences (IUPHAR database); LCav3 (GenBankTM accession no. AF484084) and yeast gene Cch1 from Schizosaccharomyces pombe (GenBankTM accession no. CAB11726) and Saccharomyces cerevisiae (GenBankTM accession no. CAA97244). Numbers at branch points represent bootstrap values based on 100 replicates in heuristic search. Phylogram branches are scaled by their length and rooted with Cch1 Ca2+ channel homologs from fungi species. C, percent amino acid similarity scores were generated from EMBOSS NEEDLE (EMBL). B, Southern blot indicates a single copy gene in the Lymnaea genome. A T-type probe hybridized to create a banding pattern (white arrows) on the blot was created from membrane transfer of genomic DNA digested with either EcoRV(a), HindIII (b), EcoRI (c), or XhoI (d). The probe contained an EcoRV restriction site, so the probe hybridized to two genomic DNA fragments digested with EcoRV. D, densitometric intensity of RT-PCR bands (illustrated in the inset) was generated from Lymnaea brain tissue.
Fig. 3A illustrates a running window of similarity between aligned LCav3 and human Cav3.3 channel protein sequences. The strongest homology is observed in the six-transmembrane segments and pore (P)-loops of each domain (Fig. 3A). Side chains of conserved negative residues lining the four P-loops contribute to a DDEE selectivity filter in T-type channels (7), including LCav3 (Fig. 3B). The pores of LCav3 and all voltage-gated Ca2+ channels also include a highly conserved aspartate adjacent to the selectivity-filter glutamate in Domain II (Fig. 3B), which may serve to attract incoming Ca2+ ions to the ion selective pore.3 One noticeably conserved region outside of the membrane-spanning domains is the gating brake present in the proximal I-II cytoplasmic linker (Fig. 3C). Comparison of LCav3 with Cav3.3 residues suggests conserved elements in the gating brake, including a putative helix-loop-helix hydrophobic core, a putative salt bridge, and potential protein-protein interaction sites facing away from the hydrophobic core (Fig. 3C).4
Expression Characteristics of LCav3 in HEK-293T Cells
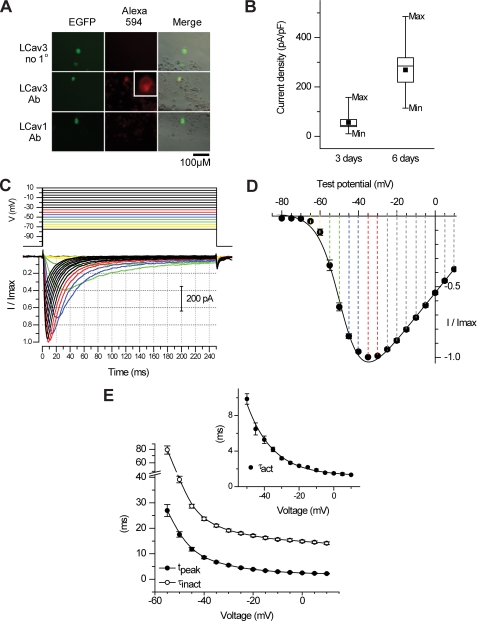
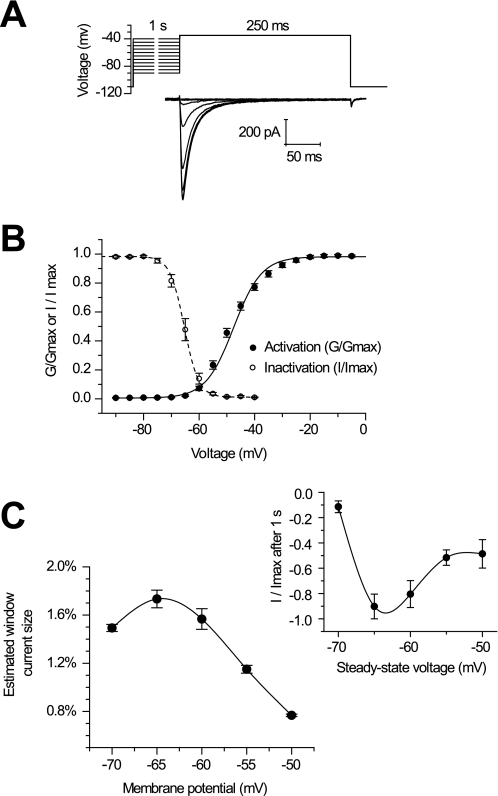
Transient transfection of LCav3 cDNA contained in pIRES2-EGFP vector reveals membrane-delimited staining of HEK-293T cells (box, Fig. 4A) with a rabbit polyclonal antibody generated against the I-II cytoplasmic linker of LCav3 (see Fig. 3A for the relative location of the epitope). Antibody staining was not apparent in LCav1-transfected cells (9) or when preimmune serum was used to detect LCav3 (Fig. 4A). Channel expression levels generally correspond to the EGFP intensity in HEK-293T cells, as would be expected with the LCav3 cDNA expressed on the same mRNA as EGFP using the bicistronic expression vector, pIRES2-EGFP. The optimal level of HEK-293T expression for electrophysiological recording (200 pA to 1.5 nA) corresponds to 3 days after transfection, whereas larger currents of up to 10 nA were possible by allowing protein expression to continue for up to 6 days (Fig. 4B). Typical transient kinetics of T-type currents are revealed in whole cell recordings of LCav3. Small voltage steps above threshold (−70 to −65 mV) are slow to activate and inactivate (requiring 10s of ms). Larger voltage steps elicit currents with progressively faster activation and inactivation kinetics that cross over each other with each successive step toward a maximal current for a voltage step of −35 mV (Fig. 4C). τ of inactivation kinetics follows the change in the time to peak and τ of activation kinetics with increasing voltage steps (Fig. 4E) (see Table 1 for a detailed comparison of biophysical parameters between Cav3 channels).
FIGURE 4.
Transient transfection of HEK-293T cells harboring the pIRES2-EGFP plasmid containing invertebrate T-type channel cDNA reveal highly abundant channels and characteristic T-type channel properties. A, membrane delimited staining of LCav3 (inset) is evident in EGFP-positive cells but only with LCav3-specific antibody and not with preimmune serum or with LCav1-transfected cells. B, the box chart indicates the current density (pA/pF (picofarads)) of LCav3 expression on 3 or 6 days after transfection. The box chart also illustrates mean, median ± 1 S.D., min/max current densities. C, sample LCav3 currents are shown in response to 5-ms voltage steps from a −110-mV holding potential. Illustrated is an ensemble of rapidly activating and inactivating Ca2+ currents where each trace “crosses over” the previous one from rest to peak, and the resulting normalized peak currents are plotted as a function of voltage step, indicating low threshold of activation (−65 mV) and maximal currents generated at a step to −35 mV (D). Current-voltage relationships were curve-fitted with an Ohmic-Boltzmann function. E, the increase in inactivation kinetics (τinact) closely follows the increasing speed at which the current approaches peak (tpeak), also reflected in the faster rate of activation, curve-fitted and represented by τact.
TABLE 1.
Comparison of biophysical parameters for recombinant LCav3 and mammalian T-type channels expressed in human cell lines
NA, not available.
| Electrophysiology | LCav3 | n | Cav3.1 | Reference | Cav3.2 | Reference | Cav3.3 | Reference |
|---|---|---|---|---|---|---|---|---|
| Activation | ||||||||
| V½ | −48.42 ± 0.34 | 5 | −44.6 ± 0.7a | 25, 49 | −42.7 ± 0.7b | 21, 24 | −40.2 ± 0.8b | 25 |
| −42.1 ± 1.1b | −43.8 ± 0.8b | |||||||
| K | 5.81 ± 0.30 | 5 | 5.8 ± 0.2 | 25, 49 | 6.0 ± 0.1 | 21, 24 | 5.8 ± .2 | 25 |
| 5.3 ± 0.2 | 6.3 ± 0.1 | |||||||
| Peak of IV (mV) | −35 | 5 | −30, −30 | 25, 49 | −30 | 21 | −25 | 25 |
| Inactivation | ||||||||
| V½ | −65.40 ± 0.15 | 7 | −77.7 ± 0.7b | 25, 49 | −77.7 ± 0.8b | 21, 24 | −68.6 ± 0.7b | 25 |
| −74.1 ± 1.6b | −78.1 ± 1.2b | |||||||
| K | 2.76 ± 0.13 | 7 | −5.0 ± 0.2b | 25 | −5.7 ± 0.2b | 21, 24 | −5.5 ± 0.5b | 25 |
| −5.3 ± 0.2b | −5.7 ± 0.1b | |||||||
| Kinetics at −20 mV | ||||||||
| τact | 2.34 ± 0.12 | 5 | 1.6 ± 0.2a | 49 | 2.9 ± 0.1a | 24 | NA | |
| τinact | 17.11 ± 0.46 | 5 | 13.9 ± 1.1a | 49 | 17.4 ± 0.9 | 24 | NA | |
| Kinetics at −30 mV | ||||||||
| τact | 3.19 ± 0.17 | 5 | 2.9 ± 0.4 | 50 | 3.8 ± 0.1a | 50 | 14 ± 1.0b | 50 |
| τinact | 19.16 ± 0.57 | 5 | 17 ± 6 | 50 | 16 ± 1 | 50 | 80 ± 5b | 50 |
| Deactivation | ||||||||
| −100 mV | 1.03 ± 0.15 | 10 | 2.6 ± 0.2b,c | 28 | 3.6 ± 0.4b,c | 28 | 1.12 ± 0.1c | 28 |
| −70 mV | 2.91 ± 0.64 | 10 | 6.2 ± 0.4b,c | 28 | 8.5 ± 1.1b,c | 28 | 2.1 ± 0.1a,c | 28 |
| Pharmacology | ||||||||
| Nickel (IC50 μm) | 300.00 ± 29.24 | 4 | 250 ± 22d | 15, 16 | 4.9 ± 2.0b,d | 15, 16 | 216 ± 9a,d | 15 |
| 12 ± 2b,d | ||||||||
| Mibefradil (IC50 μm) | 0.68 ± 0.03 | 4 | 1.2 ± 0.2d | 19 | 1.1 ± 0.2d | 9 | 1.5 ± 0.1b,d | 19 |
a p < 0.05 (one-way analysis of variance).
b p < 0.005 (one-way analysis of variance).
c 2 mm instead of 5 mm Ca2+ in extracellular solution.
d 10 mm Ba2+ in extracellular solution.
The pairing of activation and inactivation with LCav3 is typical of T-type currents and has led some to suggest that T-type channel inactivation is voltage-independent (12). The voltage sensitivity of LCav3 approximates a typical low threshold T-type current that peaks at −35 mV (Fig. 4d), although technically LCav3 is slightly lower threshold than mammalian T-types (peak between −30 and −25 mV). Channel availability at steady state was assessed after a 1-s prepulse protocol (Fig. 5A) and a 5-s prepulse protocol (data not shown), revealing a surprisingly steep and positively shifted availability curve compared with mammalian T-type channels (Fig. 5B). The combination of the very low threshold of channel activation and the large fraction of possible available channels creates a potentially large and persistent window current near the resting membrane potential of typical neurons (2) (Fig. 5C). An estimated window current was gathered by the product of available channels under steady-state conditions and the relative peak conductance. As many as 1.8% of total T-type channels may contribute to this current at −65 mV (Fig. 5C), a value at least as high as that calculated for recombinant mammalian channels gathered under similar conditions. A measure of the persistent, steady-state current amplitude was assessed after 1 s of sustained potentials held from a range of −70 to −50 mV in 5-mV increments. The largest, persistent current corresponded to the estimated maximal window current size at resting membrane potentials of −65 mV (Fig. 5c, inset).
FIGURE 5.
Invertebrate LCav3 has a large, persistent window current up to 1.8% of the total current near the resting membrane potential. A, sample current traces of maximal Cav3 currents (step to −35 mV) in response to a 1-s inactivating prepulse. B, a Boltzmann-fitted inactivation curve was generated by plotting the fraction of maximal current as a function of prepulse voltage. The fraction of maximal conductance at each voltage was plotted as an activation curve, curve-fitted with a Boltzmann function. The activation curve was derived from the current-voltage relationship minus the ohm-changes due to the driving force (illustrated in Fig 4D). C, calculation of the window currents were based on the product of the fraction of the whole cell conductance and fraction of available, non-inactivated channels at each voltage. Inset, a window current was measured at the end of a long, 1-s voltage-step. At 1 s, the majority of open channels will have been inactivated, leaving only open channels that persist under steady-state conditions, with a maximum at the resting membrane potential (−65 mV).
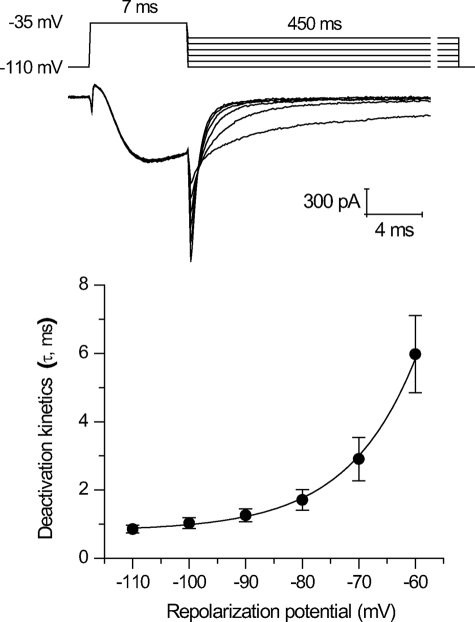
One of the characteristic features of T-type channels is a slow rate of deactivation (7). Deactivation is measured as the rate of current decay from a tail current generated by the rapid return to lower, more hyperpolarized potentials with maximally opened channels (held at −35 mV) (Fig. 6). Deactivation rates of LCav3 are fastest at hyperpolarized potentials (−110 mV) and quickly slow with depolarization steps to resting potentials (−60 mV) (Fig. 6). LCav3 fits within the faster end of the range of deactivation kinetics for mammalian T-type channels but is still manyfold slower than Cav1 and Cav2 channels. The slowness of deactivation kinetics suggests that native LCav3 currents may pass a deactivating tail current upon membrane repolarization.
FIGURE 6.
Invertebrate LCav3 slowly deactivates similar to mammalian T-type channels. Sample tail currents and curve fitting of decay rate of tail currents (τ, ms) were generated by hyperpolarizing steps between −110 and −60 mV for 450 ms from a 7-ms depolarizing step to −35 mV.
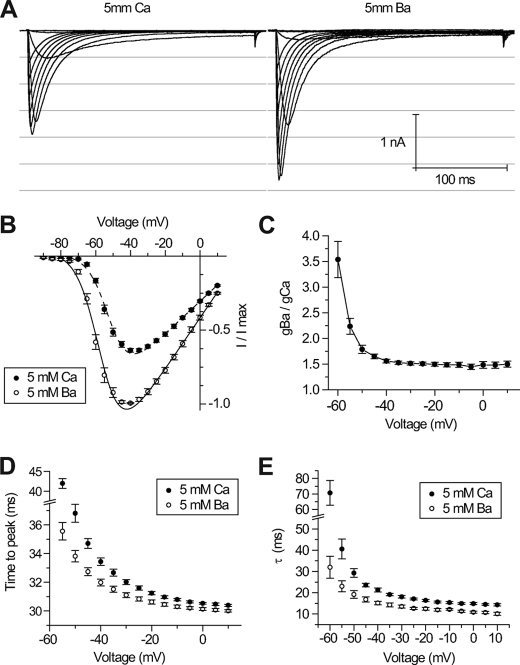
Macroscopic, native Ca2+ currents are typically equal or larger than Ba2+ currents at equimolar concentrations, although the unitary conductance is reported to be equal in high Ca2+ or Ba2+ (13). Macroscopic Ca2+ currents range from smaller, equal, or larger than Ba2+ currents for recombinant Cav3.2, Cav3.3, or Cav3.1 channels, respectively (14). Reasons for the relative differences in Ca2+ and Ba2+ permeability of different channel types are not clearly understood. LCav3 resembles Cav3.2 and other high voltage-activated snail Ca2+ channels, conducting larger amplitude whole cell Ba2+ currents than Ca2+ currents (Fig. 7A) (7). Ba2+ as a charge carrier results in a slight hyperpolarizing shift in the current-voltage relationships compared with Ca2+ (Fig. 7B), but there is still an ∼50% increase in whole cell Ba2+ conductance compared with Ca2+ in the absence of driving force changes (Fig. 7C). Kinetics are also faster when Ba2+ is the charge carrier, with faster time to peak (Fig. 7D) associated with more rapid inactivation kinetics (Fig. 7E). Ca2+-dependent inactivation typically associated with Cav1 channels is not a property of LCav3 or other T-type channels (14).
FIGURE 7.
LCav3 currents are larger and faster when Ba2+ is the charge carrier. Sample traces (A) and current-voltage relationships (B) of LCav3 currents were generated from depolarizing voltage steps from a holding potential of −110 mV while microperfusing extracellular solution containing either 5 mm Ba2+ or 5 mm Ca2+. Whole cell Ba2+ conductance was estimated to be ∼50% greater than Ca2+ conductance at all voltages (C). Kinetics of activation (time to peak current, ms) (D) and inactivation decay (tau curve fit, ms) (E) are faster when barium instead of calcium is the charge carrier.
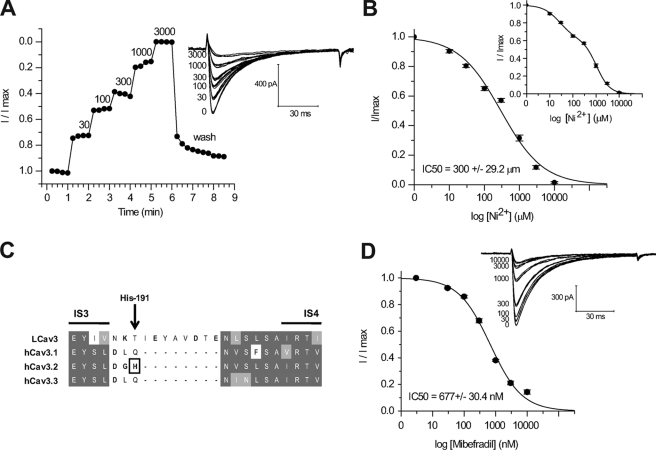
Ni2+ traditionally has been considered to be a blocker that distinguishes T-types from other channels, but only one of the three cloned mammalian T-channels, Cav3.2, is strongly inhibited by Ni2+ (15). LCav3 is approximately equally sensitive to Ni2+ as Cav3.1 and Cav3.3, with an IC50 of 300 ± 29.24 μm (Fig. 8A), but all of these T-type channels are ∼20–60-fold less sensitive than Cav3.2 (15). Lee and co-workers (16) identified that the unusual Ni2+ sensitivity of Cav3.2 critically involves His-191, imbedded in a helix-turn-helix motif known as the S3b-S4 voltage sensor paddle in Domain I (17) (Fig. 8C). Interestingly, LCav3 does not have a corresponding His-191 residue of Cav3.2, but neither does the sequence of the S3b-S4 voltage sensor paddle of LCav3 compare well with any of the Cav3 channels (Fig. 8C). LCav3 has an eight-amino acid insert in this short linker region and extra positive and negative charges compared with the mammalian Cav3 channels (Fig. 8C). The Ni2+ dose-response curve does not perfectly fit the data (Fig. 8B), but a biphasic dose-response curve does, having a high affinity IC50 of 27.25 ± 2.74 μm (38%) and a lower affinity IC50 of 1064.54 ± 79.11 μm (62%) (Fig. 8B, inset).
FIGURE 8.
Invertebrate T-type channels have similar Ni2+ and mibefradil sensitivity as mammalian T-types. A, shown is the time course of Ni2+ inhibition of normalized LCav3 peak currents (inset, representative traces). B, cumulative dose-response is illustrated, with an IC50 (300 ± 29.2 μm) value that overlaps with IC50 of Cav3.1 (304.8 ± 6.2 μm; Kang et al. (16)). Inset, a better fit illustrated with a biphasic dose-response curve is shown. C, T-type channel alignments in the region of the S3b-S4 paddle of Domain I illustrate the His-191 required for high Ni2+ sensitivity of Cav3.2 channels. LCav3 has an eight-amino acid insert with additional charged residues in the relative position of the His-191 residue in Cav3.2. D, shown is a cumulative dose-response curve of mibefradil block of LCav3 (inset, representative traces), indicating an IC50 (300 ± 29.2 μm) value that is reminiscent of the IC50 for mammalian Cav3 channels.
Mibefradil was marketed by Roche Applied Science as a drug for treatment of hypertension and angina (18) before it was withdrawn in 1998 for its potential side effects. It is a non-selective antagonist but typically has an ∼10-fold greater selectivity for T-type channels over L-type Ca2+ channels. LCav3 is in the range of sensitivity to mibefradil (680 ± 0.03 nm) as mammalian T-type channels (Fig. 8D). Caution must be heeded when directly comparing results from different studies as mibefradil is highly sensitive to the charge carrier, charge carrier concentration, and holding potential (19).
DISCUSSION
Introduction to LCav3
Here we describe the first in vitro expression characteristics of a snail homolog of mammalian T-type channels and, also remarkable, is that LCav3 is only one of two full-length cDNA sequences determined for non-mammalian T-types to date. cDNAs are assembled from predicted exons from a number of invertebrate sequenced genomes (e.g. Drosophila melanogaster Ca-α1T-RB, accession no. NM_132068), but low homology outside of the conserved transmembrane domains indicates that the predicted transcript assemblies are likely erroneous when analyzed with multiple sequence alignments of cDNAs derived from mRNA (LCav3 and C. elegans cca-1B, human Cav3.1 to Cav3.3). LCav3 codes for a 322-kDa protein of 2886 amino acids, which is the largest protein of any reported four-repeat ion channel expressed to date, including 1.25 times larger than mammalian T-type channels and 1.5 times larger than cca-1B from C. elegans, the only other reported invertebrate cDNA coding for a T-type channel. Whether T-type channels in other phylogenetic groups are this large or possibly even larger is not known.
Permeation and the DDEE Selectivity Filter
The transmembrane regions are not responsible for most of the extra mass of LCav3 and include the highly conserved, voltage-sensor domain (S1 to S4) the outer helix (S5), the P-loop, and inner helix (S6) in all four repeat domains (7). A unique DDEE selectivity filter (20) and a gating brake (21) are two trademarks of T-types that distinguish them from the Cav1 and Cav2 Ca2+ channel families. Flexible side chains of each domain harboring key glutamate residues (EEEE) contribute to the selectivity filter by extending into the permeation pathway, where they are expected to bridge Ca2+ ions as they pass through the pore of high voltage-activated Cav1 and Cav2 channels.3 A highly conserved aspartate residue upstream in the selectivity filter and adjacent to the glutamate residue in Domain II may serve to attract incoming Ca2+ ions to the ion-selective pore of all Ca2+ channels, according to modeling studies by B. Zhorov.3 T-type channels are reported to have a lower Ca2+ selectivity over monovalent cations as the estimated reversal potential is less positive than high voltage-activated Cav1 and Cav2 channels (+40 versus +60 mV) (22). Shortened carbon side chains in Domains I and II of T-types (DDEE instead of EEEE) may bridge Ca2+ ions less stringently, resulting in lower pore selectivity for Ca2+ ions in favor of faster kinetics that is typical for T-type Ca2+ channels. Interestingly, inactivation kinetic changes mirror changes in activation kinetics in T-types (12), and a modified EEDD locus alters gating properties (20).
No obvious conclusion can be drawn from differences in permeability for Ba2+ and Ca2+ ions among T-type channels. Only LCav3 and mammalian Cav3.2 channels have larger macroscopic Ba2+ than Ca2+ currents. Interestingly, greater macroscopic currents in Ba2+ over Ca2+ are a consistent feature with snail Ca2+ channels expressed in HEK-293T cells including LCav1 (9) and LCav2 (23).
Gating Brake
A gating brake shared among T-type channels is considered to prevent channel opening at hyperpolarized potentials, as nucleotide polymorphisms in patients with childhood absence epilepsy or strategically placed deletions in this region produce channels that open at even more negative potentials than typical T-type channels (24). The proximal I-II loop of LCav3 is predicted to contain the helix-loop-helix gating brake structure (21), and more distally the I-II loop has been ascribed to regulating the surface expression of T-types (25). It may be more than coincidence that the gating brake is in the equivalent position where β subunits associate with and alter the biophysical properties of high voltage-activated Cav1 and Cav2 channels as well as regulate/modulate their expression (e.g. protein folding, turnover, and membrane trafficking) (26). Indeed, invertebrate LCav3 does not require accessory β or α2δ subunits and robustly expresses in human HEK-293T cells at an efficiency that rivals the mammalian T-type channels. Continued transfection in the presence of G418 antibiotic selection has generated a number of stable HEK-293T cell lines for LCav3. A high constitutive expression of LCav3 under the strong mammalian cytomegalovirus promoter argues in favor of greater transcriptional controls for T-type channel expression in native cells compared with perhaps more post-translational checkpoints regulating the expression of Cav1 and Cav2 channels that are known to form complex, multimeric assemblies along the secretory pathway.
Overall Shared Features of T-type Channels
Scoring of the overall amino acid conservation between invertebrate and mammalian genes can lead to overestimates of the degree of structural divergence, as a sequence not under selection will drift substantially over the hundreds of millions of years separating their evolution. Comparing the in vitro expression characteristics between LCav3 and mammalian T-types suggests a structural equivalency in core regions despite the overall sequence divergence of different channels, revealing a set of quintessential properties shared by all T-types. Voltage properties are tightly regulated with fast and transient kinetics, slow deactivation, window currents produced by overlapping activation, and availability curves and channel activity limited to a narrow window of subthreshold voltages where channels are available and conducting. Also, similar drug sensitivities of LCav3 for Ni2+ ions and mibefradil suggest conserved residues in the outer pore and the aqueous permeation pathway between the selectivity filter and the aqueous, pore-lined, inner S6 helices (inverted tepee-shape) as predicted from the three-dimensional structure of crystallized K channels (27). Probing the affinity of a number of different T-type channel drugs will assist in interpreting the structural variants in the snail channel pore versus the mammalian ones.
Primitive Features in Invertebrate Channels
Invertebrate Ca2+ channels of the high voltage variety are also highly conserved in their biophysical properties. Rat Cav1.2 and snail LCav1 channels, for example, are so alike that there are no reliable biophysical features outside of drug sensitivity that separate the two channels transfected in HEK-293T cells.5 Differences outside of biophysical features appear to reflect the primitiveness of the invertebrate homologue, reminiscent of a state preceding the evolution of specializations in electromechanical coupling, such as the tetrad organization in skeletal muscle where mammalian Cav1.1 channels are directly coupled to ryanodine receptors of the sarcoplasmic reticulum (29). Invertebrate muscles lack tetrads or an equivalent Cav1.1 channel that mediates muscle contraction (29). More indirect coupling, with Ca2+ serving as a short range transmitter, is also a feature of invertebrate neurotransmission. Invertebrate Cav2 channels that are responsible for transmitter release lack a II-III loop structure containing the synaptic protein binding site of Cav2.1 and Cav2.2 channels (11) and also exhibit a synaptic organization lacking key structural proteins present in mammalian synapses (such as Bassoon and CAST) and a synaptic substructure, such as a Drosophila T-bar, which is unlike the mammalian presynaptic density (30).
T-type Channel Diversity
T-type channels are modulated through intracellular signaling cascades and are coupled to other ion channels (31), but there is little to indicate that T-types serve as instruments for electromechanical coupling in cell-type specific, multisubunit complexes in the manner of Cav1 and Cav2 channels (32). Structural diversity in the three mammalian T-type channels arose out of genomic duplication, perhaps creating some overlapping redundancy in function. Yet the presence of unique biophysical properties, tissue specificity, modulation, and putative protein-protein interactions sites suggests otherwise, indicating that the different genes may provide specialized functions in mammals. Examples that illustrate this functional divergence include the contribution to rebound burst firing in thalamocortical neurons by Cav3.1, the involvement of Cav3.2 in pain sensitivity, relaxation of coronary arteries and secretion of aldosterone, and the involvement of Cav3.3 in long-lasting bursts in the inferior olive and habenula served by its slower kinetics and a larger window current range compared with other T-type channels (7). Distinct, regional antibody staining within individual central neurons suggests that each gene may serve particular roles within somatic, dendritic, and perinuclear compartments (33, 34). Whether the diversity of mechanisms in mammals is contained within a single invertebrate Cav3 gene and its alternative splicing has not been explored.
Wide Range of Functions Expected for Abundant T-type Channel Transcript
Here we show that LCav3 is the most abundant Ca2+ channel transcript in the Lymnaea nervous system, and our previous analysis indicates that this reflects a transcript profile in an individual snail neuron (11). Quantitative RT-PCR of single identified respiratory VD4 neurons, measured in replicates of six neurons, indicated that LCav3 is manyfold more abundant than either LCav1 or LCav2 channel expression (11). Their abundance in invertebrates may reflect a wide range of functions associated with T-type channels such as (a) shaping nerve action potentials and pacemaking, (b) a non-electrogenic role for T-types in providing Ca2+ through window currents (2), and (c) roles in differentiating and proliferating cells (3) and (d) secretion (35). Some invertebrates also appear to have additional roles that are not served by mammalian T-type channels, such as excitation contraction coupling in jellyfish muscle cells (36) or facilitating the contraction of pharyngeal muscles in nematodes (37). Interestingly, T-type spikes can provide qualitatively different information than sodium spikes in the same invertebrate axons. Weak depolarizations initiate slow swimming via T-type spikes, whereas stronger pacemaking inputs initiate a fast escape swimming response mediated by overshooting sodium spikes in the same axons, presumably operating in the availability range outside of T-type channels (38).
Drug Binding; Nickel
LCav3 has equal (∼300 μm IC50) Ni2+ sensitivity as Cav3.1 and Cav3.3 channels. We report that the Ni2+ dose-response curve for LCav3 is biphasic, indicative of two components of drug block. A similar biphasic Ni2+ block is apparent in the dose-response data for mammalian recombinant channels (Fig. 3D in Ref. 16, Fig. 2B in Ref. 15, and Fig. 7B in Ref. 39) and in native currents (Fig. 3A in Ref. 40, Fig. 6B in Ref. 41, and Fig. 7B in Ref. 39). A biphasic response might result from two Ni2+ binding sites. Jones and co-workers suggest that Cav3.1 indeed has two binding sites, one in the outer pore and another deeper site within the pore pathway that is strongly affected by the permeant ion (42).
Unusually sensitive Ni2+ block (5–10 μm IC50) is a property of Cav3.2 channels and critically involves a His-191 residue (16) in what has been described as the S3b-S4 voltage sensor paddle for sodium channels based on the x-ray structure of potassium channels (43). More than His-191 may be critical in the S3b-S4 voltage sensor paddle as a similar high affinity cation block of Cav3.2 channels by extracellular Zn2+ involves the His-191 residue and two residues directly upstream of His-191, in particular, Asp-189 and Gly-190 (44).
The S3b-S4 is considered to carry most of the gating charge and likely drives the conformational changes required for pore opening and closing. It seems probable that Ni2+ associates with the S3b-S4 paddle motif of Cav3.2 in a manner similar to how tarantula and scorpion toxins immobilize the voltage sensor of sodium channels (17). Cav3.2 is inhibited by Ni2+ independently of voltage and is similarly blocked with Ca2+ or Ba2+ as a charge carrier, which is consistent with an inhibition by a mechanism outside the permeation pathway (42). Interestingly, LCav3 has an eight-amino acid insert in this short S3b-S4 region with extra positive and negative charges compared with Cav3 channels. The effect of the insert on Ni2+ block or voltage-gating, if any, is not known.
Other regions may also contribute to Ni2+ block. High affinity Zn2+ block in Cav3.2 channels also involves a neutral Ala-140 in IS2 that is negatively charged (Asp-140) in corresponding position of less sensitive Cav3.1 and Cav3.3 channels (44). Future chimera work may be important to evaluate whether LCav3 with a positively charged His-140 at this position influences cation block.
Drug Binding; Mibefradil
A number of new and potent T-type channel blockers are being explored, and mibefradil serves as the first T-type channel blocker that was clinically available (18). Interestingly, mibefradil block of snail LCav3 channel is in the range of potency of mammalian T-types. With doses spanning the mid-range of the IC50 (680 ± 0.03 nm), we observed that the mibefradil block of LCav3 would not readily stabilize, with accumulation of a slow but progressive block during long periods (tens of min) of continuous perfusion. We assume that this reflects a use-dependence often ascribed to mibefradil block (45). A slow time course of mibefradil block may also be explained by a reported accumulation of a hydrolyzed metabolite and more membrane-impermeant form of mibefradil (dm-mibefradil) that has an affinity for calcium channels from the cytoplasm (46). Further probing of different structures with mammalian and snail homologs will provide an opportunity for describing the high affinity drug binding in T-type channels.
Summary and Future Prospects
Expression characteristics of an invertebrate T-type channel has combined features that are reminiscent of all mammalian Cav3.1, Cav3.2, and Cav3.3 channels (see Table 1). LCav3 is 25% larger than any voltage-gated ion channel expressed to date and is the most abundantly expressed Ca2+ channel transcript in the snail nervous system. Window currents in invertebrate and mammalian T-type channels suggest a likely non-electrogenic role for T-types in providing Ca2+ for proliferating and differentiating cells and in the developing embryo. Alternative splicing of the single invertebrate gene may provide the structural diversity for shaping the window current and firing patterns catered for individual network requirements. We anticipate that the snail will provide unique perspectives for probing T-type channel physiology. Much can be learned from the simple molluscan preparation where only a single T-type channel gene is expressed in native cells and where there is relative ease in probing the physiological mechanisms in single identified cultured neurons and intact networks in the brain that underlay well described behaviors (47). An invertebrate channel also provides an opportunity to reflect on evolutionary mechanisms. Channel I as it was first described has turned out to be the most challenging Ca2+ channel to analyze since it was identified by Hagiwara et al. (5) more than 35 years ago. There is some truth in the following summary statement by Gray and Macdonald (48), reflecting on the present status of the T-type channel field: “[The] Physiologic regulation of T-type channels is simultaneously well documented and very obscure.”
Acknowledgments
We thank E. Perez-Reyes and B. Zhorov for helpful discussions, H. Vigil-Guitierrez for assistance in generating LCav3 antibodies, S. Lam for support in setting up the electrophysiological experiments, and A. N. Boone for editing the manuscript.
This work was supported by the Natural Science and Engineering Research Council (NSERC) of Canada and a NSERC Alexander Graham Bell Canada Graduate Scholarships (doctoral) (to A. Senatore).
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) AF484084.
B. Zhorov, personal communication.
E. Perez-Reyes, personal communication.
A. Senatore, S. Lam, A. N. Boone, T. F. Dawson, B. S. Zhorov, and J. D. Spafford, manuscript in preparation.
- EGFP
- enhanced green fluorescence protein
- PBS
- phosphate-buffered saline
- RT
- reverse transcription.
REFERENCES
- 1.Kim D., Song I., Keum S., Lee T., Jeong M. J., Kim S. S., McEnery M. W., Shin H. S. (2001) Neuron 31, 35–45 [DOI] [PubMed] [Google Scholar]
- 2.Chemin J., Monteil A., Briquaire C., Richard S., Perez-Reyes E., Nargeot J., Lory P. (2000) FEBS Lett. 478, 166–172 [DOI] [PubMed] [Google Scholar]
- 3.Lory P., Bidaud I., Chemin J. (2006) Cell Calcium 40, 135–146 [DOI] [PubMed] [Google Scholar]
- 4.Shin H. S., Cheong E. J., Choi S., Lee J., Na H. S. (2008) Curr. Opin. Pharmacol. 8, 33–41 [DOI] [PubMed] [Google Scholar]
- 5.Hagiwara S., Ozawa S., Sand O. (1975) J. Gen. Physiol 65, 617–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nowycky M. C., Fox A. P., Tsien R. W. (1985) Nature 316, 440–443 [DOI] [PubMed] [Google Scholar]
- 7.Perez-Reyes E. (2003) Physiol. Rev. 83, 117–161 [DOI] [PubMed] [Google Scholar]
- 8.Thomas P., Smart T. G. (2005) J. Pharmacol. Toxicol. Methods 51, 187–200 [DOI] [PubMed] [Google Scholar]
- 9.Spafford J. D., Dunn T., Smit A. B., Syed N. I., Zamponi G. W. (2006) J. Neurophysiol. 95, 42–52 [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski P., Sacchi N. (1987) Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 11.Spafford J. D., Munno D. W., Van Nierop P., Feng Z. P., Jarvis S. E., Gallin W. J., Smit A. B., Zamponi G. W., Syed N. I. (2003) J. Biol. Chem. 278, 4258–4267 [DOI] [PubMed] [Google Scholar]
- 12.Talavera K., Nilius B. (2006) Pflugers Arch. 453, 189–201 [DOI] [PubMed] [Google Scholar]
- 13.Bittner K. C., Hanck D. A. (2008) Biophys. J. 95, 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McRory J. E., Santi C. M., Hamming K. S., Mezeyova J., Sutton K. G., Baillie D. L., Stea A., Snutch T. P. (2001) J. Biol. Chem. 276, 3999–4011 [DOI] [PubMed] [Google Scholar]
- 15.Lee J. H., Gomora J. C., Cribbs L. L., Perez-Reyes E. (1999) Biophys. J. 77, 3034–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang H. W., Park J. Y., Jeong S. W., Kim J. A., Moon H. J., Perez-Reyes E., Lee J. H. (2006) J. Biol. Chem. 281, 4823–4830 [DOI] [PubMed] [Google Scholar]
- 17.Milescu M., Bosmans F., Lee S., Alabi A. A., Kim J. I., Swartz K. J. (2009) Nat. Struct. Mol. Biol. 16, 1080–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ertel S. I., Clozel J. P. (1997) Expert Opin. Investig. Drugs 6, 569–582 [DOI] [PubMed] [Google Scholar]
- 19.Martin R. L., Lee J. H., Cribbs L. L., Perez-Reyes E., Hanck D. A. (2000) J. Pharmacol. Exp. Ther. 295, 302–308 [PubMed] [Google Scholar]
- 20.Talavera K., Staes M., Janssens A., Klugbauer N., Droogmans G., Hofmann F., Nilius B. (2001) J. Biol. Chem. 276, 45628–45635 [DOI] [PubMed] [Google Scholar]
- 21.Arias-Olguín I. I., Vitko I., Fortuna M., Baumgart J. P., Sokolova S., Shumilin I. A., Van Deusen A., Soriano-García M., Gomora J. C., Perez-Reyes E. (2008) J. Biol. Chem. 283, 8136–8144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serrano J. R., Perez-Reyes E., Jones S. W. (1999) J. Gen. Physiol 114, 185–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spafford J. D., Chen L., Feng Z. P., Smit A. B., Zamponi G. W. (2003) J. Biol. Chem. 278, 21178–21187 [DOI] [PubMed] [Google Scholar]
- 24.Vitko I., Bidaud I., Arias J. M., Mezghrani A., Lory P., Perez-Reyes E. (2007) J. Neurosci. 27, 322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baumgart J. P., Vitko I., Bidaud I., Kondratskyi A., Lory P., Perez-Reyes E. (2008) PLoS One 3, e2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richards M. W., Butcher A. J., Dolphin A. C. (2004) Trends Pharmacol. Sci. 25, 626–632 [DOI] [PubMed] [Google Scholar]
- 27.Doyle D. A., Morais Cabral J., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R. (1998) Science 280, 69–77 [DOI] [PubMed] [Google Scholar]
- 28.Chemin J., Monteil A., Perez-Reyes E., Bourinet E., Nargeot J., Lory P. (2002) J. Physiol. 540, 3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Biase V., Franzini-Armstrong C. (2005) J. Cell Biol. 171, 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atwood H. L. (2006) Science 312, 1008–1009 [DOI] [PubMed] [Google Scholar]
- 31.Iftinca M. C., Zamponi G. W. (2009) Trends Pharmacol. Sci. 30, 32–40 [DOI] [PubMed] [Google Scholar]
- 32.Spafford J. D., Zamponi G. W. (2003) Curr. Opin. Neurobiol. 13, 308–314 [DOI] [PubMed] [Google Scholar]
- 33.McKay B. E., McRory J. E., Molineux M. L., Hamid J., Snutch T. P., Zamponi G. W., Turner R. W. (2006) Eur. J. Neurosci. 24, 2581–2594 [DOI] [PubMed] [Google Scholar]
- 34.Molineux M. L., McRory J. E., McKay B. E., Hamid J., Mehaffey W. H., Rehak R., Snutch T. P., Zamponi G. W., Turner R. W. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5555–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carbone E., Giancippoli A., Marcantoni A., Guido D., Carabelli V. (2006) Cell Calcium 40, 147–154 [DOI] [PubMed] [Google Scholar]
- 36.Lin Y. C., Spencer A. N. (2001) J. Exp. Biol. 204, 3717–3726 [DOI] [PubMed] [Google Scholar]
- 37.Steger K. A., Shtonda B. B., Thacker C., Snutch T. P., Avery L. (2005) J. Exp. Biol. 208, 2191–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mackie G. O., Meech R. W. (1985) Nature 313, 791–793 [DOI] [PubMed] [Google Scholar]
- 39.Perchenet L., Bénardeau A., Ertel E. A. (2000) Naunyn Schmiedebergs Arch. Pharmacol. 361, 590–599 [DOI] [PubMed] [Google Scholar]
- 40.Ferron L., Capuano V., Ruchon Y., Deroubaix E., Coulombe A., Renaud J. F. (2003) Circ. Res. 93, 1241–1248 [DOI] [PubMed] [Google Scholar]
- 41.Lalevée N., Rebsamen M. C., Barrère-Lemaire S., Perrier E., Nargeot J., Bénitah J. P., Rossier M. F. (2005) Cardiovasc. Res. 67, 216–224 [DOI] [PubMed] [Google Scholar]
- 42.Obejero-Paz C. A., Gray I. P., Jones S. W. (2008) J. Gen. Physiol. 132, 239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bosmans F., Martin-Eauclaire M. F., Swartz K. J. (2008) Nature 456, 202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang H. W., Vitko I., Lee S. S., Perez-Reyes E., Lee J. H. (2009) J. Biol. Chem. 285, 3271–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee T. S., Kaku T., Takebayashi S., Uchino T., Miyamoto S., Hadama T., Perez-Reyes E., Ono K. (2006) Pharmacology 78, 11–20 [DOI] [PubMed] [Google Scholar]
- 46.Wu S., Zhang M., Vest P. A., Bhattacharjee A., Liu L., Li M. (2000) J. Pharmacol. Exp. Ther. 292, 939–943 [PubMed] [Google Scholar]
- 47.Syed N. I., Bulloch A. G., Lukowiak K. (1990) Science 250, 282–285 [DOI] [PubMed] [Google Scholar]
- 48.Gray L. S., Macdonald T. L. (2006) Cell Calcium 40, 115–120 [DOI] [PubMed] [Google Scholar]
- 49.Shcheglovitov A., Vitko I., Bidaud I., Baumgart J. P., Navarro-Gonzalez M. F., Grayson T. H., Lory P., Hill C. E., Perez-Reyes E. (2008) FEBS Lett. 582, 3765–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gomora J. C., Murbartián J., Arias J. M., Lee J. H., Perez-Reyes E. (2002) Biophys. J. 83, 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]