Abstract
Streptococcus pneumoniae is a major cause of morbidity and mortality worldwide. The ability of this bacterium to adhere to epithelial cells is considered as an essential early step in colonization and infection. By screening a whole genome phage display library with sera from infected patients, we previously identified three antigenic fragments matching open reading frame spr0075 of the strain R6 genome. This locus encodes for an ∼120-kDa protein, herein referred to as plasminogen- and fibronectin-binding protein B (PfbB), which displays an LPXTG cell wall anchoring motif and six repetitive domains. In this study, by using isogenic pfbB-deleted mutants of the encapsulated D39 and of the unencapsulated DP1004 type 2 pneumococcal strains, we show that PfbB is involved in S. pneumoniae adherence to various epithelial respiratory tract cell lines. Our data suggest that PfbB directly mediates bacterial adhesion, because fluorescent beads coated with the recombinant PfbB sp17 fragment (encompassing one of the six repetitive domains and the C-terminal region) efficiently bound to epithelial cells. Mutants lacking PfbB bound to fibronectin and plasminogen considerably less efficiently than wild type bacteria, whereas sp17-coated beads specifically bound to both of these substrates. Taken together, our data suggest that, by directly interacting with fibronectin, PfbB significantly increases the ability of S. pneumoniae to adhere to human epithelial cells.
Keywords: Extracellular Matrix/Fibronectin, Organisms/Bacteria, Protein/Adhesion, Antigen, Protein Motifs, Bacterial Adherence, Plasminogen, Pneumococcus
Introduction
Although it is frequently found as a harmless colonizer of the human upper respiratory tract, Streptococcus pneumoniae (the pneumococcus) can cause local infections, such as otitis, as well as invasive life-threatening diseases, such as sepsis and meningitis (1). Pneumococci cause at least 1–2 million deaths worldwide every year, mostly as a result of community-acquired pneumonia (2). One of the most promising strategies to control pneumococcal diseases is targeting the colonization factors that promote pathogen adhesion to human tissues (3). Although knowledge of the mechanisms leading to pneumococcal colonization is still limited, it appears that a critical step in this process is the binding of human host proteins by a complex array of bacterial adhesins (3, 4). Choline-binding proteins, such as PspC and PsaA, which are noncovalently attached to the cell wall, are likely involved in the adherence to mucosal cells. PspC, for example, binds to the polymeric immunoglobulin receptor of respiratory epithelial cells and may be at least partially responsible also for transcytosis across the human mucosa (5, 6). PsaA, a metal-binding lipoprotein, has been recently reported to bind to nasopharyngeal cells through an interaction with E-cadherin (7). Moreover, as shown for other Gram-positive bacteria, two different types of pili were recently implicated in the adherence of pneumococci to respiratory cells (8–10).
The ability to bind to host fibronectin (Fn)3 is a characteristic shared by many pathogens, especially by Gram-positive cocci, and is considered as a critical early step in the infection process (11). Fn is a large glycoprotein present in soluble form (e.g. in plasma, cerebrospinal, and amniotic fluids) or in insoluble form on the cell surface, in the extracellular matrix, and in basement membranes. Fn, whose amino acid sequence is highly conserved among vertebrates, is involved in a number of essential biological processes, including embryogenesis and wound healing (11). Therefore, targeting of Fn is considered a basic strategy by which invading pathogens exploit essential host processes to establish or disseminate infection (12).
Although pneumococci strongly bind Fn (13), the molecular mechanisms governing this interaction are as yet little understood. PavA is one of the proteins involved in this process, because pavA mutants show decreased ability to bind to Fn (14). Although PavA is homologous to Fn-binding proteins of other pathogens (e.g. Fbp54 of Streptococcus pyogenes or FbpA of Listeria monocytogenes), it is likely that PavA does not bind directly to Fn but rather modulates the surface expression of other adhesins that are directly responsible for this interaction (15). Recently, a protein designated as PfbA (standing for plasmin- and fibronectin-binding protein A) was reported to bind to Fn, plasminogen, plasmin, and human serum albumin (16). A mutant strain lacking this protein had reduced ability to adhere to lung and laryngeal cell lines compared with the parental strain.
By screening of a whole genome display library, we have previously identified an ∼120-kDa immunogenic protein (encoded by open reading frame spr0075 in the annotated genome of the serotype 2 R6 strain) displaying an LPXTG cell wall anchoring motif and six repetitive domains (17). In this study, we show that this protein, herein referred to as plasminogen- and fibronectin-binding protein B or PfbB, has a significant role in mediating pneumococcal adhesion to human respiratory epithelial cells. We also show that some recombinant fragments of PfbB can directly bind plasminogen and Fn.
EXPERIMENTAL PROCEDURES
Bacterial Strains
The pneumococcal strains used in this study were the encapsulated serotype 2 D39 strain (18) and its unencapsulated derivative DP1004 (19). The FP228 strain, a DP1004 mutant deleted for pfbB (spr0075), has been described previously (17). The FP242 strain, a D39 pfbB-deleted mutant, was constructed by transformation of D39 with a cassette amplified from FP228 genomic DNA using the external primers 0075_1 (CTTCAGCAGGTCAAGACCATGT) and 0075_4 (CAGTTAAATCCAGCATTTCTT) (17). Kanamycin (500 μg/ml) was used as a selection agent. All bacteria were grown at 37 °C in Todd-Hewitt broth (Oxoid). When necessary, kanamycin was added to a final concentration of 500 μg/ml.
Production of PfbB Recombinant Fragments and Antisera
The sp4 and sp17 recombinant PfbB fragments were produced as described previously (17). Briefly, DNA inserts from λ phage clones SP-cl.4 and SP-cl.17 (17) were subcloned into the bacterial expression vector pGEX-SN to produce, respectively, pGEX-SNsp4 and pGEX-SNsp17 that allow the expression of recombinant proteins as fusions to glutathione S-transferase (GST). Similarly, to produce the sp17 fragments RD6 (amino acids 899–1053 of the PfbB protein) and C terminus (amino acids 1054–1133), the corresponding DNA sequences were amplified from pGEX-SNsp17 and cloned into pGEX-SN. After induction of the fusion proteins, these were purified from the cytoplasm of bacterial cells by affinity chromatography (17). Recombinant GST, to be used as a control, was also produced using the same procedures. In some experiments, the GST tag was removed enzymatically using factor Xa (Promega) according to the manufacturer's instructions, followed by removal of free GST by affinity chromatography. Mouse antisera were produced by immunizing 6-week-old specific pathogen-free CD1 mice (Charles River Laboratories, Italia) by the intraperitoneal injection of recombinant sp17-GST or GST (50 μg) in complete Freund's adjuvant on day 0 and in incomplete Freund's adjuvant on days 14 and 28. The use of complete Freund adjuvant in the first immunization was justified by our previous observations that high titered sera were more consistently obtained with this adjuvant, as compared with other less “inflammatory” adjuvants such as alum. However, care was taken to minimize discomfort to the animals by injecting a low volume of the emulsion (0.1 ml containing 0.05 mg of mycobacteria) and by using sterile solutions and techniques to prepare it. Under these conditions, no significant abdominal distension or complications at the injection site were observed throughout the experimental period. To obtain anti-pneumococcal immune sera, to be used as positive controls, an additional group of mice was immunized with a choline-binding protein-enriched fraction designated as CCR6. This mixture was obtained from strain DP1004 cells grown to the early exponential phase (A600 = 0.2), washed with phosphate-buffered saline (PBS; pH 7.2), and incubated in the presence of 2% choline chloride (Sigma) at 20 °C for 10 min. The supernatant was dialyzed, concentrated, and used as described above to immunize mice using 50 μg (total protein content) for each immunization. All immunizations were conducted at the animal facilities of the Metchnikoff Department of the University of Messina according to the European Union guidelines for the handling of laboratory animals and were approved by the relevant national authority (Istituto Superiore di Sanità).
Flow Cytometry Immunofluorescence Analysis
S. pneumoniae strains grown to the early log phase (A600 = 0.2) were harvested by centrifugation, washed three times with PBS, and blocked for 20 min at 20 °C with PBS containing 2% fetal calf serum (FCS). Mouse antisera were diluted 1:100 in FCS-supplemented PBS (PBS/FCS) and incubated with bacterial cells for 40 min at 4 °C. Phycoerythrin-conjugated goat anti-mouse IgG (Jackson ImmunoResearch), diluted 1:50 was then added to the cells and incubated at 4 °C for an additional 30 min. Bacteria were then washed and analyzed with an LSR Flow Cytometer using the CellQuest software (both from BD Biosciences).
Bacterial Adhesion Assays
The human respiratory cell lines A549 (alveolar), Chang (conjunctival), and Hep-2 (laryngeal) were grown in Dulbecco's modified Eagle's medium with 10% FCS in 7% CO2 and dispensed into 24-well plates at a density of 1 × 105/ml. The cells were cultured for 24 h before the adherence assay. Before use, the monolayers were washed twice with PBS. Bacteria were grown to the early log phase (A600 = 0.2), washed, resuspended in Dulbecco's modified Eagle's medium without FCS, and applied to the monolayers at a multiplicity of infection of 1:20. For adherence assays, infected monolayers were incubated for 1 h at 37 °C and washed three times to remove nonadherent bacteria. After the addition 1 ml of cold H2O and gentle scraping, cell lysates were serially diluted and plated in triplicate onto tryptic soy agar plates supplemented with 3% defibrinated sheep blood (Oxoid) for the enumeration of CFUs. Adherence results were expressed as the percentage of CFUs recovered in lysates relative to the CFUs initially added to the monolayers. Pneumococcal adherence to A549 cells was also assessed microscopically. Experimental conditions were as detailed above, except that cells were grown on glass coverslips, and infected monolayers were not lysed but rather fixed with paraformaldehyde (3.7% in PBS for 10 min at 4 °C) and permeabilized with PBS/Triton X-100 (0.25% for 10 min). After blocking with PBS/bovine serum albumin for 1 h at 20 °C, the monolayers were exposed to anti-CCR6 mouse serum (diluted 1:400) followed by fluorescein isothiocyanate-conjugated goat anti-mouse IgG diluted 1:1000 (Sigma). TRITC-conjugated phalloidin (Sigma) was used at a 1:80 dilution for cytosolic F-actin staining. 4′,6-Diamidino-2-phenylindole (Molecular Probes) was used for nuclear staining according to the manufacturer's instructions. At least 300 cells were counted using a fluorescence microscope (Axio Observer) equipped with a structured illumination apparatus (Apotome) and with the Axiovision software (all from Carl Zeiss). Results were expressed as the number of bacteria per cell. Under the conditions described here for the adherence assays (specifically at 1 h after the addition of bacteria), all cell-associated pneumococci were localized to the cell surface with no detectable internalization, as assessed in preliminary immunofluorescence microscopy experiments using permeabilized and nonpermeabilized cells (data not shown).
Coupling of Microspheres with Recombinant Proteins
Fluorescent beads (Fluoresbrite YG 1.00-μm microspheres, Polysciences) were conjugated with recombinant PfbB fragments (or with GST, as a control) according to the manufacturer's instructions using a protein concentration of 300 μg/ml. The amount of protein coupled on beads was calculated by subtracting the amount of protein present in the supernatant after adsorption. Care was taken to use in each experiment beads with similar amounts of coupled proteins.
Adhesion of Microspheres to Cells
A549 cells were prepared as described above under “bacterial adhesion assays,” and protein-coupled beads were added in serum-free medium at a concentration of 108 beads/ml. After 30 min of incubation at 37 °C, monolayers were washed three times, and attached beads were counted by using a fluorescent microscope. At least 300 cells were counted. For competitive inhibition binding assays, the monolayers were incubated with increasing concentrations of soluble sp17-GST or GST for 30 min and washed, and the assay was performed as described above.
Adhesion of Microspheres or Bacteria to Immobilized Human Proteins
For the microsphere adhesion assay, silane-treated 18-mm2 glass coverslips were incubated overnight at 4 °C with the proteins indicated below (10 μg/ml in PBS), blocked with PBS supplemented with 2% casein for 1 h at 20 °C, and exposed to 108 beads in 1 ml of FCS-free PBS. After incubation at 20 °C overnight, the slides were washed and observed under a fluorescent microscope. To examine the binding of pneumococci to human proteins, similarly prepared coverslips were exposed to bacteria. Briefly, pneumococci were grown to the early log phase (A600 = 0.2), washed, and resuspended in PBS without serum and applied to the coverslips (1 × 105 CFU/ml). Slides were then Gram-stained and observed under a bright field microscope. Results were expressed as number of particles per field of vision at the indicated magnification. At least 20 different fields per slide were counted. The following substrates (all from Sigma) were used: human fibronectin; human serum albumin; human collagen type 1; human chondroitin sulfate B; human plasminogen, and bovine serum albumin.
RESULTS
PfbB Protein Is Expressed on the Pneumococcal Surface
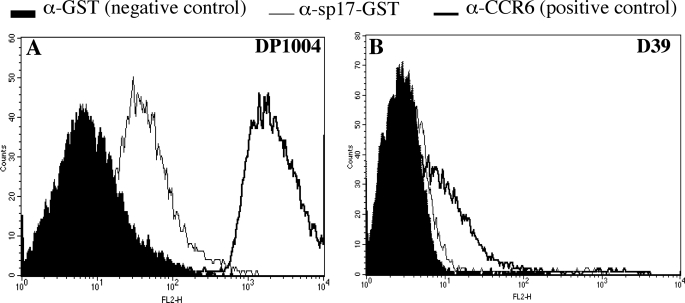
By using a whole genome phage display library, we have previously identified several antigenic pneumococcal fragments based on their ability to bind antibodies in convalescent sera. Three of these fragments matched the sequence of open reading frame spr0075 in the R6 strain genome. The protein encoded by spr0075, herein referred to as PfbB, is predicted to contain a putative signal peptide, six adjoining repeated regions (each composed of a 150–152-amino acid-long domain), and an LPXTG motif, characteristic of surface proteins that are covalently bound to peptidoglycan (Fig. 1). To assess whether PfbB is indeed expressed on the bacterial surface, we expressed the recombinant sp17 fragment of PfbB (Fig. 1) and used it to immunize mice. Fig. 2A shows that sera from mice immunized with sp17 fused to GST, but not sera from mice immunized with the GST control, bound to the surface of the unencapsulated strain DP1004. Anti-sp17-GST sera also bound weakly to the surface of the encapsulated D39 strain (Fig. 2B). Weaker binding to the encapsulated strain relative to the unencapsulated one was not surprising, because it is known that the polysaccharide capsule can mask surface expression of cell wall-linked proteins (20). As expected, anti-sp17 sera did not bind the pfbB-deleted mutants FP228 and FP242 (data not shown). This evidence indicated that PfbB is expressed on the surface of both encapsulated and unencapsulated strains.
FIGURE 1.
Schematic representation of protein PfbB encoded by the spr0075 open reading frame of the R6 strain genome. A, predicted amino acid sequence of PfbB. RD1–6, repeat domains 1–6. The arrow indicates the cleavage site predicted by the PSORT program. The LPXTG cell wall anchoring motif near the C terminus is underlined, and the sequences of the sp4 and sp17 fragments are in boldface. B, structure of PfbB primary sequence. N, N terminus; SP, signal peptide; RD1–6, repeat domains 1–6. The white and dark gray areas indicate, respectively, the N- and C-terminal sequences located outside of the repeat domains 1–6. The boundaries of the sp4 and sp17 PfbB fragments are also indicated.
FIGURE 2.
Presence of PfbB on the bacterial surface as assessed by immunofluorescence flow cytometry analysis of the unencapsulated DP1004 strain (A) and of the encapsulated D39 strain (B). Bacteria were exposed to mouse antisera raised against GST (α-GST, used as a negative control), against a choline-binding protein-enriched fraction (α-CCR6, used as a positive control), or against the sp17 fragment of PfbB fused to GST (sp17-GST). Antibody binding was detected with phycoerythrin-conjugated goat anti-mouse IgG.
PfbB Is an Adhesin of S. pneumoniae
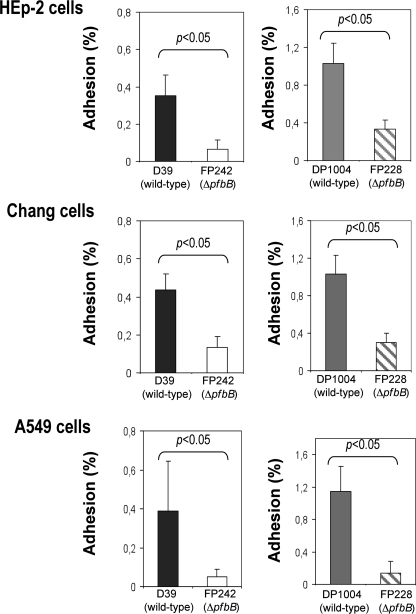
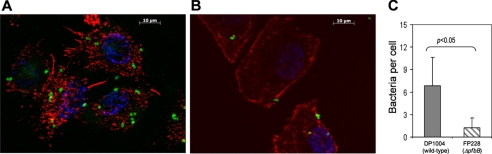
To investigate whether PfbB is involved in adherence to human cells, we compared the adhesion capabilities of DP1004 and D39 with those of the corresponding pfbB-deleted mutants. In these experiments, the numbers of adhering bacteria were determined using three different human epithelial cell lines (namely, A549, Chang and Hep-2, derived from lung, conjunctiva, and larynx tissues, respectively). As expected, the unencapsulated DP1004 strain (Fig. 3, right panels) adhered to the cells more efficiently than the encapsulated parental D39 strain (Fig. 3, left panels), in agreement with the known ability of the capsule to mask adhesins in pneumococci (4, 20). Notably, the pfbB-deleted strains adhered less efficiently than the corresponding parental strains to all tested cell lines. For example, the adherence of the pfbB-deleted FP228 strain was 68–88% lower than that observed with the parental DP1004 strain. Interestingly, even in the presence of the capsule, PfbB was required for efficient adherence, because adherence of the pfbB-deleted FP242 strain was significantly reduced compared with that of the wild type D39 strain. The poor adherence of pneumococci in the absence of PfbB was further confirmed by microscopic analysis (Fig. 4). Altogether, these findings suggested that PfbB plays a significant role in the ability of S. pneumoniae to adhere to human epithelial cells and that this effect is not masked by the presence of a polysaccharide capsule.
FIGURE 3.
Role of PfbB in adherence of encapsulated and unencapsulated pneumococci to human epithelial cell lines. The left panels show the adherence of the D39 strain (encapsulated) and of its isogenic ΔpfbB mutant (FP242). The right panels show the adherence of the DP1004 strain (unencapsulated) and of its isogenic ΔpfbB mutant (FP228). The number of adherent bacteria was determined by counting CFUs in bacterial lysates as described under “Experimental Procedures.” Data show means ± S.D. of three independent experiments conducted in triplicate. Statistical analysis was performed using Student's t test.
FIGURE 4.
Role of PfbB in adherence to A549 cells, as evidenced by microscopic analysis. Cells grown on coverslips were incubated with S. pneumoniae strain DP1004 (A) and its isogenic ΔpfbB mutant strain FP228 (B). Bacteria were labeled green using mouse anti-pneumococcal antibodies followed by fluorescein isothiocyanate-goat anti-mouse IgG. A549 cells were labeled with TRITC-phalloidin for cytosolic F-actin staining and by 4′,6-diamidino-2-phenylindole for nuclear staining. C, adherence to A549 cells of strain DP1004 and its isogenic ΔpfbB mutant strain FP228, as determined by microscopic counts. Data show means ± S.D. of three independent experiments conducted in duplicate. Statistical analysis was performed using Student's t test.
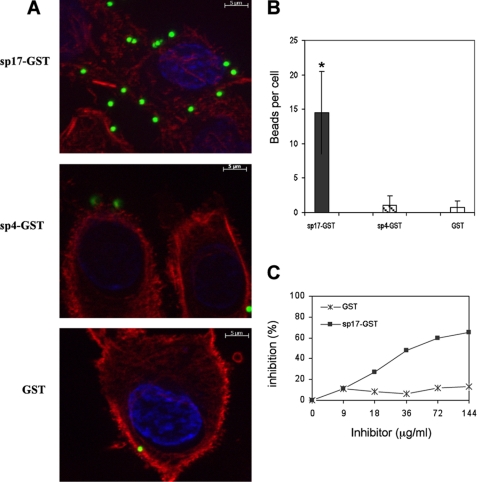
The above data did not clarify whether PfbB is directly involved in adhesion or merely participates in this process by modulating, for example, the expression of “true” adhesins. To discriminate between these two possibilities, we evaluated the ability of two recombinant PfbB fragments to bind to epithelial cells. As depicted in Fig. 5, beads coated with sp17-GST, in contrast to beads coated with sp4-GST or GST, strongly adhered to A549 alveolar epithelial cells. Moreover, adherence of sp17-GST-coated beads was inhibited in a dose-dependent manner by pretreatment of cells with soluble sp17-GST, but not with GST, thus further confirming the specificity of sp17 binding (Fig. 5C). These data suggested that PfbB directly binds to the surface of human epithelial cells.
FIGURE 5.
Direct binding of PfbB fragments to A549 cells. Fluorescent latex microspheres were coated with PfbB fragments expressed as GST fusion proteins (sp17-GST or sp4-GST) or with GST. After incubation with A549 cells, adhering particles were counted microscopically (A and B). Columns in B show means ± S.D. of three independent experiments. *, significantly different (p < 0.05) from GST-coated beads, as assessed by Student's t test. C, competitive inhibition of binding of sp17-GST-coated particles to A549 cells by cell pretreatment with soluble sp17-GST or GST, used as a control. Shown are data from one representative experiment of three producing similar results.
Binding of the sp17 Fragment of PfbB to Human Proteins
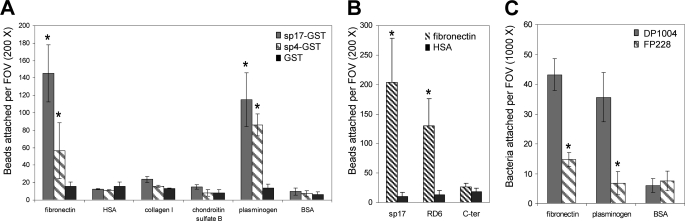
The above data indicated that PfbB is expressed on the surface of S. pneumoniae and directly interacts with host molecules. To determine whether this protein can interact with human proteins, we evaluated the ability of the sp17 and sp4 fragments to bind several extracellular matrix or plasma components, including chondroitin sulfate B, collagen, Fn, plasminogen, and albumin. To this end, we measured binding of the fragments to substrates immobilized on glass coverslips. Fig. 6A shows that beads coated with sp17-GST or sp4-GST, but not with GST, bound to immobilized Fn or plasminogen. Neither sp4- nor sp17-coated beads bound to chondroitin sulfate B, collagen, or human serum albumin. These data indicated that both of the PfbB fragments examined adhered to immobilized plasminogen or Fn, although there was a tendency for the sp17 fragment to bind more efficiently than the sp4 fragment to Fn.
FIGURE 6.
Binding of PfbB fragments (A and B) or S. pneumoniae (C) to coverslips coated with human proteins. A, fluorescent latex microspheres were coated with the sp17 or sp4 PfbB fragments expressed recombinantly as GST fusion proteins (sp17-GST or sp4-GST) or with GST. After incubation with protein-sensitized coverslips, adhering particles were counted microscopically. BSA, bovine serum albumin. B, fluorescent latex microspheres were coated with sp17 or with two sp17 fragments (RD6 and C terminus (C-ter)) after enzymatic removal of the GST tag. After incubation with coverslips sensitized with fibronectin or with human serum albumin (HSA), adhering particles were counted microscopically. C, coverslips sensitized with the indicated proteins were incubated with strain DP1004 and its isogenic ΔpfbB mutant strain FP228. Bacterial binding was detected after Gram staining of coverslips. Results were expressed as particles per field of vision (FOV) at the indicated magnification and represent means ± S.D. of three (A and C) or six (B) independent experiments. *, significantly different (p < 0.05) from GST-coated beads (A), HSA-coated beads (B), or wild type strain DP1004 (C) as assessed by Student's t test.
Because sp17 differs from sp4 for containing an 80-amino acid-long sequence located at the C terminus of PfbB outside of the repeat domain region (Fig. 1), it was of interest to ascertain whether this C-terminal region played a significant role in the binding of sp17 to immobilized Fn. To this end, we produced two separate fragments encompassing the whole length of sp17 and consisting of, respectively, the RD6 domain (amino acids 899–1053 of PfbB) and the C terminus (C-terminal amino acids 1054–1133). Next, we assessed binding to immobilized Fn of beads coated with these fragments, in comparison with sp17-coated beads. To avoid possible interfering effects of the relatively bulky GST tag, in these experiments we used polypeptides from which GST had been enzymatically removed. Fig. 6B shows that the RD6 domain could bind to Fn, although the C-terminal portion showed no binding. There was a tendency for sp17 to bind to Fn more efficiently than RD6 (Fig. 6B), but this difference was not statistically significant. These data suggest a model whereby repeat domains are sufficient for specific binding and thus appear mainly responsible for the interaction between PfbB and Fn.
Finally, we investigated whether PfbB contributed significantly to the ability of bacteria to adhere to immobilized Fn or plasminogen. To this end, we compared the DP1004 strain with its pfbB-defective mutant (FP228) for binding to these substrates. Fig. 6B shows that wild type pneumococci strongly bound to Fn and plasminogen but not to bovine serum albumin, used as a control. Notably, binding to fibronectin and plasminogen was markedly reduced in the strain lacking PfbB. These data suggested that PfbB significantly contributes to the overall activity of pneumococci to bind Fn and plasminogen.
DISCUSSION
It is well established that binding to cell surface Fn is a fundamental step in the ability of several Gram-positive bacterial pathogens to adhere to host epithelial barriers and to invade them (3, 11). Considerable insight into the molecular mechanisms of bacteria-Fn interactions has come from studies on Fn-binding proteins of S. pyogenes and Staphylococcus aureus, although comparatively little is known of the mechanisms and functional significance of Fn binding by pneumococci (11). This study describes the ability of a surface protein, designated as PfbB, to promote Fn binding of pneumococci, as well as their adherence to human epithelial cells. PfbB, which was previously identified based on its ability to induce specific antibodies during pneumococcal infections (17), contains a typical LPXTG motif for peptidoglycan attachment at its C terminus, and six repeats of 150–152 amino acid residues. Although orthologs of this protein are present in all of the sequenced genomes of S. pneumoniae strains, the number of repeats may vary. For example, SP0082, the TIGR4 strain ortholog of PfbB, contains only four of such repeats (21). A detailed bioinformatics analysis of SP0082 was performed by Bumbaca et al. (22), who recombinantly expressed the repeat domain and described its ability to interact with Fn, although high concentrations (>100 μg/ml) of the recombinant domain were needed to detect some degree of binding. Because Δsp0082 mutants were not generated by these authors, the functional role of this protein or its contribution to the overall Fn binding activity of pneumococci could not be discerned from their data. In this study, we showed that pneumococci adhered to immobilized Fn with a considerably lower efficiency in the absence of PfbB. Notably, we also found that PfbB significantly contributed to the ability of S. pneumoniae to adhere to human cells. For example, using the alveolar A549 cell line, the adherence of the pfbB-defective mutant was reduced by ∼90%, as compared with the parental unencapsulated DP1004 strain. Genetic evidence was complemented biochemically by showing that sp17, a recombinant PfbB fragment, bound efficiently to A549 cells and to immobilized Fn.
It is well known that the presence of a thick capsule can hinder pneumococcal adherence, due to a “masking” effects on surface adhesins (4, 20). Accordingly, we found that the encapsulated D39 strain adhered to epithelial cells severalfold less efficiently than its unencapsulated derivatives. Thus, it was of interest to ascertain whether PfbB played a role in adherence even in the presence of a capsule. Our data indicate that this was indeed the case, because the PfbB-defective isogenic mutant of the encapsulated D39 strain also showed significantly decreased adherence compared with the wild type parental strain. This agreed with the detectable, albeit weakly reactive, presence of PfbB on the D39 surface, as determined by immunofluorescence flow cytometry.
The occurrence of repetitive domains in PfbB is reminiscent of Fn-binding proteins of other bacteria, such as those of S. aureus, Streptococcus dysgalactiae, and S. pyogenes (11). Although, in the latter adhesins, the primary sites of Fn interaction have been localized to the repeat domains, it is now evident that regions outside of these domains may also contribute to high affinity interactions (11). We investigated here whether the C-terminal portion of PfbB, located outside of the repeat domain region, can directly interact with Fn. This, however, did not seem to be the case. In contrast, a single recombinant repeat domain was able to bind to immobilized Fn. Collectively, our data are compatible with a model whereby PfbB binding to Fn is primarily mediated by the repeat domains, although an involvement of the C-terminal region in this process cannot be categorically excluded.
Although PfbB appears to significantly contribute to the ability of pneumococci to bind Fn and to adhere to epithelial cells, other adhesins may also play an important role in this process. It is well known that adherence and colonization to mucosal surfaces requires the coordinated action of several bacterial components. Like PfbB, the recently described PfbA binds Fn and plasminogen and is expressed on the surface of the R6 strain, an unencapsulated derivative of the D39 strain (16). In the same study, a ΔpfbA mutant strain lost ∼50% of the ability of the parental R6 strain to bind to epithelial cells. At variance with PfbB, PfbA is devoid of repetitive domains and displays a 19-amino acid-long region homologous to Fn-type III repeat, which may be responsible for interactions with host Fn type I repeats. Therefore, it is possible that by using different mechanisms to interact with Fn, PfbB and PfbA cooperate in enabling S. pneumoniae to bind to this molecule. It should be noted that in our study, as well in the one by Yamaguchi et al. (16), strains were used that are devoid of the pili that have been recently described in pneumococci (8–10). Indeed, because these structures may be crucial in mediating the adherence to epithelial cells of a significant proportion of S. pneumoniae strains, it remains to be determined if and to what extent PfbB or PfbA contribute to the adherence of piliated strains.
We found here that PfbB fragments, particularly sp17, bind to plasminogen, in addition to Fn. Moreover, PfbB appeared to significantly contribute to the ability of whole bacteria to bind plasminogen, as shown by experiments using a pfbB-deleted mutant. Such ability is considered an important virulence factor because, by this mechanism, pneumococci may acquire potent proteolytic activity, which would enable them to degrade the extracellular matrix and fibrin and thereby to disseminate within the body (4, 23, 24). It was recently proposed that pneumococci covered with active plasmin degrade intercellular junction proteins and migrate, by this process, through epithelial barriers using a pericellular route (24). Moreover, plasminogen could promote pneumococcal adherence to epithelial cells, because this protein is present not only in plasma but also (albeit at considerably lower concentrations) in bronchoalveolar fluid, from which it could be captured by pneumococci and used to interact with the plasminogen receptors expressed by epithelial cells (24). Further studies are underway to evaluate this possibility. The best characterized plasmin(-ogen) binding factors in pneumococci are surface glyceraldehyde-3-phosphate dehydrogenase (25) and enolase (26). The latter enzyme displays a nonameric peptide, which is directly responsible for interaction with plasminogen (27). This peptide, however, is not present in PfbB. Therefore, future studies on the mechanisms of plasminogen binding by this molecule, as well as on its functional significance, may provide new insights into the pathogenesis of pneumococcal infections.
In conclusion, in this work we have described some functional properties of a surface pneumococcal protein capable of interacting with host components, particularly Fn. It has been clearly demonstrated that immunization with the several Fn-binding proteins of S. pyogenes, such as Sfb1 (28), Fbp54 (29), and FbaA (30), can protect mice against infection with this pathogen. Therefore, the identification of surface-exposed Fn-binding proteins of pneumococci, such as the one described here, may be potentially useful in the development of protein vaccines and/or of novel therapeutic approaches against S. pneumoniae.
Acknowledgments
We are deeply grateful to Gianni Pozzi and Marco R. Oggioni for helpful discussions.
This work was supported in part by grants from the MIUR PRIN/COFIN 2005 Research Program of Italy.
- Fn
- fibronectin
- FCS
- fetal calf serum
- PBS
- phosphate-buffered saline
- TRITC
- tetramethylrhodamine isothiocyanate
- CFU
- colony-forming unit
- GST
- glutathione S-transferase.
REFERENCES
- 1.Sinha A., Levine O., Knoll M. D., Muhib F., Lieu T. A. (2007) Lancet 369, 389–396 [DOI] [PubMed] [Google Scholar]
- 2.File T. M. (2003) Lancet 362, 1991–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kline K. A., Fälker S., Dahlberg S., Normark S., Henriques-Normark B. (2009) Cell Host Microbe 5, 580–592 [DOI] [PubMed] [Google Scholar]
- 4.Hammerschmidt S. (2006) Curr. Opin. Microbiol. 9, 12–20 [DOI] [PubMed] [Google Scholar]
- 5.Hammerschmidt S., Tillig M. P., Wolff S., Vaerman J. P., Chhatwal G. S. (2000) Mol. Microbiol. 36, 726–736 [DOI] [PubMed] [Google Scholar]
- 6.Zhang J. R., Mostov K. E., Lamm M. E., Nanno M., Shimida S., Ohwaki M., Tuomanen E. (2000) Cell 102, 827–837 [DOI] [PubMed] [Google Scholar]
- 7.Anderton J. M., Rajam G., Romero-Steiner S., Summer S., Kowalczyk A. P., Carlone G. M., Sampson J. S., Ades E. W. (2007) Microb. Pathog. 42, 225–236 [DOI] [PubMed] [Google Scholar]
- 8.Bagnoli F., Moschioni M., Donati C., Dimitrovska V., Ferlenghi I., Facciotti C., Muzzi A., Giusti F., Emolo C., Sinisi A., Hilleringmann M., Pansegrau W., Censini S., Rappuoli R., Covacci A., Masignani V., Barocchi M. A. (2008) J. Bacteriol. 190, 5480–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barocchi M. A., Ries J., Zogaj X., Hemsley C., Albiger B., Kanth A., Dahlberg S., Fernebro J., Moschioni M., Masignani V., Hultenby K., Taddei A. R., Beiter K., Wartha F., von Euler A., Covacci A., Holden D. W., Normark S., Rappuoli R., Henriques-Normark B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2857–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson A. L., Ries J., Bagnoli F., Dahlberg S., Fälker S., Rounioja S., Tschöp J., Morfeldt E., Ferlenghi I., Hilleringmann M., Holden D. W., Rappuoli R., Normark S., Barocchi M. A., Henriques-Normark B. (2007) Mol. Microbiol. 66, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwarz-Linek U., Höök M., Potts J. R. (2004) Mol. Microbiol. 52, 631–641 [DOI] [PubMed] [Google Scholar]
- 12.Knodler L. A., Celli J., Finlay B. B. (2001) Nat. Rev. Mol. Cell Biol. 2, 578–588 [DOI] [PubMed] [Google Scholar]
- 13.van der Flier M., Chhun N., Wizemann T. M., Min J., McCarthy J. B., Tuomanen E. I. (1995) Infect. Immun. 63, 4317–4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes A. R., McNab R., Millsap K. W., Rohde M., Hammerschmidt S., Mawdsley J. L., Jenkinson H. F. (2001) Mol. Microbiol. 41, 1395–1408 [DOI] [PubMed] [Google Scholar]
- 15.Pracht D., Elm C., Gerber J., Bergmann S., Rohde M., Seiler M., Kim K. S., Jenkinson H. F., Nau R., Hammerschmidt S. (2005) Infect. Immun. 73, 2680–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaguchi M., Terao Y., Mori Y., Hamada S., Kawabata S. (2008) J. Biol. Chem. 283, 36272–36279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beghetto E., Gargano N., Ricci S., Garufi G., Peppoloni S., Montagnani F., Oggioni M., Pozzi G., Felici F. (2006) FEMS Microbiol. Lett. 262, 14–21 [DOI] [PubMed] [Google Scholar]
- 18.Iannelli F., Pearce B. J., Pozzi G. (1999) J. Bacteriol. 181, 2652–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shoemaker N. B., Guild W. R. (1974) Mol. Gen. Genet. 128, 283–290 [DOI] [PubMed] [Google Scholar]
- 20.Bootsma H. J., Egmont-Petersen M., Hermans P. W. (2007) Infect. Immun. 75, 5489–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tettelin H., Nelson K. E., Paulsen I. T., Eisen J. A., Read T. D., Peterson S., Heidelberg J., DeBoy R. T., Haft D. H., Dodson R. J., Durkin A. S., Gwinn M., Kolonay J. F., Nelson W. C., Peterson J. D., Umayam L. A., White O., Salzberg S. L., Lewis M. R., Radune D., Holtzapple E., Khouri H., Wolf A. M., Utterback T. R., Hansen C. L., McDonald L. A., Feldblyum T. V., Angiuoli S., Dickinson T., Hickey E. K., Holt I. E., Loftus B. J., Yang F., Smith H. O., Venter J. C., Dougherty B. A., Morrison D. A., Hollingshead S. K., Fraser C. M. (2001) Science 293, 498–506 [DOI] [PubMed] [Google Scholar]
- 22.Bumbaca D., Littlejohn J. E., Nayakanti H., Rigden D. J., Galperin M. Y., Jedrzejas M. J. (2004) OMICS 8, 341–356 [DOI] [PubMed] [Google Scholar]
- 23.Attali C., Durmort C., Vernet T., Di Guilmi A. M. (2008) Infect. Immun. 76, 5350–5356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Attali C., Frolet C., Durmort C., Offant J., Vernet T., Di Guilmi A. M. (2008) Infect. Immun. 76, 466–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergmann S., Rohde M., Hammerschmidt S. (2004) Infect. Immun. 72, 2416–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergmann S., Rohde M., Chhatwal G. S., Hammerschmidt S. (2001) Mol. Microbiol. 40, 1273–1287 [DOI] [PubMed] [Google Scholar]
- 27.Bergmann S., Rohde M., Preissner K. T., Hammerschmidt S. (2005) Thromb. Haemost. 94, 304–311 [DOI] [PubMed] [Google Scholar]
- 28.Guzmán C. A., Talay S. R., Molinari G., Medina E., Chhatwal G. S. (1999) J. Infect. Dis. 179, 901–906 [DOI] [PubMed] [Google Scholar]
- 29.Kawabata S., Kunitomo E., Terao Y., Nakagawa I., Kikuchi K., Totsuka K., Hamada S. (2001) Infect. Immun. 69, 924–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terao Y., Okamoto S., Kataoka K., Hamada S., Kawabata S. (2005) J. Infect. Dis. 192, 2081–2091 [DOI] [PubMed] [Google Scholar]